Abstract
The genus Clavibacter comprises one species and five subspecies of plant-pathogenic bacteria, four of which are classified as quarantine organisms due to the high economic threat they pose. Clavibacter michiganensis subsp. michiganensis is one of the most important pathogens of tomato, but the recommended diagnostic tools are not satisfactory due to false-negative and/or -positive results. To provide a robust analysis of the genetic relatedness among a worldwide collection of C. michiganensis subsp. michiganensis strains, relatives (strains from the four other C. michiganensis subspecies), and nonpathogenic Clavibacter-like strains isolated from tomato, we performed multilocus sequence-based analysis and typing (MLSA and MLST) based on six housekeeping genes (atpD, dnaK, gyrB, ppK, recA, and rpoB). We compared this “framework” with phenotypic and genotypic characteristics such as pathogenicity on tomato, reaction to two antisera by immunofluorescence and to five PCR identification tests, and the presence of four genes encoding the main C. michiganensis subsp. michiganensis pathogenicity determinants. We showed that C. michiganensis subsp. michiganensis is monophyletic and is distinct from its closest taxonomic neighbors. The nonpathogenic Clavibacter-like strains were identified as C. michiganensis using 16S rRNA gene sequencing. These strains, while cross-reacting with C. michiganensis subsp. michiganensis identification tools, are phylogenetically distinct from the pathogenic strains but belong to the C. michiganensis clade. C. michiganensis subsp. michiganensis clonal complexes linked strains from highly diverse geographical origins and also strains isolated over long periods of time in the same location. This illustrates the importance of seed transmission in the worldwide dispersion of this pathogen and its survival and adaptation abilities in a new environment once introduced.
INTRODUCTION
Management of bacterial plant diseases is usually restricted to prophylaxis, given the limited and inefficient chemical options available. As numerous plant-pathogenic bacteria are seed borne, control methods are essentially based on seed health testing, which also allows for control of quarantine organisms. Strict seed sanitation measures are thus required to control the introduction and spread of pathogens in disease-free areas. The diagnostic procedures are based on detection and identification tools that have to be specific, sensitive, and fast. Specificity is one of the key factors in the design of identification tools, because it plays in concert with sensitivity. Increasing the specificity of identification tools calls for a thorough knowledge of target diversity. Hence, a well-established phylogeny is often a prerequisite for the setup of powerful identification tests.
Clavibacter michiganensis subsp. michiganensis is the causal agent of bacterial canker of tomato (Solanum lycopersicum). It is one of the most important bacterial pathogens in tomato-producing areas worldwide. The economic threat posed by this disease and the difficulties encountered in attempts to control its spread have led to the inclusion of this pathogen in the quarantine list in the European Union (19; http://www.eppo.org/QUARANTINE/listA2.htm) as well as in other countries (18). This pathogen is seed borne, and infected seeds are often considered the major inoculum source for long-distance spread and the cause of bacterial canker outbreaks (13, 29, 76). Therefore, extensive efforts have been made to develop sensitive and reliable assays for detection of C. michiganensis subsp. michiganensis in tomato seeds and plants (7, 29, 38). The detection methods currently recommended (19; http://www.worldseed.org/cms/medias/file/TradeIssues/PhytosanitaryMatters/SeedHealthTesting/ISHI-Veg/Tomato_Cmm_010811.pdf) for seed health assays are based on plating seed extracts on semiselective media, selecting strains based on colony morphology, and using confirmatory identification tools such as pathogenicity tests, PCR with specific primers (17, 67), and immunofluorescence (23). Other detection assays include the enzyme-linked immunosorbent assay (ELISA) (17), immunomagnetic separation (14), and bio-PCR (29). Tests aimed at identifying the presence of the four main pathogenicity determinants, CelA, ChpC, Pat-1, and PpaA, of C. michiganensis subsp. michiganensis were also designed. The chpC and ppaA genes are located in the chp subregion of the chromosomal pathogenicity island (PI) of C. michiganensis subsp. michiganensis. chpC encodes a protease belonging to serine protease family S1A, and ppaA encodes a chymotrypsin-related serine protease (40, 41). The two plasmid-borne genes celA and pat-1 encode an endo-β-1,4-glucanase and a serine protease of the chymotrypsine type, respectively (17, 35).
The current identification methods for C. michiganensis subsp. michiganensis produce unreliable results for some strains compared to pathogenicity assays. False-negative results have been published and arose from the use of these different tools (17, 29, 37, 40, 44). For example, two tomato-virulent strains exhibiting an atypical dry colony phenotype on solid media did not react with an otherwise highly specific monoclonal antibody (MAb) (Cmm1; 38). Southern hybridization and PCR of the pat-1 gene coding for a serine protease required for pathogenicity also led to false-negative results (37, 40). False-positive results have been reported using ELISA and phage typing and were due to cross-reactions with both nonvirulent C. michiganensis subsp. michiganensis strains and other coryneform bacteria (17). PCR-based detection of two genes encoding determinants of C. michiganensis subsp. michiganensis pathogenicity that are plasmid borne or located on the chromosome (celA and ppaA, respectively) identified all tested pathogenic strains but also all six or one of the six nonpathogenic tested strains, respectively (40).
Nonvirulent counterparts to virulent strains exist for many plant-pathogenic bacteria, as for human pathogens (75). Nonvirulent strains of C. michiganensis subsp. michiganensis arose in natural populations sampled in greenhouses and may have two causes. The existence of nonvirulent strains of C. michiganensis subsp. michiganensis was first linked to the instability of plasmids encoding virulence genes that are required for symptom expression. The two main plasmids of C. michiganensis subsp. michiganensis (pCM1 and pCM2) can be lost under stress conditions (i.e., temperature above 30°C), leading to reduced virulence or nonvirulent phenotypes (54). Bacteria are still able to colonize tomato but are unable to cause disease as a consequence of the loss of celA and/or pat-1. The other explanation is that partial deletion of the chromosomal pathogenicity island of C. michiganensis subsp. michiganensis may cause the emergence of avirulent strains in the field. In this case, bacteria are poor colonizers and although the strains carry the plasmid-borne pathogenicity genes, their in planta population sizes are too low to induce disease (40).
Elucidation of population genetics and taxonomic and evolutionary relatedness among strains is based on the analysis of multiple core genes and the clustering patterns of the strains (26, 32). These approaches, called multilocus sequence analysis (MLSA) and multilocus sequence typing (MLST), have been successfully used to revisit the taxonomy of very closely related strains (1, 7, 31, 48, 59) or more distantly related strains (7, 50, 58). Both methods depend on the sequencing of multiple (usually four to eight) housekeeping genes, i.e., genes conferring a basic metabolic function. MLSA relies on the comparison of partial DNA sequences of each gene or of concatenated sequences among strains, while MLST is based on the analysis of the combination of alleles at each locus, defining a sequence type (ST). MLSA provides a framework for species definition and allows the identification of species by electronic taxonomy (8), while MLST usually allows strains to be distinguished below the species level. MLSA and MLST studies have been conducted on diverse Gram-negative and Gram-positive bacteria; however, no full MLSA study on any Gram-positive plant-pathogenic bacteria has been published so far.
The objectives of this study were to analyze the genetic relatedness among the members of a worldwide collection of C. michiganensis subsp. michiganensis strains, their relatives (strains from the four other C. michiganensis subspecies), and nonpathogenic Clavibacter-like strains isolated from tomato and to compare this “framework” with the phenotypic characteristics of the strains. This allowed us to evaluate the specificity and the sensitivity of the currently recommended identification methods for C. michiganensis subsp. michiganensis. We characterized about 200 strains for their pathogenicity on tomato and their reaction to two antisera by immunofluorescence and to five PCR tests designed for the detection and identification of this pathogen. We designed an MLSA scheme for six housekeeping genes (atpD, dnaK, gyrB, ppK, recA, and rpoB). Using a representative subcollection of 88 C. michiganensis subsp. michiganensis strains and phylogenetically related taxa, we evaluated its robustness with respect to defining phylogenetic clusters and unraveling the molecular evolution of strains. We used MLST to decipher genetic relationships among strains in order to gain knowledge on C. michiganensis subsp. michiganensis epidemiology.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Two collections of strains were used in this study. The first collection (the A-collection) is made of 141 C. michiganensis subsp. michiganensis and 56 Clavibacter-like strains isolated from tomato seeds and plants (total of 197 strains) that were provided by the French Collection of Plant-associated Bacteria (CFBP), the National Collection of Plant Pathogenic Bacteria (United Kingdom), Anses-LSV (France), GEVES (France), Naktuinbouw (The Netherlands), NL Plant Directorate (The Netherlands), ILVO (Belgium), Syngenta, Enza Zaden, Anne Alvarez's laboratory collection (United States), Vilmorin, Rijk Zwaan, and DGBBG (Belgium). The list of strains and their descriptions are available upon request. For this study, these strains were thoroughly characterized for pathogenicity by immunofluorescence and by PCR diagnostic tests.
A second collection (the B-collection) was established for a thorough phylogenetic characterization of some strains of the A-collection. This B-collection (Table 1) contained (i) 69 C. michiganensis subsp. michiganensis strains representing diversity in terms of geographical origin, host of isolation, pathogenicity, and reactions to the above-mentioned identification tests; (ii) 6 nonpathogenic Clavibacter-like strains isolated from tomato seeds, illustrating the various types of cross-reactions with detection tools that we observed; (iii) 1 Clavibacter-like strain isolated from maize to be identified, and (iv) a set of 12 strains representing the other four subspecies of C. michiganensis. One outgroup strain (Rathayibacter iranicus) was added to this selection to root phylogenetic trees. Strains from the B-collection are all deposited at the CIRM-CFBP (International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; http://www.angers.inra.fr/cfbp), and other strains are maintained in the laboratory collection of GEVES. Cultures were stored after lyophilization and/or in a −80°C freezer in 40% glycerol. They were checked for purity and routinely cultivated on YPGA (yeast extract, 7 g liter−1; peptone, 7 g liter−1; glucose, 7 g liter−1; agar, 15 g liter−1, pH at 6.5) for 2 to 4 days at 28°C. Growth of each strain was monitored on SCMfast (sucrose, 10 g liter−1; K2HPO4, 2 g liter−1; KH2PO4, 0.5 g liter−1; H3BO3, 1.5 g liter−1; yeast extract, 2 g liter−1; Mg2SO4 · 7H2O, 0.25 g liter−1; agar, 18 g liter−1; trimethoprim, 80 mg liter−1) and CMM1tris (sucrose, 10 g liter−1; Trizma base, 3.32 g liter−1; Tris HCl, 11.44 g liter−1; Mg2SO4 · 7H2O, 0.25 g liter−1; LiCl, 5 g liter−1; yeast extract, 2 g liter−1; NH4Cl, 1 g liter−1; casein hydrolysate, 4 g liter−1; agar, 15 g liter−1).
Table 1.
List of the 88 strains from the B-collection used in phylogenetic analysis and results of their polyphasic characterizationa
| CFBP code | C. michiganensis subsp. | Country of origin | Host | Yr of isolation | Plant pathogenicityb | IFc |
Result of PCR-based tests for indicated primer(s)d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRI | Loewe | ZTO 55/56 | Cmm F/R | PSA 4/R | PSA R/8 | Ptssk | ppaA F/R | chpC F/R | PCF 3/5 | P 5/6 | ||||||
| 5 | michiganensis | France | Solanum lycopersicum | 1956 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1460 | michiganensis | France | S. lycopersicum | 1974 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1461 | michiganensis | France | S. lycopersicum | 1974 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1462 | michiganensis | France | S. lycopersicum | 1975 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1463 | michiganensis | France | S. lycopersicum | 1975 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1464 | michiganensis | France | S. lycopersicum | 1975 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1465 | michiganensis | France | S. lycopersicum | 1975 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1714 | michiganensis | France | S. lycopersicum | 1975 | + | + | + | + | + | + | + | + | + | + | + | + |
| 1940 | michiganensis | Spain | S. lycopersicum | 1978 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2108 | michiganensis | France | S. lycopersicum | 1981 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2492 | michiganensis | Algeria | S. lycopersicum | 1985 | +/− | + | + | + | + | + | + | + | + | + | + | + |
| 2493 | michiganensis | Algeria | S. lycopersicum | 1985 | + | + | + | + | + | + | + | + | + | + | + | − |
| 2494 | michiganensis | Algeria | S. lycopersicum | 1984 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2495 | michiganensis | Algeria | S. lycopersicum | 1985 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2496 | michiganensis | Algeria | S. lycopersicum | 1978 | + | + | + | + | + | + | + | + | + | + | + | − |
| 2497 | michiganensis | Algeria | S. lycopersicum | 1978 | +/− | + | + | + | + | + | + | + | + | + | + | − |
| 2498 | michiganensis | Algeria | S. lycopersicum | 1984 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2499 | michiganensis | Algeria | S. nigrum | 1984 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2500 | michiganensis | Algeria | S. nigrum | 1984 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2501 | michiganensis | Algeria | Capsicum annum | 1984 | + | + | + | + | + | + | + | + | + | + | + | − |
| 4999T | michiganensis | Hungary | S. lycopersicum | 1957 | + | + | + | + | + | + | + | + | + | + | + | + |
| 5842 | michiganensis | Brazil | C. annum | 1993 | + | + | + | + | + | + | + | + | + | + | + | + |
| 5843 | michiganensis | Brazil | S. lycopersicum | 1994 | + | + | + | + | + | + | + | + | + | + | − | + |
| 6885 | michiganensis | France | S. lycopersicum | 2004 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7158 | michiganensis | New Zealand | S. lycopersicum | 1968 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7309 | michiganensis | United States | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7310 | michiganensis | United States | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7311 | michiganensis | Morocco | S. lycopersicum | 1989 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7312 | michiganensis | China | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7313 | michiganensis | United States | S. lycopersicum | 2002 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7314 | michiganensis | United States | S. lycopersicum | 2002 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7315 | michiganensis | United States | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7316 | michiganensis | United States | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7317 | michiganensis | France | naf | 2001 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7318 | michiganensis | na | na | 2001 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7439 | michiganensis | na | na | 1992 | + | + | + | + | + | + | + | + | + | + | − | + |
| 7444 | michiganensis | na | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7449 | michiganensis | France | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7464 | michiganensis | Morocco | S. lycopersicum | na | + | + | + | + | + | + | + | + | + | + | + | + |
| 7471 | michiganensis | France | S. lycopersicum | 1997 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7478 | michiganensis | Switzerland | S. lycopersicum | 2007 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7487 | michiganensis | France | S. lycopersicum | 2008 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7488 | michiganensis | France | S. lycopersicum | 2008 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7504 | michiganensis | France | S. lycopersicum | 2009 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7506 | michiganensis | na | na | 2009 | + | + | + | + | + | + | + | + | + | + | + | − |
| 7507 | michiganensis | France | S. lycopersicum | 2010 | + | + | + | + | + | + | + | + | + | + | − | + |
| 7508 | michiganensis | France | S. lycopersicum | 2009 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7551 | michiganensis | Switzerland | S. lycopersicum | 2007 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7555 | michiganensis | Slovenia | S. lycopersicum | 2001 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7559 | michiganensis | na | na | na | + | + | + | + | + | + | + | + | + | + | + | + |
| 7562 | michiganensis | Portugal | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7567 | michiganensis | na | S. lycopersicum | na | +/− | + | + | + | + | + | + | + | + | + | + | − |
| 7568 | michiganensis | United States | S. lycopersicum | 2000 | + | + | + | + | + | + | + | + | + | + | + | − |
| 7569 | michiganensis | na | S. lycopersicum | na | +/− | + | + | + | + | + | + | + | − | − | + | + |
| 7572 | michiganensis | na | S. lycopersicum | 2002 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7574 | michiganensis | na | S. lycopersicum | na | +/− | + | + | + | + | + | + | + | + | + | + | + |
| 7578 | michiganensis | na | S. lycopersicum | na | + | + | + | + | + | + | + | + | + | + | + | − |
| 7584 | michiganensis | The Netherlands | S. lycopersicum | 2008 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7586 | michiganensis | Belgium | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7589 | michiganensis | Belgium | S. lycopersicum | 1984 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7590 | michiganensis | Taiwan | S. lycopersicum | 1988 | (+/−) | + | + | + | + | + | + | + | − | + | + | + |
| 7591 | michiganensis | Taiwan | S. lycopersicum | 1988 | +/− | + | + | + | + | + | + | + | − | + | + | + |
| 7594 | michiganensis | Belgium | S. lycopersicum | 2008 | +/− | + | + | + | + | + | + | + | + | + | + | + |
| 7597 | michiganensis | Belgium | S. lycopersicum | 2008 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7599 | michiganensis | France | S. lycopersicum | 2007 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7606 | michiganensis | The Netherlands | S. lycopersicum | 1998 | + | + | + | + | + | + | + | + | + | + | + | + |
| NCPPB1064 | michiganensis | Italy | S. lycopersicum | 1961 | + | + | + | + | + | + | + | + | + | + | + | + |
| NCPPB2034 | michiganensis | South Africa | S. lycopersicum | 1992 | +/− | + | + | + | + | + | + | + | + | + | + | + |
| NCPPB382 | michiganensis | na | S. lycopersicum | 1956 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7492 | saprophyte | India | S. lycopersicum | 2000 | − | + | + | + | + | − | + | − | − | − | − | − |
| 7495 | saprophyte | Chile | S. lycopersicum | 2007 | − | − | − | + | + | − | − | − | − | − | − | − |
| 7500 | saprophyte | na | S. lycopersicum | 1999 | − | + | + | + | + | +fe | − | − | − | − | − | − |
| 7505 | saprophyte | na | S. lycopersicum | 2009 | − | + | + | + | + | + | − | − | − | − | − | − |
| 7575 | saprophyte | na | S. lycopersicum | na | − | + | + | + | + | +f | − | − | − | − | − | − |
| 7576 | saprophyte | na | S. lycopersicum | 1997 | − | + | + | − | − | − | − | − | − | − | + | − |
| 7577 | unknown | na | Zea mays | na | − | − | + | − | − | − | − | − | − | − | − | − |
| 2404T | insidious | United States | M. sativa | 2404 | − | + | + | − | − | +f | − | − | +f | +f | + | − |
| 6488 | insidiosus | Czech Republic | Medicago sativa | 1998 | na | na | na | na | na | na | na | na | − | − | + | − |
| 6492 | insidious | Czech Republic | M. sativa | 2000 | − | + | + | − | − | +f | − | − | − | − | + | − |
| 2405T | nebraskensis | United States | Zea mays | 1971 | − | + | + | − | − | +f | − | − | − | − | − | − |
| 3521 | nebraskensis | United States | Z. mays | 1979 | na | na | na | na | na | na | na | na | − | − | − | − |
| 7553 | nebraskensis | United States | Z. mays | 1985 | − | + | + | − | − | +f | − | − | − | − | − | − |
| 2049T | sepedonicus | Canada | S. tuberosum | 1968 | − | − | + | − | − | − | − | − | − | − | − | − |
| 3559 | sepedonicus | France | S. tuberosum | 1993 | − | +ag | +a | − | − | +f | − | − | − | + | − | − |
| 3560 | sepedonicus | Argentina | S. tuberosum | 1977 | na | na | na | na | na | na | na | na | − | − | − | − |
| 3494 | tessellarius | na | Triticum aestivum | 1978 | na | na | na | na | na | na | na | na | − | − | − | − |
| 3496T | tessellarius | na | T. aestivum | 1978 | − | + | + | − | − | − | − | − | − | − | − | − |
| 3499 | tessellarius | na | T. aestivum | 1977 | na | na | na | na | na | na | na | na | − | − | − | − |
| 807 | Rathayibacter iranicus | Iran | Triticum aestivum | 1966 | na | na | na | na | na | na | na | na | − | − | − | − |
Strains were tested for pathogenicity on tomato, reactions to specific antisera in immunofluorescence, and production of specific signals with four PCR-based identification tests and four tests allowing the amplification of pathogenicity determinants.
Pathogenicity on tomato plants using ISF-recommended test. +, strain-induced canker and wilting of the plant parts above the area of inoculation; +/−, the strain induced a canker without any associated wilting of the plant parts above the area of inoculation; (+/−), tiny canker and no wilting of the plant parts above the area of inoculation; −, no visible reaction.
Results of immunofluorescence (IF) reaction tests run with two antisera.
Primer codes (reference, source, or associated gene): ZTO 55/56 (62); Cmm F/R (Van Betteray, unpublished); PSA 4/R (59); PSA R/8 (cf. ISF method); Ptssk (6); Chpc F/R, chpC; ppaA F/R, ppaA; PFC3/5, celA CB domain; P5/P6, pat-1 (42).
f, faint bands that upon sequencing corresponded exactly to the target sequences.
na, not available.
a, atypical reaction.
Pathogenicity and HR tests.
The pathogenicity of strains was tested in accordance with the International Seed Federation (ISF) method (http://www.worldseed.org/cms/medias/file/TradeIssues/PhytosanitaryMatters/SeedHealthTesting/ISHI-Veg/Tomato_Cmm_010811.pdf). Briefly, tomatoes (cv. Marmande) at the two-leaf stage were inoculated by piercing the main stem between the cotyledons and the first true leaf with a toothpick dipped in a fresh colony. Plants were incubated in a growth chamber at 28°C during 21 days. Strains inducing canker and wilting were considered fully pathogenic and were scored positive (+). Strains inducing canker at the inoculation point without any wilting of the plant parts above it were considered pathogenic but were differentiated from the former ones and scored doubtful (±). Strains with an absence of symptoms 21 days after inoculation was scored negative (−). Three plants were inoculated per strain. The C. michiganensis subsp. michiganensis type strain (CFBP4999) was used as the positive control and water as the negative control. The pathogenicity was tested twice independently. The same pathogenicity test was done on tomatoes of cv. Heinz and cv. Amely for strains of the B-collection that did not induce any symptoms on tomato cv. Marmande. These strains were also tested twice for a hypersensitive response (HR) on tobacco (Nicotiana benthamiana and N. tabaccum) leaves using the classical blunt-end syringe method and a 1 × 108 CFU/ml bacterial suspension in sterile distilled water. Plant inoculations were carried out under conditions of quarantine regulation at GEVES, Angers, France.
Immunological assays.
Immunofluorescence analysis was performed with two anti-C. michiganensis subsp. michiganensis polyclonal antibodies (Plant Research International, Wageningen, The Netherlands, and Loewe). Strain suspensions adjusted to 1 × 106 and 1 × 105 CFU/ml in sterile distilled water were deposited on 40-μl glass slide wells and fixed with alcohol. Immunofluorescence was performed using the French reference method (3).
DNA extraction.
Suspensions made from fresh cultures (overnight growth at 28°C under conditions of agitation in YP broth [yeast extract, 7 g liter−1; peptone, 7 g liter−1; pH 7.2]) were used for DNA extraction using a DNeasy blood and tissue kit (Qiagen). The quality and quantity of DNA were spectrophotometrically evaluated and adjusted (Nanodrop ND-100; Nanodrop Technologies). The extracted DNAs were then separated into aliquots and stored at −20°C.
PCR-based assays.
The PCR primers for C. michiganensis subsp. michiganensis identification were ZTO55/56 (A. Rijlaarsdam, B. Woudt, G. Simons, H. Koenraadt, J. Oosterhof, M. Asma, P. Buddiger, P. Roorda, V. Grimault, and J. de Koning, presented at the EPPO Meeting, Noordwijkerhout, Netherlands, 19 to 22 April 2004), Cmm F/R (B. Van Betteray, unpublished data), PSA 4/R (61), PSA R/8 (a modification of PSA 4/R), and Ptssk (S. M. H. Berendsen, H. Koenraadt, B. Woudt, and J. Oosterhof, presented at the APS-IPPC Meeting, Honolulu, HI, 6 to 10 August 2011). The Cmm F/R primers (Cmm-F, TGAGCGGGAGGATGACC; Cmm-R, GGTCCTCGTGCTCACCCTGC) allowed amplification of a fragment of 380 bp by a PCR done in a 20-μl volume containing 200 μM deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 0.1 μM (each) primer, 0.05 U μl−1 of GoTaq Flexi DNA polymerase (Promega) (final concentrations), and 5 μl of a boiled bacterial suspension (1 × 107 CFU ml−1). PCR conditions were 5 min at 94°C followed by 35 cycles of 30 s at 94°C, 60 s at 60°C, and 1 min at 72°C and ended with 10 min at 72°C. The PSA R/8 primers are described in the details of the ISF method provided online (http://www.worldseed.org/cms/medias/file/TradeIssues/PhytosanitaryMatters/SeedHealthTesting/ISHI-Veg/Tomato_Cmm_010811.pdf). PSA R/8 and Ptssk primers were used following the ISF methodology. ZTO55/56 (A. Rijlaarsdam et al., EPPO Meeting, Noordwijkerhout, Netherlands) and PSA 4/R (61) were used as previously described. Primers described by Kleitman and colleagues (40) were used to detect four genes involved in C. michiganensis subsp. michiganensis pathogenicity: chpC, ppaA, celA, and pat-1. PCRs were carried out in a total volume of 20 μl containing 1× GoTaq buffer (Promega), 125 μM (each) dNTP, 0.25 μM (each) primer, 0.4 U of GoTaq polymerase, and 5 μl of DNA suspension at 4 ng μl−1. All PCRs were performed with the following cycling conditions: an initial denaturation step at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s, and a final extension step at 72°C for 5 min. Amplification products were separated on a 1.5% agarose gel in 1× Tris-acetate-EDTA (TAE).
Amplification of the 16S rRNA gene was performed using A1 (5′-GAGTTTGATCATGGCTCAG-3′) and B6 (5′-TTGCGGGACTTAACCCAACAT-3′) primers (47), which complement bases 9 to 27 and bases 1082 to 1102 (Escherichia coli 16S rRNA gene sequence numbering), respectively. PCR was performed using 50-μl reaction mixtures and 1× GoTaq buffer (Promega), 2 mM MgCl2, 225 μM dNTP, 0.64 μM (each) primer, and 1 U of GoTaq polymerase in a PE9600 thermocycler (Applied Biosystems). The amplification program included denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 1 min, and extension to 72°C for 2 min, and a final extension step at 72°C for 3 min. PCR products were sequenced with reverse and forward primers at the Biogenouest platform (Nantes, France).
Housekeeping gene sequencing.
Primers for partial sequencing of six housekeeping genes (atpD [ATP synthase β chain], dnaK [70-kDa heat shock protein], gyrB [DNA gyrase β subunit], ppk [polyphosphate kinase], recA [recombinase A], and rpoB [ARN polymerase β subunit]) were designed (Table 2) from the genomic sequence of C. michiganensis subsp. michiganensis NCPPB382 (25). In case of amplification difficulties with these primer sets, alternative primer sets were designed to amplify (i) the gyrB gene of CFBP7310, CFBP7568, and CFBP7504 on the basis of the gyrB sequences of Leifsonia xyli subsp. xyli (NC_006087), C. michiganensis subsp. michiganensis (AM711867) genomes, and the partial gyrB sequence of Curtobacterium flaccumfaciens pv. poinsettiae (AM410841), (ii) the atpD gene of CFBP807 on the basis of Leifsonia xyli subsp. xyli (NC_006087) and C. michiganensis subsp. michiganensis (AM711867) atpD sequences, and (iii) the ppk gene of CFBP807 on the basis of Leifsonia xyli subsp. xyli (NC_006087), C. michiganensis subsp. michiganensis (AM711867) genomes, and the partial ppk gene of Rathayibacter iranicus (AM410841). PCR amplifications were performed in a 50-μl reaction mixture containing 1× GoTaq buffer (Promega), 200 μM dNTP, 0.5 μM (each) primer, 0.4 U of GoTaq polymerase, and 5 ng of template genomic DNA in an Applied Biosystems thermocycler with an initial denaturation at 94°C for 5 min, 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at a gene-specific temperature (Table 2), and extension for 1 min at 72°C, and a final extension for 10 min at 72°C. Purity and yield of PCR products were checked by running an 8-μl reaction mixture in 1.2% agarose gels and poststaining using ethidium bromide. The remaining PCR products were sequenced with reverse and forward primers at the Biogenouest platform.
Table 2.
Primers for housekeeping gene amplification and sequencing
| Target | Primer code | Sequence (5′→3′) | Tma (°C) | Fragment size (bp) |
|---|---|---|---|---|
| atpD | atpdF | CGGTCTACAACGCCCTCAAGA | 60 | 697 |
| atpdR | TGCGTGAAGCGGAAGATGTTG | |||
| atpDb | atpD2F | GACATCGAGTTCCCGCAC | 55 | 1,104 |
| atpD2R | CGATGATCTCCTGGAGCTCCTTGT | |||
| dnaK | dnakF | GCTCGTGCAGTAGGAATCG | 59 | 704 |
| dnakR | CTTGGCGATCTGTCGTTCGAGAC | |||
| gyrB | gyrbF | GGGGTCGGCAGCTCCGTCGTGAAC | 60 | 909 |
| gyrbR | TGGCAGTCCTTGAGCTTGCCAG | |||
| gyrBc | gyrb2F | GGCCGCGGCATCCCGGT | 60 | 1,160 |
| gyrb2R | ACGTTGAGGATCTTGCCGCG | |||
| ppk | ppkF | GAGAACCTCATCCAGGCCCT | 60 | 604 |
| ppkR | CGAGCTTGCAGTGGGTCTTGAG | |||
| ppkb | ppk2F | GGACGAGACCGAGAACCTGATCAAG | 60 | 674 |
| ppk2R | CGGTGCCGATGTGGGAGTAGTG | |||
| recA | recaF | GACCGCGCTCGCACAGATCGACCG | 60 | 724 |
| recaR | GCCATCTTGTTCTTGGACGACCTTG | |||
| rpoB | rpobF | ACGGTGACCGACTGCTTG | 60 | 662 |
| rpobR | TCAACTCGTTCGGCTTCATCGA |
Tm, melting temperature.
A specific primer set was designed to amplify the atpD and ppk genes of CFBP807.
A specific primer set was designed to amplify the gyrB gene of CFBP7310, CFBP7568, and CFBP7504.
Sequence acquisition and alignment.
Forward and reverse nucleotide sequences were edited, assembled, translated, and aligned using Geneious Pro 4.8.5 software to obtain high-quality sequences. 16S rRNA gene sequence data were compared to those referenced in the NCBI database by using BLAST (database of November 2011) with default parameters. For housekeeping gene fragments, multiple alignments were manually edited using the BioEdit program (30). Amino acid alignments were transposed back to the nucleotide sequence level to gain a codon-based alignment (30). Sequences were concatenated following the alphabetic order of the genes, ending in a sequence of 3,633 bp (bp 1 to 561 for atpD, 562 to 1140 for dnaK, 1141 to 1884 for gyrB, 1885 to 2289 for ppk, 2290 to 2883 for recA, and 2884 to 3633 for rpoD). The GenBank accession numbers for the partial sequences used in this study are listed below.
Data analysis.
Haplotype (Hap) numbers, haplotype diversity (Hd) values (56), nucleotidic diversities (θπ and θw) (56, 79), and neutrality estimates (Tajima's D and Fu and Li's D* and F*) (24, 73) were estimated for each of the six genes for each population using DnaSP, version 4.0 (66). The neutrality estimates give information about the evolutionary forces operating on a particular gene. Under conditions of neutrality, the expected value of these estimates is “0”; for diversifying selected genes, the expected value is positive; and under conditions of purifying selection, the expected value is negative.
The Nei and Gojobori (57) method was used to evaluate the synonymous/nonsynonymous substitution (Ka/Ks) ratios by the MEGA 4.1 program. The detection of potential recombinant sequences and identification of likely parental sequences were carried out by using a set of seven nonparametric detection methods implemented in RDP version 3.38 (52): RDP (51), Geneconv (60), MaxChi (71), Chimaera (62), BootScan (53), SiScan (27), and 3Seq (9). The analysis was performed with default settings for the different detection methods, and the Bonferroni-corrected P value cutoff was set at 0.05. Recombination events were accepted when detected with at least three out seven detection methods. The Web-based service GARD (genetic algorithm for recombination detection) (42) was also used to detect and locate recombination breakpoints. Split decomposition analyses were performed with SplitsTree4 V4.6 (34), which is available at http://www.splitstree.org/, using the Neighbor-Net algorithm. Split decomposition is a parsimony method that permits a tree-like network structure if conflicting phylogeny signals are detected in the data set (34). Intragenic recombination was estimated by split decomposition of individual genes, and the total recombination (inter- and intragenic) was estimated by using the concatenated sequences.
Phylogenetic analyses were performed on individual gene sequences as well as on the data set of concatenated sequences. Strain CFBP807 of Rathayibacter iranicus was used to root trees. R. iranicus is one of the closest genera of C. michiganensis based on 16S rRNA gene sequencing (43). Neighbor-joining (NJ) trees were generated with the Neighbor program from Mega 5 (74) by using the Jukes-Cantor distance methods. The model of evolution for maximum likelihood (ML) analysis was determined using Modeltest 3.7 in Paup. Both the hierarchical likelihood ratio test (hLRT) and the standard Akaike Information Criterion (AIC) were used to evaluate the model scores. Phylogenetic trees and bootstrap values for the nucleotide and amino acid sequences of each gene fragment and of concatenated sequences were obtained by the PhyML (28) method using TOPALi program version 2.5 (55), available at http://www.topali.org/. Bootstrap analyses were done with 1,000 replicates for NJ and ML analysis. Trees were generated with Mega 5. The Shimodara-Hasegawa test (69) implemented in the DNAML program from PHYLIP (22) was used to test whether the tree topologies based on each locus fell within the same confidence limits.
MLST analysis.
Each unique sequence of a gene was assigned an allele number, and the combination of allele numbers for each isolate defined the sequence type (ST). ST were grouped into clonal complexes (CC) using eBURST V3 (72). The program identified the putative ancestral genotype, which is the ST with the most single-locus variants.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the partial sequences used in this study are as follows: for atpD, JX889733 to JX889821; for dnaK, JX889911 to JX889999; for gyrB, JX890000 to JX890088; for ppk, JX890089 to JX890177; for recA, JX890178 to JX890266; and for rpoB, JX889822 to JX889910.
RESULTS
Most of the tested identification tools showed cross-reactions with Clavibacter-like strains that were not pathogenic on tomato.
A large majority (129/141) of the C. michiganensis subsp. michiganensis strains induced both canker and wilting on tomato plants. These strains reacted positively with both antisera used, and expected signals were obtained with the five PCR-based identification tests (Table 3). Less than 10% of the C. michiganensis subsp. michiganensis strains (12 of 141 strains) induced canker only at the inoculation point, without any associated wilting of the plant parts above that point. These strains were nevertheless correctly identified based on the five PCR-based identification methods and both antisera used in immunofluorescence analysis. Only one strain (CFBP7590) induced weak symptoms on tomato: a reduced canker (less than 1 cm long) at the point of inoculation but no wilting of plant parts above that point. The presence of the bacterium was monitored in the inoculated plant, and isolations showed that the strain was confined to the main stem and was at a lower concentration than the control CFBP7572 (data not shown). CFBP7590 was not isolated from leaf petiole, indicating a poor ability to colonize the host. Based on the results of immunofluorescence and PCR tests, this strain should be identified as C. michiganensis subsp. michiganensis.
Table 3.
Characterization of a worldwide collection of 197 C. michiganensis subsp. michiganensis and Clavibacter-like strains isolated from tomato seedsa
| No. of strains | Plant pathogenicityb | IF |
Result of PCR-based tests for indicated primer(s)c |
|||||
|---|---|---|---|---|---|---|---|---|
| PRI | Loewe | ZTO 55/59 | Cmm F/R | PSA 4/R | PSA R/8 | Ptssk | ||
| 129 | + | + | + | + | + | + | + | + |
| 11 | +/− | + | + | + | + | + | + | + |
| 1 | (+/−) | + | + | + | + | + | + | + |
| 1 | − | + | + | + | + | + | − | − |
| 1 | − | + | + | + | + | − | + | − |
| 3 | − | + | + | + | + | − | − | − |
| 5 | − | + | + | + | + | + (f) | − | − |
| 2 | − | + | + | − | − | +(f) | + | − |
| 4 | − | + | + | − (1 f) | − | − | − | − |
| 4 | − | + | + | − | − | + (f) | − | − |
| 2 | − | − | + | − | − | − (1 f) | − | − |
| 2 | − | − | − | + | + | − | − | − |
| 32 | − | − | − | − | − | − (1 f) | − | − |
Clavibacter-like colonies were selected based on their ability to grow on selective media and produce typical Clavibacter-like morphology. Strains were tested for their ability to induce canker and wilting on tomato plants, to react to specific antisera in immunofluorescence, and to produce specific signals with PCR-based identification tests.
Pathogenicity on tomato plants determined using ISF-recommended test. +, strain induced canker and wilting of the plant parts above the area of inoculation, +/−: the strain induced a canker without any associated wilting of the plant parts above the area of inoculation, (+/−): tiny canker and no wilting of the plant parts above the area of inoculation.
The remaining 56 strains were isolated from tomato seeds and plant parts and presented a Clavibacter-like morphology on SCMfast and CMM1tris media (data not shown). These 56 strains did not induce any symptoms on tomato (cv. Marmande). For more than half of these Clavibacter-like strains, there were no reactions with the antisera and no signals with the identification primers. However, 25 nonpathogenic Clavibacter-like strains reacted with the antisera and/or gave signals for some of the identification primers, illustrating false-positive reactions for these tests. While they were nonpathogenic on tomato, 20 and 22 strains reacted positively with Plant Research International (PRI) and Loewe antisera, respectively. ZTO55/56 and Cmm F/R primer pairs allowed the amplification of a correct signal for 11 nonpathogenic strains (Table 3). PSA 4/R primers allowed some false-positive reactions with 15 nonpathogenic strains, but in most (13/15) cases the amplicons were not as strong as those of the positive controls, and faint signals were observed (Fig. 1). Three nonpathogenic strains gave a positive signal with PSA R/8 using Platinum Taq polymerase. No cross-reactions using Ptssk primers and any of these 56 nonpathogenic strains were monitored.
Fig 1.
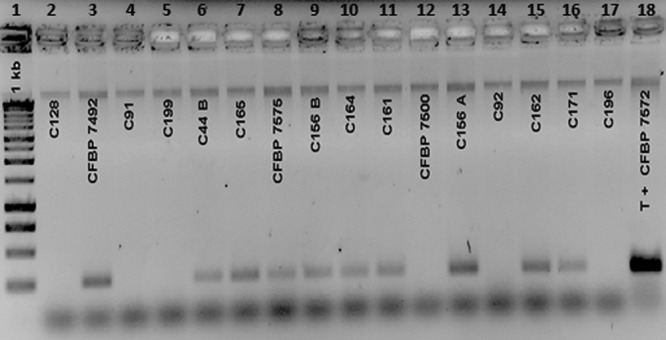
Gel photograph showing faint cross-reactions observed with PSA 4/R primers (61) for some nonpathogenic Clavibacter-like strains isolated from tomato seeds. Lane 1, 1-kb DNA ladder; lanes 2, 4, 5, 12, 14, and 17, nonpathogenic strains that did not react with PSA 4/R primers; lanes 3, 6 to 11, 13, 15, and 16, faint signals obtained for nonpathogenic strains with PSA 4/R primers. The positive control (lane 18) was DNA from CFBP7572. The three nonpathogenic CFBP7492, CFBP7500, and CFBP7500 strains were included in the B-collection.
Genes coding for the four main pathogenicity determinants were not amplified from any pathogenic C. michiganensis subsp. michiganensis strain.
Based on these results of pathogenicity and identification tests, we selected 69 C. michiganensis subsp. michiganensis strains and 6 nonpathogenic Clavibacter-like strains from tomato seeds to represent the diversity we observed. We confirmed the absence of pathogenicity of these six Clavibacter-like strains by using the same pathogenicity test on two other tomato cultivars (cv. Amely and cv. Heinz) and by the absence of HR after infiltration of N. tabaccum and N. benthamiana leaves (data not shown). These six strains did not induce any symptoms on tomatoes or any HR on tobacco; they behaved as saprophytes. The presence of genes coding the four main pathogenicity determinants was monitored for each of the 88 strains of the B-collection (Table 1). The four pathogenicity determinants (the ppaA, chpcC, celA, and pat-1 genes) were detected in the majority (51/69) of the pathogenic C. michiganensis subsp. michiganensis strains. However, the pat-1 gene was not detected in 6 pathogenic C. michiganensis subsp. michiganensis strains, and 4 strains did not harbor the celA gene. Among the pathogenic strains, 9 strains induced only canker and no wilting of the aerial plant part, and 4 of them harbored these 4 genes; 2 strains lacked pat-1, 1 lacked chpC, and the ppaA gene was not amplified in 3 of the 9 weakly aggressive strains. Among the four pathogenicity determinants, only celA was identified in 1 of the 6 saprophytes we tested. Hence, the presence of chpC and/or ppaA was a good but not absolute indicator of C. michiganensis subsp. michiganensis pathogenicity on tomato. Nevertheless, note that the chpC and ppaA genes were identified in one C. michiganensis subsp. insidiosus strain, celA in the three tested C. michiganensis subsp. insidiosus strains, and chpC in one C. michiganensis subsp. sepedonicus strain.
The identification of nonpathogenic Clavibacter-like strains isolated from tomato seeds was confirmed using partial 16S rRNA gene sequencing.
An internal fragment of the 16S rRNA gene was amplified from the 6 saprophytic strains and the Clavibacter-like strain isolated from maize (CFBP7577). Their 16S rRNA partial gene sequences shared more than 99% nucleotidic identity with C. michiganensis on 100% of query coverage for fragments 625 to 966 bp long, except for CFBP7492, for which the fragment was 433 bp long (Table 4). The Clavibacter-like strain isolated from maize (CFBP7577) was also identified as belonging to C. michiganensis (99% identity on 100% of query coverage for a 972-bp-long fragment) (Table 4).
Table 4.
Identification of unidentified isolates based on 16S rRNA gene sequencing
| Strain code | Result of NCBI Blast searches |
||||
|---|---|---|---|---|---|
| Identification | Accession code | Total score | Query coverage (%) | Maximum identification (%) | |
| CFBP7492 | C. michiganensis | HE608962.1 | 782 | 100 | 99 |
| C. michiganensis subsp. insidiosus | GQ332310.1 | 782 | 100 | 99 | |
| C. michiganensis subsp. nebraskensis | U96182.1 | 782 | 100 | 99 | |
| C. michiganensis subsp. michiganensis | HQ144242.1 | 776 | 100 | 99 | |
| CFBP7495 | C. michiganensis | HE608962.1 | 1,262 | 100 | 99 |
| C. michiganensis subsp. insidiosus | GQ332308.1 | 1,256 | 100 | 99 | |
| C. michiganensis subsp. tessellarius | AM410693.1 | 1,256 | 100 | 99 | |
| CFBP7505 | C. michiganensis | JN603288.1 | 1,371 | 100 | 99 |
| C. michiganensis subsp. insidiosus | GQ332308.1 | 1,365 | 100 | 99 | |
| C. michiganensis subsp. tessellarius | AM410693.1 | 1,365 | 100 | 99 | |
| CFBP7500 | C. michiganensis | HE608962.1 | 1,432 | 100 | 100 |
| C. michiganensis subsp. insidiosus | GQ332308.1 | 1,426 | 100 | 99 | |
| C. michiganensis subsp. tessellarius | AM410693.1 | 1,426 | 100 | 99 | |
| C. michiganensis subsp. michiganensis | JN603288.1 | 1,421 | 100 | 99 | |
| CFBP7576 | C. michiganensis | HE608962.1 | 1,375 | 100 | 100 |
| C. michiganensis subsp. insidiosus | GQ332308.1 | 1,339 | 100 | 99 | |
| C. michiganensis subsp. tessellarius | AM410693.1 | 1,339 | 100 | 99 | |
| C. michiganensis subsp. michiganensis | JN603281.1 | 1,334 | 100 | 99 | |
| CFBP7577 | C. michiganensis partial 16S | HE608962.1 | 1,784 | 100 | 100 |
| C. michiganensis subsp. nebraskensis | AM410697.1 | 1,779 | 100 | 99 | |
The C. michiganensis subsp. michiganensis subspecies was monophyletic and was phylogenetically distinct from the saprophytes.
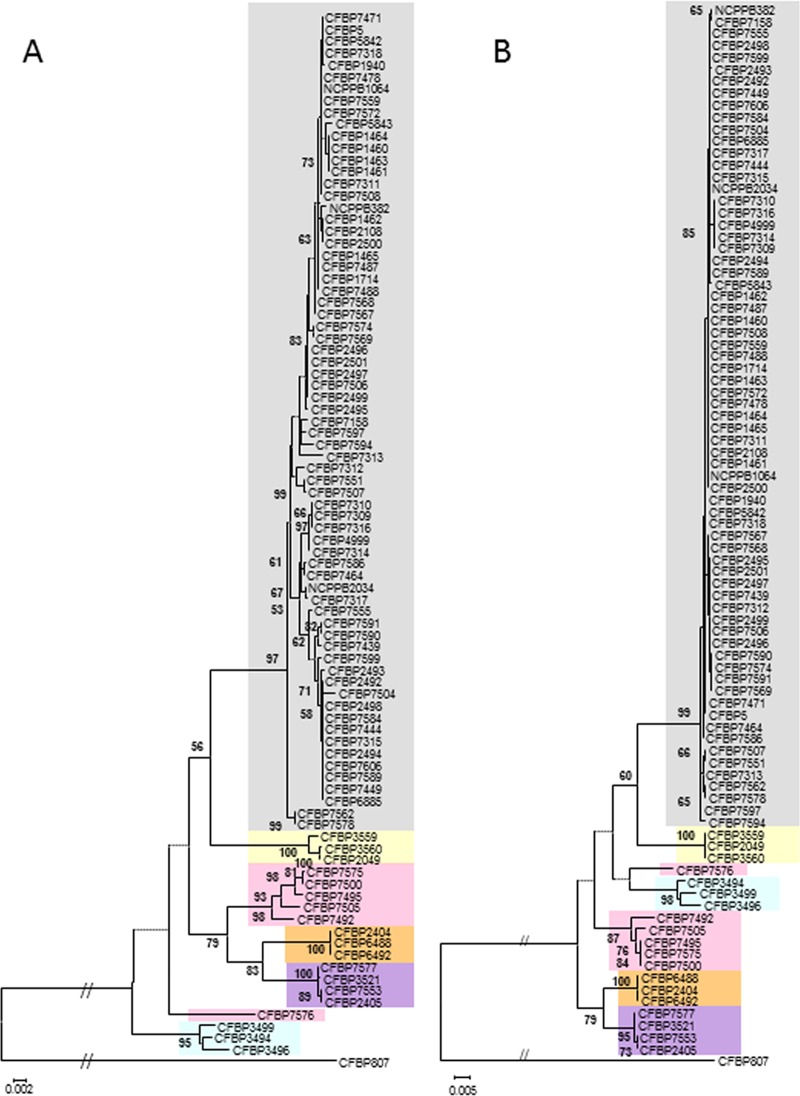
The phylogenetic tree based on the data set of concatenated sequences of six housekeeping gene fragments presented a phylogenetic history strongly supported by high bootstrap values that clearly differentiated C. michiganensis subsp. michiganensis from the four other subspecies and from the saprophytes within C. michiganensis (Fig. 2). The strains from the four subspecies were separated into four distinct clusters together with their respective type strains. Five of the six saprophytes clustered together and formed a monophyletic group. The nucleotidic distances separating all these groups were highly similar (Fig. 2), indicating that the group with the nonpathogens could be considered a new subspecies within C. michiganensis. One saprophytic strain (CFBP7576) did not cluster with the five other saprophytes; its position remains to be clarified, as it was not supported by strong bootstrap values. C. michiganensis subsp. sepedonicus strains appeared as a group closely related to C. michiganensis subsp. michiganensis. The saprophyte group was phylogenetically close to C. michiganensis subsp. insidiosus and C. michiganensis subsp. nebraskensis, while C. michiganensis subsp. tessellarius strains formed the group outlying furthest. Altogether, the genetic diversity within C. michiganensis strains was low, as indicated by the low level of polymorphism in the analyzed loci (313 polymorphic sites per 3,555 sites analyzed and an overall nucleotide diversity [θπ] of 0.01314; Table 5). The percentages of polymorphic sites were 7.66, 5.03, 12.77, 9.40, 10.77, 5.62, and 8.80 for atpD, dnaK, gyrB, ppk, recA, rpoB, and the data set of concatenated sequences, respectively.
Fig 2.
Maximum likelihood trees based on concatenated partial sequences of (A) atpD, dnaK, gyrB, ppk, recA, and rpoB and (B) gyrB and recA. Bootstrap scores (1,000 replicates) are displayed at each node. Strains from each C. michiganensis subspecies (insidiosus, michiganensis, nebraskensis, sepedonicus, and tesselarius) of C. michiganensis are highlighted in orange, gray, purple, yellow, and blue, respectively. Saprophytes are highlighted in pink.
Table 5.
Sequence variation at the six loci among strains of the B-collection (88 strains)
| Locus | No. of sitesa | GC% | Sb | Hapc | Hdd | θπe | θWf | Tajima's Dg | Fu & Li's Dg | Fu's Fg |
|---|---|---|---|---|---|---|---|---|---|---|
| atpD | 561 | 0.681 | 43 | 17 | 0.794 | 0.01153 | 0.01589 | −0.87776 | 1.63438* | 0.76536 |
| dnaK | 576 | 0.693 | 29 | 20 | 0.841 | 0.0059 | 0.00997 | −1.25588 | −1.70693 | −1.8353 |
| gyrB | 744 | 0.661 | 95 | 18 | 0.825 | 0.01785 | 0.02742 | −1.16528 | −0.04185 | −0.60965 |
| ppk | 564 | 0.688 | 53 | 21 | 0.888 | 0.01721 | 0.01896 | −0.29895 | −0.03642 | 1.028 |
| recA | 594 | 0.7 | 64 | 17 | 0.752 | 0.01786 | 0.02501 | −0.94167 | −0.14782 | −0.5668 |
| rpoB | 516 | 0.651 | 29 | 16 | 0.779 | 0.00624 | 0.01152 | −1.41461 | 0.96484 | 0.06075 |
| Concath | 3.594 | 0.679 | 313 | 48 | 0.972 | 0.01313 | 0.01872 | −1.0211 | 0.1261 | 0.855 |
Data represent the number of analyzed sites.
S, number of polymorphic sites.
Hap, number of haplotypes.
Hd, level of haplotype diversity (54).
θπ, level of nucleotide diversity (54).
θW, level of nucleotide diversity from S (78).
Data represent the results of neutrality tests performed using the method of Tajima (73) and Fu and Li (24) (Tajima's D, Fu and Li's D*, and Fu's F*) and associated P values (*, P < 0.05).
Concat, data set of concatenated sequences.
The phylogenetic trees built with the ML algorithm for each of the six loci did not all depict the same phylogenetic history (see Fig. S1 in the supplemental material). C. michiganensis subsp. michiganensis appeared as a monophyletic group strongly supported by high bootstrap values and clearly separated from the four other subspecies of C. michiganensis and saprophytes on the basis of atpD, dnaK, gyrB, recA, and rpoB partial gene sequences. Better support of the nodes was found in gyrB, recA, and rpoB trees. C. michiganensis subsp. michiganensis strains were dispersed in several groups on the basis of ppk sequences. The results of a Shimodeira-Hasegawa test performed on individual gene sequences and on the data set of concatenated sequences (Table 6) showed that all trees were significantly incongruent with each other but were not significantly different from the tree based on the data set of concatenated sequences, except for rpoB, for which the probability was at the threshold value (P = 0.046). Hence, these genes did not exhibit the same evolutionary history, but the tree based on the data set of concatenated sequences did not contradict the information brought by each gene. To define a minimal MLSA scheme, a tree based on the concatenated gyrB and recA sequences was built. This tree gave an image congruent with the previous one (Fig. 2). Note that the genes studied here were not positively selected, as indicated by the neutrality estimates (Table 5). This was confirmed by the Ka/Ks ratios for the six loci. The values ranged from 0.041 (for recA) to 0.335 (for ppk). All loci were polymorphic, and the number of polymorphic sites ranged from 29 for dnaK and rpoB, the least polymorphic loci, to 95 for gyrB, the most polymorphic locus. The number of alleles at each locus ranged from 16 for rpoB to 21 for ppk (Table 5).
Table 6.
P values determined using the Shimodaira-Hasegawa test of tree topologies run on each of the maximum likelihood trees based on the 6 loci and the data set of concatenated sequences
| Locus |
P value |
||||||
|---|---|---|---|---|---|---|---|
| atpD | dnaK | gyrB | ppk | recA | rpoB | Concata | |
| atpD | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | |
| dnaK | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| gyrB | 0.000 | 0.001 | 0.000 | 0.003 | 0.000 | 0.000 | |
| ppk | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| recA | 0.000 | 0.001 | 0.003 | 0.000 | 0.000 | 0.000 | |
| rpoB | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Concat | 0.207 | 0.371 | 0.316 | 0.220 | 0.309 | 0.046 | |
Data represent the results determined with a maximum likelihood tree designed with the data set of concatenated sequences.
Little evidence of homologous recombination was found in the data set.
To further investigate relationships among strains, we constructed phylogenetic networks using the Neighbor-Net algorithm to highlight conflicting signals in the gene sequence data, which would suggest exchange or acquisition of genetic material among C. michiganensis strains (Fig. 3). In a phylogenetic network, alternative phylogenies are represented by parallelograms. The more reticulation there is in a network, the more conflicting signals exist in the sample, possibly due to exchange of genetic material. A relevant reticulation was found connecting strains within each locus. However, ppk and dnaK clearly showed many more reticulations than the other loci. These split graphs highlighted the alleles for atpD shared between one saprophyte and 36 C. michiganensis subsp. michiganensis strains grouped in one ST (Fig. 3; see also Table S1 in the supplemental material). In a similar way, alleles for rpoB shared among the three C. insidiosus strains and one C. sepedonicus strain, and between one saprophyte and one C. michiganensis subsp. michiganensis strains, were highlighted.
Fig 3.
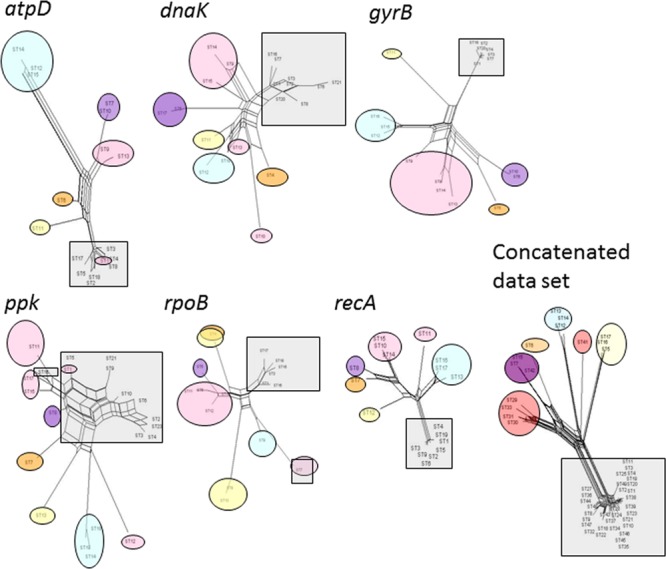
Split graphs of multilocus sequence analysis of the B collection of strains of each sequence type (ST) for the six loci (atpD, dnaK, gyrB, ppk, rpoB, and recA) and the data set of the concatenated sequences. The designation at each of the leaves indicates the ST number. See Table S1 in the supplemental material for strain designations and ST correspondences. Strains from each subspecies (insidiosus, michiganensis, nebraskensis, sepedonicus, and tesselarius) of C. michiganensis are highlighted in orange, gray, purple, yellow, and blue, respectively. Saprophytes are highlighted in pink.
The split graph built on the data set of the concatenated sequences showed very few reticulations, indicating that recombination should not have contributed predominantly to genetic evolution within C. michiganensis (Fig. 3). This was confirmed by three other tests aimed at evaluating the importance of recombination in the genetic evolution of these strains. Using GARD software, only one breakpoint (at 204 bp) was predicted within atpD. Using RDP, no significant recombination event was detected in the data set. One event was detected between positions 8 and 182 on the atpD gene fragment by 3Seq. It involved ST9 as a major parent on ST14, ST12, and ST15. These events were not confirmed by any other method in the RDP package. We then estimated the relative contributions of recombination and point mutation to sequence diversity (r/m ratio) and diversification of the population using the method of Feil et al. (21). The r/m ratio was 0.027:1 on the concatenated gene fragments. No recombination imports were detected in ppk and recA gene fragments, which contained 53 and 64 point mutations, respectively. Both dnaK and rpoB showed 1 recombination event per 27 point mutations (r/m, 0.037:1), and r/m ratios were 3:37 and 3:86 for atpD and gyrB, respectively.
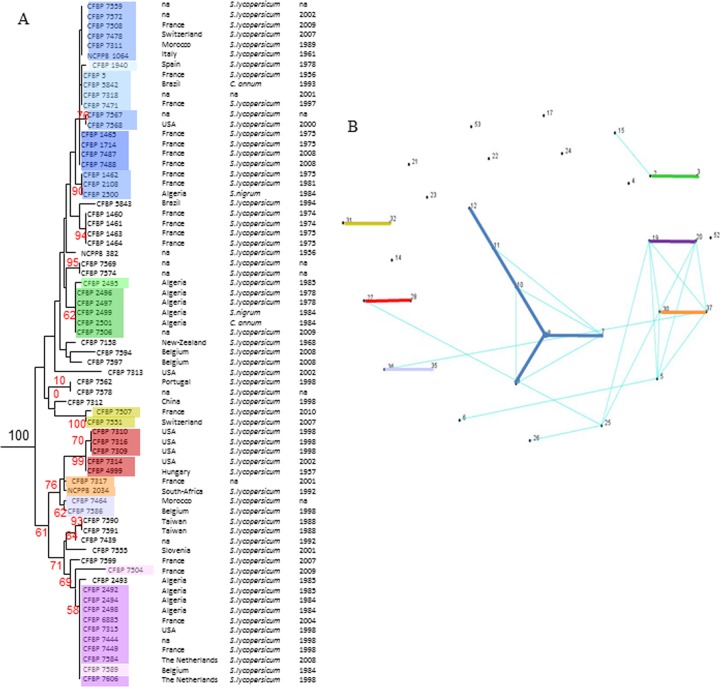
MLST analysis highlighted the relatedness of strains from different geographical origins and isolated over a long period of time.
A total of 48 sequence types (ST) was obtained for the B-collection, but only 32 concerned C. michiganensis subsp. michiganensis strains (see Table S1 in the supplemental material). The B-collection gathers 69 C. michiganensis subsp. michiganensis strains in a collection of 88 strains representing the known diversity within C. michiganensis. Thirty-three ST were represented by a single isolate, and the largest ST grouped 10 strains. Using the eBurst algorithm, we were able to identify seven clonal complexes (CC) within the C. michiganensis subsp. michiganensis strains of the B-collection. These CC linked single-locus variants. Six CC linked 2 ST, and one linked 6 ST. All other ST were singletons (Fig. 4). Altogether, these 7 CC clustered 49 strains representing 71% of the C. michiganensis subsp. michiganensis strains of the B-collection. In these CC, strains isolated in different countries, even different continents, over a long period of time were grouped. For example, in the clonal complex linking ST7-ST8-ST9-ST10-ST11 and ST12, we observed strains that were isolated in Algeria, Brazil, France, Italy, Spain, Switzerland, and the United States between 1961 and 2009 (Fig. 4).
Fig 4.
Relatedness and patterns of evolutionary descent among isolates with similar genotypes. (A) Sub-neighbor-joining tree focused on C. michiganensis subsp. michiganensis strains with place, host, and year of isolation. Rectangles of the same color cluster strains from the same ST and decreasing intensity illustrate increasing distances from the founding ST. (B) Graph from an e-Burst analysis showing ST linked by single-locus variations (plain colored lines) and double-locus variation (turquoise line).
Nearly 80% of the C. michiganensis subsp. michiganensis strains were linked into four CC according to the results of analysis of double-locus variants. The pattern of CC1 and CC6 was not changed, and that of CC2 was enlarged, with links to CC4, CC5, CC7, ST5, ST6, ST25, and ST26. CC3 was enlarged with a link to ST15. The 9 remaining ST were singletons representing 14 strains isolated in diverse locations. Only 20% of C. michiganensis subsp. michiganensis strains were considered genetically isolated.
The MLSA scheme allowed an unidentified strain to be allocated to a clade.
Strain CFBP7577 was isolated from maize and was identified as belonging to C. michiganensis based on 16S rRNA gene sequencing. Based on the sequences of the six housekeeping gene fragments, this strain clustered in the same group with the three C. michiganensis subsp. nebraskensis strains included in this study. Hence, CFBP7577 clustered with the pathotype strain of michiganensis subsp. nebraskensis (CFBP2405), which is a pathogen of maize. This strain produced acid from sorbitol but not from mannitol (data not shown).
DISCUSSION
MLSA has deciphered the phylogenetic relationships for numerous bacteria. Based on sequence analysis of several housekeeping genes, this technique has been developed for species delineation and has also been used for infraspecies phylogenies (20, 26, 31, 32, 49, 63, 82). The advantage of MLSA is that phylogenetic relationships of large sets of strains can be analyzed with better portability than any previous genotyping technique such as DNA/DNA hybridization, amplified fragment length polymorphism (AFLP), or repetitive sequence-based PCR (rep-PCR). It has also proven valuable for assessing phylogenetic relationships within complex species such as Stenotrophomonas maltophilia (36), Agrobacterium tumefaciens species complex (4, 12), and Ralstonia solanacearum (11). The MLSA scheme we designed is based on six housekeeping genes routinely used for phylogenetic studies of Gram-positive bacteria belonging to Microbacterium, Aureobacterium (63), Lactobacillus (59), and Gram-negative bacteria (20, 58, 80, 82). This MLSA scheme revealed a robust phylogenetic structure within Clavibacter, with each subspecies clustering in differentiated clades. It also highlighted an unknown part of the diversity within this genus. Indeed, five nonpathogenic C. michiganensis strains isolated from tomato seeds formed one well-defined cluster whereas another strain clustered separately and apart from the five well-known subspecies within Clavibacter. At the least, one novel subspecies should be defined within C. michiganensis that would encompass these five nonpathogenic strains. Analysis of more nonpathogenic strains should reveal if their phylogenetic diversity is high or limited to two groups. The genetic diversity within C. michiganensis was limited. Overall, the percentages of variable sites (5.0% to 12.8%) can be considered low to medium and comparable to those of X. campestris (1.8% to 10.2%) (20). They are, however, far higher than what is observed for Myxococcus xanthus (0.7% to 2.4%) (77) but considerably lower than those observed for Helicobacter pylori (19.8% to 23.7%) (45).
Incongruence between individual gene phylogenies may highlight past recombination events or different impacts of mutation on gene evolution. Split-tree analysis revealed that recombination may have preferentially affected dnaK and ppk. However, no signs of recombination could be found with the dedicated software (RDP package and Guard) for any genes. Moreover, the r/m values inferred using the data set (0.027:1) were very low relative to the range of r/m ratios so far reported for other plant-pathogenic bacteria: for X. campestris, 1.5:1 (20); and for Xylella fastidiosa, 0.46:1 (68). In many Gram-negative plant-pathogenic bacteria, recombination is thought to contribute more than mutation to the diversification of strains (10, 11, 16, 20, 70, 81). This did not seem to be the case for C. michiganensis subsp. michiganensis, for which the impact of mutation was greater than the impact of recombination. Nevertheless, a few alleles were shared between strains belonging to different subspecies (between C. michiganensis subsp. michiganensis and the nonpathogenic strain clade and between C. michiganensis subsp. insidiosus and C. michiganensis subsp. sepedonicus). These recombination events involved some nonpathogenic Clavibacter-like strains and illustrate the consequences of niche sharing for bacterial genetic evolution.
The genes gyrB and recA appeared to provide robust phylogenies that were congruent with the ones based on the data set of the concatenated sequences. These genes did not display any signs of evolution involving recombination. The analysis confirmed the results of previous studies reporting a robust use of gyrB as a phylogenetic marker in the genus Clavibacter (83) and the use of recA for differentiation of the five subspecies of C. michiganensis (78). Both the gyrB and recA loci were sufficiently discriminating to identify infraspecies sequence variation. We recommend using these two genes for accurate and robust identification of strains belonging to Clavibacter and for phylogenetic positioning of unknown strains. This strategy prevents mispositioning strains due to recombination events that may affect only part of the genome and hence be nonvisible if only one gene is used. As proposed by Bishop et al. (8), results of MLSA could be used for in silico taxonomic assignment of strains, provided the database is hosted in a dedicated website. Sequences obtained are made available through GenBank and for MLST analysis through PAMDB databases (2).
We used 16S rRNA partial gene sequencing to identify strains at the species level, as the genus Clavibacter has currently only one species. Indeed, a cutoff of 97% similarity in the 16S rRNA gene is often used to group strains into operational taxonomic units, but strains that are more than 97% similar in their 16S rRNA gene sequences can still be different species according to the DNA-DNA hybridization criterion (65). The seven Clavibacter-like strains were identified as C. michiganensis and, using the proposed MLSA scheme, positioned within the known diversity of this genus. Nonpathogenic strains formed a novel clade that is positioned between the current subspecies and that did not enlarge the boundaries of this species. Moreover, MLSA appeared to be an adequate method to allocate strains to the correct subspecies, as observed for the Clavibacter-like strain isolated from maize. This allocation corroborated biochemical characteristics of this strain known to be specific for the C. michiganensis subspecies nebraskensis: production of acid from sorbitol but not from mannitol. The colony morphology of this strain was also typical of C. michiganensis subsp. nebraskensis strains (33). Based on rep-PCR profiles, Louws et al. (44) could not differentiate avirulent C. michiganensis subsp. michiganensis strains from virulent ones; thus, MLSA provides better resolution for correct allocation of Clavibacter strains into subspecies.
The analysis of the evolutionary genetic diversity of large collections of strains requires discriminative, high-throughput techniques able to identify strain types and provide reproducible results. MLST, based on allele analysis of several housekeeping genes, has proven valuable for molecular epidemiology studies of various pathogens (5, 46, 64). Despite the worldwide distribution and the major economic importance of C. michiganensis subsp. michiganensis, a quarantine organism, no evaluation of this popular typing technique for population structure analyses had yet been conducted. We used MLST to unravel the evolutionary dynamics, i.e., the different strain evolution characteristics, within C. michiganensis subsp. michiganensis. Compared to Stenotrophomonas maltophilia, the number of alleles we obtained per locus was low (from 16 to 21 alleles on loci of 516 to 744 sites and from 38 to 53 alleles on loci of 444 to 558 sites for C. michiganensis subsp. michiganensis and S. maltophilia [36], respectively). The total number of sequence types we obtained for C. michiganensis subsp. michiganensis in this collection was limited (49 ST for 88 strains typed on 6 loci representing 3,555 sites). MLST revealed the existence of clonal groups and of singletons within C. michiganensis subsp. michiganensis. Most (70%) of the C. michiganensis subsp. michiganensis strains collected were linked by single variations. If one considers double-locus variations, the proportion of linked strains increases to 80%. Singletons usually grouped one strain. In one case, four strains composed a singleton. They were isolated in the same location in two consecutive years.
MLST provided information indicating that both seed transmission and maintenance in the environment are capacities shared by strains that are epidemiologically fit. Indeed, CC grouped strains that were epidemiologically fitter than singletons as they dispersed and were regularly isolated from outbreaks. Most ST and, obviously, the clonal groups linking ST were not related to the geographical origin of strains or the year of their isolation. In contrast, MLST revealed that genetically similar or closely related strains were isolated in different continents and over long periods of time. This information is coherent with the known main means of dispersal of this pathogen, i.e., via contaminated seeds (18, 74). The international seed trade contributed to the dispersal of epidemiologically efficient strains in the main tomato-growing areas where outbreaks occurred and from which strains were collected. Also highlighted is the importance of local inocula in disease outbreak. Isolating genetically similar strains from the same country over more than 30 years reflects the fact that these strains were propagated on susceptible hosts or survived saprophytically in the same location. Survival over such long periods of time could not be attributed only to a survival in association with seeds, because this survival duration is far longer than that of the viability of tomato seeds, which is less than 10 years. It appears that the same ST isolated over a long period of time within the same country corresponds to the dispersal of a strain after its introduction rather than to different introduction events, taking into account that, at least in Europe, this pathogen is under quarantine and hence the importation of tomato seeds in Europe is subject to inspections and analyses. Recently, a study indicated that 21 strains individually isolated from different locations in Turkey were genetically different based on inter-simple sequence repeat–PCR (ISSR-PCR), suggesting that they correspond to different introduction events (6), whereas in Canary Island, 54 strains isolated from 2002 to 2007 could not be differentiated based on randomly amplified polymorphic DNA-PCR (RAPD-PCR), BOX-PCR, and AFLP. de Leon and collaborators (15) suggested that the bacterium was introduced once into the Canary Islands from only one origin. In Japan, C. michiganensis subsp. michiganensis haplotypes were maintained over years within different greenhouses and differed among greenhouses, suggesting that they originated from the previous C. michiganensis subsp. michiganensis population in each greenhouse. It has been found that disbudding and defoliation contribute to secondary spread in greenhouse (39). It would be of interest to evaluate the robustness of the proposed MLST scheme using such collections.
While it has long been known that C. michiganensis subsp. michiganensis strains could lose or show reduced virulence along with curing of one or its two plasmids or deletion of (part of) the genetic island dedicated to pathogenicity, we show here that under natural conditions, nonpathogenic Clavibacter-like strains that were isolated from tomato niches do belong to C. michiganensis but have diverged from a common ancestor shared with the five known subspecies, forming a distinct clade. Our results confirm those of Kleitman and colleagues (40), who showed on the basis of 16S rRNA primers, Gram staining, and ELISA that the nonvirulent strains belong to C. michiganensis. The saprophytic strains cross-reacted with most of the detection tools designed to specifically detect C. michiganensis subsp. michiganensis, illustrating their proximity in various genetic fragments and surface antigens. We observed that the saprophytes differed from the pathogens from a phylogenetic point of view. None of the saprophytes shared the same ST with C. michiganensis subsp. michiganensis strains; however, some alleles were shared between saprophytes and C. michiganensis subsp. michiganensis, essentially for atpD and rpoB. These saprophytes were not pathogenic to tomato (three cultivars were tested) and did not induce any HR on N. benthamiana or N. tabaccum, and most of them generally did not harbor determinants of C. michiganensis subsp. michiganensis pathogenicity; hence, we consider these results to be demonstrations of a lack of pathogenic abilities. It should be emphasized that the genus Clavibacter encompasses plant-pathogenic bacteria and also bacteria that share the same habitat as pathogens but have engaged in a saprophytic lifestyle.
Some C. michiganensis subsp. michiganensis isolates induced only canker and no wilting of the plant part above the inoculation point. This was due to poor colonization abilities, which limit dispersal of the strain in the plant. We illustrated this point for one strain with very low pathogenicity. This strain was restricted to the main stem and could not colonize secondary ramifications and leaf petioles. Its multiplication in the main stem was limited in comparison with the multiplication of a fully aggressive strain. This is in agreement with previous results (40). We show here that these weakly pathogenic strains are interspersed in the phylogenetic tree among fully aggressive C. michiganensis subsp. michiganensis strains. They even share the same ST as fully aggressive strains. However, the four main pathogenicity determinants described in C. michiganensis subsp. michiganensis (40) were identified in half of these weakly aggressive strains, based on PCR tests. It may be argued that these determinants could be nonfunctional in these strains or indicate that as-yet-unknown pathogenicity determinants were absent in these strains. The weakly pathogenic strains were different from the nonpathogenic strains isolated from the same niche, but they were shown to be phylogenetically distant and they generally did not harbor the main pathogenicity determinants.
We compared five identification tests designed for C. michiganensis subsp. michiganensis using a large worldwide collection made of C. michiganensis subsp. michiganensis and nonpathogenic Clavibacter spp. While none of the five tests generated false-negative results, only one test produced no false-positive result. The Ptssk test (Berendsen et al., APS-IPPC Meeting) produced results that totally matched pathogenicity test results. It also identified strains that had a weak pathogenicity, but it did not identify saprophytic Clavibacter sp. strains. It is hence a highly reliable tool for identification of C. michiganensis subsp. michiganensis.
In conclusion, using MLSA/MLST approaches coupled with a thorough analysis of in planta pathogenicity, the presence of pathogenicity determinants, and diversity through the use of seven detection tools, C. michiganensis subsp. michiganensis was identified as a monophyletic clade, clearly separated from its closest phylogenetic neighbors and from nonpathogenic Clavibacter strains that share the same habitat and interfere in C. michiganensis subsp. michiganensis detection, leading to false positives. These saprophytic C. michiganensis strains clustered in one main clade. Based on DNA similarities, these saprophytic C. michiganensis strains formed at least one other new subspecies. The C. michiganensis subsp. michiganensis collection that was established for this study encompasses strains that appeared to belong to several clonal groups. These clonal groups exhibited different rates of epidemiological success. Some clones dispersed over long distances and were isolated over long periods of time, illustrating the efficacy of seed dispersal and powerful survival strategies. We provide a reduced MLSA scheme that is useful for positioning any new isolate and an MLST scheme allowing efficient typing of C. michiganensis subsp. michiganensis for epidemiological surveys.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially funded by the French Ministry of Agriculture, AOP Tomates et Concombres de France, the French federation of vegetable plantlet growers (SF3P), through CLAVITOM project “Gestion de Clavibacter michiganensis subsp. michiganensis, un enjeu sanitaire majeur pour la production de tomate en France.”
We acknowledge Marisa Ferreira and Armelle Darrasse for critically reviewing the manuscript. We thank CIRM-CFBP (INRA, France) for strain conservation and supply.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ah-You N, et al. 2009. Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas. Int. J. Syst. Evol. Microbiol. 59:306–318 [DOI] [PubMed] [Google Scholar]
- 2. Almeida NF, et al. 2010. PAMDB, a multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology 100:208–215 [DOI] [PubMed] [Google Scholar]
- 3. Anonymous 2006. Semences de tomate. Détection de Clavibacter michiganensis subsp. michiganensis (agent du chancre bactérien) par immunofluorescence indirecte. Méthode LNPV BH/06/01 version a. Ministère de l'Agriculture et la Pêche. Direction Générale de l'Alimentation, Paris, France [Google Scholar]
- 4. Aujoulat F, et al. 2011. Multilocus sequence-based analysis delineates a clonal population of Agrobacterium (Rhizobium) radiobacter (Agrobacterium tumefaciens) of human origin. J. Bacteriol. 193:2608–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baldwin A, et al. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baysal O, et al. 2011. Clavibacter michiganensis subsp. michiganensis: tracking strains using their genetic differentiations by ISSR markers in Southern Turkey. Physiol. Mol. Plant Pathol. 75:113–119 [Google Scholar]
- 7. Biggerstaff CM, Gleason ML, Braun EJ. 2000. Refinement of a nondestructive tomato seed assay for Clavibacter michiganensis subsp. michiganensis using seed fiber. Seed Sci. Technol. 28:261–269 [Google Scholar]
- 8. Bishop CJ, et al. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7:3 doi:10.1186/1741-7007-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bull CT, et al. 2011. Multilocus sequence typing of Pseudomonas syringae sensu lato confirms previously described genomospecies and permits rapid identification of P. syringae pv. coriandricola and P. syringae pv. apii causing bacterial leaf spot on parsley. Phytopathology 101:847–858 [DOI] [PubMed] [Google Scholar]
- 11. Castillo JA, Greenberg JT. 2007. Evolutionary dynamics of Ralstonia solanacearum. Appl. Environ. Microbiol. 73:1225–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costechareyre D, et al. 2010. Rapid and efficient identification of Agrobacterium species by recA allele analysis. Microb. Ecol. 60:862–872 [DOI] [PubMed] [Google Scholar]
- 13. Davis MJ, Gillaspie AG, Vidaver AK, Harris RW. 1984. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34:107–117 [Google Scholar]
- 14. de León L, Siverio F, Rodriguez A. 2006. Detection of Clavibacter michiganensis subsp. michiganensis in tomato seeds using immunomagnetic separation. J. Microbiol. Methods 67:141–149 [DOI] [PubMed] [Google Scholar]
- 15. de Leon L, Rodriguez A, Llop P, Lopez MM, Siverio F. 2009. Comparative study of genetic diversity of Clavibacter michiganensis subsp. michiganensis isolates from the Canary Islands by RAPD-PCR, BOX-PCR and AFLP. Plant Pathol. 58:862–871 [Google Scholar]
- 16. Doroghazi JR, Buckley DH. 2010. Widespread homologous recombination within and between Streptomyces species. ISME J. 4:1136–1143 [DOI] [PubMed] [Google Scholar]
- 17. Dreier J, Bermpohl A, Eichenlaub R. 1995. Southern hybridization and PCR for specific detection of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Phytopathology 85:462–468 [Google Scholar]
- 18. Eichenlaub R, Garteman K-H. 2011. The Clavibacter michiganensis subspecies: molecular investigation of gram positive bacterial plant pathogens. Annu. Rev. Phytopathol. 49:445–464 [DOI] [PubMed] [Google Scholar]
- 19. European and Mediterranean Plant Protection Organization (EPPO) 2005. PM7/421. Clavibacter michiganensis subsp. michiganensis. OEPP/EPPO Bull. 35:271–273 [Google Scholar]
- 20. Fargier E, Fischer-Le Saux M, Manceau C. 2011. A multilocus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer-attacking pathovars of this species. Syst. Appl. Microbiol. 34:156–165 [DOI] [PubMed] [Google Scholar]
- 21. Feil EJ, Maiden MCJ, Achtman M, Spratt BG. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496–1502 [DOI] [PubMed] [Google Scholar]
- 22. Felsenstein J. 2005. PHYLIP Phylogeny Inference Package. Version 3.63. Department of Genetics, University of Washington, Seattle, WA [Google Scholar]
- 23. Franken AAJM, Kamminga GC, Snyders W, Van Der Zouwen PS, Birnbaum YE. 1993. Detection of Clavibacter michiganensis ssp. michiganensis in tomato seeds by immunofluorescence microscopy and dilution plating. Neth. J. Plant Pathol. 99:125–137 [Google Scholar]
- 24. Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gartemann KH, et al. 2008. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190:2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gevers D, et al. 2005. Re-evaluating bacterial species. Nat. Microbiol. Rev. 3:733–739 [DOI] [PubMed] [Google Scholar]
- 27. Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582 [DOI] [PubMed] [Google Scholar]
- 28. Guindon S, et al. 2010. New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 29. Hadas R, Kritzman G, Klietman F, Gefen T, Manulis S. 2005. Comparison of extraction procedures and determination of the detection threshold for Clavibacter michiganensis subsp. michiganensis in tomato seeds. Plant Pathol. 54:643–649 [Google Scholar]
- 30. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 31. Hanage WP, et al. 2005. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187:6223–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanage WP, Fraser C, Spratt BG. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holt JG, Krieg NR, Sneath PA, Staley JT, Williams ST. 1994. Group 20. Irregular nonsporing gram-positive rods, p 571–572 In Bergey's manual of determinative bacteriology (9th ed). Lippincott Williams & Wilkins, Philadelphia, PA: ISBN 0-683-00603-7 [Google Scholar]
- 34. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 35. Jahr H, Dreier J, Meletzus D, Bahro R, Eichenlaub R. 2000. The endo-β-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant Microbe Interact. 13:703–714 [DOI] [PubMed] [Google Scholar]
- 36. Kaiser S, Biehler K, Jonas D. 2009. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 191:2934–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaneshiro WS, Alvarez AM. 2001. Specificity of PCR and ELISA assays for hypovirulent and avirulent Clavibacter michiganensis subsp. michiganensis. Phytopathology 91:S46 [Google Scholar]
- 38. Kaneshiro WS, Mizumoto CY, Alvarez AM. 2006. Differentiation of Clavibacter michiganensis subsp. michiganensis from seed-borne saprophytes using ELISA, Biolog and 16S rDNA sequencing. Eur. J. Plant Pathol. 116:45–56 [Google Scholar]
- 39. Kawaguchi A, Tanina K, Inoue K. 2010. Molecular typing and spread of Clavibacter michiganensis subsp. michiganensis in greenhouses in Japan. Plant Pathol. 59:76–83 [Google Scholar]
- 40. Kleitman F, et al. 2008. Characterization of a Clavibacter michiganensis subsp. michiganensis population in Israel. Eur. J. Plant Pathol. 121:463–475 [Google Scholar]
- 41. Kokoskova B, Mraz I, Janse JD, Fousek J, Jerabkova R. 2005. Reliability of diagnostic techniques for determination of Clavibacter michiganensis subsp. sepedonicus. J. Plant Dis. Prot. 112:1–16 [Google Scholar]
- 42. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098 [DOI] [PubMed] [Google Scholar]
- 43. Lee IM, Bartoszyk IM, Gundersen DE, Mogen B, Davis RE. 1997. Nested PCR for ultrasensitive detection of the potato ring rot bacterium, Clavibacter michiganensis subsp. sepedonicus. Appl. Environ. Microbiol. 63:2625–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louws FJ, et al. 1998. rep-PCR-mediated genomic fingerprinting: a rapid and effective method to identify Clavibactermichiganensis. Phytopathology 88:862–868 [DOI] [PubMed] [Google Scholar]
- 45. Maggi Solcà N, Bernasconi MV, Valsangiacomo C, Van Doorn LJ, Piffaretti JC. 2001. Population genetics of Helicobacter pylori in the southern part of Switzerland analysed by sequencing of four housekeeping genes atpD, glnA, scoB and recA), and by vacA, cagA, iceA and IS605 genotyping. Microbiology 147:1693–1707 [DOI] [PubMed] [Google Scholar]
- 46. Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Manceau C, Horvais A. 1997. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl. Environ. Microbiol. 63:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcelletti S, Ferrante P, Scortichini M. 2010. Multilocus sequence typing reveals relevant genetic variation and different evolutionary dynamics among strains of Xanthomonas arboricola pv. juglandis. Diversity 2:1205–1222 [Google Scholar]
- 49. Martens M, et al. 2007. Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 57:489–503 [DOI] [PubMed] [Google Scholar]
- 50. Martens M, et al. 2008. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 58:200–214 [DOI] [PubMed] [Google Scholar]
- 51. Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563 [DOI] [PubMed] [Google Scholar]
- 52. Martin DP, Williamson C, Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 53. Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21:98–102 [DOI] [PubMed] [Google Scholar]
- 54. Meletzus D, Bermpohl A, Dreier J, Eichenlaub R. 1993. Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175:2131–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Milne I, et al. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 24:126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nei M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY [Google Scholar]
- 57. Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 58. Nzoué A, et al. 2009. Multilocus sequence analysis of bradyrhizobia isolated from Aeschynomene species in Senegal. Syst. Appl. Microbiol. 32:400–412 [DOI] [PubMed] [Google Scholar]
- 59. Oh PL, et al. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4:377–387 [DOI] [PubMed] [Google Scholar]
- 60. Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 61. Pastrik K, Rainey F. 1999. Identification and differentiation of Clavibacter michiganensis subspecies by polymerase chain reaction-based techniques. J. Phytopathol. 147:687–693 [Google Scholar]
- 62. Posada D, Crandall K. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 24:13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Richert K, Brambilla E, Stackebrandt E. 2007. The phylogenetic significance of peptidoglycan types: molecular analysis of the genera Microbacterium and Aureobacterium based upon sequence comparison of gyrB, rpoB, recA and ppk and 16SrRNA genes. Syst. Appl. Microbiol. 30:102–108 [DOI] [PubMed] [Google Scholar]
- 64. Romano S, et al. 2009. Multilocus sequence typing supports the hypothesis that Ochrobactrum anthropi displays a human-associated subpopulation. BMC Microbiol. 9:267 doi:10.1186/1471-2180-9-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rossello-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39–67 [DOI] [PubMed] [Google Scholar]
- 66. Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 [DOI] [PubMed] [Google Scholar]
- 67. Santos MS, Cruz L, Norskov P, Rasmussen OF. 1997. A rapid and sensitive detection of Clavibacter michiganensis subsp. michiganensis in tomato seeds by polymerase chain reaction. Seed Sci. Technol. 25:581–584 [Google Scholar]
- 68. Scally M, Schuenzel EL, Stouthamer R, Nunney L. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 71:8491–8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114–1116 [Google Scholar]
- 70. Silva C, Vinuesa P, Eguiarte LE, Souza V, Martinez-Romero E. 2005. Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol. Ecol. 14:4033–4050 [DOI] [PubMed] [Google Scholar]
- 71. Smith JM. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 72. Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129–134 [DOI] [PubMed] [Google Scholar]
- 73. Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tamura K, et al. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 76. Tsiantos J. 1987. Transmission of Corynebacterium michiganense pv. michiganense by seeds. J. Phytopathol. 119:142–146 [Google Scholar]
- 77. Vos M, Velicer GJ. 2006. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl. Environ. Microbiol. 72:3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Waleron M, Waleron K, Kamasa J, Przewodowski W, Lojkowska E. 2011. Polymorphism analysis of housekeeping genes for identification and differentiation of Clavibacter michiganensis subspecies. Eur. J. Plant Pathol. 131:341–354 [Google Scholar]
- 79. Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7:256–276 [DOI] [PubMed] [Google Scholar]
- 80. Whatmore AM, Perrett LL, MacMillan AP. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7:34 doi:10.1186/1471-2180-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yan S, et al. 2008. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Appl. Environ. Microbiol. 74:3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Young JM, Park DC, Shearman H, Fargier E. 2008. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 31:366–377 [DOI] [PubMed] [Google Scholar]
- 83. Zaluga J, et al. 2011. GyrB sequence analysis and MALDI-TOF MS as identification tools for plant pathogenic Clavibacter. Syst. Appl. Microbiol. 34:400–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.