Abstract
Ochratoxin A (OTA), a mycotoxin produced by Aspergillus and Penicillium species, is composed of a dihydroisocoumarin ring linked to phenylalanine, and its biosynthetic pathway has not yet been completely elucidated. Most of the knowledge regarding the genetic and enzymatic aspects of OTA biosynthesis has been elucidated in Penicillium species. In Aspergillus species, only pks genes involved in the initial steps of the pathway have been partially characterized. In our study, the inactivation of a gene encoding a nonribosomal peptide synthetase (NRPS) in OTA-producing A. carbonarius ITEM 5010 has eliminated the ability of this fungus to produce OTA. This is the first report on the involvement of an nrps gene product in OTA biosynthetic pathway in an Aspergillus species. The absence of OTA and ochratoxin α, the isocoumaric derivative of OTA, and the concomitant increase of ochratoxin β, the dechloro analog of ochratoxin α, were observed in the liquid culture of transformed strain. The data provide the first evidence that the enzymatic step adding phenylalanine to polyketide dihydroisocoumarin precedes the chlorination step to form OTA in A. carbonarius and that ochratoxin α is a product of hydrolysis of OTA, giving an interesting new insight into the biosynthetic pathway of the toxin.
INTRODUCTION
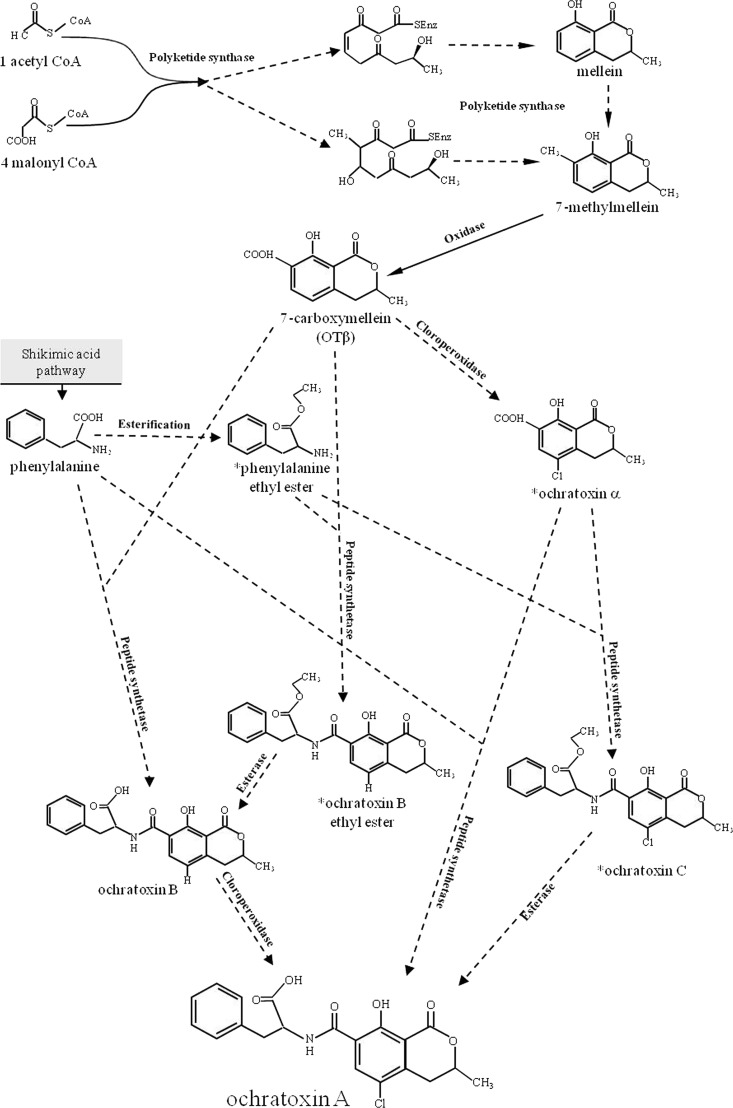
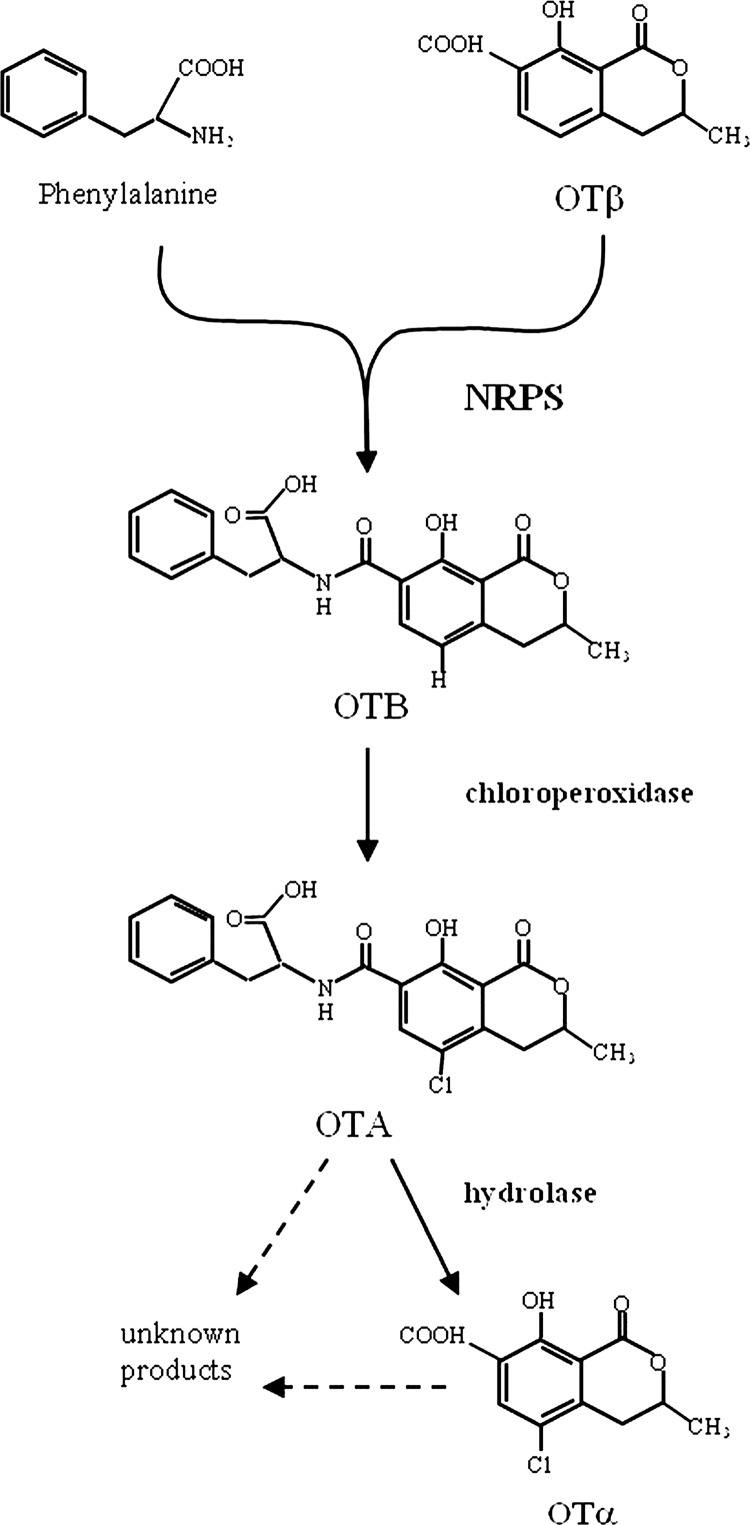
Ochratoxin A (OTA), initially described as a toxic metabolite of Aspergillus ochraceus (53), is a secondary metabolite produced by Aspergillus and Penicillium species. The economic impact of this mycotoxin is significant due to the fact that it is found as a contaminant in a wide variety of food commodities. In particular, Penicillium verrucosum is mainly responsible for OTA contamination of cereal-based products, Penicillium nordicum is responsible for contamination of some dry-cured foods, Aspergillus ochraceus, Aspergillus carbonarius, Aspergillus westerdijkiae, and Aspergillus steynii are responsible for contamination of coffee, cocoa, spices, and dried fruits, while contamination of grapes, grape juice, dried vine fruits, must, and wine is due mainly to A. carbonarius and, to a lesser extent, to Aspergillus niger “aggregate” species (21, 35, 36, 44, 48, 52). Ochratoxin A contamination of grapes constitutes a serious health and economic problem not only in southern European countries, but also in other areas of the world with Mediterranean-type climates (7, 9, 38, 44). Ochratoxin A is a potent nephrotoxin that also displays hepatotoxic, teratogenic, immunosuppressive, and carcinogenic effects (1, 17, 18, 19, 46, 47); it has been classified as a class 2B human carcinogen by the International Agency for Research on Cancer (IARC) (30). At present, wine is considered, after cereals, the second major source of OTA, and strict European Union regulation has been established, setting maximum levels for OTA of 2 μg kg−1 in wine, musts, and grape juice and 10 μg kg−1 for dried vine fruits (Commission regulation no. 1881/2006). Structurally, OTA comprises a dihydrocoumarin moiety linked to a molecule of l-β-phenylalanine (Phe), derived from the shikimic acid pathway, by an amide bond. Several related compounds were also reported to occur in OTA-producing organism cultures, such as the dechlorinated analog ochratoxin B (OTB), the isocoumarin nucleus of OTA ochratoxin α (ΟΤα) and its dechlorinated analogue ochratoxin β (ΟΤβ)—which are not linked to phenylalanine, methyl and ethyl esters, including ochratoxin C (OTC), which is an ethyl ester derivative of OTA—and several amino acid analogues (39, 40, 57). Unlike other important mycotoxins, the biosynthesis pathway of OTA has not yet been completely elucidated in detail. However, it is clear that the pathway involves some crucial steps, such as the biosynthesis of the isocoumarin group through the catalyzing action of a polyketide synthase (PKS), its ligation with the amino acid phenylalanine through the carboxyl group in a reaction catalyzed by a peptide synthetase, and the chlorination step, but the order of the reactions is not yet well defined. In this regard, several schemes have been proposed that have resulted from different studies. Huff and Hamilton (29) suggested a role in the polyketide synthesis for mellein, which would be carboxylated to OTβ and then transformed through a chloroperoxidase to OTα. According to their hypothesis, in the subsequent step, OTα would be phosphorylated and linked to the ethyl ester of phenylalanine to generate OTC, and then a deesterification would lead to the final product OTA (Fig. 1). Precursor feeding experiments with A. ochraceus, carried out by Harris and Mantle (27), suggested that one, possibly dominant, biosynthetic pathway involves the passage from OTβ to OTα and then to OTA, with a chlorinating step prior to the ligation of the polyketide (OTα) to phenylalanine, without OTC and phenylalanine ethyl ester intermediates. Since they could not rule out the role of OTB, they also maintain a possible alternative pathway in which the formation of OTA goes through the synthesis from OTβ to OTB, but in this case, a biosynthetic role for OTα, which occurs naturally, could not be explained. All of the hypothetical pathways for OTA biosynthesis are summarized in Fig. 1, in which the esterification or not of phenylalanine and the consequent presence of OTB ethyl ester with an intermediary role are also contemplated.
Fig 1.
Scheme showing all the different hypotheses of the OTA biosynthesis pathway according to the literature data available so far (27, 29). *, hypothetical intermediary compounds.
Most of the molecular aspects of OTA biosynthesis have been elucidated in Penicillium species. In P. nordicum, a putative OTA biosynthetic cluster has been identified containing biosynthetic genes encoding (i) a PKS (otapksPN), (ii) a nonribosomal peptide synthetase (NRPS) (otanpsPN) putatively responsible for the formation of the peptide bond between the polyketide and the phenylalanine, (iii) a gene (otachlPN) with some homology to chlorinating enzymes, thought to be involved in the chlorination step, and (iv) a gene (otatraPN) with high homology to a transporter protein hypothesized to be involved in OTA export (25, 31). The orthologs of these genes have also been identified for the closely related species P. verrucosum, but have not yet been identified in the major OTA-producing aspergilli. So far, in Aspergillus spp., specifically in A. ochraceus (41) and in A. westerdijkiae (6), only the pks genes involved in the initial steps of the pathway have been characterized. In A. carbonarius, an economically and scientifically interesting species due to its predominant role in the contamination of grapes, the presence of different pks genes was investigated (5). In our previous work, we identified an additional pks gene in the same species whose expression pattern showed a correlation to OTA production under permissive conditions (22). In order to identify genes and proteins related to OTA biosynthesis in A. carbonarius, other studies were performed based on differential expression profiles, but no OTA homologous genes were found (10, 13, 14).
Recently, the genome sequence of A. carbonarius strain ITEM 5010, which is capable of producing up to 7.5 mg of OTA per liter of inductive culture medium (43), was generated by the United States Department of Energy's Joint Genome Institute (http://jgi.doe.gov/carbonarius/). Similar to many other ascomycetes so far sequenced, the A. carbonarius genome contains a large number of pks and nrps genes. These genes encode complex and multifunctional proteins involved in the biosynthesis of most of the fungal secondary metabolites (11, 12, 32). In particular, a new pks gene (protein identification [PI] no. 173482) was identified which showed a similarity of about 70% at the nucleotide level and 74% at the amino acid level to the predicted OTA cluster pks gene (XP001397313) of the OTA-producing A. niger strain CBS 513.88 (42). The percentages of identity to A. ochraceus (AY272043) and A. westerdijkiae (AY583209) OTA PKS partial sequences were 63.7% and 37%, respectively, whereas an amino acid identity of 38.7% was observed to the PKS sequence previously identified in A. carbonarius (22). Furthermore, it showed an identity of 30.4% to the partial sequence of P. nordicum OTA PKS (AY557343).
Interestingly, A. niger strain ATCC 1015, which does not produce OTA, has a 21-kb deletion in this predicted biosynthetic cluster (4). Similar to most characterized mycotoxins, OTA biosynthetic genes in A. carbonarius are expected to be arranged in a cluster. The genomic region, located by the OTA putative pks gene in this fungus, was investigated, leading to the identification of a gene encoding an NRPS hypothesized to catalyze the ligation of phenylalanine to the polyketide group. The gene arrangement of putative OTA clusters in different fungal species is shown in Fig. S1 in the supplemental material.
In this work, we present the inactivation of the gene in A. carbonarius designated AcOTAnrps, which completely inhibited the production of OTA, OTB, and OTα. The principal secondary metabolites involved in OTA biosynthetic pathway were identified in wild-type and transformant strains by high-resolution mass spectrometry (HRMS). This result indicates for the first time the involvement of an NRPS in the OTA biosynthetic pathway of an Aspergillus species. The absence of OTA, OTB, and OTα and the concomitant increase of OTβ concentration in the culture of ΔAcOTAnrps strain confirm the hypothesis that the bond between the phenylalanine and the polyketide dihydroisocoumarin, catalyzed by the synthetase, precedes the chlorination step, clarifying the order of reactions in the OTA biosynthetic pathway. Moreover, we present evidence that the non-OTA-producing ΔAcOTAnrps strain keeps the capability to degrade OTA in OTα.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The A. carbonarius strain used in this study is the wild-type strain ITEM 5010 from the Agri-Food Toxigenic Fungi Culture Collection of the Institute of Sciences of Food Production, CNR, Bari, Italy (www.ispa.cnr.it/Collection). The other two strains used in this study were generated by the wild-type strain and are indicated as KB1039 (ΔkusA) and KB1041 (ΔkusA ΔAcOTAnrps). The mutant strains were grown on minimal medium supplemented with hygromycin (100 μg ml−1) and/or glufosinate (Basta; bioWorld, Dublin, OH) (1 mg ml−1) for selection of transformants.
To check the production of OTA and other secondary metabolites, conidia (105 spores in 100 μl of water) were inoculated into Erlenmeyer flasks containing 20 ml of minimal medium (6 g/liter NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4 · 7H2O, 1.52 g/liter KH2PO4 [pH 6.5], 10 g/liter glucose, 2 ml/liter Hutner's trace elements). Incubation was carried out at 25°C in the dark under stationary conditions for 14 days. After 7 and 14 days, the fungal biomass was collected, filtered on Whatman paper no. 4 (Whatman International, Ltd., Maidstone, United Kingdom), and weighed. Sporulation and pigmentation of fungal colonies were evaluated by visual observation. Liquid cultures were collected and analyzed for OTA and OTα content by high-pressure liquid chromatography with fluorescence detection (HPLC-FLD). Liquid cultures of transformants KB1039 and KB1041 were analyzed by HPLC-HRMS to confirm the results of HPLC-FLD and to identify other metabolites belonging to the OTA biosynthetic pathway.
For kinetic studies, the KB1041 strain was grown for 25 days at 25°C in 100 ml of minimal medium supplemented with OTA (1.2 μg ml−1 [3 μM]). As a control, the same amount of OTA was added to minimal medium not inoculated with the fungus. Every day, 1 ml of liquid culture was collected from each replicate and stored at −20°C until the HPLC-FLD determination of OTA and OTα content. Triplicate cultures were prepared and analyzed for each experiment.
Deletion of the AcOTAnrps gene in A. carbonarius.
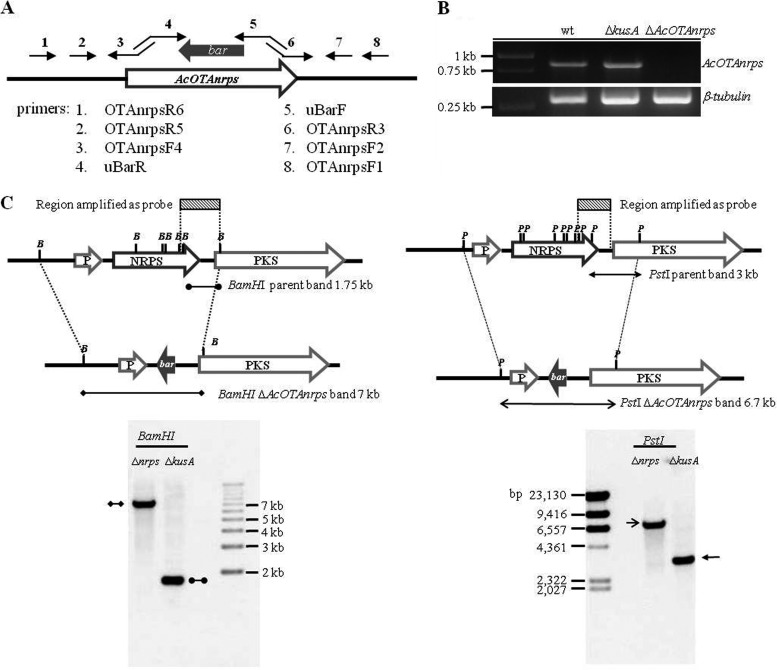
A deletion cassette for AcOTAnrps was designed and transformed into strain KB1039 of A. carbonarius ITEM 5010 lacking the kusA gene, which is required for nonhomologous end joining (NHEJ) double-strand-break repair of DNA, to facilitate homologous integration (M. Praseuth, A. Gallo, S. E. Baker, J. R. Collett, C. C. C. Wang, and K. S. Bruno, submitted for publication). The deletion cassette was generated by a commonly used PCR approach (58) in which sequences flanking the coding region of the AcOTAnrps gene were fused to the bar gene encoding glufosinate resistance. The primers used to create the deletion cassette are listed in Table 1. In particular, two ∼1,200-bp fragments, one upstream and one downstream of the targeted gene, were amplified from the genomic A. carbonarius DNA with the primer pairs OTAnrpsR6/OTAnrpsF4 and OTAnrpsR3/OTAnrpsF1, respectively. A fusion PCR attached the two fragments to the bar resistance marker, which was amplified from the plasmid pCB1530 (Fungal Genetics Stock Center, Kansas City, MO) by using the primer pair uBarR/uBarF, which have extension sequences overlapping extension sequences of primers OTAnrpsF4 and OTAnrpsR3. The joined product was amplified with the nested primers OTAnrpsR5 and OTAnrpsF2 to obtain the construct, which was transformed into protoplasts prepared from strain KB1039 (Fig. 2A). Colonies were selected for stable integration of the cassette on minimal medium supplemented with 1 mg/ml glufosinate (bioWorld, Dublin, OH). The transformants were first screened for correct integration by diagnostic PCR using the primers uBarR and OTAnrpsF1. This pair of primers can only amplify a fragment from transformants that have integrated the bar gene at the targeted locus. All three of the obtained transformants tested positive for proper integration. To further screen for the deletion, we amplified the entire locus using primer OTAnrpsR6 and OTAnrpsF1. These primers are outside the construct that was used to transform the parent strain. The ΔkusA parental strain had an amplicon larger than 8 kb, and the transformants had an amplicon of about 3.4 kb (see Fig. S2 in the supplemental material). These matched the predicted sizes and were consistent with the predicted changes that should have occurred at that locus. A final confirmation of the gene deletion was made by Southern blotting, by using standard methods for DNA electrophoresis and blotting (37). Southern analysis was performed by digesting the genomic DNA of the strains with BamHI and PstI and by hybridization with a 1,586-bp probe, amplified with primers AcNRPS-2rev and OTAnrpsF1. The probe was labeled and developed using a North2South chemiluminescent detection kit (Thermo Fisher Scientific). The blot was imaged using a Kodak MR2000 equipped with a charge-coupled device (CCD) camera. Markers were visualized for size determination on the blot by loading either the Invitrogen 1-kb-plus DNA size marker or HindIII-digested lambda DNA into the outside lanes. These bands were detected using labeled lambda DNA.
Table 1.
Primers used in this study
| Primer name | Primer sequence (5′→3′) |
|---|---|
| OTAnrpsF1 | 5′-AATCTCCGACAGCTGGAAAA-3′ |
| OTAnrpsF2 | 5′-CTTTGGGCAGAGGTTTGAAG-3′ |
| OTAnrpsR3 | 5′-TGACCTCCACTAGCTCCAGCGCAATCCGGCTTTACTTTCA-3′ |
| OTAnrpsF4 | 5′-AATAGAGTAGATGCCGACCGGTGGAAAGTGGGAAATGGTG-3′ |
| OTAnrps5R | 5′-GGGAATGTCTTCTGCCTTGA-3′ |
| OTAnrps6R | 5′-CTGACTGCGTAGCCTCTCCT-3′ |
| uBarF | 5′-GCTGGAGCTAGTGGAGGTCAGTCGACAGAAGATGATATTG-3′ |
| uBarR | 5′-CGGTCGGCATCTACTCTATTGTCGACCTAAATCTCGGTGA-3′ |
| AcNRPS-1for | 5′-TCATCTCCGACGAGGAAC-3′ |
| AcNRPS-2rev | 5′-CAAAGGAATCCTCGTCACT-3′ |
Fig 2.
(A) Schematic representation of the AcOTAnrps gene deletion construct. (B) RT-PCR profile of AcOTAnrps gene expression in the wild-type, ΔkusA, and ΔAcOTAnrps strains grown on a medium permissive for OTA. β-Tubulin was used as a control. (C) Map of the AcOTAnrps locus in parental and ΔAcOTAnrps strains of A. carbonarius and Southern blot hybridizations from genomic DNA digested with BamHI and PstI. The restriction sites BamHI and PstI are indicated with B and P, respectively.
Nucleic acid extraction, cDNA synthesis, and RT-PCR.
DNA was isolated using the Fungal DNA miniprep kit (E.Z.N.A., Omega Bio-Tek, Inc., Doraville, GA) according to the manufacturer's protocol. Total RNA was extracted from frozen mycelium pulverized in liquid nitrogen using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA samples were treated with RNase-free DNase I (Qiagen) to eliminate possible trace amounts of contaminating DNA. RNA aliquots were preserved at −80°C. First-strand cDNA was synthesized using about 1.5 μg of total RNA, oligo(dT)18 primer, and SuperScript III reverse transcriptase (Invitrogen, San Diego, CA) according to the manufacturer's protocol.
Reverse transcription (RT)-PCR was used to analyze the presence of nrps transcript in the mycelium of wild-type strain ITEM 5010 and the mutant strains KB1039 and KB1041. The primer pair AcNRPS-1for/2rev (Table 1) was used under the following conditions: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 50 s, and 72°C for 50 s, and then a final extension step of 72°C for 5 min. The primers Bt2a/Bt2b (26) were used to monitor β-tubulin gene expression as endogenous control. These primers span three introns, which also allowed checking for DNA contamination.
Chemical analyses of liquid cultures by HPLC-FLD and HPLC-HRMS.
For OTA and OTα determination in liquid cultures of the three strains of A. carbonarius ITEM 5010 (wild type, KB1039, and KB1041), aliquots of liquid cultures were filtered through a 0.22-μm-pore filter (Sartorius AG, Goettingen, Germany), and 100 μl was injected into the HPLC-FLD apparatus. Direct injection of liquid culture was possible since no interfering peaks eluted at retention times of OTα and OTA. The limit of detection of this method for OTA and OTα was 0.1 ng/ml. This methodology was also used to measure OTA and OTα in liquid cultures for the kinetics study of OTA degradation by the ΔAcOTAnrps strain. Samples containing high OTA concentrations exceeding the calibration range of the OTA standard were appropriately diluted with the HPLC mobile phase and reanalyzed.
To confirm the lack of OTA production by A. carbonarius KB1041 at low concentration, aliquots of liquid cultures were purified and concentrated through OchraTest immunoaffinity (IMA) columns (Vicam L.P., Watertown, MA) prior to HPLC-FLD determination. In particular, 5 ml of liquid culture was diluted with a solution containing 1% polyethylene glycol–5% NaHCO3 and loaded onto the IMA column, which was eluted at 1 ml/min by discarding the eluate. The column was then washed with 5 ml washing solution containing 2.5% NaCl, 0.5% NaHCO3, and 5 ml pure water. The column was eluted with 2 ml methanol that was collected in a vial, evaporated to dryness at 50°C under a nitrogen stream, and reconstituted with 250 μl of the HPLC mobile phase. Fifty microliters of purified extract, equivalent to 1 ml of liquid culture, was injected into the HPLC-FLD apparatus. The limit of detection of this method for OTA was 0.005 ng/ml. OTα could not be measured by this method because the IMA column has a poor retention for this compound, which is lost during sample loading on the column.
The HPLC-FLD apparatus was an 1100 series liquid chromatography (LC) system comprising a binary pump, an autosampler, a fluorescence detector (excitation wavelength, 333 nm; emission wavelength, 460 nm) from Agilent Technologies (Waldbronn, Germany). The column was a Waters Symmetry C18 column (150 mm by 4.6 mm, 5-μm particles) (Waters, Milford, MA), preceded by a 0.5-μm Rheodyne guard filter. The mobile phase was an isocratic mixture of acetonitrile and water (45:55) containing 1% acetic acid. The flow rate of the mobile phase was 1 ml/min.
Standard solutions of OTA and OTα were purchased from Romer Labs Diagnostic GmbH (Tulln, Austria), calibration solutions were prepared in HPLC mobile phase, and calibration curves were prepared in the range of 0.1 to 100 ng/ml.
Twenty microliters of filtered liquid cultures of A. carbonarius KB1039 (ΔkusA) and KB1041 (ΔkusA ΔAcOTAnrps) were also analyzed by HPLC-HRMS for the identification of 7-methylmellein (7-MM), OTβ, OTα, phenylalanine (Phe), ethyl ester of Phe, ethyl ester of OTB, OTB, OTC, and OTA. HPLC-HRMS retention times and HR mass data of the metabolites are shown in Table 2. The column was a Kinetex C18 column (100 mm by 2.10 mm, 2.6 μm). The mobile phase was a multistep gradient of water (eluent A) and methanol (eluent B), both containing 0.5% acetic acid and 1 mM ammonium acetate. A gradient elution was performed by changing the mobile phase composition as follows. After 3 min at 20% eluent B, the proportion was set at 40% and then linearly increased to 63% in 35 min and kept constant for 11 min. The column was reequilibrated with 20% eluent B for 10 min prior to the successive injection (34). HPLC-HRMS analyses were performed on a benchtop single-stage mass spectrometer (Exactive) equipped with a heated electrospray ion source (HESI II) (Thermo Fisher Scientific, Bremen, Germany), coupled to an Accela HPLC system (Thermo Fisher Scientific, San Jose, CA). The HESI II interface was used in the positive-ion mode, and the scan range was 50.2 to 1,003.0 m/z, with a resolution power of 100,000 FWHM (full width at half maximum). Other settings were as follows: sheath and auxiliary gas flow rates, 30 and 10 arbitrary units, respectively; sweep gas, 0 arbitrary units; capillary temperature, 300°C; capillary voltage, 4 kV. The Xcalibur software (version 2.1.0, Thermo Fisher Scientific) was used for data acquisition and processing.
Table 2.
Summary of HPLC-HRMS retention times and HR mass data of OTA and other metabolites identified in cultures of wild-type and ΔAcOTAnrps strains
| Compound | Retention time (min) | Mass (Da) |
Error (mDa) | Formula | |
|---|---|---|---|---|---|
| Measured | Monoisotopic | ||||
| Phe | 2.64 | 165.0782 | 165.0789 | −0.7 | C9H11NO2 |
| OTB ethyl ester | —a | — | 399.1682 | — | C22H25NO6 |
| OTβ | 5.89 | 222.0516 | 222.0528 | −1.2 | C11H10O5 |
| Phe ethyl ester | 6.11 | 193.1093 | 193.1103 | −1.0 | C11H15NO2 |
| 7-MM | 7.61 | 192.0777 | 192.0786 | −0.9 | C11H12O3 |
| OTα | 8.00 | 256.0132 | 256.0138 | −0.6 | C11H9ClO5 |
| OTC | — | — | 431.1136 | — | C22H22ClNO6 |
| OTB | 18.98 | 369.1201 | 369.1212 | −1.1 | C20H19NO6 |
| OTA | 25.27 | 403.0807 | 403.0823 | −1.6 | C20H18ClNO6 |
—, OTB ethyl ester and OTC were not present in cultures of either wild-type or ΔAcOTAnrps strains.
Statistical analysis.
Statistical analysis was performed by using the GraphPad Instat software (Instat, San Diego, CA). Data were subjected to the paired t test (two-tailed P value) to determine the statistical differences when two groups of data were compared. One-way analysis of variance (ANOVA) with posttest (standard parametric methods) was used when three groups of data were compared.
RESULTS
Identification and inactivation of the AcOTAnrps gene in A. carbonarius.
In the genome of A. carbonarius ITEM 5010, we identified a 5,691-bp gene, designated AcOTAnrps, encoding a nonribosomal peptide synthetase (PI no. 132610), and located at about 900 nucleotides (nt) upstream of a pks gene. The NRPS protein is predicted to contain 1,875 amino acid (aa) residues and is characterized by one module with the typical three core domains and an additional last adenylation domain. The predicted amino acid sequence displays ∼60% identity to the nrps gene product of the predicted OTA cluster in A. niger (42) and a low similarity (11% identity) to the OTA NRPS in P. nordicum (AY534879).
A deletion cassette for AcOTAnrps was designed and transformed into strain KB1039 of A. carbonarius ITEM 5010, lacking the kusA gene, which is required for NHEJ double-strand-break repair of DNA. Many species of filamentous fungi lacking the NHEJ process show a much higher percentage of targeted gene deletion, and the ΔkusA mutant strain was created for this aim in a previous study (Praseuth et al., submitted). Three transformants positive by PCR screening for the integration of the bar resistance marker at the AcOTAnrps locus were obtained (see Fig. S2 in the supplemental material). One of them was chosen for subsequent analyses and was designated KB1041. A Southern blot hybridization, performed on the genomic DNA of the wild-type and mutant strains digested with BamHI and PstI, confirmed the complete deletion of AcOTAnrps gene in KB1041: hybridizing bands of 1.75 kb and 3.0 kb with BamHI and PstI digestions, respectively, indicated the endogenous gene, whereas a homologous integration of the deletion cassette resulted in a band of ∼7.1 kb when DNA was digested with BamHI and of 6.7 kb when DNA was digested with PstI (Fig. 2C). The RT-PCR analysis shown in Fig. 2B confirmed the lack of expression of the AcOTAnrps gene in mutant strain KB1041, when the wild type and the two mutant strains were grown in an OTA permissive medium.
Production of OTA, OTα, and other secondary metabolites in mutant and wild-type strains of A. carbonarius.
Both A. carbonarius mutant strains KB1039 (ΔkusA) and KB1041 (ΔkusA ΔAcOTAnrps) and wild-type strain ITEM 5010 were inoculated into minimal medium and incubated for 7 and 14 days at 25°C in the dark. During the incubation period, no difference in fungal growth, sporulation, and pigment production was observed. In particular, at 7 days the mean biomass weights (triplicate measurements) were 0.94 ± 0.13, 0.82 ± 0.14, and 0.94 ± 0.21 g for the wild-type, ΔkusA, and ΔAcOTAnrps strains, respectively, and no statistical difference was observed between them (P = 0.6295). Similar results were obtained at 14 days (P = 0.6815). Cultures were analyzed for the presence of OTA and OTα by HPLC-FLD. Cultures of A. carbonarius ITEM 5010 and mutant strains KB1039 and KB1041 were also analyzed for 7-MM, OTβ, the OTB ethyl ester, Phe, Phe ethyl ester, OTα, OTB, OTC, and OTA by HPLC-HRMS. The simultaneous identification of these metabolites in liquid cultures was possible through use of the Orbitrap mass analyzer, which enables high mass resolution (up to 150,000), high mass accuracy, large dynamic range, and a high mass/charge range (28).
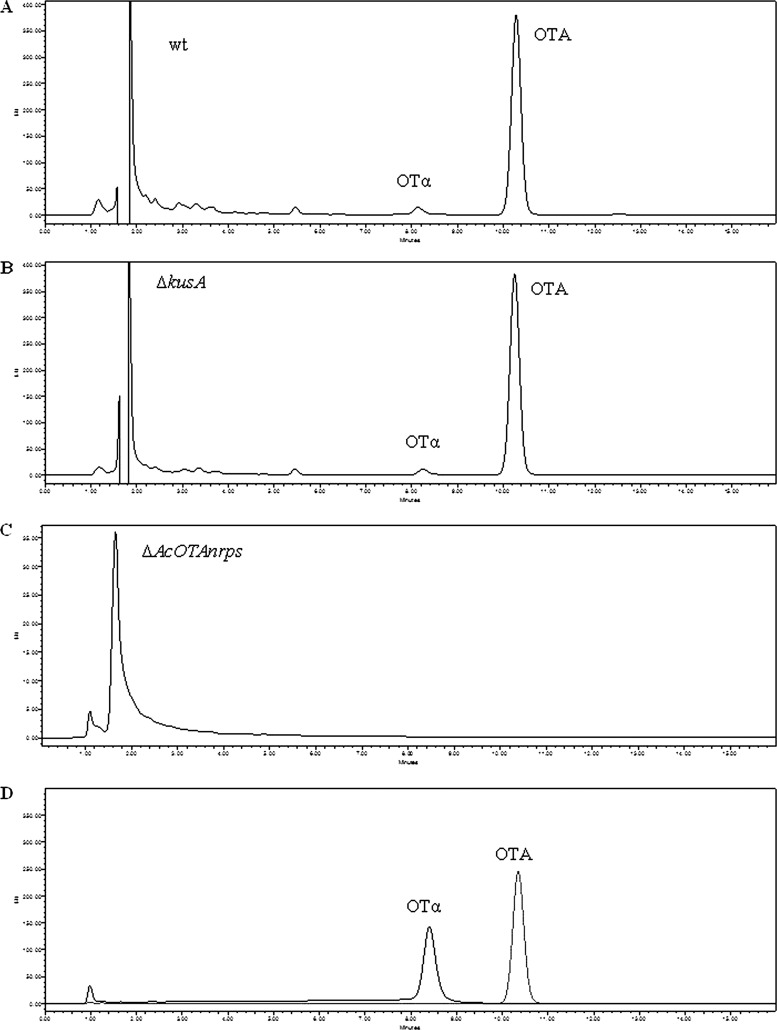
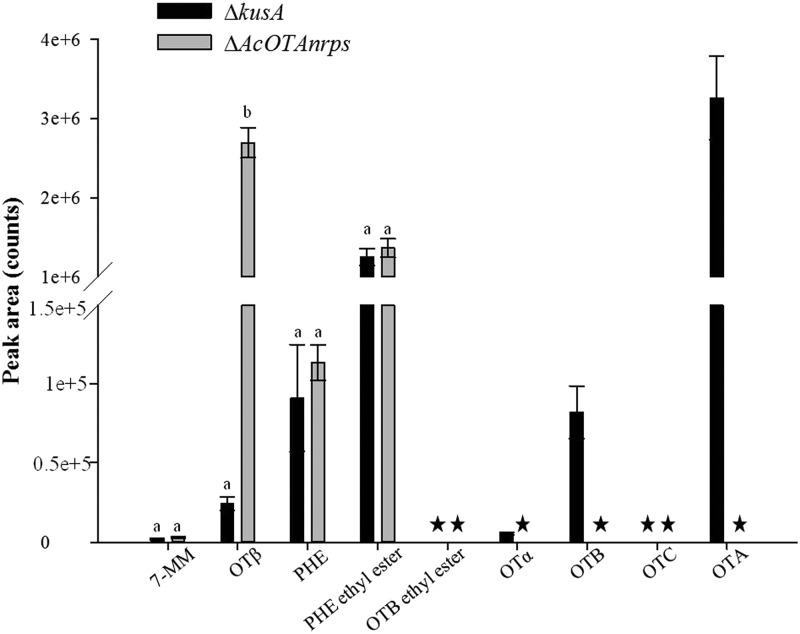
HPLC-FLD analyses showed that OTA and OTα were detected in the cultures of wild-type and KB1039 strains, the latter with the kusA gene deleted (Fig. 3A and B). In particular, after 7 days, mean values of OTA and OTα in the wild-type strain culture were 66.7 and 3.2 ng/ml, respectively. In the strain KB1039 lacking in the kusA gene, after 7 days, the mean concentrations of OTA and OTα were 54.7 and 2.8 ng/ml, respectively. No statistically significant differences were observed for OTA (P = 0.1103) and OTα (P = 0.2245) among the wild-type and ΔkusA strains. No statistically significant differences were also observed between the two strains after 14 days of growth (OTA, 79.3 and 62.6 ng/ml, P = 0.1594, and OTα, 5.1 and 4.1 ng/ml, P = 0.4663, in the wild-type and ΔkusA strains, respectively). The HPLC-FLD analyses showed the same metabolite profile for cultures of wild-type strain ITEM 5010 and mutant strain KB1039 (ΔkusA), both producing OTA and OTα. In cultures of mutant strain KB1041, lacking the kusA and AcOTAnrps genes, neither OTA nor OTα was detected, with a limit of detection (LOD) of 0.1 ng/ml. The lack of OTA production by the mutant strain KB1041 was also confirmed when liquid cultures were purified and concentrated with IMA columns, which permitted the lowering of the LOD from 0.1 ng/ml to 0.005 ng/ml (Fig. 3C). The absence of OTA and OTα in liquid cultures of KB1041 was further confirmed by HPLC-HRMS. Comparison of HPLC-HRMS metabolite profiles between mutant strains KB1039 and KB1041 in triplicate experiments showed the following major differences among these strains: OTA, OTB, and OTα which occurred in relevant amounts in the culture of mutant strain KB1039, lacking the kusA gene, were not present in mutant strain KB1041, lacking the kusA and AcOTAnrps genes. Other pieces of evidence from HRMS analysis were a clear and significant increase of OTβ (P = 0.0018) in the culture of mutant strain KB1041 compared to mutant strain KB1039 and the presence of 7-MM, Phe, and ethyl ester of Phe in cultures of both strains. Neither ethyl ester of OTA (OTC), nor ethyl ester of OTB, a hypothetical intermediary before OTB formation, was detected in either cultures (Fig. 4).
Fig 3.
HPLC-FLD chromatograms of OTA and OTα. (A) Culture medium of wild-type A. carbonarius; (B) culture medium of ΔkusA mutant KB1039; (C) culture medium of ΔAcOTAnrps mutant KB1041 purified by IMA column before HPLC-FLD analysis; (D) OTA and OTα standards. Retention times: OTA, 10.35 min; OTα, 8.41 min.
Fig 4.
Metabolites supposed to be involved in OTA biosynthesis and identified by HPLC-HRMS in cultures of ΔkusA (KB1039) and ΔAcOTAnrps (KB1041) strains incubated in triplicate for 7 days. *, not detected. Values are means ± standard errors. Different letters indicate statistical differences within pairs of data (P < 0.005).
Kinetics of OTA degradation by wild-type and ΔAcOTAnrps mutant strains of A. carbonarius.
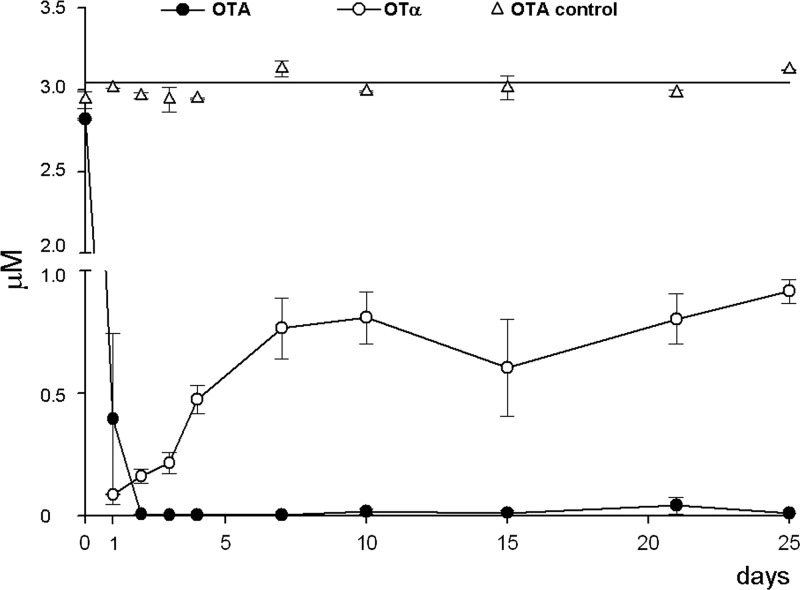
The A. carbonarius strain lacking in functional AcOTAnrps was tested for its ability to degrade OTA. To examine the kinetics of OTA degradation, toxin was added in 100 ml of MM culture (3 μM) prior to inoculum of the fungus, and the presence of OTA and OTα was monitored for 25 days in a 1-ml sample taken every day.
As shown in Fig. 5, the measured concentrations of OTA in control medium (no fungus) and at time zero of medium inoculated with fungus were the same (about 3 μM), and no OTα was detected in either culture. The initial amount of OTA measured in the medium was degraded almost completely (>99%) after 2 days of incubation. As a result of OTA degradation, OTα appeared in the medium already 1 day after the addition of OTA. The formation of OTα could be the result of OTA hydrolysis produced by the mutant strain. In particular, the OTα concentration rose from 0.09 μM after 1 day to a maximum of about 0.8 μM (27% of the OTA concentration measured at time zero) already after 7 days and was constant until the end of the experiment. In the uninoculated control, the concentration of OTA remained constant over the time course.
Fig 5.
Kinetics of OTA degradation and OTα formation by the ΔAcOTAnrps (KB1041) mutant during 25 days of incubation in minimal medium spiked with 3 μM OTA. The concentration of OTA remained unchanged in control medium over the entire duration of the experiments. Values are means ± standard errors.
DISCUSSION
Ochratoxin A is a chlorinated polyketide molecule containing the amino acid phenylalanine. According to the molecular structure of OTA, several enzymatic activities are required for the biosynthetic process, such as a polyketide synthase for the synthesis of the polyketide dihydroisocoumarin, a chlorinating enzyme, and also a peptide synthetase, which is necessary to link the phenylalanine to the dihydroisocoumarin ring. However, the order in which these enzymes act in the biosynthetic pathway has, until this study, not yet been clarified. Most studies of OTA biosynthetic genes have been focused on pks genes, which are involved in the first steps of the pathway. These pks genes have been shown experimentally to have a role in OTA biosynthesis of different fungal species of the genera Aspergillus and Penicillium (24). In Penicillium, other OTA production genes have been identified and investigated; an nrps gene, encoding a nonribosomal peptide synthetase, was identified to be responsible for the formation of the peptide bond between the polyketide dihydroisocoumarin and the amino acid phenylalanine (31).
Nonribosomal peptide synthetases are multimodular enzymes present in large numbers in the genomes of filamentous fungi. Both PKSs and NRPSs constitute a class of multifunctional proteins involved in the synthesis of a multitude of secondary metabolites with a wide range of biological activities. A great number of natural products of microbial origin are biosynthetically derived from amino acids and carboxylic acid moieties. The synthesis of these mixed peptide-polyketide natural products may be catalyzed by a hybrid NRPS-PKS system involving direct interactions between NRPS and PKS modules, or the peptide-polyketide backbone is assembled without a direct functional hybridization between NRPS and PKS proteins, which act individually (16). In particular, NRPSs have been proposed to operate via a thiotemplate mechanism in a manner independent of the ribosome. They exhibit a modular structure, with one module being a semiautonomous unit that recognizes, activates, and modifies a single residue of the final peptide by means of three typical domains: (i) the adenylation domain (A), which is responsible for substrate specificity and activation; (ii) the peptidyl carrier domain (P), which covalently binds the substrate to the enzymes via a thioester linkage; and (iii) the condensation domain (C), which catalyzes peptide bond formation (20, 50, 56). Our analyses of the recently sequenced A. carbonarius genome revealed the presence of about 13 complete nrps genes, one of which was identified as being in the OTA cluster based on homology to the predicted OTA biosynthetic cluster of A. niger (42). The gene we designated AcOTAnrps is about 900 nt upstream of a pks gene, and they are transcribed in the same direction, differently from P. nordicum, in which the OTA pks and nrps genes are transcribed in the opposite direction (31). AcOTAnrps encodes a protein (PI no. 132610) with 1,876 aa, characterized by one module with the typical three core domains and an additional last adenylation domain (see Fig. S3 in the supplemental material). Monomodular NRPS-related enzymes involved in secondary metabolites have so far not been well characterized (55), but the complete module identified in the AcOTAnrps product would be sufficient for ligation of the isocoumarin group to the amino acid phenylalanine through the carboxylic group in the OTA molecule. We confirmed this hypothesis by gene inactivation. The resulting strain of A. carbonarius lacking the nrps gene was incapable of OTA production, proving that the AcOTAnrps gene is involved in the biosynthetic pathway of OTA.
This is the first report about the identification in Aspergillus species of an nrps gene involved in OTA biosynthesis. Moreover, our attention was focused on the gene encoding a peptide synthetase with the aim of elucidating the order of the reactions in the biosynthetic pathway, which is still not completely defined. For this reason, the presence of OTα also was monitored during the analysis of OTA production by the mutant strain. Ochratoxin α is the chlorinated isocoumaric derivative of ΟΤΑ not linked to phenylalanine, which is identified naturally and usually detected in the cultures of OTA-producing species. We observed its presence parallel to the production of OTA in cultures of the A. carbonarius wild-type and ΔkusA strains analyzed after 7 and 14 days of incubation. The role of OTα in the biosynthetic pathway was not clear. According to the predominant hypothesis of Huff and Hamilton (29), it is derived from the chlorination of OTβ by a chloroperoxidase and then linked to the ethyl ester of phenylalanine via a synthetase to generate OTC, which is finally deesterified to produce OTA. Harris and Mantle (27) suggested that the conversion of OTα to OTA is the main biosynthetic pathway, with the chlorinating step being intermediate and prior to ligation of the polyketide to the phenylalanine molecule. However, their feeding experiments have shown that chlorination of OTB also can give rise to OTA, leading to an alternative pathway that entails the conversion of OTβ to OTB and then the chlorination to OTA, with the chlorinating step as the last reaction and without the direct involvement of OTα, even though the latter was demonstrated to occur naturally. One of our initial hypotheses was that the inactivation of the nrps gene, which encodes the peptide synthetase responsible of the ligation of phenylalanine moiety to the polyketide, could aid in clarifying this question. In the cultures of the ΔAcOTAnrps strain, which was unable to produce OTA, the absence of OTα was observed, contrary to the hypothesis involving chlorination of the intermediate OTα; in this hypothesis, accumulation of OTα would result from a block at the phenylalanine addition step. The absence of OTA and OTα in the cultures of the mutant strain was also confirmed by HPLC-HRMS analysis. Other related metabolites were analyzed by HRMS, and among them, the ethyl esters of OTA (OTC) and of OTB were not detected in the cultures of both producing and mutant strains (Fig. 4). This result suggests that probably phenylalanine and not its esterified form is involved in the bond with the carboxy group of OTβ. The hypothesis of esterification of phenylalanine in the biosynthesis pathway was proposed by Huff and Hamilton (29) for its predicted protective role of the phenylalanine carboxy group in the binding reaction leading to OTC. However, in their subsequent study, Harris and Mantle (27) found no evidence for the intermediate role of OTC.
Moreover, from our HRMS analysis, the absence of OTB and, at the same time, a statistically significant (P = 0.0018) increase in OTβ, accumulating in the medium as a precursor of OTB as a consequence of nrps inactivation, confirm the hypothesis that the step of ligation of phenylalanine to polyketide catalyzed by an NRPS most occurs prior to the chlorination step (Fig. 6). Furthermore, this finding is corroborated by the studies of Geisen et al. (25) on the expression kinetics of the known OTA-related genes in P. nordicum, in correlation with the OTA biosynthesis. They observed a consecutive occurrence of expression optima of the various genes, which indicated the enzymatic step of NRPS occurred before the chlorination step, with the latter step being the last reaction just before OTA excretion. According to these authors, this putative order of reactions agrees with a possible self-protection mechanism of the producing organism from the toxic OTA at the completion of biosynthesis.
Fig 6.
New schematic representation of the last steps of the OTA biosynthetic pathway as proposed in this work.
One of the historical arguments against this hypothesis of the biosynthetic pathway has been the difficulty to explain the natural occurrence of OTα in fungal cultures if it is not directly involved in biosynthesis. However, several reports have now been published about the presence of OTα as a product of OTA biodegradation by different microorganisms, among which are various Aspergillus species (2, 8, 54). The amide bond between OTα and phenylalanine in OTA is hydrolyzed by several different enzymes, including a carboxypeptidase A (15), lipase (51), commercial proteases, and as recently determined, a metalloprotease isolated in A. niger (3).
In the light of the hypothesis we present in this work regarding the OTA biosynthetic pathway, the presence of OTα in the OTA-producing A. carbonarius strain culture is most likely due to the biodegradation activity of the fungus and is independent of OTA biosynthesis. This is also confirmed by the fact that the mutant strain, lacking the AcOTAnrps gene and therefore unable to produce the mycotoxin, retains the capability to degrade OTA. Ochratoxin A was quickly and completely degraded, with only 14% remaining in culture after only 1 day and with less than 1% remaining after 2 days of incubation with exogenous OTA. OTα was detected after only 1 day, and the amount of OTα that had accumulated in culture at the end of incubation was equivalent to 27% of the initial amount of OTA, suggesting that OTA could be degraded to other metabolites in addition to OTα (23) or that OTα could be degraded further. The degradation process appeared faster and more significant than those observed in other studies in which the kinetics of nonochratoxigenic Aspergillus species were analyzed (2, 8). From the results of this experiment, it was not possible to have an indication of whether the hydrolysis activity occurred in the medium due to an enzyme excreted out of the cell or if externally applied OTA was absorbed into the cell where the degradation process would have taken place. Previous studies have indicated that OTA is a substrate for different kinds of transporters facilitating not only the efflux of compounds but also cellular uptake (49). Actually, contradictory findings have been reported about the excretion of OTA-degrading enzymes by different fungal species. Abrunhosa and Venancio (3) purified the metalloprotease responsible for OTA hydrolysis from the culture medium of A. niger, after separation of the fungal cell, whereas Peteri et al. (45) reported that the enzyme responsible of OTA degradation in Phaffia rhodozyma was not excreted, because the simple fermented broth was unable to degrade OTA. A specific OTA degradation activity of the producing strain could be supposed as the strategy of a self-protection mechanism. Also in the biosynthesis of trichothecenes by Fusaria, a mechanism of self-defense for producers was found, related to the acetylation of the trichothecene ring owing to the tri101 gene of the biosynthetic cluster (33). Additional investigation of the OTA biosynthetic cluster of A. carbonarius would be worthwhile, with the aim of finding a specific degradation gene product for a better explanation of mycotoxin biosynthesis and in view of its potential application.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by the Italian Ministry of Economy and Finance (MEF) Project, CISIA-Conoscenze Integrate per la Sostenibilità e l'Innovazione del made in Italy Agroalimentare, and by the private-public initiative Molecular Biodiversity Laboratory, partially funded by the Italian Ministry of University and Research (MIUR), Fondo FAR Legge 297/1999 Art. 12/lab project grant DM19410. The genome sequencing of Aspergillus carbonarius was conducted by the U.S. Department of Energy Joint Genome Institute and supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. We appreciate the DOE Office of the Biomass Program support for the research conducted at the Pacific Northwest National Laboratory.
We thank Filomena Epifani, Giuseppe Panzarini, and Roberto Schena for valuable technical assistance.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abouzied MM, et al. 2002. Ochratoxin A concentrations in food and feed from a region with Balkan endemic nephropathy. Food Addit. Contam. 19:755–764 [DOI] [PubMed] [Google Scholar]
- 2. Abrunhosa L, Serra R, Venancio A. 2002. Biodegradation of ochratoxin A by fungi isolate from grapes. J. Agric. Food Chem. 50:7493–7496 [DOI] [PubMed] [Google Scholar]
- 3. Abrunhosa L, Venancio EA. 2007. Isolation and purification of an enzyme hydrolyzing ochratoxin A from Aspergillus niger. Biotechnol. Lett. 29:1909–1914 [DOI] [PubMed] [Google Scholar]
- 4. Andersen MR, et al. 2011. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 21:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atoui A, Dao HP, Mathieu F, Lebrihi A. 2006. Amplification and diversity analysis of ketosynthase domains of putative polyketide synthase genes in Aspergillus ochraceus and Aspergillus carbonarius producers of ochratoxin A. Mol. Nutr. Food Res. 50:488–493 [DOI] [PubMed] [Google Scholar]
- 6. Bacha N, Atoui A, Mathieu F, Liboz T, Lebrihi A. 2009. Aspergillus westerdijkiae polyketide synthase gene “aoks1” is involved in the biosynthesis of ochratoxin A. Fungal Genet. Biol. 46:77–84 [DOI] [PubMed] [Google Scholar]
- 7. Battilani P, Magan N, Logrieco A. 2006. European research on ochratoxin A in grapes and wine. Int. J. Food Microbiol. 111(Suppl 1):S2–S4 [DOI] [PubMed] [Google Scholar]
- 8. Bejaoui H, Mathieu F, Taillandier P, Lebrihi A. 2006. Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes: a potential means of ochratoxin A decontamination in grape juice and musts. FEMS Microbiol. Lett. 255:203–208 [DOI] [PubMed] [Google Scholar]
- 9. Belli N, et al. 2004. Occurrence of ochratoxin A and toxigenic potential of fungal isolates from Spanish grapes. J. Sci. Food Agric. 84:541–546 [Google Scholar]
- 10. Botton A, et al. 2008. A cDNA-AFLP approach to study ochratoxin A production in Aspergillus carbonarius. Int. J. Food Microbiol. 127:105–115 [DOI] [PubMed] [Google Scholar]
- 11. Chiang YM, Oakley BR, Keller NP, Wang CC. 2010. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biotechnol. 86:1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cramer RA, et al. 2006. Phylogenomic analysis of non-ribosomal peptide synthetase in the genus Aspergillus. Gene 383:24–32 [DOI] [PubMed] [Google Scholar]
- 13. Crespo-Sempere A, Gonzales-Candela L, Martinez-Culebras PV. 2010. Genes differentially expressed by Aspergillus carbonarius strains under ochratoxin A producing conditions. Int. J. Food Microbiol. 142:170–179 [DOI] [PubMed] [Google Scholar]
- 14. Crespo-Sempere A, Gil JV, Martinez-Culebras PV. 2011. Proteome analysis of the fungus Aspergillus carbonarius under ochratoxin A producing conditions. Int. J. Food Microbiol. 147:162–169 [DOI] [PubMed] [Google Scholar]
- 15. Deberghes P, et al. 1995. Detoxification of ochratoxin A, a food contaminant: prevention of growth of Aspergillus ochraceus and its production of ochratoxin A. Mycotox. Res. 11:37–47 [DOI] [PubMed] [Google Scholar]
- 16. Du L, Sanchez C, Shen B. 2001. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 3:78–95 [DOI] [PubMed] [Google Scholar]
- 17. EFSA 2006. Opinion of the scientific panel on contaminants in the food chain on a request from the commission related to ochratoxin A in food. EFSA J. 365:1–56 [Google Scholar]
- 18. EFSA Panel on Contaminants in the Food Chain 2010. Statement on recent scientific information on the toxicity of ochratoxin A. EFSA J. 8:1626 doi:10.293/j.efsa.2010.1626 [Google Scholar]
- 19. FAO/WHO 2001. Safety evaluation of certain mycotoxins in food. Fifty-Sixth Meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA)—WHO Food Additives series 47. FAO food and nutrition paper 74. Food and Agriculture Organization, World Health Organization, Geneva, Switzerland [Google Scholar]
- 20. Finking R, Marahiel MA. 2004. Biosynthesis of non ribosomal peptides. Annu. Rev. Microbiol. 58:453–488 [DOI] [PubMed] [Google Scholar]
- 21. Frisvad JC, et al. 2011. Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS One 6:e23496 doi:10.1371/journal.pone.0023496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallo A, et al. 2009. Characterisation of a pks gene which is expressed during ochratoxin A production by Aspergillus carbonarius. Int. J. Food Microbiol. 129:8–15 [DOI] [PubMed] [Google Scholar]
- 23. Galtier P, Alvineire M. 1976. In vitro transformation of ochratoxin A by animal microbioal floras. Ann. Rech. Vet. 7:91–98 [PubMed] [Google Scholar]
- 24. Geisen R, Schmidt-Heydt M. 2009. Physiological and molecular aspects of ochratoxin A biosynthesis, p 353–376 In Anke T, Weber D. (ed), Physiology and genetics. The Mycota XV. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 25. Geisen R, Schmidt-Heydt M, Karolewiez A. 2006. A gene cluster of the ochratoxin A biosynthetic genes in Penicillium. Mycotox. Res. 22:134–141 [DOI] [PubMed] [Google Scholar]
- 26. Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris JP, Mantle PG. 2001. Biosynthesis of ochratoxins by Aspergillus ochraceus. Phytochemistry 558:709–716 [DOI] [PubMed] [Google Scholar]
- 28. Hu Q, et al. 2005. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40:430–443 [DOI] [PubMed] [Google Scholar]
- 29. Huff WE, Hamilton PB. 1979. Mycotoxins—their biosynthesis in fungi: ochratoxins—metabolites of combined pathways. J. Food Prot. 42:815–820 [DOI] [PubMed] [Google Scholar]
- 30. IARC 1993. Some naturally occurring substances, food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC monographs on the evaluation of carcinogenic risks to humans, vol. 56 IARC, Lyon, France [Google Scholar]
- 31. Karolewiez A, Geisen R. 2005. Cloning a part of the ochratoxin A biosynthetic gene cluster of Penicillium nordicum and characterization of the ochratoxin polyketide synthase gene. Syst. Appl. Microbiol. 28:588–595 [DOI] [PubMed] [Google Scholar]
- 32. Keller NP, Turner G, Bennet JW. 2005. Fungal secondary metabolism-from biochemistry to genomics. Nat. Rev. Microbiol. 3:937–947 [DOI] [PubMed] [Google Scholar]
- 33. Kimura M, Shingu Y, Yoneyama K, Yamaguchi I. 1998. Features of Tri101, the trichothecene 3-O-acetyltransferase gene, related to the self defense mechanism in Fusarium graminearum. Biosci. Biotechnol. Biochem. 62:1033–1036 [DOI] [PubMed] [Google Scholar]
- 34. Lattanzio VMT, Solfrizzo M, Visconti A. 2007. Simultaneous determination of aflatoxins, ochratoxin A and Fusarium toxins in maize by liquid chromatography/tandem mass spectrometry after multitoxin immunoaffinity cleanup. Rapid Commun. Mass Spectrom. 221:3253–3261 [DOI] [PubMed] [Google Scholar]
- 35. Lund F, Frisvad JC. 2003. Penicillium verrucosum in cereals indicates production of ochratoxin A. J. Appl. Microbiol. 95:1117–1123 [DOI] [PubMed] [Google Scholar]
- 36. Magnoli C, et al. 2007. Ochratoxin A and Aspergillus section Nigri in peanut seeds at different months of storage in Cordoba, Argentina. Int. J. Food Microbiol. 119:213–218 [DOI] [PubMed] [Google Scholar]
- 37. Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Martinez-Culebras PV, et al. 2009. Molecular characterization of the black Aspergillus isolates responsible for ochratoxin A contamination in grapes and wine in relation to taxonomy of Aspergillus section Nigri. Int. J. Food Microbiol. 132:33–41 [DOI] [PubMed] [Google Scholar]
- 39. Moss MO. 1996. Mode of formation of ochratoxin A. Food Addit. Contam. 13:5–9 [PubMed] [Google Scholar]
- 40. Moss MO. 1998. Recent studies of mycotoxins. Symp. Ser. Soc. Appl. Microbiol. 84:62S–76S [DOI] [PubMed] [Google Scholar]
- 41. O'Callaghan J, Caddick MX, Dobson ADW. 2003. A polyketide synthase gene required for ochratoxin A biosynthesis in Aspergillus ochraceus. Microbiology 149:3485–3491 [DOI] [PubMed] [Google Scholar]
- 42. Pel HJ, et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221–231 [DOI] [PubMed] [Google Scholar]
- 43. Perrone G, et al. 2006. Ochratoxin A production and amplified fragment length polymorphism analysis of Aspergillus carbonarius, Aspergillus tubingensis, and Aspergillus niger strains isolated from grapes in Italy. Appl. Environ. Microbiol. 72:680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perrone G, et al. 2007. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 59:53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peteri Z, Téren J, Vagvölgyi C, Varga J. 2007. Ochratoxin degradation and adsorption caused by astaxanthin-producing yeasts. Food Microbiol. 24:205–210 [DOI] [PubMed] [Google Scholar]
- 46. Petzinger E, Ziegler K. 2000. Ochratoxin A from a toxicological perspective. J. Vet. Pharmacol. Ther. 23:91–98 [DOI] [PubMed] [Google Scholar]
- 47. Pfohl-Leszkowicz A, Manderville RA. 2007. Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 51:61–99 [DOI] [PubMed] [Google Scholar]
- 48. Sanchez-Hervas M, Gil JV, Bisbal F, Ramon D, Martinez-Culebras PV. 2008. Mycobiota and mycotoxin producing fungi from cocoa beans. Int. J. Food Microbiol. 125:336–340 [DOI] [PubMed] [Google Scholar]
- 49. Schrickx J, Lekatarau Y, Fink-Gremmels J. 2006. Ochratoxin A secretion by ATP-dependent transporters in Caco-2 cells. Arch. Toxicol. 80:243–249 [DOI] [PubMed] [Google Scholar]
- 50. Schwarzer D, Finking R, Marahiel MA. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275–287 [DOI] [PubMed] [Google Scholar]
- 51. Stander MA, Bornscheuer UT, Henke E, Steyn PS. 2000. Screening of commercial hydrolases for the degradation of ochratoxin A. J. Agric. Food Chem. 48:5736–5739 [DOI] [PubMed] [Google Scholar]
- 52. Taniwaki MH, Pitt JI, Teixeira AA, Iamanaka BT. 2003. The source of ochratoxin A in Brazilian coffee and its formation in relation to processing methods. Int. J. Food Microbiol. 82:173–179 [DOI] [PubMed] [Google Scholar]
- 53. Van der Merwe KJ, Steyn PS, Fourie L. 1965. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 205:1112–1113 [DOI] [PubMed] [Google Scholar]
- 54. Varga J, Rigo K, Teren J. 2000. Degradation of ochratoxin A by Aspergillus species. Int. J. Food Microbiol. 59:1–7 [DOI] [PubMed] [Google Scholar]
- 55. von Dohren H. 2009. A survey of nonribosomal peptide synthetase (NRPS) genes in Aspergillus nidulans. Fungal Genet. Biol. 46:S45–S52 [DOI] [PubMed] [Google Scholar]
- 56. Weber T, Marahiel MA. 2001. Exploring the domain structure of modular nonribosomal peptide synthetase. Structure 9:R3–R9 [DOI] [PubMed] [Google Scholar]
- 57. Xiao H, Marquard RR, Frohlich AA, Ling YZ. 1995. Synthesis and structural elucidation of analogues of ochratoxin A. J. Agric. Food Chem. 43:524–530 [Google Scholar]
- 58. Yu JH, et al. 2004. Double joint PCR: a PCR based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.