Abstract
The Helicobacter pylori outer membrane protein HopZ is regulated by a phase-variable CT repeat and occurs in two distinct allelic variants. Whole-genome comparisons of isolates from one human volunteer recently provided evidence for in vivo selection for the hopZ ON status. We explored the frequency of sequence variation in hopZ during acute and chronic human infection and studied the association of hopZ with the phylogeographic population structure of H. pylori. hopZ ON variants were cultured from 24 out of 33 volunteers challenged with the hopZ OFF strain BCS 100. Transmission of H. pylori within families was also frequently associated with a status change of hopZ. In contrast, hopZ sequences obtained from 26 sets of sequential isolates from chronically infected individuals showed no changes of status, suggesting that the hopZ status selected during early infection is subsequently stable. Mutations leading to amino acid changes in HopZ occurred more frequently in ON than in OFF status isolates during chronic infection, indicating that sequence changes are more likely the result of positive selection in ON isolates than of a loss of negative selection pressure in OFF isolates. Analysis of 63 isolates from chronically infected individuals revealed no significant correlation of hopZ status with chronic atrophic gastritis. hopZ sequences were obtained from a globally representative collection of 54 H. pylori strains. All H. pylori populations contained hopZ-positive isolates. The data suggest that hopZ has been acquired and split into the two variants before the human migration out of Africa.
INTRODUCTION
The human gastric pathogen Helicobacter pylori infects approximately 50% of the world population (55). All infected individuals develop chronic active gastritis, which can give rise to more severe long-term effects such as ulcer disease, chronic atrophic gastritis, gastric adenocarcinoma, and gastric lymphoma of the mucosa-associated lymphoid tissue (MALT). The factors that determine the long-term outcome of H. pylori infection remain largely unknown; however, the course of infection likely depends on the complex interplay of bacterial virulence factors, environmental influences, and the host immune response. Several bacterial factors have been associated with virulence and disease outcome; these include the cag pathogenicity island (cagPAI) and the vacuolating cytotoxin VacA (9, 13, 47). Another property that has frequently been suggested to be decisive in infection, mucosal inflammation, and disease outcome is adhesion of bacteria to host tissue, which is mediated by multiple bacterial adhesins and their interactions with host ligands (8, 15, 19, 26, 27, 36).
H. pylori possesses a large repertoire of outer membrane proteins (OMPs) encoded by a family of paralogous genes (57). The largest subgroup of OMPs is the Hop group, encoded by 21 genes (2). The complement of OMP genes varies between strains (2, 24, 30), and several hop genes (e.g., hopZ, oipA, sabA, sabB, babA, babB, and babC) can be regulated by phase-variable dinucleotide repeats in their 5′ coding region (3, 11, 52, 57, 59). The Hop family includes the two best-studied adhesins of H. pylori, the Lewis b blood group antigen-binding adhesin BabA (25) and the sialyl Lewis x antigen-binding adhesin SabA (38), as well as additional proteins that have been implicated in cell or tissue adhesion (14, 46, 48).
One of the Hop family proteins whose pathobiology is less well understood is HopZ (HP0009/JHP0007). HopZ has been reported to be involved in adhesion to gastric epithelial cells (48, 61); a host receptor for HopZ has not yet been identified. hopZ is phase variable due to a CT dinucleotide repeat in its signal sequence-encoding region (48, 61). Evidence from both human infection and animal models suggests that HopZ plays an important role during infection. We have recently reported a strong selection for strains expressing hopZ (hopZ ON) during an experimental human infection (31). We noted that 17 out of 18 reisolates cultured from a volunteer who had been challenged with the hopZ OFF H. pylori strain BCS 100 were hopZ ON. In atpb4-tox176 mice, which show parietal cell depletion and are thus a model system for chronic atrophic gastritis, the ability to express HopZ provided bacteria with a fitness advantage (20).
hopZ has been described to occur in two allelic variants, HopZ-I and HopZ-II (48). In addition to its regulation via the phase-variable repeat, hopZ has also been reported to be regulated at the transcriptional level in response to changing pH (41) and to contact with host cells (20). In this study, we systematically investigated phase and sequence variability in hopZ during acute and chronic infection of humans and after transmission, as well as the association of hopZ with the phylogeny of H. pylori.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For a complete list of bacterial isolates, see Table S1 in the supplemental material. Twenty-six groups of sequential isolates, 18 isolates from members of 6 families, 55 strains belonging to 7 phylogeographic populations and 11 subpopulations, and 50 additional strains obtained from the Kalixanda study, a large population-based study performed in northern Sweden, were analyzed for their hopZ sequences (4, 12, 33, 37, 54, 56). Twenty-two additionally available isolates from patient Kx345 from the Kalixanda study and 56 isolates from a vaccine challenge study were analyzed for the phase-variability-mediating repeat (1). H. pylori strains NCTC11637, 8A3 (31), a reisolate from a human infection with strain BCS 100 (1, 22), and N6 (6, 18) were used for functional analyses and generation of hopZ mutants. Bacteria were cultured from frozen stocks on blood agar plates (Columbia agar base II; Oxoid, Wesel, Germany) supplemented with 10% horse blood (Oxoid, Wesel, Germany) and antibiotics (10 mg/liter vancomycin, 3.2 mg/liter polymyxin B, 5 mg/liter trimethoprim, and 4 mg/liter amphotericin B); plates for mutant strains carrying the kanamycin resistance cassette were additionally supplemented with kanamycin (20 mg/liter). Plates were incubated either in an incubator at 5% O2, 10% CO2, and 85% N2 at 37°C or in airtight jars (Oxoid, Wesel, Germany) containing Anaerocult C gas-generating bags (Merck, Darmstadt, Germany).
For cultivation in liquid culture, brain heart infusion broth (BHI; Oxoid, Wesel, Germany) was supplemented with yeast extract (2.5 g/liter), 10% heat-inactivated horse serum, and antibiotics (for concentrations, see above). Bacteria grown on plates were resuspended in the medium, and the culture was adjusted to a starting optical density at 600 nm (OD600) of 0.1. Cultures were incubated for 16 h with shaking in jars (Oxoid, Wesel, Germany) containing Anaerocult C gas-generating bags (Merck, Darmstadt, Germany). For the analysis of protein expression, BHI broth supplemented with yeast (pH ∼7) was adjusted with HCl to pH 6 and pH 5. The medium was then sterile filtered, and horse serum and antibiotics were added.
Escherichia coli strain DH5α (23) was used for DNA cloning, and strain ER2566 (NEB, Ipswich, MA) was used for protein expression. E. coli was grown on LB plates or, for protein expression, in LB broth (Lennox L broth; Invitrogen GmbH, Karlsruhe, Germany) supplemented with ampicillin (200 mg/liter) and kanamycin (20 mg/liter) as required.
DNA methods.
Bacterial DNA was extracted from bacteria grown on plates using the QIAamp DNA purification kit (Qiagen, Hilden, Germany) or the Amplicor respiratory specimen preparation kit (Roche Diagnostics, Mannheim, Germany). PCRs were performed according to standard protocols. PCR products were purified using the PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Sequencing of hopZ was performed by amplifying the HopZ-encoding open reading frame (ORF) and its flanking genomic region with oligonucleotides HPdelta_hopZ_1 and HPdelta_hopZ_2 and by subsequent sequencing with internal oligonucleotides (see Table S2 in the supplemental material). For the amplification of the babB locus (J99), oligonucleotide AHp-AS (24) was used in combination with oligonucleotide babLocusB-rev (see Table S2). For 22 additional isolates from patient Kx345 from the Kalixanda study and 56 additional reisolates from the vaccine challenge study, only the DNA segment containing the phase-variability-mediating dinucleotide repeat was amplified from genomic DNA and sequenced with oligonucleotides HPhopZ_for2 and HPhopZ_rev or H1_omp1_for and H1_omp1_rev, respectively (see Table S2). cagA and vacA typing was performed as previously described (10).
Sequence analysis.
Assembly of sequences and sequence analyses as well as the generation of phylogenetic trees and calculation of similarity values were performed using Bionumerics 6.01 (Applied Maths, Sint-Martens-Latem, Belgium). Prediction of signal sequence cleavage sites was performed using SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/) (7). SignalP3.0 was applied for the hopZ sequence from strain NCTC11637, and a probable signal sequence was predicted to be located between amino acids 18 and 19 (LNA-ED [leucine, asparagine, alanine-glutamic acid, aspartic acid]). The glutamic acid residue was then considered the first amino acid in the mature protein, and the amino acid positions of the mature protein were used for further analyses.
Generation of isogenic mutants.
Mutagenesis was performed by a PCR- and ligation-based approach without cloning as previously described (53). In short, hopZ-flanking genomic regions were amplified from genomic DNA of the parental strains NCTC11637, 8A3, and N6 with oligonucleotides HPdelta_hopZ_1, HPdelta_hopZ_3C, HPdelta_hopZ_2, and HPdelta_hopZ_4 (see Table S2 in the supplemental material). A kanamycin resistance cassette was amplified from the plasmid pILL600 (34) with primers Km8_for_BamHI and Km9_rev_NdeI (see Table S2). The resulting PCR products were digested with NdeI and BglII and with NdeI and BamHI, respectively, and ligated subsequently. The ligation product was used as the template for a PCR with primers HPdelta_hopZ_1 and HPdelta_hopZ_2. The product containing the hopZ-flanking regions homologous to the genome of the parental strain flanking the kanamycin resistance cassette, in the same orientation as the hopZ gene in the genome of the parental strains, was gel purified and either used directly for transformation or cloned into pUC18 (62) using the PstI restriction sites. Kanamycin-resistant clones obtained after natural transformation of the respective wild-type H. pylori strains were isolated, and the correct integration of the cassette into the target gene was verified by PCRs before further characterization of the mutants. The kanamycin cassette from pILL600 used to create the allelic disruptions contains a strong promoter and is inserted in the same direction as the target gene, minimizing the risk of polar effects (50). In addition, the two genes located immediately downstream of hopZ are transcribed in the opposite direction, making a polar effect of the inactivation of hopZ on downstream genes unlikely.
Expression and purification of HopZ and generation of antisera.
A hopZ fragment was amplified from PCR products of strains NCTC11637 (allelic variant HopZ-I) and N6 (allelic variant HopZ-II) with oligonucleotides HopZ_F_expr_SapI_rev and HopZ_F_expr_NdeI_for_B and oligonucleotides HopZ_F_expr_N6_NdeI_for and HopZ_Fexpr_N6_SapI_rev, respectively. Specific HopZ gene fragments were used in order to avoid the expression of insoluble parts of the protein. In addition, partial sequences of hopZ were carefully selected for the expression cloning, in order to allow generation of antisera with high specificity for HopZ and not for other members of the H. pylori hop family (2), since parts of the hop genes closely resemble each other. The amplified gene segments were subsequently cloned via the NdeI and SapI restriction sites into the pTWIN1 vector of the impact expression system (NEB, Ipswich, MA), generating a C-terminal fusion construct (see Table S2 in the supplemental material). Partial HopZ proteins were expressed and purified from the insoluble fraction under denaturing conditions in the presence of 8 M urea. Partial renaturation was performed by dialysis to 2 M urea, and the HopZ fragment was subsequently purified according to the manufacturer's instructions using chitin beads and dithiothreitol (DTT) for cleavage of the HopZ fragment from the intein tag. Purified partial HopZ proteins were then used for the generation of rabbit antisera by intradermal injection (Biogenes, Berlin, Germany). The rabbit preimmune sera showed some reaction with H. pylori proteins (in particular, bands corresponding to a mass of ∼49 and ∼82 kDa; see Fig. S1) that were also visible as nonspecific bands in Western blot analyses performed with the final antisera raised against HopZ-I and HopZ-II. The identity of the additional bands recognized by the sera is not known, but it is likely that they represent conserved proteins of proteobacteria that the rabbits had been in contact with before the immunization. Preimmune sera and final antisera also did not recognize the major Hop proteins BabA or BabB (see Fig. S1). Identification of an ∼74-kDa band as HopZ (corresponding to the predicted molecular mass of mature HopZ after cleavage of the signal peptide) was confirmed by comparisons with isogenic hopZ mutants for all three H. pylori wild-type strains used in this study (26695, J99, and N6).
Analysis of HopZ protein expression in H. pylori.
H. pylori strains were grown on plates for up to 48 h and resuspended in phosphate-buffered saline (PBS), or samples were taken directly from liquid cultures. Bacterial numbers were adjusted by means of OD600 measurements, and samples were collected by centrifugation at 5,000 × g and 4°C for 10 min. Bacterial pellets were resuspended in SDS-PAGE sample buffer, boiled for 10 min, and analyzed via SDS-PAGE and subsequent Western blotting.
Protein methods.
Standard methods were used for protein separation and detection (35, 58). For protein separation, 8% SDS-PAGE was used. As secondary antibodies, horseradish peroxidase-conjugated goat anti-rabbit heavy and light IgG (Dianova, Hamburg, Germany) was used in a dilution of 1:20,000. The blots were incubated for 1 min with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) and exposed to Amersham Hyperfilm ECL (GE Healthcare, Little Chalfont, United Kingdom) films.
RESULTS
HopZ status in isolates obtained from an experimental human infection during a vaccine trial.
We had previously observed strong selection for the hopZ ON status in one human volunteer (volunteer 008) who had received the H. pylori challenge strain BCS 100 (31). While 16 single-colony purified clones isolated from BCS 100 were all hopZ OFF, 16 out of 17 single-colony isolates cultured from the volunteer at 6 and 10 weeks after infection were hopZ ON (31). Such selection might either be a rare event, restricted to a few individuals, or occur frequently. We therefore determined the hopZ status from 56 additional reisolates obtained from 32 additional volunteers who took part in the same study and were all initially infected with BCS 100 and reendoscoped 3 months postinfection. In 38 out of the 56 reisolates, hopZ had switched to ON status. Twenty-three of the additional 32 volunteers (72%) carried at least one hopZ ON reisolate, and in 20 of these, both reisolates tested were hopZ ON (Fig. 1; see also Table S3 in the supplemental material).
Fig 1.
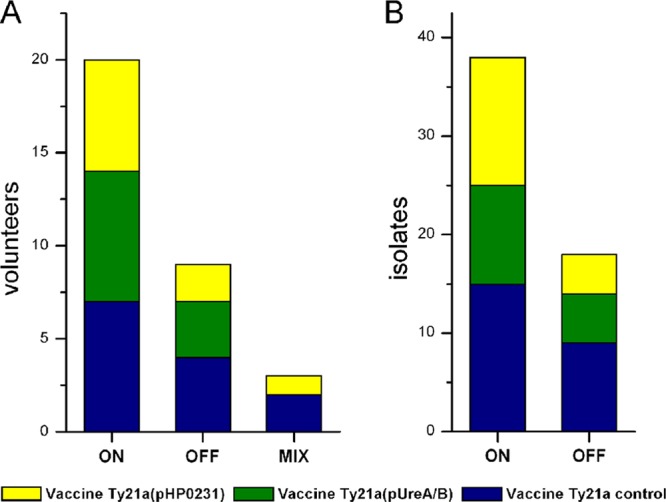
hopZ status of H. pylori reisolates from volunteers participating in the vaccine challenge study. hopZ status was predicted from the sequenced CT dinucleotide repeat in the 5′ region of the gene. (A) Numbers of volunteers where all available H. pylori reisolates were in ON (CT × 7), where all were in OFF (CT × 6), or where one of the isolates was in ON and one was in OFF status (“mix”). (B) Numbers of reisolates in ON or OFF status.
hopZ variability in phylogeographic populations of H. pylori.
We next studied the sequence variability of hopZ in a phylogeographic context. hopZ sequences were obtained from a globally representative collection of 54 H. pylori strains belonging to 7 different populations and 11 subpopulations as defined by structure analysis of haplotypes generated by multilocus sequence analysis (16, 37, 43) (Table 1). The proportion of hopZ ON to OFF strains varied among phylogeographic populations. All but two subpopulations contained both ON and OFF status isolates. Only strains assigned to the hspAfrica2SA and hspAmerind subpopulations were exclusively hopZ OFF (Table 1).
Table 1.
HopZ status and allele type in H. pylori isolates belonging to different phylogeographic populations and subpopulationsa
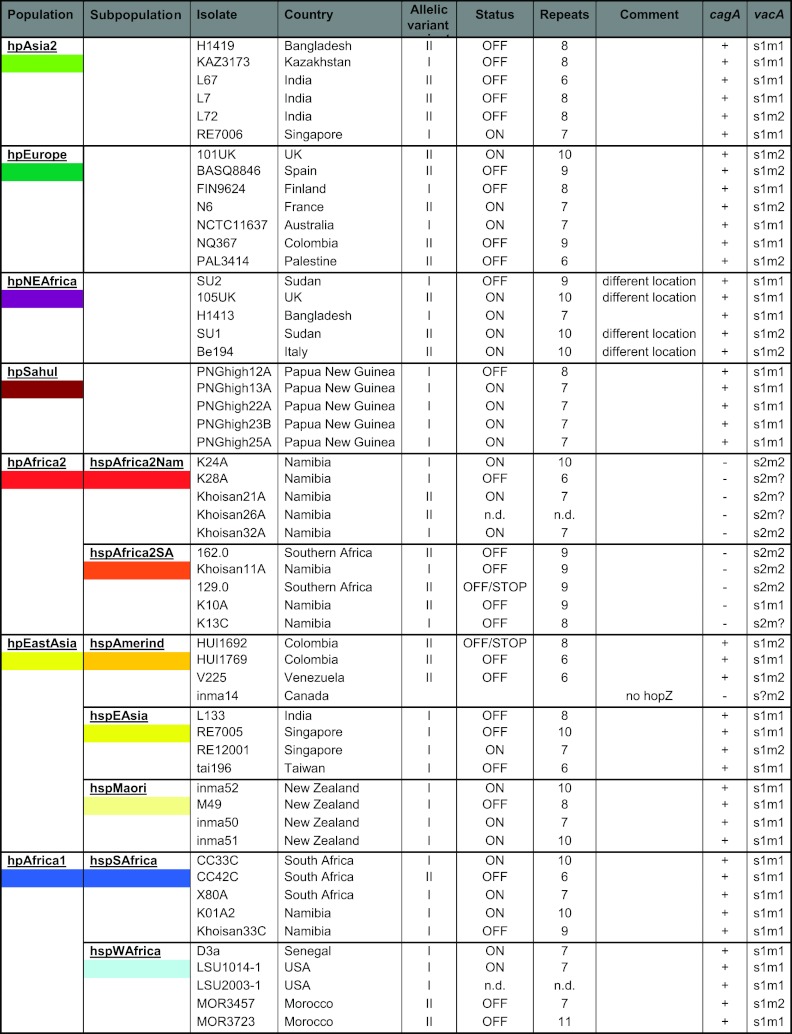
n.d., no data.
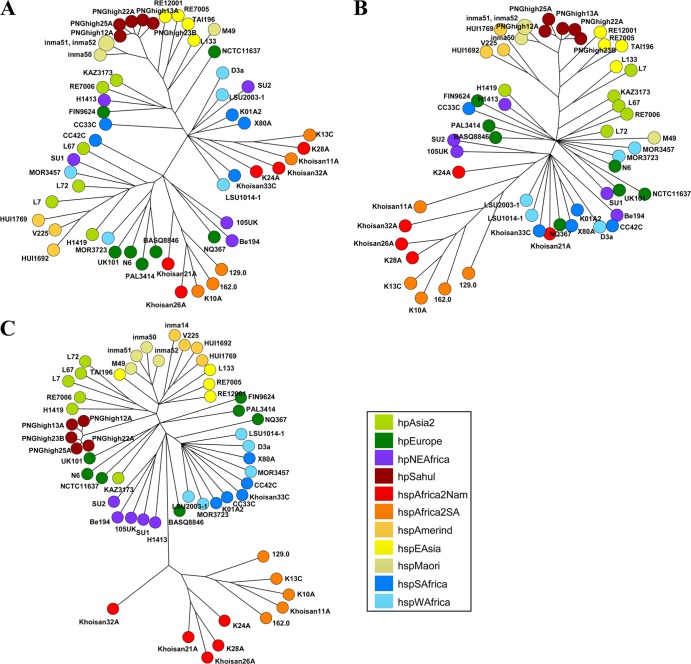
Phylogenetic analysis of the hopZ sequences revealed two major branches (100% bootstrap support) separating sequences containing the previously described two allelic variants HopZ-I and HopZ-II. Within the two branches, sequences from strains of the same phylogeographic population frequently clustered together (Fig. 2A). When only a C-terminal part of the hopZ gene that did not include the allele-specific segment was analyzed, the split into two major branches disappeared, and the correlation of hopZ sequence clustering with phylogeographic populations became even more apparent (Fig. 2B).
Fig 2.
Phylogenetic analysis based on full-length hopZ sequences (A), sequences encoding the C-terminal part of HopZ (B), or concatenated multilocus sequence typing (MLST) fragments (C) of 54 (A and B) or 55 (C) isolates belonging to 7 H. pylori populations. Neighbor-joining trees based on nucleotide sequences (no correction) are depicted as majority summary trees as generated with Bionumerics.
The frequency of the two allelic variants differed between populations. For example, strains belonging to hspMaori and hpSahul contained only allelic variant I (Fig. 2 and Table 1; see also Table S4 in the supplemental material). Strains from the hspAmerind subpopulation contained allelic variant II or lacked hopZ altogether (one strain). We also analyzed the data for possible correlations between the status and allelic variant of hopZ and the two best-studied virulence factors of H. pylori, cagA and vacA. We could not detect any statistically significant correlation of either factor with hopZ status or between the allelic variant of hopZ and carriage of the cagA gene. We did, however, detect a correlation of the allelic variant of hopZ with the vacA m type (hopZ allelic variant I with vacA m1 and hopZ allelic variant II with vacA m2) (χ2 test, P < 0.01).
For four of five isolates belonging to the H. pylori population hpNEAfrica, we observed much smaller PCR products when amplifying the genomic region usually containing hopZ, and sequencing revealed an empty site for these isolates. hopZ was identified at a different genomic location in these strains, namely, the locus where babB is found in strain J99 (Table 1).
Expression and regulation of hopZ in three different H. pylori strains.
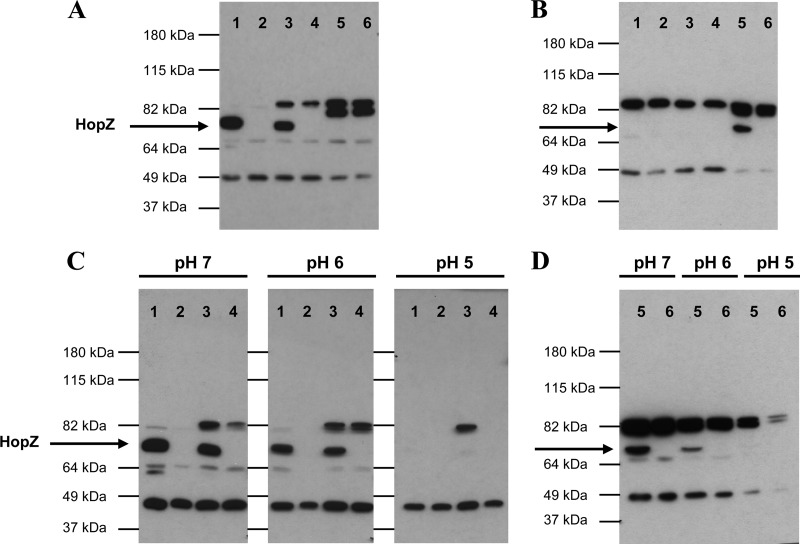
In order to analyze functional properties of hopZ, we generated isogenic mutants in three different H. pylori strains, NCTC11637, N6 (18), and 8A3, the hopZ ON reisolate of BCS 100 whose genome was completely sequenced (1, 22, 31). NCTC11637 and 8A3 carry allelic variant I, while N6 carries allelic variant II. In order to study protein expression, polyclonal antisera were raised against both variant proteins and used in Western blot analyses. An ∼74-kDa band consistent with the predicted molecular mass of HopZ was detected in strains NCTC11637, 8A3, and N6 (Fig. 3A and B, lanes 1, 3, and 5), and the identification of this band as HopZ was confirmed by comparison with isogenic hopZ mutants constructed for all three wild-type strains, which all lacked the corresponding bands (Fig. 3A and B, lanes 2, 4, and 6). Each of the two antisera was variant specific, detecting only the specific HopZ variant against which it had been raised (Fig. 3A and B; see also Fig. S1 in the supplemental material). Antisera were subsequently also used to confirm for selected pairs of isolates that a hopZ switch identified by sequence analysis resulted in corresponding changes of protein expression (see Fig. S2).
Fig 3.
Detection of HopZ from H. pylori grown under various conditions using two different antisera with specificity for allelic variants. Bacterial strains NCTC11637 (lanes 1), NCTC11637ΔhopZ (lanes 2), 8A3 (lanes 3), 8A3 ΔhopZ (lanes 4), N6 (lanes 5), and N6 ΔhopZ (lanes 6) were grown on plates (A and B) or in liquid culture at different pH values (pH 5, 6, and 7) (C and D) for 16 h and lysed for SDS-PAGE analysis. Immunoblotting assays were performed with antisera generated against a HopZ fragment cloned from either strain NCTC11637 (allelic variant I) (A and C) or strain N6 (allelic variant II) (B and D).
Previous reports, based on microarray data, have concluded that the transcription of hopZ is differentially regulated in response to changing pH (41), but regulation has not been ascertained for the HopZ protein. We cultivated the mutants and parental H. pylori strains in BHI medium (pH 7) and media adjusted to pH 5 and pH 6 for 16 h and analyzed the strains for HopZ protein by Western blotting. For all three strains tested, HopZ expression was decreased at pH 6 and almost completely abrogated at pH 5 (Fig. 3C and D). To rule out the possibility that this reduction of HopZ expression might be due to a selection of hopZ OFF variants, the hopZ status was reassessed after culture at pH 5, showing that all strains were still hopZ ON (data not shown).
HopZ status in H. pylori isolates from patients with chronic atrophic gastritis versus subjects with nonatrophic gastritis.
Since hopZ has previously been shown to confer a selective advantage in a mouse model of chronic atrophic gastritis (20), we asked whether the hopZ status might correlate with the development of chronic atrophic gastritis in humans. We sequenced hopZ in 63 isolates from the Kalixanda study, a large population-based study of peptic ulcer disease performed in northern Sweden (4, 54). These isolates included 31 H. pylori strains from patients with different stages of atrophy as well as 32 control isolates obtained from individuals with nonatrophic gastritis. hopZ was in ON status in 19 out of 32 strains (59%) from subjects with nonatrophic gastritis and in 20 out of 31 strains (65%) from atrophic gastritis samples. Thus, there was no association of hopZ status with the presence of atrophic gastritis in our study population. In this population, allelic variant I occurred more frequently than did allelic variant II (41 versus 22 isolates). There was also no significant association between gastric histopathology and hopZ status when data were analyzed separately for the two allelic variants (see Fig. S3 and Table S5 in the supplemental material).
We next analyzed the data for a correlation between the H. pylori hopZ status and the “inflammatory capacity” of the bacteria. As a scoring system for inflammatory capacity, we used the average of the scores of the infiltration by lymphocytes, plasma cells, and granulocytes and subtracted the score of the Helicobacter density, such that a low bacterial density associated with a high degree of inflammation would yield a value of >0. As depicted in Fig. S3B in the supplemental material, we observed a trend toward a higher proportion of isolates carrying hopZ in the ON status in strains with a high inflammatory capacity (>0); the observed difference was, however, not significant.
hopZ microevolution during chronic infection.
As the data from the vaccine trial had shown frequent selection for the hopZ ON status during the early phase of infection in humans, we next sought to quantify the frequency of hopZ switching during chronic infection by analyzing paired sequential isolates. We sequenced hopZ in 26 pairs of sequential isolates obtained at intervals ranging from 3 months to 4 years and in two additional follow-up isolates with a time interval of 16 years after the initial biopsy. In these sequential isolates, we observed very few repeat length changes in the phase-variability-mediating CT dinucleotide repeat and no single change of hopZ status. Fourteen sets of isolates contained hopZ in ON status, while 12 sets of isolates were in OFF status.
We next obtained the entire nucleotide sequences of the hopZ coding regions from the same sets of sequential isolates. Sequence differences in hopZ were observed in 18 of the 26 pairs, while eight pairs had identical hopZ sequences. The observed sequence differences included changes in the phase-variable repeat without an impact on status, one or multiple isolated single nucleotide polymorphisms, and clusters of nucleotide polymorphisms (polymorphisms with a distance of less than 200 bp), indicative of recombination events. Overall, the sequence comparisons for pairs with time intervals up to 4 years showed a sequence similarity ranging from 98.4% to 100%. A particularly striking case is strain pair NQ392/4060 (16 years), where a recombination event exchanged an allele II fragment for an allele I fragment and reduced the overall sequence similarity between the two isolates to 94%.
The number of amino acid changes was correlated with the switch status of the gene, with more changes occurring at the amino acid level in a gene in ON status than in a gene in OFF status (Fig. 4; see also Table S6 in the supplemental material). For this analysis, only the amino acid sequences of the predicted mature proteins were used in order to analyze differences regardless of switch status of the gene, as the phase-variability-mediating repeat forms part of the signal sequence and is thus not present in the predicted mature protein. The only gene containing a frameshift in the coding region of hopZ not caused by the CT repeat was excluded from this analysis, as well as the 16-year follow-up isolates. In the remaining set of sequential isolates, the genes in the ON status had acquired mutations resulting in 0.25 amino acid changes per month of infection on average, while the OFF status isolates had acquired only 0.01 amino acid changes in the predicted mature HopZ per month (Welch's t test for unequal variances, P < 0.02).
Fig 4.
Graphical representation of pairwise comparisons of hopZ sequences of sequential isolates. Gene sequences and derived amino acid sequences of the predicted mature proteins regardless of status. Sequences of the respective first isolates were used as references. White blocks represent identical sequence in an isolate pair, while black vertical lines indicate positions of polymorphisms or amino acid exchanges that occur in the second isolate in comparison to the first within the pair of sequential isolates. Insertion and deletion events were also counted as polymorphic sites and are thus also indicated as black vertical lines. The hatched box for isolate pair Kx1039 indicates a frameshift caused by a 1-nucleotide indel in isolate Kx1039_0 causing a premature stop codon. Numbers on the right side of the figure indicate the presence of allelic variant I or II. Happlots were generated using the program Happlot with manual corrections in the case of the nucleotide sequences, and amino acid sequences happlots were generated manually.
The observed stability of the gene status as well as the occurrence of sequence changes over time during chronic infection was independent from disease progression in the included strain pairs obtained from individuals who displayed either stable gastritis (Kx208, Kx573, Kx1165, and Kx1259), stable atrophic gastritis (Kx1167), regression of atrophy (Kx424 and Kx1377), or disease progression (Kx201, Kx345, Kx439, Kx1039, Kx1172, and Kx438) (see Table S6 in the supplemental material).
In order to assess the stability of hopZ status during chronic infection and within one stomach, 24 single-colony isolates from the corpus of one patient's stomach obtained at two different time points with an interval of 4 years were analyzed. All the single colonies showed an identical repeat region of 7 CT dinucleotide copies and were thus uniformly hopZ ON (Kx345). This result indicated that the phase status of hopZ within one stomach can be remarkably stable.
In this collection of sequential isolates, the amino acid sequence of allelic variant II changed less frequently than did the sequence of allelic variant I. This difference was statistically significant (Welch's t test for unequal variances, P < 0.05); the comparison was, however, limited by the small number of pairs with allelic variant II (five), compared to 21 pairs of allelic variant I.
HopZ status changes after intrafamilial transmission.
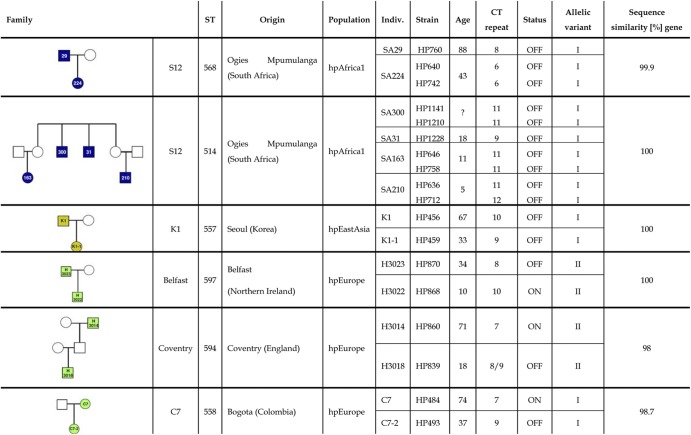
Since the results obtained with reisolates from the vaccine challenge trial suggested that switching of the hopZ status might occur frequently in the early phase of H. pylori infection, we tested the hopZ status of highly related H. pylori strains isolated from members of the same family. Due to the high allelic diversity of H. pylori, strains with identical sequence types (STs; identical sequences at seven housekeeping gene loci) occur very rarely in unrelated individuals and are widely accepted as evidence of recent transmission. We analyzed hopZ sequences from seven ST-identical pairs of strains from family members (51). In contrast to the paired isolates from chronically infected individuals, changes in the CT dinucleotide repeat were frequent, leading to changes of hopZ status in three out of the seven assumed intrafamilial transmission events (Table 2). In three cases of transmission in family S12 from Ogies (Mpumulanga), all isolates contained hopZ in OFF status, as well as in one case of intrafamilial transmission from South Korea (Table 2).
Table 2.
Comparison of hopZ sequences in isolates with identical sequence types from family membersa
Indiv., individual.
In agreement with the results reported in the previous paragraph, we again observed very few changes in hopZ sequences with a stable OFF status, while hopZ sequences were more variable, with sequence similarities ranging from 98 to 100% when one of the isolates was in ON status.
DISCUSSION
The Hop family of Helicobacter outer membrane proteins harbors multiple proteins that mediate the interaction of H. pylori with its host. The high degree of sequence similarity between the paralogous genes and their metastability due to phase-variable sequences may be reasons why relatively little information is still available about most Hop proteins. Building on our recent observation of selection for an active hopZ gene in one human volunteer who was challenged with a hopZ OFF H. pylori strain (31), this study explored the sequence variation of hopZ both during human infection and in a phylogeographic context.
hopZ phase variation in vivo.
The majority of volunteers challenged with the hopZ OFF strain BCS 100 yielded hopZ ON H. pylori in one or both biopsy specimens when they were reendoscoped 3 months later. This high frequency of selection of a hopZ ON genotype was observed by analysis of one single colony isolate per biopsy specimen. Sampling of additional biopsy specimens or multiple clones per biopsy specimen might have resulted in an even higher rate of detection of hopZ ON strains. In striking contrast to this selection during early infection, the status of hopZ (ON or OFF) was found to be remarkably stable during chronic infection. No single change of hopZ status was observed for 26 pairs of sequential isolates with a cumulated separation time of 1,335 months. This permits calculation of an upper bound of the 99% confidence interval for the rate of switching as 0.0476 per year. The difference between the switching frequencies observed during acute infection of volunteers and those observed in chronically infected individuals was highly significant (P = 1.38 × 10−37).
Additionally, we observed changes in hopZ status in 3 out of 7 cases of intrafamilial transmissions, further supporting our hypothesis that HopZ plays a role in the initial colonization by H. pylori, at least in some individuals. It is possible that transmission from one individual to another can also be accompanied by a switch from hopZ ON to OFF status. In fact, in one of the presumed intrafamilial transmission events (Coventry family, Table 2), the older family member carried a hopZ ON strain while the strain from the younger family member was hopZ OFF, suggesting an ON-to-OFF switch when the strain was transmitted to the younger family member. However, the direction of transmission is only assumed, so that there is currently no clear-cut evidence of ON-to-OFF switches upon transmission. Taken together, we propose that HopZ plays a role predominantly during the initial phase of infection of a new individual, which can select for a different hopZ status. After establishment of chronic infection, the hopZ status is very stable.
We cannot exclude intraindividual variation of hopZ status within one stomach, as has been shown for CagA EPIYA repeat status (28). However, we analyzed multiple single-colony isolates from one stomach for volunteer 008 (31) and patient Kx345 (this study) and did not observe such intraindividual variation.
Sequence variation in hopZ in phylogeographic context.
All H. pylori populations and subpopulations analyzed in this study were found to contain hopZ-carrying strains (see Table S4 in the supplemental material). The only strain lacking hopZ belonged to the hspAmerind population, for which the lack of hopZ in some strains was already previously described (30, 32).
The neighbor-joining tree based on the hopZ sequences revealed a bipartite split reflecting the previously described two allelic variants of hopZ (48). Within the two major branches, further subclusters roughly correlated with the phylogeographic populations (Fig. 2). The correlation between hopZ phylogeny and the phylogeographic populations was even stronger when the tree was based only on a C-terminal part of the predicted protein that does not contain the allele-specific fragment. This indicates that this part of the hopZ gene has coevolved with the rest of the H. pylori genome for a long time and has likely been acquired before the cag pathogenicity island (which is lacking from the hpAfrica2 population).
We note that the two distinct allelic variants also occurred in almost all H. pylori populations and subpopulations, suggesting that they have existed concurrently since before the split of H. pylori into two main superlineages (42). Remarkably, two subpopulations, hspMaori and hpSahul, carried only allelic variant I (Table 1). The observation of only one allelic variant in the hspMaori subpopulation might be explained by strong human population bottlenecks which reduced diversity of hopZ, as the genetic diversity within the hspMaori sample has already been shown to be extremely low (17).
There was no correlation of the presence of the cagPAI with the hopZ allelic variant. We detected a significant correlation of the allelic variant of hopZ with the vacA m type. This association could be due to different frequencies of vacA m types in different phylogeographic populations (Table 1). Alternatively, this observation might reflect a functional interaction between hopZ and vacA, a hypothesis that needs further investigation.
Functional implications of variation in hopZ.
The precise functional role of hopZ in the infectious process has so far not been identified. The gene was not found in a screening for essential genes in a gerbil infection model (29). Additionally it has been shown to be nonessential in different mouse models on the basis of isogenic mutants (20, 60). This is supported by our observation that some strains lack a functional hopZ gene. Unfortunately, despite intensive efforts, we could not reproduce the previously reported role of HopZ in gastric cell adhesion (48), limiting our possibilities for functional analyses. We hypothesize that subtle differences in experimental conditions (e.g., with respect to the culture of AGS cells) may be responsible for these discrepancies.
Regardless of its precise biological function during the infectious process, our results point to a role of HopZ during initial or early infection, a situation that has been characterized to be accompanied by a transient hypochlorhydria (21, 39, 44, 45, 49). This represents a link to the status of chronic atrophic gastritis, which is also accompanied by a disturbed acid secretion and in which hopZ has been shown to confer a selective advantage in a mouse model (20). This role of hopZ during a situation of perturbed acid secretion is in line with its high expression at near neutrality and a reduction of HopZ protein levels already at mildly acidic pH, which is consistent with previously published microarray studies (41).
In the set of sequential isolates analyzed in this study, we detected a significantly higher frequency of nonsynonymous sequence changes with effects on the amino acid sequence of the mature HopZ protein in genes in ON status than in genes in OFF status, indicating that the observed variability in HopZ among sequentially isolated strains is more likely due to positive or diversifying selection than to degeneration of the gene in OFF status. This could reflect processes of adaptation to interaction partners of the individual host, as has been described for the adhesin BabA (5), and/or could be the result of immune pressure, consistent with the observation that hopZ contains one of the 50 most immunogenic H. pylori epitopes, as identified during the generation of a comprehensive antigenic profile (40).
Supplementary Material
ACKNOWLEDGMENTS
We thank Lars Agreus, Tom Storskrubb, Jukka Ronkainen, and Pertti Aro for providing the Kalixanda strains and Thomas Borén for kindly providing BabA and BabB antisera. We are very grateful to Xavier Didelot for help with the statistical evaluation of switching frequencies in acutely versus chronically infected individuals.
This study was supported by grant ERA-NET PathoGenoMics3 (HELDIVPAT) from the German Ministry of Education and Research (BMBF) and SFB 900/A1 from the German Research Foundation (DFG). Further support was provided by the DFG as a Ph.D. stipend to L.K. in the framework of the International Research Training Group IRTG 1273 and the Hannover Biomedical Research School (HBRS).
Footnotes
Published ahead of print 1 October 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Aebischer T, et al. 2008. Correlation of T cell response and bacterial clearance in human volunteers challenged with Helicobacter pylori revealed by randomised controlled vaccination with Ty21a-based Salmonella vaccines. Gut 57:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alm RA, et al. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alm RA, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180 [DOI] [PubMed] [Google Scholar]
- 4. Aro P, et al. 2004. Valid symptom reporting at upper endoscopy in a random sample of the Swedish adult general population: the Kalixanda study. Scand. J. Gastroenterol. 39:1280–1288 [DOI] [PubMed] [Google Scholar]
- 5. Aspholm-Hurtig M, et al. 2004. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305:519–522 [DOI] [PubMed] [Google Scholar]
- 6. Behrens W, Bonig T, Suerbaum S, Josenhans C. 2012. Genome sequence of Helicobacter pylori hpEurope strain N6. J. Bacteriol. 194:3725–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 8. Borén T, Falk P, Roth KA, Larson G, Normark S. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892–1895 [DOI] [PubMed] [Google Scholar]
- 9. Censini S, et al. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U. S. A. 93:14648–14653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chattopadhyay S, et al. 2004. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. J. Clin. Microbiol. 42:2821–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colbeck JC, Hansen LM, Fong JM, Solnick JV. 2006. Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect. Immun. 74:4375–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Correa P, et al. 2000. Chemoprevention of gastric dysplasia: randomized trial of antioxidant supplements and anti-Helicobacter pylori therapy. J. Natl. Cancer Inst. 92:1881–1888 [DOI] [PubMed] [Google Scholar]
- 13. Cover TL, Blanke SR. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320–332 [DOI] [PubMed] [Google Scholar]
- 14. Dossumbekova A, et al. 2006. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: genetic and functional genomic analysis of hopH gene polymorphisms. J. Infect. Dis. 194:1346–1355 [DOI] [PubMed] [Google Scholar]
- 15. Falk P, et al. 1993. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proc. Natl. Acad. Sci. U. S. A. 90:2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falush D, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585 [DOI] [PubMed] [Google Scholar]
- 18. Ferrero RL, Cussac V, Courcoux P, Labigne A. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerhard M, et al. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. U. S. A. 96:12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giannakis M, et al. 2009. Response of gastric epithelial progenitors to Helicobacter pylori isolates obtained from Swedish patients with chronic atrophic gastritis. J. Biol. Chem. 284:30383–30394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graham DY, Alpert LC, Smith JL, Yoshimura HH. 1988. Iatrogenic Campylobacter pylori infection is a cause of epidemic achlorhydria. Am. J. Gastroenterol. 83:974–980 [PubMed] [Google Scholar]
- 22. Graham DY, et al. 2004. Challenge model for Helicobacter pylori infection in human volunteers. Gut 53:1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 24. Hennig EE, Allen JM, Cover TL. 2006. Multiple chromosomal loci for the babA gene in Helicobacter pylori. Infect. Immun. 74:3046–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ilver D, et al. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–377 [DOI] [PubMed] [Google Scholar]
- 26. Ilver D, Barone S, Mercati D, Lupetti P, Telford JL. 2004. Helicobacter pylori toxin VacA is transferred to host cells via a novel contact-dependent mechanism. Cell. Microbiol. 6:167–174 [DOI] [PubMed] [Google Scholar]
- 27. Ishijima N, et al. 2011. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 286:25256–25264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karlsson A, Ryberg A, Nosouhi DM, Borch K, Monstein HJ. 2012. Variation in number of cagA EPIYA-C phosphorylation motifs between cultured Helicobacter pylori and biopsy strain DNA. Infect. Genet. Evol. 12:175–179 [DOI] [PubMed] [Google Scholar]
- 29. Kavermann H, et al. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawai M, et al. 2011. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 11:104 doi:10.1186/1471-2180-11-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kennemann L, et al. 2011. Helicobacter pylori genome evolution during human infection. Proc. Natl. Acad. Sci. U. S. A. 108:5033–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kersulyte D, et al. 2010. Helicobacter pylori from Peruvian Amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One 5:e15076 doi:10.1371/journal.pone.0015076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraft C, et al. 2006. Genomic changes during chronic Helicobacter pylori infection. J. Bacteriol. 188:249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Labigne-Roussel A, Courcoux P, Tompkins L. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 36. Linden SK, et al. 2009. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 5:e1000617 doi:10.1371/journal.ppat.1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Linz B, et al. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahdavi J, et al. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marshall BJ, Armstrong JA, McGechie DB, Glancy RJ. 1985. Attempt to fulfil Koch's postulates for pyloric Campylobacter. Med. J. Aust. 142:436–439 [DOI] [PubMed] [Google Scholar]
- 40. Meinke A, et al. 2009. Composition of the ANTIGENome of Helicobacter pylori defined by human serum antibodies. Vaccine 27:3251–3259 [DOI] [PubMed] [Google Scholar]
- 41. Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moodley Y, et al. 2012. Age of the association between Helicobacter pylori and man. PLoS Pathog. 8:e1002693 doi:10.1371/journal.ppat.1002693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moodley Y, et al. 2009. The peopling of the Pacific from a bacterial perspective. Science 323:527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morris A, Nicholson G. 1987. Ingestion of Campylobacter pyloridis causes gastritis and raised fasting gastric pH. Am. J. Gastroenterol. 82:192–199 [PubMed] [Google Scholar]
- 45. Morris AJ, Ali MR, Nicholson GI, Perez-Perez GI, Blaser MJ. 1991. Long-term follow-up of voluntary ingestion of Helicobacter pylori. Ann. Intern. Med. 114:662–663 [DOI] [PubMed] [Google Scholar]
- 46. Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537–1548 [DOI] [PubMed] [Google Scholar]
- 47. Olbermann P, et al. 2010. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 6:e1001069 doi:10.1371/journal.pgen.1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramsey EJ, et al. 1979. Epidemic gastritis with hypochlorhydria. Gastroenterology 76:1449–1457 [PubMed] [Google Scholar]
- 50. Schmitz A, Josenhans C, Suerbaum S. 1997. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J. Bacteriol. 179:987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwarz S, et al. 2008. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 4:e1000180 doi:10.1371/journal.ppat.1000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. 2004. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 101:2106–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sterzenbach T, et al. 2008. The role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J. Bacteriol. 190:6398–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Storskrubb T, et al. 2005. A negative Helicobacter pylori serology test is more reliable for exclusion of premalignant gastric conditions than a negative test for current H. pylori infection: a report on histology and H. pylori detection in the general adult population. Scand. J. Gastroenterol. 40:302–311 [DOI] [PubMed] [Google Scholar]
- 55. Suerbaum S, Michetti P. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175–1186 [DOI] [PubMed] [Google Scholar]
- 56. Taylor NS, et al. 1995. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J. Clin. Microbiol. 33:918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 58. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamaoka Y. 2008. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J. Med. Microbiol. 57:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yamaoka Y, et al. 2002. Helicobacter pylori infection in mice: role of outer membrane proteins in colonization and inflammation. Gastroenterology 123:1992–2004 [DOI] [PubMed] [Google Scholar]
- 61. Yamaoka Y, et al. 2004. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology 126:1030–1043 [DOI] [PubMed] [Google Scholar]
- 62. Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.