Abstract
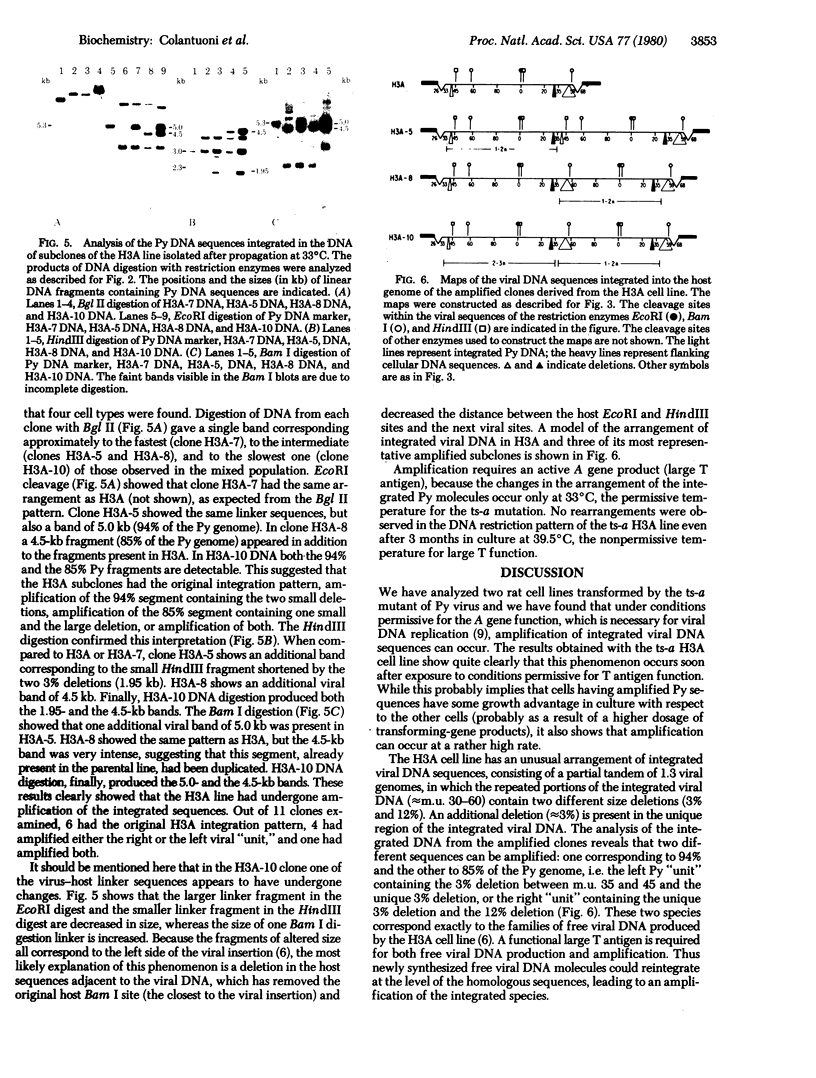
Polyoma virus (Py) transformation of rat cells requires integration of viral genomes into the host DNA, which generally occurs in a partial or full head-to-tail tandem arrangement. The instability of this structure was previously demonstrated by the high rate of loss of integrated Py genomes in the presence of viral large tumor (T) antigen. We now show that integrated Py DNA sequences can also undergo amplification. We studied two rat cell lines transformed by the ts-a Py mutant, which codes for a thermolabile large T antigen. In a derivative of the ts-a H6A cell line, we have observed loss of full-length Py DNA molecules from the integrated tandem ("curing"), accompanied by the creation of new tandem repeats of two segments of viral DNA corresponding to 38% and 10% of the viral genome, each containing the origin of DNA replication. In the ts-a H3A cell line, which contains an integrated partial tandem of about 1.3 viral genomes with three distinct deletions, propagation at 33 degrees C resulted in the generation of full tandem repeats of a 94% Py DNA "unit" (including two 3% deletions), an 85% "unit" (including a 3% and the 12% deletion), or both. Amplification of integrated viral DNA was not observed in cells propagated at 39.5 degrees C, the nonpermissive temperature for large T antigen function. Amplification of integrated Py DNA sequences thus requires an active large T antigen and can generate a full tandem of integrated viral DNA molecules long after the initial integration event.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Basilico C., Zouzias D., Della-Valle G., Gattoni S., Colantuoni V., Fenton R., Dailey L. Integration and excision of polyoma virus genomes. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):611–620. doi: 10.1101/sqb.1980.044.01.064. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Hourcade D., Dressler D., Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. B., Cozzarelli N. R., Deutscher M. P., Lehman I. R., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXII. Replication of duplex deoxyribonucleic acid by polymerase at a single strand break. J Biol Chem. 1970 Jan 10;245(1):39–45. [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lania L., Griffiths M., Cooke B., Ito Y., Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (Hr-t) mutant. Cell. 1979 Nov;18(3):793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- Peterson H. M., Laughnan J. R. INTRACHROMOSOMAL EXCHANGE AT THE BAR LOCUS IN DROSOPHILA. Proc Natl Acad Sci U S A. 1963 Jul;50(1):126–133. doi: 10.1073/pnas.50.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad I., Zouzias D., Basilico C. State of the viral DNA in rat cells transformed by polyoma virus. I. Virus rescue and the presence of nonintergrated viral DNA molecules. J Virol. 1976 May;18(2):436–444. doi: 10.1128/jvi.18.2.436-444.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Silver J., Schaffhausen B., Benjamin T. Tumor antigens induced by nontransforming mutants of polyoma virus. Cell. 1978 Oct;15(2):485–496. doi: 10.1016/0092-8674(78)90018-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Unequal mitotic sister chromatin exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1272–1276. doi: 10.1073/pnas.71.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Zouzias D., Prasad I., Basilico C. State of the viral DNA in rat cells transformed by polyma virus. II. Identification of the cells containing nonintegrated viral DNA and the effect of viral mutations. J Virol. 1977 Oct;24(1):142–150. doi: 10.1128/jvi.24.1.142-150.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]