Abstract
The emergence of new pathogens and the exploitation of novel pathogenic niches by bacteria typically require the horizontal transfer of virulence factors and subsequent adaptation—a “fine-tuning” process—for the successful incorporation of these factors into the microbe's genome. The function of newly acquired virulence factors may be hindered by the expression of genes already present in the bacterium. Occasionally, certain genes must be inactivated or deleted for full expression of the pathogen phenotype to occur. These genes are known as antivirulence genes (AVGs). Originally identified in Shigella, AVGs have improved our understanding of pathogen evolution and provided a novel approach to drug and vaccine development. In this review, we revisit the AVG definition and update the list of known AVGs, which now includes genes from pathogens such as Salmonella, Yersinia pestis, and the virulent Francisella tularensis subspecies. AVGs encompass a wide variety of different roles within the microbe, including genes involved in metabolism, biofilm synthesis, lipopolysaccharide modification, and host vasoconstriction. More recently, the use of one of these AVGs (lpxL) as a potential vaccine candidate highlights the practical application of studying AVG inactivation in microbial pathogens.
INTRODUCTION
All species evolve over time. The evolution or acquisition of new genes enhances an organism's ability to adapt within novel niches and ultimately augments the organism's fitness. In the case of microbial species, exposure to new environments, competition with other species for limited resources, and the need to evade predators and/or host immune defenses all contribute to selective pressures that determine which organism—and, more importantly, which set of genes—will endure. The rise of antimicrobial resistance is a classic example of pathoadaptation. The horizontal transfer of antibiotic-resistance genes on plasmids or pathogenicity islands provides many modern-day pathogens an edge in survival within the clinical setting.
Gene loss can be just as critical to microbial survival as gene acquisition, although less attention has been given to this facet of the evolutionary process. Most commonly, loss-of-function gene mutations result from bacterial adaptation to a more specific niche. As certain gene products or pathways become superfluous in this new environment, neutral mutations are allowed to accumulate in unnecessary genes with negligible consequences on bacterial fitness. In the earliest stage of this reductive evolution, organisms start to accumulate pseudogenes in unnecessary pathways, although they still retain the majority of the genes necessary for a free-living bacterium. Next, at an intermediate stage in reductive evolution, all or many of the superfluous genes become inactivated but the nonfunctional remnants of these genes still persist. At this stage, the expression of such pseudogenes may already be in a state of erosion. The intermediate stage of reductive evolution can be observed in specific niche-adapted organisms known to boast a high frequency of pseudogenes, such as Shigella flexneri, Salmonella enterica, and Yersinia pestis (11, 39, 54). Over time, regions no longer encoding functional genes may be gradually eliminated from a bacterial genome (37). At the final stage of reductive evolution, bacteria possess considerably smaller genomes than their predecessors and few pseudogenes, indicating that they are reaching the end of this evolutionary pathway (29). Such final stage organisms include endosymbionts and obligate intracellular pathogens, such as Buchnera, Mycobacterium leprae, and Chlamydia trachomatis, which have adapted to scavenge key nutrients from their hosts rather than directly synthesize substrates and have accordingly lost numerous biosynthetic pathways (8, 30, 47, 48).
A second evolutionary force driving gene loss also occurs in microbial pathogens. The concept of antagonistic pleiotropy proposes that a gene whose expression was advantageous in one environment may be detrimental in another environment (10). Consequently, niche adaptation requires selection for and against traits to optimize pathogen fitness in the new environment. As virulence is a critical factor in the continued survival of host-restricted pathogens, the expression of any gene that interferes with virulence will be detrimental to the pathogen's fitness. Adaptation to any newly acquired virulence factors may require selection against these detrimental genes in order to maintain an organism's fitness. This concept is known as antivirulence (31). An antivirulence gene (AVG) is a gene whose expression in a pathogen is incompatible with the virulence of that pathogen (see Table 2). Therefore, an AVG must be inactivated, deleted, or differentially regulated to prevent its expression from interfering with the pathogen's virulence.
Table 2.
Definition of an AVG
| Question | Answer |
|---|---|
| What is an AVG? | An AVG is a gene whose expression in a pathogen is incompatible with the virulence of that pathogen. In the pathogen, an AVG is inactivated, deleted, or differentially regulated so that it cannot interfere with the virulence of that organism. |
| What is not an AVG? | Suppressors. Certain genes, when inactivated under experimental conditions or determined to be nonfunctional in a limited number of natural isolates, can lead to an increase in virulence, a phenomenon commonly known as hypervirulence. |
| Genes not functional in the ancestral species. AVGs must have been present in the ancestral species before being inactivated or lost. As the ancestral strain or species may not always be available for study, the nearest related extant species may be acceptable for comparison. For example, Shigella species arose from an ancestral E. coli strain and Y. pestis diverged from Y. pseudotuberculosis. Pseudogene formation (and, consequently, AVG formation) in Shigella and Y. pestis may be studied by comparing their genomes or phenotypes to the corresponding extant species (E. coli and Y. pseudotuberculosis, respectively). | |
| Genes that have undergone genetic decay due to close association with the known AVG(s). Only that gene which causes the antivirulence phenotype will be considered the AVG. Once that gene has been lost, other associated genes may become superfluous in the absence of a fully functional system and may therefore also decay. This process is part of reductive evolution. |
In this review, we highlight the AVGs that have been described since the last comprehensive review (31) and present a list of known AVGs (Table 1). These include not only AVGs that were lost as a result of commensal-to-pathogen evolution, such as in the case of Escherichia coli to Shigella, but also during pathogen-to-pathogen evolution, as in the case of Yersinia pseudotuberculosis to Yersinia pestis. While we limit our discussion to AVG discoveries in bacterial species, antivirulence has the potential to occur in other pathogens, such as parasites, although the genetic complexities of these organisms make AVGs more difficult to identify.
Table 1.
Known AVGs
| Pathogen(s) | Gene(s) | Process(es) inhibited by presence of functional gene(s) | Reference(s) |
|---|---|---|---|
| Shigella/EIEC | nadA/nadB | T3SS secretion | 41, 42 |
| Shigella/EIEC | cadA | PMN transepithelial migration, ShET1/ShET2 enterotoxin activity, phagolysosome escape | 9, 18, 32, 33 |
| Shigella | speG | Oxidative stress survival | 4 |
| Shigella | ompT | Stability of IcsA | 3 |
| Salmonella | lacI | Expression of SPI-2 genes | 16 |
| Burkholderia pseudomallei | araA-araH | Virulence in golden Syrian hamster model | 36 |
| Yersinia pestis | rcsA, nghA | Biofilm formation, biofilm stability | 14, 15, 49 |
| Yersinia pestis | lpxL | Protection from host immune response | 35 |
| Francisella tularensis subsp. tularensis and holarctica | pepO | Systemic spread of pathogen | 21 |
WHAT IS (AND WHAT IS NOT) AN AVG
Before reviewing the most recent additions to the AVG list, we must first correct some common errors made in the classification of such genes (Table 2). First, virulence suppressors or regulators are not AVGs. The term “hypervirulent” is often used to describe a strain that exhibits a significant increase in virulence compared to either the parental wild-type strain or related strains within the same species. Such an increase in virulence may be due to the functional loss of one or more genes, and this phenomenon has been documented in wild-type strains or discovered through genetic manipulation of strains within the laboratory (3, 26, 44). However, such genes are still functional in either the majority of wild-type strains within the same species or, in the case of lab-induced hypervirulence, in the parent wild-type strain and therefore do not fit our criteria for AVGs. Instead, these genes are best defined as regulators of virulence or virulence suppressors.
Second, AVGs must have originally been both widely present and functional in the ancestral species. ompT, previously characterized as an AVG in Shigella, presents an example of the difficulties in applying this criterion (38). OmpT, a surface protease, degrades the bacterial protein IcsA, consequently inhibiting Shigella cell-to-cell spread. ompT is situated within the DLP12 cryptic lambdoid-like prophage, just downstream of the gene envY. This prophage is carried by most, but not all, lineages of E. coli (109/144 strains in the EcoCyc database carry ompT; data retrieved on 11 July 2012, EcoCyc version 16.1) (27). None of the sequenced Shigella strains carry the DLP12 cryptic prophage at this position, so we cannot say with certainty that all or indeed any of the ancestral Shigella strains ever had, and subsequently lost, ompT. Although we tentatively label ompT as a Shigella AVG, the inclusion of ompT in this category remains debatable.
There are other obvious difficulties in applying this second criterion. In certain cases, we simply will not have an extant species available to study. In the case of host-restricted Shigella, which is postulated to have arisen from multiple lineages within free-living commensal E. coli, evaluation of nutrient requirements within the host, conserved metabolic pathways, and readily accessible genome sequences make the identification of putative AVG targets more straightforward. In the absence of a reference species to compare to the pathogen of interest, it becomes much more challenging to differentiate putative AVGs from those that were lost due to reductive evolution.
Finally, any gene that has undergone genetic decay as a result of close association with the AVG will not be considered an AVG in its own right. cadA, for example, is the AVG linked to absence of the lysine decarboxylase system in Shigella. The closely associated cadB gene, which encodes a lysine-cadaverine antiporter, is also inactivated in nearly all species. However, the presence of cadB does not appear to have any effect on virulence alone. With the loss of functional cadA, the lysine decarboxylase system in Shigella was lost, and in the absence of any selective pressure to maintain a functional cadB, mutations were allowed to accumulate in this gene. However, note that in certain pathways or systems, more than one gene may be involved in the antivirulence phenotype. For example, both nadA and nadB of Shigella encode enzymes that catalyze the synthesis of quinolinic acid, a small-molecule inhibitor of Shigella virulence (42). If either gene is inactivated, the pathway to quinolinic acid synthesis is lost, so both nadA and nadB are considered AVGs for Shigella.
THE REPERTOIRE OF AVGs IN BACTERIAL PATHOGENS
Shigella, enteroinvasive E. coli (EIEC), enterohemorrhagic E. coli (EHEC), and Shiga toxin-producing E. coli (STEC).
Shigella likely arose 35,000 to 270,000 years ago from multiple ancestral E. coli lineages (40). Although Shigella retains a separate genus and species classification because of its medical significance, more recent phylogenetic analyses suggest that this pathogen is actually part of the E. coli pathovar (40).
During its evolution from an extracellular resident of the mammalian colon to an intracellular pathogen, Shigella acquired a large 220-kb virulence plasmid which harbors the genes required for successful invasion, replication, and dissemination inside host cells, in addition to the induction of the host inflammatory response that is critical to the bacterium's life cycle (46). As the majority of Shigella virulence factors are plasmid encoded and the rest are localized to distinct pathogenicity islands, it has been suggested that Shigella evolved from commensal E. coli strains. In addition to gene acquisition, Shigella accumulated a plethora of pseudogenes. On average, each strain appears to have lost the functionality of approximately 200 genes (55). As E. coli remains the paradigm species for bacterial research and Shigella has so recently evolved, there is a unique opportunity to study both reductive evolution and antivirulence in Shigella.
EIEC, one of the five classical pathogenic E. coli subtypes, shares a particularly close relationship with Shigella. EIEC is a nonmotile facultative intracellular pathogen, harbors the same 220-kb virulence plasmid as Shigella, and is usually grouped with Shigella as a single pathovar within E. coli (28). Only a few metabolic differences, including mucate and acetate production, separate Shigella and EIEC. Many AVGs of Shigella, such as those responsible for lysine decarboxylase activity and quinolinic acid synthesis, have also been mutated in or lost from EIEC (6, 41). A previous review focused on the loss of two AVG-encoded proteins by Shigella and EIEC, i.e., the surface protease encoded by ompT and the lysine decarboxylase enzyme encoded by cadA (31). No new publications have addressed ompT, so we will not discuss that particular AVG further. Since the last review, several additional AVGs have been identified in Shigella, including nadA/nadB and speG (4, 42) (Fig. 1).
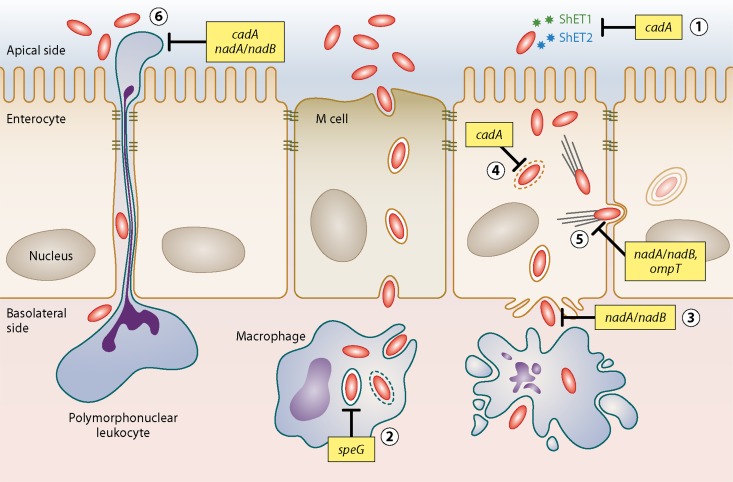
Fig 1.
Inhibition of pathogenesis in Shigella. Pathogenesis phenotypes interrupted by Shigella AVGs. The product of the lysine decarboxylase reaction, cadaverine, inhibits ShET1/ShET2 enterotoxin activity (part 1), phagosome escape (part 4), and PMN transepithelial migration (part 6). Another small molecule, quinolinic acid, is the product of the nadA/nadB enzymatic reactions and inhibits both Shigella invasion (part 3) and intracellular spread (part 5). Inactivation of speG, which encodes the spermidine acetyltransferase, allows spermidine to accumulate within the phagosome and ultimately promotes bacterial survival in the macrophage (part 2). OmpT, an outer membrane protease, cleaves IcsA from the bacterial surface, preventing actin tail polymerization and inhibiting cell-to-cell spread (part 5).
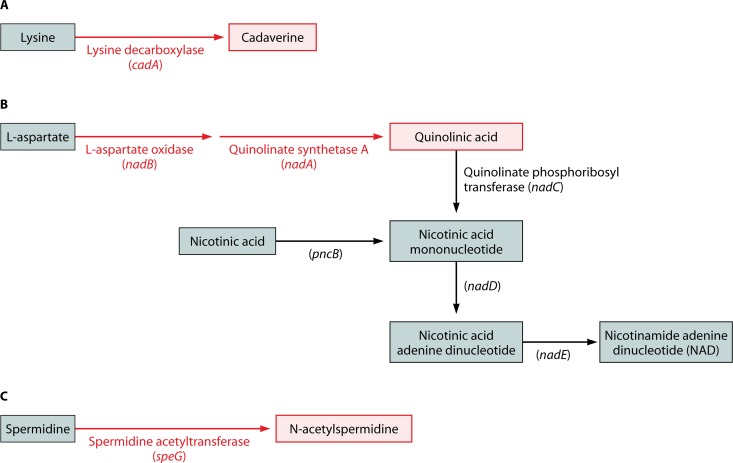
The lysine decarboxylase enzyme converts the amino acid lysine to the polyamine cadaverine. This exchange functions as one of several acid resistance systems in E. coli. In contrast, a functional lysine decarboxylase system has been uniformly lost by all Shigella species (1) (Fig. 2A).This system consists of two operons, cadBA, encoding a lysine/cadaverine antiporter and a lysine decarboxylase enzyme, respectively, and cadC, the transcriptional activator of cadBA. Absence of lysine decarboxylase activity is one of the classic hallmarks that distinguish Shigella/EIEC from other E. coli species in the clinical laboratory. A wide variety of different mutations or deletions have resulted in the loss of cadA, and often cadB and cadC, by all known Shigella/EIEC species (9). The obvious convergence of these different Shigella strains is indicative of strong selective pressure against the cadA locus.
Fig 2.
Metabolic pathways lost in Shigella. Compounds or enzymes still present in Shigella are marked in black; those that have been lost are marked in red. (A) Lysine decarboxylation. (B) Biosynthetic and salvage NAD pathways. (C) Spermidine metabolism.
The expression of functional cadA in wild-type Shigella or exposure to physiologically relevant levels of the exogenous end product, cadaverine, results in the disruption of ShET1/ShET2 enterotoxin activity, inhibition of polymorphonuclear leukocyte (PMN) transepithelial migration, and blockage of bacterial phagosome escape, all of which are critical contributors to Shigella virulence inside the host (18, 32, 33). The mechanism(s) of action of cadaverine with regard to the inhibition of Shigella virulence, although under investigation, remains unknown.
Although approximately 90% of E. coli species encode a functional lysine decarboxylase system, the cad operon has also been inactivated in or lost by certain strains of EHEC and STEC. The reconstitution of a functional lysine decarboxylase system in these strains decreases their ability to adhere to host cells (52, 53). In these studies, the increased adherence seen in cadA-negative strains appears to be the result of increased expression of the outer membrane adhesin intimin, although the exact mechanism of action is not yet clear. This phenomenon, while not considered antivirulence, suggests that the lysine decarboxylase system plays an important role not only in acid tolerance in E. coli but also in the regulation of numerous virulence factors of both Shigella and E. coli. Although cadA is currently not inactivated across all EHEC and STEC strains, which precludes the classification of cadA as an AVG in these organisms, these events may define a case of evolutionary transition. If selective pressure to lose this locus continues, the cadA gene may eventually be considered an AVG in these pathogens.
In order for E. coli to synthesize nicotinic acid mononucleotide, a precursor of the essential coenzyme NAD, the enzymes l-aspartate oxidase (encoded by nadB) and quinolinate synthetase A (encoded by nadA) are required. In contrast, Shigella has a strict nutritional requirement for nicotinic acid for growth on minimal media, suggesting that Shigella relies on a salvage pathway for NAD (20). When the sequences of genes required for de novo NAD synthesis in E. coli were compared to the Shigella genome, it quickly became apparent that all Shigella species have inactivated nadA, nadB, or both genes (41, 42). These inactivations take the form of multiple amino acid substitutions, complete or partial deletions of nadA or nadB, or insertion sequence (IS) elements interrupting either or both genes. The wide variety of different genetic alterations utilized by Shigella to inactivate these genes suggests that there was strong selective pressure to lose this part of the NAD pathway. To compensate, Shigella bypasses this early block in the pathway by importing exogenous nicotinic acid and converting it to the NAD precursor nicotinic acid mononucleotide through the actions of nicotinate phosphoribosyltransferase (42) (Fig. 2B).
Prunier et al. showed that one of the intermediates in the de novo synthesis pathway, quinolinic acid, attenuates both Shigella invasion and intracellular dissemination, in addition to blocking PMN transepithelial migration (42). The secretion of Shigella type III secretion system (T3SS) effectors, such as IpaB and IpaC, is greatly decreased in the presence of quinolinic acid, suggesting that inadequate secretion of Shigella effectors most likely contributes to these phenotypes. Interestingly, this inhibition is limited to the Shigella T3SS. Other organisms with a T3SS, such as Salmonella enterica, enteropathogenic E. coli, or Y. enterocolitica, do not exhibit virulence inhibition in the presence of quinolinic acid. The exact mechanism of quinolinic acid inhibition upon Shigella virulence is not yet known.
Finally, the ability to acetylate another polyamine, spermidine, has also been lost in the evolution of E. coli to Shigella (4). Barbagallo and colleagues utilized a particularly novel approach in an attempt to locate new Shigella AVGs. Working on the hypothesis that the transcriptional activators of the major virulence plasmid, such as VirF, might also incidentally activate other genes on the E. coli chromosome, they conducted a global transcriptional analysis to identify VirF-activated E. coli genes and then determined whether or not those genes were correspondingly inactivated in Shigella. Genes that were turned on in E. coli by VirF but were functionally lost in Shigella were identified as putative AVGs. One of the genes identified via this method was speG, which encodes a spermidine acetyltransferase that transfers an acetyl group onto the polyamine spermidine to produce N-acetylspermidine (Fig. 2C).
speG is inactivated in all Shigella lineages through a startling number of missense mutations, IS insertions, and complete deletions of speG (4). Unlike loss of the lysine decarboxylase system, loss of speG was not designed to prevent the synthesis of an inhibitory end product. Instead, Shigella likely inactivated or lost this enzyme because the pathogen actually requires increased amounts of the substrate, spermidine, for intracellular survival. Therefore, the blockage of any pathway that metabolizes this polyamine would be favored by the emerging pathogen.
In wild-type strains from each of the four Shigella species (S. flexneri, S. boydii, S. sonnei, and S. dysenteriae), absence of functional SpeG prevents spermidine metabolism and, correspondingly, spermidine levels are significantly higher in wild-type Shigella than in wild-type E. coli. When the active speG gene in E. coli is replaced with the inactive speG gene from S. dysenteriae, spermidine levels rise to the levels seen in the wild-type Shigella strains (4). Increased spermidine levels correlate with increased survival in response to oxidative stress, such as exposure to hydrogen peroxide in macrophages, which is particularly critical to this microbe's pathogenesis. In the early stages of infection, Shigella is taken up by macrophages, where it survives and eventually escapes the unfavorable conditions of the phagosome, which include reactive oxygen species such as superoxide and hydrogen peroxide. Ultimately, the pathogen induces pyroptosis of the host macrophage to invade neighboring epithelial cells (50).
In a mouse macrophage model, wild-type Shigella is able to outcompete the same Shigella strain expressing the active speG gene from E. coli (4). Taken together, these data suggest that the accumulation of spermidine may be critical for Shigella intracellular survival in vivo, and loss of a functional speG gene was crucial for Shigella pathoadaptation to its new host niche. Although the mechanism for this survival is not yet clear, Barbagallo and colleagues (4) determined that in the presence of increased spermidine, the expression of katG also rose. The hydroperoxidase encoded by katG is critical for antioxidant defense in bacteria. Its expression is mediated by the OxyR stress response which is induced in the presence of polyamines such as spermidine. Thus, spermidine-mediated induction of OxyR may account for the increased macrophage survival seen in wild-type Shigella. Taken together, these data support the identity of speG as an AVG in Shigella.
SALMONELLA
E. coli and Salmonella diverged from a common ancestor approximately 100 million years ago (12). The genus Salmonella comprises two main species: Salmonella enterica, which includes the serovars Typhi, Typhimurium, and Enteritidis; and Salmonella bongori, a closely related species that infects cold-blooded animals. It has been postulated that these two species diverged approximately 25 to 40 million years ago and that subsequent development of S. enterica ultimately required the acquisition of new virulence factors by lateral gene transfer (5). Among these, S. enterica obtained SPI-2, a chromosomally located pathogenicity island that contains the genes necessary for intracellular survival and replication.
One of the characteristics that distinguish E. coli and Salmonella is the ability of E. coli to ferment lactose, while Salmonella is traditionally thought of as a nonfermenter. Paradoxically, both organisms thrive in the human gut, where dietary lactose is readily available as an energy source. It is unknown if the ancestral lineage that gave rise to both E. coli and Salmonella was a lactose fermenter or if this ability was obtained only by E. coli after the divergence of these two organisms.
In E. coli, the lac system contains four genes, three of which are located in an operon, i.e., lacZ (β-galactosidase), lacY (lactose permease), and lacA (transacetylase). The fourth gene, lacI, encodes the lac operon repressor and negatively regulates the system under lactose-depleted conditions.
Although the majority of Salmonella strains are unable to ferment lactose, there have been isolated reports of strains that can do so (34). Upon closer inspection of the lactose-fermenting Salmonella strains, it was determined that the functional lacZYA genes in these particular strains are likely carried on transmissible plasmids rather than in the chromosome (16). Furthermore, the vast majority of these strains lack lacI. The only known S. enterica strain (ST-2) that synthesizes a functional LacI protein has decreased repression efficiency (17). S. bongori also appears to be undergoing loss of the lac system; of the four genes, only lacI and lacZ are present, and transcriptional analysis suggests that lacI, at least, is a pseudogene (16).
Eswarappa and colleagues introduced a fully functional lacI gene into Salmonella to study its effect on pathogenicity. When lacI is expressed in S. enterica lacking a functional lac operon, bacterial virulence is significantly reduced in a murine typhoid fever model compared to that in the wild-type parent (16). Moreover, although the bacteria expressing lacI are able to invade as well as their wild-type parent, they lack the ability to proliferate inside murine macrophages, suggesting that the expression of lacI interferes with postinvasion events. A microarray analysis revealed that several genes of the SPI-2 pathogenicity island, which harbors a T3SS thought to be critical in the assembly of the Salmonella-containing vacuole, are downregulated in the presence of functional lacI. These downregulated genes include ssaK, which encodes a T3SS apparatus protein; sseB, which encodes a T3SS effector that makes up part of the SPI-2 translocon; and spiC, which encodes a T3SS effector essential for the SPI-2-mediated secretion of SseB, SseC, and SseD (24, 56).
Although the exact mechanism of action has yet to be elucidated, the ability of LacI to bind operator sequences is not required for the AVG phenotype, suggesting that LacI does not function as a direct repressor to downregulate these genes. Instead, it is possible that LacI may inhibit the expression of these genes via protein-protein interactions with other transcription factors. The functional consequence of the downregulation of these T3SS genes for bacteria residing in the macrophage vacuole is decreased survival, which ultimately marks lacI as an AVG of Salmonella.
YERSINIA
Yersinia pestis, the agent of bubonic and pneumonic plague, exploits a flea vector for its mammalian host transmission cycle. In contrast, the closely related species Y. pseudotuberculosis, which is a much milder enteric pathogen in humans, is transmitted through the fecal-oral route and (like Y. pestis) preferentially infects lymphoid tissue. Despite drastic differences in the mode of transmission, these species share at least 97% genetic identity, and an ancestral Y. pseudotuberculosis strain is believed to have given rise to Y. pestis (1, 7). Intriguingly, the evolution of Y. pseudotuberculosis to Y. pestis has affected only a few genes (22). The acquisition of new genes by Y. pestis has been well documented and includes the horizontal transfer of two plasmids, pPCP1 and pMT1, which are vital for bacterial dissemination and flea vector survival, respectively (57).
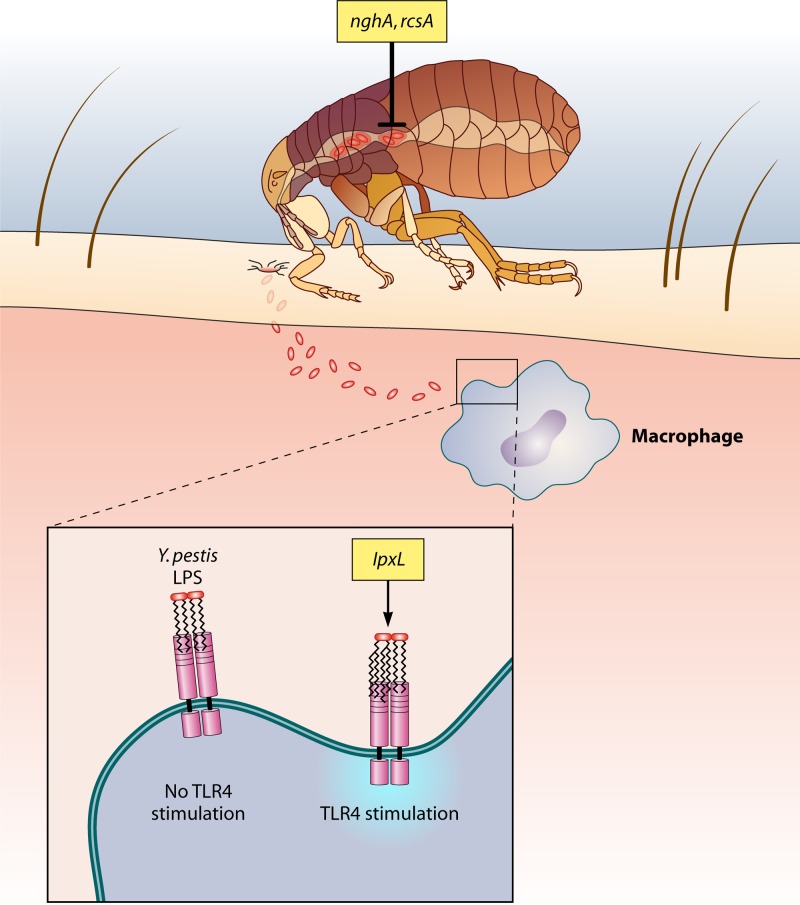
Loss-of-function mutations have also played a significant role in the evolution of Y. pestis from Y. pseudotuberculosis. Approximately 200 genes are inactivated in Y. pestis, according to a genome comparison with Y. pseudotuberculosis (7). Notably, Y. pestis lost genes whose products repress biofilm synthesis (rcsA) and enhance biofilm degradation (nghA), underscoring the importance of biofilm stability in this pathogen's lifestyle. Furthermore, Y. pestis has also lost lpxL, a gene that encodes an acyltransferase that modifies bacterial lipopolysaccharide (LPS). In the absence of this protein, the bacteria are unable to stimulate host Toll-like receptor 4 (TLR4), which plays a critical role in pathogen evasion of the host immune response (Fig. 3).
Fig 3.
Inhibition of pathogenesis in Yersinia. nghA and rcsA encode proteins that inhibit biofilm formation in Yersinia: NghA directly degrades formed biofilm, and RcsA increases the repression ability of RcsB, an inhibitor of biofilm formation. Inactivation of these genes allows Yersinia to form a biofilm on the proventriculus of the flea, enabling bacterial transmission. LpxL mediates hexa-acetylation of lipid A on bacterial LPS, thus activating TLR4 and stimulating the host immune response to this pathogen. Loss of lpxL in all sequenced Y. pestis isolates leads to increased pathogen evasion of host innate immune defenses.
Y. pestis must generate a specialized hms-dependent biofilm over the spines of the proventriculus of the flea in order to optimize the low efficiency of transmission from flea to mammalian host (22). Even in the preferred vector, Xenopsylla cheopis, the transmission rate of infected fleas only reaches around 50% and biofilm formation across the proventriculus is considered a requirement for even this relatively low rate (22). Not only does an extracellular matrix permit the bacteria to aggregate and adhere to midgut epithelial cells, the biofilm also blocks ingested food from entering, ultimately starving the flea. As the flea attempts, unsuccessfully, time and time again to feed and sate its appetite, the frequency of new host bites rises and, correspondingly, the probability of bacterial transmission increases. Not surprisingly, blocked fleas transmit Y. pestis at a much higher rate than their unblocked counterparts do (23).
Because of the critical role biofilm formation plays in Y. pestis transmission, genes that function in biofilm breakdown or negatively regulate biofilm synthesis would conflict with the ability of Y. pestis to pass from vector to host. Two biofilm-associated genes—rcsA and nghA—are inactivated in all known Y. pestis strains and fit the criteria for Y. pestis AVGs (14, 49).
rcsA is a negative regulator of biofilm synthesis (49). RcsA, an accessory protein for the Rcs histidine kinase bacterial phosphorelay system, increases the repression activity of RcsB, a DNA-binding protein involved in the negative regulation of biofilm synthesis genes. Loss of functional RcsA decreases the binding stability and repressor activity of RcsB, which is unable to fully repress biofilm synthesis independently.
In 2008, Sun and colleagues determined that expressing the functional Y. pseudotuberculosis rcsA gene in a wild-type Y. pestis background not only abolished in vitro biofilm formation but also significantly decreased flea blockage in vivo (49). In contrast, when the reciprocal experiment was performed and the functional rcsA gene in Y. pseudotuberculosis was replaced with the Y. pestis pseudogene, the resulting strain was able to form biofilms in a C. elegans model, unlike its wild-type parent. The presence of a functional RcsA protein inhibits biofilm formation, which would be incompatible with the Y pestis lifestyle in the flea. rcsA, therefore, is an AVG for Y. pestis.
The most common inactivating mutation of rcsA is a 30-bp duplication insertion midway through the reading frame that not only has been verified in the main subspecies, Y. pestis subsp. pestis, but is also suspected to be prevalent in all other subspecies (Y. pestis subsp. caucasica, altaica, hissarica, and ulegeica) (15). This result suggests that inactivation of rcsA likely occurred early in Y. pestis evolution, possibly as a result of considerable selective pressure upon the bacteria to inactivate rcsA and allow for biofilm formation in order to enhance transmission from the flea. The only other known mutation of rcsA in Y. pestis occurs in the strain Antiqua, which lacks the characteristic 30-bp insertion but instead carries a 1.2-kb putative transposase gene inserted midway through the reading frame. Although the functionality of this interrupted Antiqua rcsA has not yet been experimentally determined, it is probably inactive.
The loss of a biofilm degradation mechanism also appears to contribute to the ability of Y. pestis to form biofilms in the flea. Y. pestis extracellular matrix formation in the midgut of the flea is dependent on the expression of hms genes, which synthesize a poly-β-1,6-N-acetyl-d-glucosamine biofilm (14). nghA (formerly chb) encodes an active glycosyl hydrolase in Y. pseudotuberculosis. In one of the Y. pestis strains tested by Erickson and colleagues, no β-hexosaminidase activity was detected, suggesting that the Y. pestis nghA protein is nonfunctional. An 11-bp deletion early in the Y. pestis nghA gene leads to a premature stop codon, likely resulting in a truncated and nonfunctional protein product. This inactivating mutation appears to be conserved in all Y. pestis lineages, while Y. pseudotuberculosis strains retain full-length nghA.
When a functional nghA gene from Y. pseudotuberculosis is expressed in Y. pestis, the resulting product is able to cleave β-linked N-acetylglucosamine residues from the HMS-dependent extracellular matrix, ultimately destabilizing formed biofilms (14). Correspondingly, although this strain colonizes the midgut, similar to Y. pseudotuberculosis, biofilm formation on the flea proventriculus is markedly decreased. These experiments suggest that Y. pestis has adapted to its flea vector by selecting against the gene responsible for degradation of the extracellular matrix.
In addition to differences in the ability to form biofilms, Y. pestis and Y. pseudotuberculosis also exhibit another striking dissimilarity: Y. pestis lacks the ability to convert its LPS lipid A from tetra-acylated to hexa-acylated (35). Hexa-acylation, a posttranslational modification of the lipid A subunit of bacterial LPS, is critical to provoking a potent immune response through interactions with CD14 and TLR4 on the host cell surface. Because of this deficit in lipid A hexa-acylation, infection with Y. pestis induces only a weak innate immune response and ultimately gives the bacteria a direct survival advantage in vivo. In contrast, Y. pseudotuberculosis displays hexa-acylated lipid A at the host temperature of 37°C and produces an LPS capable of activating TLR4 (45).
Hexa-acylation of lipid A is carried out by two proteins, LpxL and LpxP. LpxL is a lauroyl acyltransferase that attaches secondary acyl chains to the tetra-acylated lipid A subunit (43). The lpxL gene is completely absent from all sequenced strains of Y. pestis (35). LpxP, an acyltransferase produced by Y. pestis, is active exclusively under cold shock conditions and cannot rescue hexa-acylation in an lpxL mutant at normal host temperatures (43).
When Montminy and colleagues expressed a functional lpxL gene from E. coli in a wild-type Y. pestis strain, the bacteria strongly activated human peripheral blood mononuclear cells compared to the wild-type strain, as evidenced by an increase in tumor necrosis factor (TNF), interleukin-6 (IL-6), and IL-8 from these host cells (35). In a mouse model, the wild-type strain predictably resulted in 100% mortality, while the strain expressing lpxL did not result in any noticeable signs of disease. Mice infected with the latter strain had significantly lower spleen bacterial titers, in addition to increases in TNF levels and the appearance of liver microabscesses, indicative of a robust immune response. This host protection was TLR4 dependent, as TLR4-deficient mice infected with the strain expressing lpxL succumbed to infection. Conclusively, it was demonstrated that when the lipid A of Y. pestis undergoes hexa-acylation, the immune system is able to effectively recognize and mount an appropriate immune response against the infection. To optimize pathogenesis and increase evasion of host immune responses, Y. pestis deleted lpxL, which joins rcsA and nghA as a Y. pestis AVG.
Montminy and colleagues proposed that this new AVG might be a potential vaccine candidate and successfully tested this hypothesis in a mouse model of infection. Mice infected with the Y. pestis strain expressing lpxL were completely protected against a challenge with wild-type Y. pestis at least 40 days postinoculation (35). This utilization of an AVG demonstrates the powerful potential for practical application of such studies and further illustrates the significance of continuing to search for other AVGs in bacterial pathogens.
FRANCISELLA TULARENSIS
The genus Francisella, composed of several facultative, intracellular zoonotic pathogens, is traditionally divided into two species, F. philomiragia and F. tularensis (51). F. philomiragia is a pathogen of muskrats and fish, though it may rarely cause disease in immunocompromised humans. Both the virulence and the transmission of this organism are poorly understood (25). F. tularensis is further split into four main subspecies, F. tularensis subsp. tularensis, holarctica, mediasiatica, and novicida.
F. tularensis subsp. tularensis (also known as F. tularensis type A), the most virulent subspecies of this subset, infects a broad mammalian host range and is most commonly spread via a wide variety of arthropod vectors, although less common routes of infection may include aerosolization and direct contact with infected animal meat (13). In humans, infection manifests itself as a potentially fatal ulceroglandular or pneumonic tularemia. Strains of F. tularensis subsp. holarctica (also known as F. tularensis type B) also infect healthy individuals but are only moderately virulent compared to strains of F. tularensis subsp. tularensis. F. tularensis subsp. mediasiatica and F. tularensis subsp. novicida, in contrast, typically occur only as human pathogens in immunocompromised individuals. On the basis of phylogenetic analysis, F. tularensis subsp. novicida likely branched first from an ancestral Francisella lineage, followed by F. tularensis subsp. tularensis and F. tularensis subsp. mediasiatica, which likely diverged at around the same time. F. tularensis subsp. holarctica, which branched last, is thought to be the most recent subspecies to emerge (51).
F. tularensis subsp. novicida PepO is an M13 zinc metalloprotease that cleaves the neuropeptide met-enkephalin in vitro (21). This secreted metalloprotease is an orthologue of the mammalian enzyme ECE-1, which cleaves proendothelin into the potent vasoconstrictor endothelin. PepO proteins from other bacterial species, including those of the genera Streptococcus and Porphyromonas, have been shown to mimic the activity of the host vasoconstrictor and may, in fact, represent lateral gene transfer between eukaryotes and prokaryotes (2, 19). Strains of F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, and F. tularensis subsp. mediasiatica have all lost the ability to secrete an active PepO (21).
pepO is secreted from F. tularensis subsp. novicida via a modified type 4 pilus (T4P), a system that in Francisella is postulated to act as a means of secreting proteins rather than as an actual pilus structure (21). When pilC, an inner membrane protein essential for T4P assembly, is deleted from F. tularensis subsp. novicida, the resulting mutant has significantly higher lethality in a mouse model of infection than its wild-type parent does. The bacterial burden in the spleen of mice infected with the mutant is correspondingly higher, leading to the hypothesis that bacterial spread from the initial site of infection is hindered. Because of the predicted function of PepO, a pepO mutant was constructed and tested in the mouse model to determine if T4P-secreted PepO was responsible for inhibiting bacterial spread. Strikingly, like the pilC mutant, the F. tularensis subsp. novicida pepO deletion mutant displays enhanced lethality in the mouse model, compared to that of the wild-type parent, in addition to a greater bacterial load in the spleen (21). In a separate model of disease in which mice are exposed to aerosolized wild-type or pepO mutant strains, PMN influx into the lung is increased in response to mutant but not wild-type infection. These results suggest that increased vasoconstriction occurs in the presence of active PepO, as found in wild-type strains of F. tularensis subsp. novicida, ultimately restricting both bacterial and PMN spreading. In contrast, loss of functional PepO in the more virulent Francisella subspecies likely allows bacterial dissemination and increased pathogenicity.
Loss or inactivation of pepO in F. tularensis subsp. tularensis and holarctica stems from multiple mutation events. In F. tularensis subsp. tularensis strains, IS-mediated genomic rearrangements have replaced the N-terminal secretion signal, likely resulting in an inability of these strains to secrete PepO (21). In addition, there are numerous amino acid alterations of F. tularensis subsp. tularensis PepO in comparison with active PepO from F. tularensis subsp. novicida. The effect these changes have upon protein activity is unknown. F. tularensis subsp. holarctica strains carry a different IS-mediated genomic rearrangement that also targets the N-terminal secretion signal. In addition, these strains carry a nonsense mutation partway through the gene that truncates the protein and removes the protease domain, likely rendering the protein inactive (21). F. tularensis subsp. mediasiatica appears to have undergone the same genetic rearrangement as F. tularensis subsp. holarctica. The location of the nonsense mutation differs, however, suggesting that while early rearrangement to hamper the secretion of this protein was similar in an ancestral strain, actual inactivation of the gene may have been independent for each species. Finally, pepO is absent from the only sequenced F. philomiragia genome, suggesting that either this gene has been deleted from all or certain strains of this species or pepO was never present in the ancestral progenitor strain.
The inactivation of pepO in the more lethal Francisella subspecies inhibits host vasoconstriction and permits the spread of the bacteria to systemic sites, allowing pepO to join the list of newly described AVGs.
IDENTIFICATION OF NOVEL AVGs
The isolation of putative AVGs in a pathogen's genome is not necessarily a straightforward process. An increase in the frequency of pseudogenization within an emerging pathogen could be indicative of both reductive evolution and AVG loss. Genes lost during adaptation as a consequence of reductive evolution confound the identification of putative AVGs, and it is likely that the vast majority of pseudogenes arise from reductive evolution rather than through AVG loss.
To identify putative AVG candidates, a reference species must first be defined to allow comparisons with the species of interest. E. coli, for example, would be classified as an appropriate reference species for Shigella, as would Y. pseudotuberculosis for Y. pestis. A thorough genomic analysis of all sequenced strains can be performed to identify pseudogenes that allude to the conserved inactivation of a gene or pathway across the species of interest. Different “filters” can be applied following the initial bioinformatics search, i.e., a phenotypic filter looking for conserved loss of function across all of the strains of a species, including loss of certain surface-exposed determinants or altered nutritional needs, a phylogenetic filter looking at the function of the gene/pathway in the reference species, and so on. At this stage, a thorough understanding of the pathogen, the nearest extant species, and any inactivated gene(s)/pathways is critical to pinpoint any gene(s) that should have continued to be advantageous to the pathogen following niche-specific adaptation. This is especially true where no explanation can be given for the loss of a particular gene/pathway. The lysine decarboxylase system, as an acid resistance system, could be beneficial to an organism that must survive the acidity of the stomach and host cell phagosome; why, then, have Shigella strains universally lost the function of cadA? The detrimental activity of cadA during Shigella virulence overwhelmed any benefits afforded by increased acid resistance.
Nevertheless, even this is an imperfect process; the loss of nadAB in Shigella, for example, could easily have been dismissed as an example of reductive evolution, as these genes encode enzymes in a metabolic pathway that can be bypassed through the uptake of exogenous nicotinic acid (in contrast, E. coli strains can encode both biosynthetic and scavenger pathways). Shigella, therefore, has the ability to steal exogenous nicotinic acid from the host and might have had no need for a biosynthetic pathway. In reality, the end product of the nadAB reactions, quinolinic acid, was inhibitory to Shigella virulence, arguing for positive rather than neutral selection. Until this phenotype was tested in vitro, these particular AVGs were not identified.
CONCLUDING REMARKS
Microbial evolution is the result of selection for specific traits that optimize the organism's fitness. In the case of pathogens, the acquisition of virulence traits must also correspond to the appropriate integration of those traits into the organism's current genome. This may ultimately result in inactivation of AVGs deleterious to the pathogen's new virulence factors. Such a fine-tuning process allows the pathogen to further adapt to its new host niche. It is likewise important to recognize that inactivation of AVGs can occur during the evolution of commensals to pathogens, such as E. coli to Shigella, or even during the evolution of one pathogen into another, such as Y. pseudotuberculosis to Y. pestis.
In the last few years, several new AVGs have been described in the literature, increasing not only our repertoire of known AVGs but also the number of pathogens that harbor these genes (Table 1). An impressively wide variety of different roles are attributed to AVGs in these organisms, and the current list of AVGs encompasses genes involved in metabolism, biofilm synthesis, LPS modification, lactose regulation, and protease activity, among other functions.
A more complete understanding of AVGs is vital to comprehending the evolution of pathogen virulence. More importantly, the study of antivirulence may assist in the uncovering of novel virulence targets and contribute to the development of inhibitors of those targets. The reintroduction of functional AVGs in their respective pathogens may prove effective as live-vaccine candidates, as the successful utilization of an lpxL-expressing strain of Y. pestis has recently demonstrated. Such work supports and highlights the importance of continuing the search for new AVGs.
ACKNOWLEDGMENTS
We thank Sabrina Joseph and Ana Marquez for helpful discussions regarding the manuscript.
Research on Shigella pathogenesis in the Maurelli lab is supported by grant AI24656 from the National Institute of Allergy and Infectious Diseases.
The opinions or assertions contained herein are ours and are not to be construed as official or as reflecting the views of the Department of Defense or the Uniformed Services University.
Footnotes
Published ahead of print 8 October 2012
REFERENCES
- 1. Achtman M, et al. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Awano S, et al. 1999. Sequencing, expression and biochemical characterization of the Porphyromonas gingivalis pepO gene encoding a protein homologous to human endothelin-converting enzyme. FEBS Lett. 460:139–144 [DOI] [PubMed] [Google Scholar]
- 3. Baek CH, Wang S, Roland KL, Curtiss R., III 2009. Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:1278–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbagallo M, et al. 2011. A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene. PLoS One 6:e27226 doi:10.1371/journal.pone.0027226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casalino M, Latella MC, Prosseda G, Colonna B. 2003. CadC is the preferential target of a convergent evolution during enteroinvasive Escherichia coli toward a lysine decarboxylase-defective phenotype. Infect. Immun. 71:5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chain PS, et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole ST, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011 [DOI] [PubMed] [Google Scholar]
- 9. Day WA, Jr, Fernández RE, Maurelli AT. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Day WA, Maurelli AT. 2006. Black holes and antivirulence genes: selection for gene loss as part of the evolution of bacterial pathogens, p 109–122 In Seifert HS, DiRita VJ. (ed), Evolution of microbial pathogens. ASM Press, Washington, DC [Google Scholar]
- 11. Deng W, et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doolittle RF, Feng DF, Tsang S, Cho G, Little E. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470–477 [DOI] [PubMed] [Google Scholar]
- 13. Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. 2008. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J. Bacteriol. 190:8163–8170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eroshenko GA, Vidyaeva NA, Kutyrev VV. 2010. Comparative analysis of biofilm formation by main and nonmain subspecies of Yersinia pestis strains. FEMS Immunol. Med. Microbiol. 59:513–520 [DOI] [PubMed] [Google Scholar]
- 16. Eswarappa SM, Karnam G, Nagarajan AG, Chakraborty S, Chakrovortty D. 2009. lac repressor is an antivirulence factor of Salmonella enterica: its role in the evolution of virulence in Salmonella. PLoS One 4:e5789 doi:10.1371/journal.pone.0005789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falkow S, Baron LS. 1962. Episomic element in a strain of Salmonella Typhosa. J. Bacteriol. 84:581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernandez IM, et al. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184:743–753 [DOI] [PubMed] [Google Scholar]
- 19. Froeliger EH, Oetjien J, Bond JP, Fives-Taylor P. 1999. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect. Immun. 67:5206–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gemski P, Jr, Formal SB, Baron LS. 1971. Identification of two widely separated loci conferring nicotinic acid dependence on wild-type Shigella flexneri 2a. Infect. Immun. 3:500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hager AJ, et al. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227–237 [DOI] [PubMed] [Google Scholar]
- 22. Hinnebusch BJ. 2005. The evolution of flea-borne transmission in Yersinia pestis. Curr. Issues Mol. Biol. 7:197–212 [PubMed] [Google Scholar]
- 23. Hinnebusch BJ, Perry RD, Schwan TG. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367–370 [DOI] [PubMed] [Google Scholar]
- 24. Holden DW. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161–169 [DOI] [PubMed] [Google Scholar]
- 25. Hollis DG, et al. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup Novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Y, Movahedzadeh F, Stoker NG, Coates AR. 2006. Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infect. Immun. 74:861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keseler IM, et al. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72:5080–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lerat E, Ochman H. 2004. Ψ-Φ: Exploring the outer limits of bacterial pseudogenes. Genome Res. 14:2273–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marri PR, Bannantine JP, Golding GB. 2005. Comparative genomes of metabolic pathways in Mycobacterium species: gene duplication, gene decay and lateral gene transfer. FEMS Microbiol. 30:906–925 [DOI] [PubMed] [Google Scholar]
- 31. Maurelli AT. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267:1–8 [DOI] [PubMed] [Google Scholar]
- 32. Maurelli AT, Fernández RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCormick BA, Fernandez MI, Siber AM, Maurelli AT. 1999. Inhibition of Shigella-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell. Microbiol. 1:143–155 [DOI] [PubMed] [Google Scholar]
- 34. McDonough PL, Shin SJ, Lein DH. 2000. Diagnostic and public health dilemma of lactose-fermenting Salmonella enterica serotype Typhimurium in cattle in the northeastern United States. J. Clin. Microbiol. 38:1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Montminy SW, et al. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 7:1066–1073 [DOI] [PubMed] [Google Scholar]
- 36. Moore RA, et al. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586 [DOI] [PubMed] [Google Scholar]
- 38. Nakata N, et al. 1993. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 9:459–468 [DOI] [PubMed] [Google Scholar]
- 39. Parkhill J, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 25:848–852 [DOI] [PubMed] [Google Scholar]
- 40. Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97:10567–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prunier AL, Schuch R, Fernández RE, Maurelli AT. 2007. Genetic structure of the nadA and nadB antivirulence loci in Shigella spp. J. Bacteriol. 189:6482–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prunier AL, et al. 2007. nadA and nadB of Shigella flexneri 5a are antivirulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of Shigella pathogenicity. Microbiology 153:2363–2372 [DOI] [PubMed] [Google Scholar]
- 43. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Razavi B, Apisarnthanarak A, Mundy LM. 2007. Clostridium difficile: emergence of hypervirulence and fluoroquinolone resistance. Infection 35:300–307 [DOI] [PubMed] [Google Scholar]
- 45. Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363–1373 [DOI] [PubMed] [Google Scholar]
- 46. Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21:134–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 48. Stephens RS, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 49. Sun YC, Hinnebusch BJ, Darby C. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U. S. A. 105:8097–8101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suzuki T, et al. 2007. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via IpaF and ASC in Shigella-infected macrophages. PLoS Pathog. 3:e111 doi:10.1371/journal.ppat.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Svensson K, et al. 2005. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187:3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres AG, Vazquez-Juarez RC, Tutt CB, Garcia-Gallegos JG. 2005. Pathoadaptive mutation that mediates adherence of Shiga toxin-producing Escherichia coli O111. Infect. Immun. 73:4766–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vazquez-Juarez RC, et al. 2008. CadA negatively regulates Escherichia coli O157:H7 adherence and intestinal colonization. Infect. Immun. 76:5072–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei J, et al. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang F, et al. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu XJ, et al. 2002. SpiC is required for secretion of Salmonella pathogenicity island 2 type III secretion system proteins. Cell. Microbiol. 4:531–540 [DOI] [PubMed] [Google Scholar]
- 57. Zhou D, Yang R. 2009. Molecular Darwinian evolution of virulence in Yersinia pestis. Infect. Immun. 77:2242–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]