Abstract
Archaea are widely distributed and yet are most often not the most abundant members of microbial communities. Here, we document a transition from Bacteria- to Archaea-dominated communities in microbial biofilms sampled from the Richmond Mine acid mine drainage (AMD) system (∼pH 1.0, ∼38°C) and in laboratory-cultivated biofilms. This transition occurs when chemoautotrophic microbial communities that develop at the air-solution interface sink to the sediment-solution interface and degrade under microaerobic and anaerobic conditions. The archaea identified in these sunken biofilms are from the class Thermoplasmata, and in some cases, the highly divergent ARMAN nanoarchaeal lineage. In several of the sunken biofilms, nanoarchaea comprise 10 to 25% of the community, based on fluorescent in situ hybridization and metagenomic analyses. Comparative community proteomic analyses show a persistence of bacterial proteins in sunken biofilms, but there is clear evidence for amino acid modifications due to acid hydrolysis. Given the low representation of bacterial cells in sunken biofilms based on microscopy, we infer that hydrolysis reflects proteins derived from lysed cells. For archaea, we detected ∼2,400 distinct proteins, including a subset involved in proteolysis and peptide uptake. Laboratory cultivation experiments using complex carbon substrates demonstrated anaerobic enrichment of Ferroplasma and Aplasma coupled to the reduction of ferric iron. These findings indicate dominance of acidophilic archaea in degrading biofilms and suggest that they play roles in anaerobic nutrient cycling at low pH.
INTRODUCTION
Our knowledge of archaeal contributions to the anaerobic carbon cycle continues to expand. Beyond their best-characterized role as methanogens, archaea are increasingly implicated in nutrient cycling, for example, via anaerobic methane oxidation (31) and the dicarboxylate-hydroxybutyrate cycle (32). To date, archaea have been shown to dominate in only a few environments, such as lake (34) and ocean sediments (43), and in some “extreme” environments, such as those characterized by high salinity (44) or in acidic hot springs (33). Understanding the factors that enrich for archaea and the physiological processes that sustain them in situ remain important questions.
In metal-rich, low-pH ecosystems associated with pyrite ores, several species of Archaea have been identified via clone library analysis (4, 10, 11, 51, 58), metagenomic sequencing (6, 59), and isolation (24, 29). Archaea have also been identified as members of consortia used in bioleaching systems for metal recovery (46). Arguably, the best-studied low-pH, high-iron environment is the Richmond Mine AMD system at Iron Mountain, CA. In the Richmond Mine and in other acidic metal-rich systems, archaea are typically found in relatively low abundance (4, 38, 39). The archaea in the system belong to two distinct lineages of the Euryarcheota: the class Thermoplasmata—which includes Ferroplasma species as well as the “alphabet plasmas” referred to as Aplasma through Iplasma (4)—and a deeply branching clade referred to as ARMAN (6, 13). Within the Thermoplasmata belongs the order Thermoplasmatales, which includes the genera Thermoplasma, Ferroplasma, Picrophilus, and Thermogymnomonas. Iplasma is the most divergent of the AMD plasmas and is almost certainly not in the order Thermoplasmatales; it may, in fact, represent a separate class (A. P. Yelton, L. Comolli, C. Castelle, N. Justice, B. C. Thomas, and J. F. Banfield, unpublished data). Metagenomic sequencing has allowed for nearly complete reconstruction of genomes for Ferroplasma, four Thermoplasmata, and four ARMAN Archaea (5, 59, 62). Among the AMD plasmas, genomic analyses consistently indicate facultatively anaerobic and heterotrophic lifestyles (59; Yelton et al., unpublished). Furthermore, isolates from the Thermoplasmatales lineage, including Thermoplasma volcanium, Thermoplasma acidophilum, and Ferroplasma spp., have been characterized as facultatively anaerobic heterotrophs (20, 53).
Generally, biofilms beginning to grow at the air-solution interface in the Richmond Mine are devoid of archaea, but archaeal populations become more abundant with increasing biofilm age and thickness (61). We present here evidence that, in contrast to floating biofilms, archaea dominate the submerged suboxic biofilms and, as such, may be key players in carbon and nutrient cycling in AMD environments.
MATERIALS AND METHODS
Site description and sample collection.
Biofilm samples were collected underground within the Richmond Mine at Iron Mountain, CA. Temperature was measured in situ, and the pH was determined shortly after sample collection. Samples for metagenomic analysis were frozen on site on dry ice, whereas those for cultivation experiments were maintained on ice water during transport to the laboratory, where they were used immediately for inoculation experiments. At the time of sampling, biofilm thickness was estimated visually to indicate growth (developmental) stage. Based on prior direct measurements, early growth stage biofilms are typically <30 μm thick, whereas late growth stage biofilms can be up to 200 μm thick (61).
Microscopy.
Environmental samples were fixed and stained with lineage-specific fluorescent in situ hybridization (FISH) probes to determine archaeal and bacterial relative abundances, as described previously (9). Fixed cells were counted manually on a Leica DMRX epifluorescence microscope at ×630 magnification. At least five fields of view and >1,000 cells were counted for each sample. FISH cell counts were used to calculate species percentages from total cell counts obtained using DAPI (4′,6′-diamidino-2-phenylindole). Probes used in the present study are listed in Table 1.
Table 1.
FISH probes
| Probe | E. coli position | Sequence (5′–3′) | Target | % Formamide | Reference |
|---|---|---|---|---|---|
| ARC915 | 915–934 | GTGCTCCCCCGCCAATTCCT | Domain Archaea | 35 | 2 |
| EUB338 mix | 338–355 | GCTGCCTCCCGTAGGAGT | Domain Bacteria | 35 | 1, 14 |
| 338–355 | GCWGCCACCCGTAGGTGT | Domain Bacteria | 35 | 1, 14 | |
| SUL230 | 230–247 | TAATGGGCCGCGRGCYCC | Sulfobacillus | 9a | |
| FER656 | 656–674 | CGTTTAACCTCACCCGATC | Thermoplasmata | 25 | 24 |
| LF655 | 655–673 | CGCTTCCCTCTCCCAGCCT | Leptospirillum groups I, II, and III | 35 | 9 |
| ARMAN mix | 331–350 | GTCTCAGTACCCTTCTGGGG | ARMAN | 20 | 5 |
| 329–348 | CCGYAGTGCTAGGGTCCTTC | ARMAN | 20 | 5 | |
| 424–442 | GGCAAAAGTTCCCTTCCGG | ARMAN | 20 | 5 | |
| 927–944 | ACCCGTTTTTGTGCTCCC | ARMAN | 20 | 5 |
That is, adapted from reference 9.
16S rRNA sequencing and analysis.
For all DNA extractions, ∼1 g of frozen biofilm was washed twice with 0.5 ml of cold phosphate-buffered saline (pH 1.2) before being resuspended in 500 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and 500 μl of STEP buffer (0.5% sodium dodecyl sulfate, 50 mM Tris-HCl [pH 7.5] 400 mM EDTA, 1 mg of proteinase K/ml, 0.5% Sarkosyl). The samples were cycled three times between liquid nitrogen and a 50°C water bath to facilitate lysis. After the last cycle, an additional 500 μl of STEP buffer was added, and the sample was left at 50°C for 20 min. The samples were extracted three times with 1 volume each of a 25:24:1 phenol-chloroform-isoamyl alcohol mix (Fisher BioReagents, Pittsburgh, PA) using a 15-ml Phase Lock Gel tube (5 Prime, Gaithersburg, MD). The aqueous layer was precipitated with 1 volume of cold isopropanol and 0.1 volume of cold sodium acetate (3 M; pH 5.2). Pellets were washed with 1 ml of cold ethanol and resuspended in TE buffer.
For clone library construction, 16S rRNA gene sequences were amplified by PCR in mixtures containing 1× PCR Master Mix (Promega, Madison, WI), ∼40 ng of template DNA, 300 mM concentrations (each) of forward and reverse primers, and 0.5 mg of bovine serum albumin (New England BioLabs, Ipswich, MA)/ml. The primers for the Bacteria-specific libraries were 27F (5′-AGAGTTTGATCMTGGCTCAG) and 1492R (5′-TACGGYTACCTTGTTACGACTT) (15) and for the Archaea-specific libraries were 522F (5′-GGYAAGACSGGTGSCAGC) and 1354R (5′-GCGRTTACTASGGAWTCC), designed to bind to both archaea of the Thermoplasmatales and the ARMAN. Reactions were incubated in a DNA Engine Dyad thermocycler (MJ Research, Quebec, Canada) for 94°C (2 min), followed by 30 cycles of 94°C for 45 s, 53°C for 45 s, and 72°C for 90 s, with a final extension of 72°C for 10 min. PCR products were purified and cloned, as described previously (10). Sequencing was carried out using the same amplification primers as used in PCRs on a 3730XL DNA Analyzer (Applied Biosystems, Carlsbad, CA). Sequences were assembled with Phred/Phrap software, and chimeric sequences were screened using Mallard (3). Sequences were clustered using UClust (23) at 97% percent identity. Sequences were aligned with MAFFT (37) and curated with MEGA (57). Trees were constructed using the maximum-likelihood algorithm in MEGA using the general time reversible model. Bootstrap values were calculated from 1,000 iterations.
Metagenomic sequencing and analysis.
For metagenomic sequencing, DNA was extracted from samples, as described above. Samples were treated with 10 U of RNase (New England BioLabs)/ml for 15 min at room temperature to improve sequencing quality. DNA from both samples was sequenced with 454 FLX Titanium (454 Life Sciences, Branford, CT), according to the manufacturer's protocols, with the exception that fragment libraries were quantified with digital PCR rather than with titrations (60). The data were quality controlled by removing reads with at least one ambiguous base. Artificial duplicate reads formed as a result of the 454 FLX Titanium sequencing method were removed, as described previously (16, 30). For analysis of the community composition, reads over 100 nucleotides in length were compared by BLAST to genomes of organisms from the Richmond Mine recovered by prior metagenomic studies (6, 59). Only matches with >98% identity were reported, and the number of hits for each species was normalized by genome size.
Protein analysis.
Whole-cell fractions of proteins were extracted from frozen biofilms (∼500 mg) via sonication and trichloroacetic acid precipitation, as previously described (18). Briefly, the cells were washed with H2SO4 (pH 1.1) to remove residual iron, and then the pellets were resuspended in 6 ml of 20 mM Tris-SO4 (pH 8.0) and sonicated on ice using a microtip sonicator. Next, 5 ml of 0.4 M Na2CO3 (pH 11) was added, and nonlysed cells and polymers were removed by centrifugation at 6,000 × g for 20 min. The protein was precipitated using trichloroacetic acid (1:10) for at least 3 h, centrifuged (14,000 × g for 10 min), and washed with cold methanol. The protein was resuspended in 6 M guanidine–10 mM dithiothreitol, and enzymatically digested with trypsin (Promega, Madison, WI). The peptides were separated using Nano-2D-LC for 24 h (strong cation exchange/reversed phase) and analyzed by tandem mass spectrometry on a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA), as described previously (18, 42). At least two technical replicates were run for each sample. Unmodified peptides were identified by using the SEQUEST algorithm (26) to search tandem mass spectra against the predicted protein database amdv1_reads_11212008_arman_Biofilm_AMD_CoreDB_04232008_B constructed from previously published community genomic data sets (6, 59). The SEQUEST search results were filtered and sorted with the DTASelect algorithm (56). False-positive rates for this method of analysis have been determined to be between 1 and 5% (18, 45). The database is available online (http://compbio.ornl.gov/amd_gtl_ms_results/databases/).
Normalized spectral abundance factors (NSAF) were calculated as described previously (27, 42). For clustering analysis of Gplasma proteins, organism-specific NSAF values (orgNSAF) were calculated by normalizing NSAF values to the total detected spectral counts from that organism, after which they were arcsin transformed, mean centered, and scaled as described previously (42). Only samples with >150 Gplasma proteins were used in order to prevent bias from zeros during normalization. Hierarchical clustering was carried out in a Multiple Experiment Viewer (49, 50) using a Pearson correlation coefficient and average linkage. A Fisher exact test was performed in R (47) to compare enrichment of functional categories in the group of proteins enriched in sunken samples versus enrichment among other proteins in the data set. Functional categories were defined using the Clusters of Orthologous Groups (COG) database.
Deamidation analysis.
Tandem mass spectra were searched again for deamidated peptides with the SEQUEST algorithm by considering the dynamic modification of deamidation for all glutamine (Q) and asparagine (N) residues. DTASelect output files were analyzed with custom Ruby scripts to calculate the spectral counts of deamidated Q and deamidated N residues, as well as the total spectral counts of Q and N residues in identified peptides. These spectral counts were summed for each organism or domain. The deamidation frequencies of Archaea and Bacteria were calculated as the percentages of the run-averaged spectral counts of deamidated Q or N over the total spectral counts of Q or N, respectively, in each domain. In order to ensure that deamidation percentages were calculated from a representative number of peptides, runs in which a single peptide contributed more than 50% of the total deamidation percentage for either archaeal or bacterial domains were not included in the analysis. By searching the reversed database (i.e., all peptide sequences reversed), false-discovery rates of deamidated peptides (∼2%) and deamidated spectra (∼3%) were identified. For analysis of differences of deamidation between growth stages and domains, data were analyzed with a two-way analysis of variance (ANOVA) in R (47) after verification that the assumptions of normality and homoscedasticity were met.
Cultivation of aerobic biofilms.
The floating biofilm was cultivated in laboratory bioreactors as previously described (7, 8). After growing to a thickness consistent with late-growth-stage biofilms, the biofilm was dislodged from the reactor walls and submerged at the bottom of the reactor chamber under ∼2 cm of slowly flowing AMD solution. After 7 days, sunken biofilm samples were taken and either immediately fixed for FISH or frozen for later proteomic analyses.
Cultivation of anaerobic biofilms.
To maximize diversity for anaerobic community culturing, inocula were derived from a variety of floating and sunken biofilm samples collected from several locations within the Richmond Mine (Redding, CA). Approximately 5-ml aliquots of the mixed inocula were added to replicate 120-ml serum vials filled with 50 ml of nitrogen-degassed ferric-9kBR medium containing 100 mM Fe2(SO4)3, 1 mM (NH4)2SO4, 0.5 mM KCl, 0.050 mM K2HPO4, 5 mM MgSO4·7H2O, 0.2 mM CaSO4, and trace minerals (8), all adjusted to pH 1.1 with H2SO4.
Triplicate cultures were supplemented with one of each of the following organic substrates: glycolate, formate, lactate, acetate, and betaine (all at 10 mM; Sigma-Aldrich), Casamino Acids (0.02%, acid-hydrolyzed casein; Becton Dickinson, East Rutherford, NJ), or peptone (0.02%, enzymatic digest of protein; Becton Dickinson). In addition, some cultures were supplemented with naturally derived biofilm substrate obtained by lyophilization of a mature floating biofilm from the Richmond Mine, followed by redissolution into sterile deionized H2O and sonication for 5 min with a microtip sonicator. This natural substrate was added to cultures at a concentration of 0.05% (wt/vol). The sterility of this substrate was monitored via DNA extractions and microscopy in uninoculated controls. Replicates of all cultures were cultivated with or without 0.01% yeast extract. A 10% volume of each culture was transferred three times into 5-ml volumes of fresh medium every 3 to 4 weeks and transferred into 40 ml in the fourth and final transfer. Ferrozine assays of the dissolved iron concentration were performed, and DNA was extracted from cultures on the fourth transfer 28 days after inoculation. The ferrozine assays were done as previously described (55), and DNA extractions were carried out as described above.
RESULTS
Sunken biofilm sampling.
Three sunken biofilms were collected from the 5-way site (February 2008, pH 0.98, 38°C), and three were collected from the UBA site (June 2009, pH 1.1, 38°C) (see the location map in Fig. S1 in the supplemental material). At the 5-way site, we sampled a mature, thick (∼0.1-mm) flexible biofilm floating on a ∼5-cm-deep flowing AMD solution (floating growth stage 2 [GS2], Fig. 1A). In addition, we collected three fragile sunken biofilms of similar thickness that were stratified underneath the floating biofilm and suspended close to the stream base. We labeled these biofilms “5-way–Sunken1,” reflecting its location immediately below the floating GS2 biofilm; “5-way–Sunken2,” the biofilm a few millimeters below 5-way–Sunken1; and “5-way–Sunken3,” the biofilm resting on the stream channel (Fig. 1A). Fragile (easily disintegrated) sunken biofilm samples collected from the UBA site (UBA-Sunken1, UBA-Sunken2, and UBA-Sunken3) were recovered from three different regions of a slowly draining AMD pool about 25 cm deep. All of the samples collected for the present study, and those obtained in previous studies, are listed in Table S1 in the supplemental material.
Fig 1.

(A) Schematic showing the 5-way sampling site and the stratification of the sunken samples. (B) Picture taken of the 5-way sampling site showing the floating biofilm on top, with the sunken biofilm visible beneath.
FISH microscopy of natural sunken biofilms.
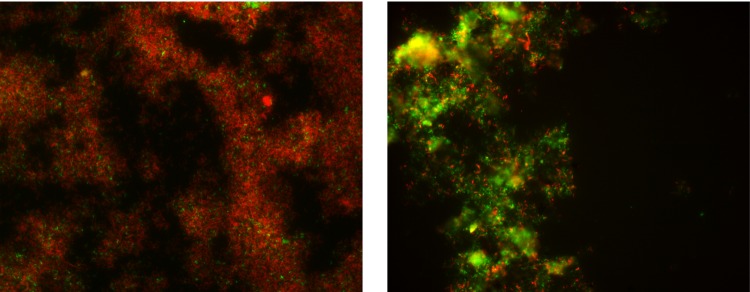
FISH was used to determine the relative proportion of Bacteria to Archaea in sunken biofilms (Fig. 2; see Table S2 in the supplemental material). In contrast to previously characterized early-growth-stage (GS1) and late-growth-stage (GS2) floating biofilms, sunken biofilms were dominated by Archaea (Fig. 3). The relative community composition of the 5-way–Sunken3 biofilm was qualitatively similar to the other 5-way samples but with clearly reduced cell density. High autofluorescence in non-DAPI filter channels, presumably caused by degraded biofilm and minerals, prohibited quantitative analysis of this sample. In the other five sunken samples analyzed quantitatively, non-ARMAN archaeal species dominated the community. ARMAN and Sulfobacillus species were more abundant in the 5-way–Sunken samples than UBA Sunken samples (see Table S2 in the supplemental material).
Fig 2.
FISH images of 5-way–Floating GS2 (left) and 5-way–Sunken 1 biofilms for comparison of bacterial (Eubmix probe, red) and archaeal (arc915 probe, green) populations.
Fig 3.

Percentages of DAPI-stained cells detected by FISH with either bacterial (EubMix) or archaeal-specific (Arc915) probes. Error bars represent the standard deviations (SD).
Clone libraries and DNA sequencing.
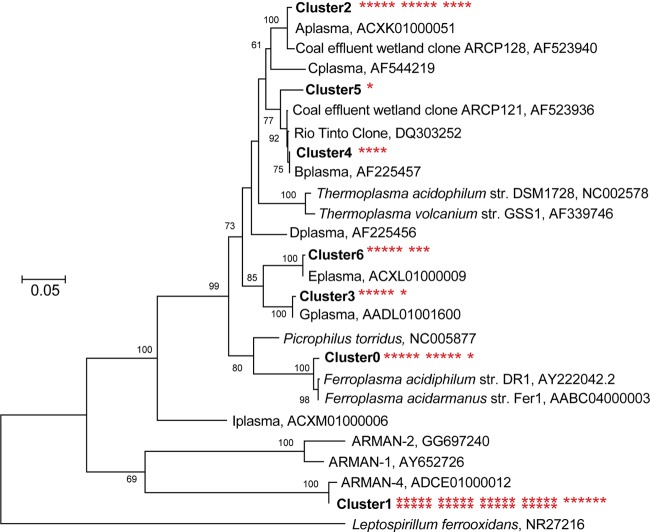
Clone libraries were constructed for 5-way–Sunken1 to better determine the phylogenetic diversity of the Archaea and Bacteria present. Forty-five of the 89 sequences obtained from 5-way–Sunken1 using Archaea-specific 16S rRNA primers belonged to the deeply branching clade known as ARMAN, a euryarchaeal lineage (5, 13) (Fig. 4). Eleven clones were closely related (>99% identity) to Ferroplasma acidarmanus, a species previously isolated from the Richmond Mine (25). The remaining clones represented Aplasma, Bplasma, Gplasma, and Eplasma (Fig. 4), a radiation of closely related Thermoplasma archaea, referred to as the “alphabet plasmas” (4, 19).
Fig 4.
Phylogenetic tree of archaeal clones generated using the maximum-likelihood method. Asterisks next to clusters identify the number of clones belonging to that cluster. Bootstrap values are shown at the respective nodes. Scale bars are equal to 0.05 changes per site.
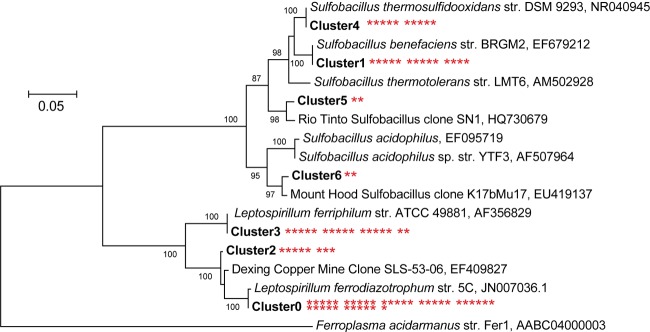
The Bacteria-specific 16S rRNA library contained 89 clones: 36 belonged to Leptospirillum ferrodiazotrophum, a member of Leptospirillum group III; 8 clones were related to, but distinct from Leptospirillum group III (3.0% divergent); 15 belonged to Leptospirillum group II (Leptospirillum ferriphilum); and the remainder to species of the genus Sulfobacillus (Fig. 5). Within the Sulfobacillus lineage, 10 sequences were Sulfobacillus thermosulfidooxidans (100% identity), 14 were Sulfobacillus benefaciens (100% identity), and 4 derived from other Sulfobacillus species.
Fig 5.
Phylogenetic tree of bacterial clones generated using the maximum-likelihood method. Asterisks next to clusters identify the number of clones belonging to that cluster. Bootstrap values are shown at the respective nodes. Scale bars are equal to 0.05 changes per site.
Proteomic comparisons of floating and sunken biofilms.
Proteomic analyses of the six sunken biofilms and the 5-way–Floating biofilm were used to analyze community structure in the context of previously sampled floating biofilms as well as to identify metabolic processes active in the sunken biofilms. The relative abundance of bacterial proteins was highest in early-growth-stage (93.0%) and late-growth-stage (84.9%) floating biofilms and was lowest in sunken biofilm samples (55.0%) (Fig. 6). Archaeal proteins were most abundant in the sunken biofilm samples (24.5%) and less abundant in the early-growth-stage (1.9%) and late-growth-stage (7.1%) floating biofilms.
Fig 6.

Bar graph showing average percentage of proteome composition for floating early growth stage (GS1), floating late growth stage (GS2), and sunken biofilms. The percentages are based on normalized spectral abundance factors. “Other” includes unassigned proteins, viral proteins, and plasmid-associated proteins. Error bars represent the SD.
The proteomes of the 5-way–Floating biofilm and the three stratified sunken biofilms showed a striking increase in the relative abundance of archaeal proteins with increasing depth, 5-way–Sunken1 contained relatively more archaeal protein (7.3%) than 5-way–Floating (1.9%). Below 5-way–Sunken1, a large increase in the relative abundance of the Archaea was found in 5-way–Sunken2 (35.8% Archaea), with a similar relative abundance in 5-way–Sunken3 (35.9% Archaea) (Fig. 7).
Fig 7.

Percent of proteome composition for 5-way samples. The percentages are based on normalized spectral abundance factors.
Overall, 2,334 distinct archaeal proteins were identified across all samples analyzed (see Table S3 in the supplemental material). Of those, 545 proteins were only identified in sunken samples, 761 in floating samples, and 1,128 proteins were identified in both. Proteins detected represented many major metabolic pathways, such as tricarboxylic acid (TCA) cycle, glycolysis, fatty acid oxidation, and electron transport. Furthermore, many proteins associated with the acquisition and breakdown of organic carbon were detected, including peptide transporters, sugar transporters, peptidases, and extracellular glucoamylases (see Table S4 in the supplemental material).
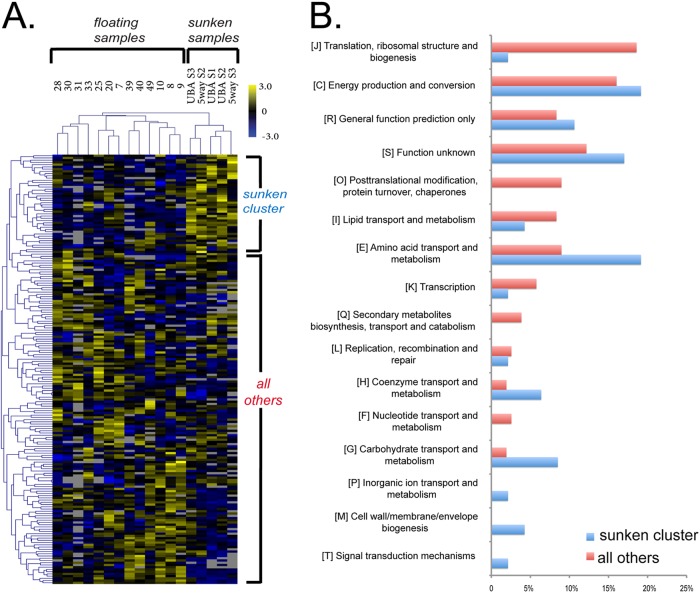
Gplasma was the only archaeal species with consistently high protein abundances across samples and growth stages, enabling proteome comparisons across growth stages by hierarchical clustering of orgNSAF values. The sunken samples formed a distinct cluster, as did a group of proteins showing relatively higher abundance in the sunken biofilms (Fig. 8A). This group of proteins had a distinct distribution of COG functional category counts (Fisher exact test, P = 0.0002), with a lower abundance of proteins involved in protein biogenesis (ribosomal) and higher abundances of proteins involved in amino acid metabolism (Fig. 8B). Elevated in abundance in the sunken biofilm protein cluster were two enzymes of the TCA cycle (malate dehydrogenase and aconitate hydratase), several enzymes associated with transformations of pyruvate (three subunits of the pyruvate dehydrogenase/2 oxoacid dehydrognease complex, a cytochrome-associate pyruvate dehydrogenase, pyruvate:ferredoxin oxidoreductase, pyruvate phosphate dikinase, and acetolactate synthase), and multiple proteins that likely participate in electron transfer reactions, including an oxidoreductase, and aldehyde dehydrogenase, and a flavoprotein (see Table S5 in the supplemental material).
Fig 8.
Hierarchical clustering of Gplasma proteins in >70% of samples using protein abundance data. Yellow is overrepresented and blue is underrepresented. The cluster of proteins uniformly overrepresented in sunken samples is indicated at the top. (B) The COG functional distribution of proteins in the sunken cluster versus the distribution of all of the other proteins on the heat-map.
454 sequencing.
The relative percentage of bacterial proteins in sunken samples was higher than what was expected based on FISH analyses, so we analyzed metagenomic sequence from a subset of samples (5-way–Sunken2 and UBA-Sunken2) as a third measure of community composition. These analyses used 50 Mb of the 454 FLX Titanium sequence data obtained for each of the UBA-Sunken2 and 5-way–Sunken2 biofilms (the read lengths averaged ∼290 bp for both samples).
BLAST comparisons of 454 sequencing reads to the genomic databases showed that the 5-way–Sunken2 and UBA-Sunken2 biofilms were dominated by archaeal species (Fig. 9). In the 5-way–Sunken2 biofilm, 98.3% of reads mapping to the database matched archaeal sequences, whereas bacterial sequences represented only 1.3% of reads. In the UBA-Sunken2 sample, archaeal species comprised 69.24% of genomic reads, whereas bacterial species made up 30.68%.
Fig 9.

Composition of 5-way–Sunken2 and UBA-Sunken2 communities based on percentages of 454 reads with BLAST matches against the genomic database (>98% identity), normalized by genome size.
Deamidation.
Compared to FISH and metagenomic analyses, proteomic measurements showed a striking overrepresentation of Bacteria in sunken communities. Although this discrepancy could be partially explained by differences in cell size (and thus protein content) between the bacterial and archaeal community members, we hypothesized that bacterial proteins may derive from cell lysates rather than living cells. It is known that glutaminyl and asparaginyl residues can undergo an acid-dependent deamidation (12, 36, 48). Thus, if our hypothesis is true, bacterial proteins in sunken communities should have more extensive deamidation than in floating communities. In contrast, archaeal proteins should have the same degree of deamidation in floating and sunken biofilms if the cells were equally viable in both communities. We measured the degree of deamidation in proteins extracted from both floating and sunken biofilms by proteomics. On average, 28.3% of the glutamines underwent deamidation in bacterial proteins from early-growth-stage biofilms, 52.5% in late-growth-stage biofilms, and 72.1% in sunken biofilms (Fig. 10A). On the other hand, glutamines in archaeal proteins were deamidated at frequencies of 17.0 and 19.0% in early- and late-growth-stage biofilms, respectively, and at 44.2% in sunken biofilms. Similarly, the frequencies of deamidation in bacterial asparagine residues were 22.2, 36.4, and 58.1% in early-growth-stage, late-growth-stage, and sunken biofilms, respectively, whereas archaeal asparagine deamidation remained relatively constant between 21.1 and 30.3% across all sample subsets (Fig. 10B). Deamidation was not limited to any particular subset of proteins and was evident for essential cytoplasmic proteins such as ribosomal proteins, TCA cycle enzymes, and glycolysis enzymes (data not shown). A two-way ANOVA was used to test for differences among deamidation frequencies between domains of life (Archaea and Bacteria) and growth stages (GS1, GS2, Sunken). Deamidation frequencies of asparagine residues differed significantly for domains [F(1,43) = 14.11, P < 0.001], as well as between growth stages [F(2,43) = 19.53, P < 0.001]. Analysis of deamidation frequencies of glutamine residues showed similar results, with significant difference among domains [F(1,44) = 52.88, P < 0.001] and between growth stages [F(2,44) = 36.97, P < 0.001]. Importantly, there was an interaction between growth stage and domain for both asparagine [F(2,43) = 6.74, P < 0.01] and glutamine [F(2,44) = 4.94, P < 0.05] residues, indicating that the two domains respond differently to increasing growth stage and submersion. A post hoc analysis using Tukey's honestly significant difference test was used to compare deamidation frequencies among growth stages and between domains for each residue (see Table S6 in the supplemental material).
Fig 10.

Deamidation of bacterial and archaeal asparagine (A) and glutamine (B) residues across growth stage 1 (GS1), growth stage 2 (GS2), and sunken samples. Error bars represent 95% confidence intervals, and italic letters correspond to groups that are not significantly different within the given bar graph by Tukey's honest significance test (P < 0.05).
Bioreactor laboratory experiments.
We submerged a laboratory-cultured biofilm in our laboratory bioreactors to test the hypothesis that a bacterial dominated floating biofilm would shift toward archaeal dominance if submerged. FISH results showed an increase in the relative abundance of Archaea after 7 days of submersion (increasing from 23.5% of the community prior to sinking to 52.0% after submersion). Bacteria populations, on the other hand, showed a marked decrease from 76.5% of the community in the floating biofilm, to 47.9% of the sunken biofilm. Proteomic data also showed an increase in the relative abundance of archaea, with archaeal proteins compromising 11.3% of the floating biofilm and 17.2% of the sunken biofilm, while bacterial proteins compromised 77.2 and 67.0% of the floating and sunken biofilms, respectively. Measures of protein deamidation showed high frequencies of deamidation for bacterial asparagine (20.15% in floating and 34.8% in sunken) and glutamine (30.6% in floating and 52.6% in sunken) residues but not for archaeal residues (between 10.2 and 12.6% for glutamine and asparagine in either floating or sunken samples) (Fig. 11).
Fig 11.
Analyses on bioreactor-cultivated floating and sunken biofilms. (A) FISH analysis of bioreactor floating and sunken biofilms. Error bars represent the SD. (B) Proteomic analysis of floating and sunken biofilms by NSAF. (C) Frequencies of glutamine deamidation and asparagine deamidation for Bacteria (blue) and Archaea (red).
Culturing.
In order to determine whether archaea are capable of carrying out anaerobic carbon oxidation in sunken biofilms, we enriched for anaerobic iron reducers using ferric sulfate media and a variety of different carbon sources. Iron was reduced to similar degrees in yeast-extract supplemented cultures of peptone, betaine, Casamino Acids, and AMD biofilm extract (see Fig. S2 in the supplemental material). Cultures without yeast extract were not viable even after the first transfer, except for native-biofilm cultures, which did not grow after the third transfer. No growth was apparent in cultures containing glycolate, formate, acetate, or lactate with or without yeast extract. Analysis of 16S rRNA sequences from cultures cultivated on peptone, betaine, Casamino Acids, and AMD biofilm were dominated by Ferroplasma acidarmanus, with Aplasma comprising 0 to 47% of the sequenced clones (see Fig. S2 in the supplemental material). We also detected no hydrogen consumption or production and no methanogenesis (data not shown).
DISCUSSION
Little is known about selection factors for Archaea or archaeal metabolism in AMD systems. We used here a combination of metagenomic, proteomic and FISH analyses to compare community structures of thin floating biofilms, thicker and higher developmental stage floating biofilms, and biofilms submerged in suboxic/anoxic environments (see Table S1 in the supplemental material). In contrast to floating biofilms that are Bacteria-dominated, we show here that field-collected communities in submerged biofilms are dominated by Archaea and that a transition from bacterial to archaeal dominance can be induced by biofilm submersion in the laboratory. Based on the identification of proteins involved in peptide transport, sugar transport, fatty acid oxidation, and extracellular protein and starch breakdown (see Table S3 in the supplemental material), it is likely that the Richmond Mine plasmas may scavenge nutrients in the form of proteins and carbohydrates, in this case derived from the decaying biofilm.
For Aplasma, Eplasma, Gplasma, and Ferroplasma, many components of the electron transport chain were identified by proteomics, including subunits of NADH dehydrogenase, Rieske Fe-S proteins, and electron transfer flavoprotein-quinone oxidoreductases, as well as other dehydrogenases and oxidoreductases that may play roles in electron transport (see Table S3 in the supplemental material). Based on this and the detection of almost all TCA cycle proteins, it seems most likely that respiration is widely used for energy generation. Interestingly, the only protein known to be involved in a terminal electron accepting process identified was a Gplasma cytochrome c oxidase identified in one of the sunken samples. The detection of terminal oxidase activity is important, given that no methods exist for direct measurement of oxygen concentrations in AMD solutions. Oxygen may be introduced upstream in flowing solutions, but its concentration is calculated to be exceedingly low (22). Based on the low-oxygen availability, we contend that an alternate electron-accepting process predominates, and ferric iron is an obvious candidate, especially given its high concentration in AMD solutions (tens of mM), the known ability of Ferroplasma to reduce ferric iron (21), and our Aplasma-Ferroplasma enrichments showing iron reduction. To date, no terminal iron-reducing proteins have been annotated in these organisms, and indeed, knowledge of enzymes involved in this process is limited for the archaeal domain (52).
Analysis of Gplasma proteins suggests a distinct metabolism in sunken biofilm compared to floating biofilm environments. The observation that, when growing in the sunken biofilms, Gplasma appears to emphasize amino acid and carbohydrate metabolism relative to protein biogenesis and posttranslational modification, may indicate greater investment in substrate oxidation than in biosynthesis. This may reflect increased energy generation by substrate oxidation to compensate for lower energy yields associated with an anaerobic respiration. Notably, the relatively high abundance in sunken samples of proteins for transformations of pyruvate (see Table S2 in the supplemental material) suggests that pyruvate may be an important node in the metabolic flux of Gplasma in this environment. The overabundance of superoxide dismutase in the sunken samples might indicate oxidative stress, although recent findings have shown high expression of this enzyme in Geobacter species carrying out iron reduction, regardless of oxygen exposure (41).
ARMAN are typically rare members of AMD biofilm communities (5). Their apparent higher abundance by FISH and metagenomic measures than by proteomic analysis is likely due to the small cell volume and perhaps lower activity of these cells (5). Although many of the ARMAN proteins identified were associated with protein biosynthesis or were of unknown function, we also detected proteins involved in fatty acid oxidation, TCA cycle, and the Embden-Meyerhoff glycolysis pathway.
As noted above, we surmise that Ferroplasma and/or Aplasma carry out chemoorganotrophic growth coupled to iron reduction. Interestingly, the native-biofilm extracts supported growth without yeast extract through three transfers, although the reasons for the failed growth in the fourth transfer are unclear (changes in the biofilm extract due to prolonged storage may have been important in this regard). The precise role of yeast extract for growth on other carbon sources is unclear, although it could function as an additional carbon source or provide micronutrients. For T. acidophilum, it has also been suggested protect against the high pH gradient (54).
Leptospirillum group II and group III are important iron oxidizers in AMD systems (17, 28, 42, 59), and Leptospirillum group III is able to fix nitrogen (59). Compared to Leptospirillum spp., Sulfobacillus spp. tend to be in lower relative abundance (4). Studies have indicated that species of Sulfobacillus are facultative anaerobes capable of both autotrophic and heterotrophic modes of growth (22, 35). Oxidation of Fe(II), S0, and sulfide minerals, anaerobic reduction of Fe(III), and utilization of yeast extract, glucose, mannose, and other carbon sources have been described for various Sulfobacillus species (22, 35). Sulfobacillus species were detected predominantly in 5-way biofilms, but we have not clearly identified ecological roles for these organisms: their broad metabolic capabilities may be indicative of a generalist ecological strategy. Currently, incomplete genomic information for Sulfobacillus species prevents widespread detection of Sulfobacillus proteins, limiting insights into their functional contributions.
Both FISH and DNA analyses showed a higher relative abundance of archaeal species than did proteomics analyses, suggesting a discrepancy between FISH, metagenomic, and proteomic measures of community composition. We attribute the discrepancy to the persistence of bacterial proteins in lysed and degrading cells, as indicated by extensive detection of acid-hydrolyzed peptides. Moreover, proteins with a high degree of solvent exposure (i.e., unfolded) are more likely to undergo deamidation (12), and low pH can also greatly accelerate this reaction (36). Given the large difference in pH between the intracellular and extracellular environments of acidophilic organisms (40), we suspect that acidophilic cells that have lost membrane integrity may show a greater degree of deamidation due to exposure of cytoplasmic proteins to low-pH solution. The difference between glutamine and asparagine deamidation frequencies is likely due to higher reactivity of glutamine residues in acidic conditions (36).
While some role for Bacteria (particularly species of Sulfobacillus) in nutrient cycling in the sunken communities cannot be ruled out, the dominance of Archaea by several measures of community composition, proteomic signature of heterotrophic growth, heterotrophic growth in anaerobic culture, and the high degree of amino acid deamidation in the Bacteria, indicate that Archaea drive nutrient cycling in suboxic and anoxic AMD environments. Complete submersion in AMD solution would appear to select against Bacteria, which may be less adapted to the low oxygen availability. The diversity of Archaea is particularly interesting and may reflect a range of ecological niches in these high-carbon, suboxic and anoxic environments.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. W. Arman (Iron Mountain Mines, Inc.) and R. Sugarek for access to the Richmond Mine and R. Carver and M. Jones for on-site assistance. We also thank Robert Butler for assistance with iron reduction assays, Kelly Wrighton for discussions, and Brett Baker for assistance with the primer design and clone library construction.
This study was supported by Genome Sciences Program in Carbon Cycling (contract DE-SC0004665), by the Systems Biology Knowledgebase (contract DE-SC0004918), and by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. App. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. App. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139–152 [DOI] [PubMed] [Google Scholar]
- 5. Baker BJ, et al. 2010. Enigmatic, ultra-small, uncultivated Archaea. Proc. Natl. Acad. Sci. U. S. A. 107:8806–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker BJ, et al. 2006. Lineages of acidophilic archaea revealed by community genomic analysis. Science 314:1933–1935 [DOI] [PubMed] [Google Scholar]
- 7. Belnap CP, et al. 2011. Quantitative proteomic analyses of the response of acidophilic microbial communities to different pH conditions. ISME J. 5:1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belnap CP, et al. 2010. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 4:520–530 [DOI] [PubMed] [Google Scholar]
- 9. Bond PL, Banfield JF. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149–161 [DOI] [PubMed] [Google Scholar]
- 10. Bond PL, Druschel GK, Banfield JF. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. App. Environ. Microbiol. 66:4962–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruneel O, et al. 2008. Archaeal diversity in a Fe-As rich acid mine drainage at Carnoules (France). Extremophiles 12:563–571 [DOI] [PubMed] [Google Scholar]
- 12. Catak S, Monard G, Aviyente V, Ruiz-Lopez MF. 2006. Reaction mechanism of deamidation of asparaginyl residues in peptides: effect of solvent molecules. J. Phys. Chem. A 110:8354–8365 [DOI] [PubMed] [Google Scholar]
- 13. Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. 2009. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 3:159–167 [DOI] [PubMed] [Google Scholar]
- 14. Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 15. DeLong EF, et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 16. Denef VJ, Banfield JF. 2012. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336:462–466 [DOI] [PubMed] [Google Scholar]
- 17. Denef VJ, Mueller RS, Banfield J. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 4:599–610 [DOI] [PubMed] [Google Scholar]
- 18. Denef VJ, et al. 2009. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ. Microbiol. 11:313–325 [DOI] [PubMed] [Google Scholar]
- 19. Dick GJ, et al. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dopson M, Baker-Austin C, Bond P. 2007. Towards determining details of anaerobic growth coupled to ferric iron reduction by the acidophilic archaeon “Ferroplasma acidarmanus” Fer1. Extremophiles 11:159–168 [DOI] [PubMed] [Google Scholar]
- 21. Dopson M, Baker-Austin C, Hind A, Bowman JP, Bond PL. 2004. Characterization of Ferroplasma isolates and Ferroplasma acidarmanus sp. nov., extreme acidophiles from acid mine drainage and industrial bioleaching environments. App. Environ. Microbiol. 70:2079–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Druschel GK, Baker BJ, Gihring TM, Banfield JF. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 24. Edwards KJ, et al. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem. Geol. 169:383–397 [Google Scholar]
- 25. Edwards KJ, Bond PL, Gihring TM, Banfield JF. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796–1799 [DOI] [PubMed] [Google Scholar]
- 26. Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 27. Florens L, et al. 2006. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goltsman DSA, et al. 2009. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (group II) and “Leptospirillum ferrodiazotrophum” (group III) bacteria in acid mine drainage biofilms. App. Environ. Microbiol. 75:4599–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Golyshina OV, et al. 2000. Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int. J. Syst. Evol. Microbiol. 50:997–1006 [DOI] [PubMed] [Google Scholar]
- 30. Gomez-Alvarez V, Teal TK, Schmidt TM. 2009. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314–1317 [DOI] [PubMed] [Google Scholar]
- 31. Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805 [DOI] [PubMed] [Google Scholar]
- 32. Huber H, et al. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U. S. A. 105:7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inskeep WP, et al. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773 doi:10.1371/journal.pone.0009773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang H, et al. 2008. Dominance of putative marine benthic Archaea in Qinghai Lake, north-western China. Environ. Microbiol. 10:2355–2367 [DOI] [PubMed] [Google Scholar]
- 35. Johnson DB, Joulian C, d'Hugues P, Hallberg KB. 2008. Sulfobacillus benefaciens sp. nov., an acidophilic facultative anaerobic Firmicute isolated from mineral bioleaching operations. Extremophiles 12:789–798 [DOI] [PubMed] [Google Scholar]
- 36. Joshi AB, Kirsch LE. 2002. The relative rates of glutamine and asparagine deamidation in glucagon fragment 22-29 under acidic conditions. J. Pharma. Sci. 91:2332–2345 [DOI] [PubMed] [Google Scholar]
- 37. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kock D, Schippers A. 2008. Quantitative microbial community analysis of three different sulfidic mine tailing dumps generating acid mine drainage. App. Environ. Microbiol. 74:5211–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macalady JL, Jones DS, Lyon EH. 2007. Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ. Microbiol. 9:1402–1414 [DOI] [PubMed] [Google Scholar]
- 40. Macalady JL, et al. 2004. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles 8:411–419 [DOI] [PubMed] [Google Scholar]
- 41. Mouser PJ, et al. 2009. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J. 3:454–465 [DOI] [PubMed] [Google Scholar]
- 42. Mueller RS, et al. 2010. Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol. Syst. Biol. 6:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. 2011. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75:361–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oren A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 39:1–7 [DOI] [PubMed] [Google Scholar]
- 45. Ram RJ, et al. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920 [PubMed] [Google Scholar]
- 46. Rawlings DE. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65–91 [DOI] [PubMed] [Google Scholar]
- 47. R Development Core Team 2010. R: a language and environment for statistical computing. R Development Core Team, Vienna, Austria [Google Scholar]
- 48. Robinson NE. 2002. Protein deamidation. Proc. Natl. Acad. Sci. U. S. A. 99:5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saeed AI, et al. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193 [DOI] [PubMed] [Google Scholar]
- 50. Saeed AI, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 51. Sanchez-Andrea I, Rodriguez N, Amils R, Luis Sanz J. 2011. Microbial diversity in anaerobic sediments at Rio Tinto, a naturally acidic environment with a high heavy metal content. App. Environ. Microbiol. 77:6085–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schroder I, Johnson E, de Vries S. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427–447 [DOI] [PubMed] [Google Scholar]
- 53. Segerer A, Langworthy TA, Stetter KO. 1988. Thermoplasma acidophilum and Thermoplasma volcanium sp. nov. from solfatara fields. Syst. Appl. Microbiol. 10:161–171 [Google Scholar]
- 54. Smith PF, Langworthy TA, Smith MR. 1975. Polypeptide nature of growth requirement in yeast extract for Thermoplasma acidophilum. J. Bacteriol. 124:884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stookey LL. 1970. Ferrozine: a new spectrophotometric reagent for iron. Anal. Chem. 42:779–781 [Google Scholar]
- 56. Tabb DL, McDonald WH, Yates JR. 2002. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tan GL, et al. 2008. Culturable and molecular phylogenetic diversity of microorganisms in an open-dumped, extremely acidic Pb/Zn mine tailings. Extremophiles 12:657–664 [DOI] [PubMed] [Google Scholar]
- 59. Tyson GW, et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 60. White RA, Blainey PC, Fan HC, Quake SR. 2009. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics 10:116 doi:10.1186/1471-2164-10-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wilmes P, et al. 2009. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3:266–270 [DOI] [PubMed] [Google Scholar]
- 62. Yelton AP, et al. 2011. A semi-quantitative, synteny-based method to improve functional predictions for hypothetical and poorly annotated bacterial and archaeal genes PLoS Comput. Biol. 7:e1002230 doi:10.1371/journal.pcbi.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.