Abstract
Herpes simplex virus 1 (HSV-1) is a common pathogen infecting the majority of people worldwide at some stage in their lives. The early host response to viral infection is initiated by the cells of the innate immune response, including macrophages. Here, we have characterized the secretome of HSV-1-infected human primary macrophages using high-throughput quantitative proteomics. We identified and quantified 516 distinct human proteins with high confidence from the macrophage secretome upon HSV-1 infection, and the secretion of 411 proteins was >2-fold increased upon beta interferon (IFN-β) priming and/or HSV-1 infection. Bioinformatics analysis of the secretome data revealed that most of the secreted proteins were intracellular, and almost 80% of the proteins whose secretion increased more than 2-fold were known exosomal proteins. This strongly suggests that nonclassical, vesicle-mediated protein secretion is activated in IFN-β-primed and HSV-1-infected macrophages. Proteins related to immune and inflammatory responses, interferon-induced proteins, and endogenous danger signal proteins were efficiently secreted upon IFN-β priming and HSV-1 infection. The secreted IFN-induced proteins include interferon-induced tetratricopeptide protein 2 (IFIT2), IFIT3, signal transducer and activator of transcription 1 (STAT1), and myxovirus resistance protein A (MxA), implicating that these proteins also have important extracellular antiviral functions. Proinflammatory cytokine interleukin-1β was not released by HSV-1-infected macrophages, demonstrating that HSV-1 can antagonize inflammasome function. In conclusion, our results provide a global view of the secretome of HSV-1-infected macrophages, revealing host factors possibly having a role in antiviral defense.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a common pathogen infecting the majority of people worldwide at some stage in their lives (42). It is an icosahedral, enveloped, nuclear-replicating, double-stranded DNA virus belonging to the neurotropic Alphaherpesvirinae subfamily (36). The site of primary infection by HSV-1 is usually the oral mucosa, from where it spreads to infect nearby sensory neurons. In the sensory neurons it forms a lifelong infection, staying unnoticed in a latent state, from which it is periodically activated to a lytic state. HSV-1 can cause diseases such as cold sores, keratitis, and encephalitis and can be lethal to individuals with a compromised immune system (47).
HSV-1 infection is recognized by the pattern recognition receptors (PRRs) of the innate immune system (27). Macrophages express multiple PRRs, and they have been shown to play a critical role in early host response against HSV-1 infection (13). HSV-1 infection activates macrophages, which leads to the production and secretion of different chemokines, such as CXC motif chemokine 10 (CXCL10), C-C motif chemokine 2 (CCL2), CCL3, and cytokines, including tumor necrosis factor alpha and type I and type III interferons (IFNs) (21, 22). Type I IFNs are an important group of cytokines in host defense against viral infection (4). They activate leukocytes and induce the upregulation of interferon-stimulated genes (ISGs) (38) that are crucial for the anti-HSV response. In addition, we have previously shown that type I IFNs enhance the HSV-1-induced chemokine response in human macrophages (22). Furthermore, Cotter and coworkers have demonstrated that intact type I signaling is required for HSV-1-induced cytokine production (6). In addition to chemokines and cytokines, macrophages also secrete other proteins that play a role in the early antiviral response (5, 16, 34).
Secreted proteins are an important group of molecules estimated to be encoded by approximately 10% of the human genome (28). Secreted proteins can reflect a broad variety of different conditions of the cell, and therefore, they can be used as biomarkers or drug targets. These proteins can be released to the extracellular space through various mechanisms, including classical secretion involving vesicular migration from the endoplasmic reticulum to the Golgi apparatus, through nonclassical vesicle-mediated mechanisms, and also through shedding from the surface of living cells. The nonclassically secreted proteins normally localize in the cytoplasm or the nucleus, where they mediate well-characterized intracellular functions (24). In the presence of a specific external stimulus, they are released from cells to mediate extracellular functions distinct from their original intracellular function. A large number of nonclassically secreted proteins include cytokines, growth factors, and other molecules with important signaling roles in physiological processes such as inflammation. A prototype of an unconventionally secreted protein is proinflammatory cytokine interleukin-1β (IL-1β). It is first synthesized as pro-IL-1β, and this proform has to be cleaved by caspase-1 in order for it to be secreted. Caspase-1 in turn is activated in a molecular platform called the inflammasome (18). It has been suggested that inflammasomes and active caspase-1 mediate activation of unconventional protein secretion (8, 12).
The focus of previous studies on HSV-1-induced protein secretion from macrophages has been on chemokines and cytokines (21, 22), and no large-scale studies on HSV-1-induced protein secretion have been published. Currently, mass spectrometry (MS)-based proteomics make it possible to identify and quantify thousands of proteins from cellular samples, including the analysis of the global pattern of secreted proteins, the secretome (16, 17, 34, 45, 48). Here, we have characterized the secretome of HSV-1-infected human primary macrophages using high-throughput quantitative MS-based proteomics combined with bioinformatics to discover novel secreted host factors involved in antiviral defense.
MATERIALS AND METHODS
Cell culture and sample preparation.
Leukocyte-rich buffy coats obtained from healthy blood donors were purchased from the Finnish Red Cross blood transfusion service (Helsinki, Finland). Peripheral blood mononuclear cells were isolated from leukocyte-rich buffy coats as previously described (31). In brief, human peripheral blood mononuclear cells were collected by density gradient centrifugation over Ficoll-Paque Plus (GE Healthcare) in Leucosep tubes (Greiner Bio-One GmbH). A mononuclear cell layer containing monocytes was collected and washed with Dulbecco's phosphate-buffered saline (DPBS) (Gibco), and the monocyte count was determined with a Coulter AcT Diff hematology analyzer (Beckman Coulter Corp). Cells were plated at a density of 1.4 × 106 monocytes/6-well plate well (Falcon Multiwell; Nunc/Thermo Fisher Scientific) in RPMI 1640 medium (Sigma-Aldrich) supplemented with 2 mM l-glutamine and antibiotics consisting of 50 U/ml of penicillin and 50 μg/ml streptomycin (both from Invitrogen Life Technologies). Cells were allowed to attach for 45 min, and then unattached cells were washed away with DPBS. Monocytes were left to differentiate into macrophages in macrophage serum-free medium (SFM; Gibco) supplemented with 10 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Biosource International Inc., Camarillo, CA) and antibiotics. The medium was changed every 2 days. After 7 days of culture, the resulting macrophages were CD14 positive (data not shown) and used in experiments.
Differentiated macrophages were left untreated or primed with 200 IU/ml recombinant beta interferon (IFN-β; Betaferon; Bayer Schering Pharma) in macrophage SFM (supplemented as described above) for 4 h. After this, cells were infected with the wild-type KOS strain of HSV-1 (a gift from Jesper Melchjorsen, Aarhus, Denmark) at a multiplicity of infection (MOI) of 1 or were left uninfected. After 2 h of infection, the cell culture medium was discarded, cells were washed three times with DPBS and fresh RPMI 1640 (supplemented as described above and with 10 mM HEPES) was added. At 18 h postinfection (hpi), cell culture supernatants were collected and pooled. One 6-well plate of cells, with 2 wells containing cells from each of three different donors, was used for each stimulation. A volume of 400 μl of the supernatant was taken for Luminex assay analysis. The remaining 5.5 ml of supernatants was concentrated using Amicon Ultra-15 (Millipore) centrifugal filter devices with a 10,000-nominal-molecular-weight limit (NMWL) to a final volume of 180 to 250 μl, and the proteins were purified using a 2-D Clean-Up kit (GE Healthcare/Amersham) according to the manufacturer's instructions.
Total cell lysates were prepared from the macrophages. Cells from the three donors were washed and detached on ice in DPBS, pooled, and lysed in cell lysis buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 50 mM NaF, and 1% Triton X-100 supplemented with protease inhibitor cocktail (Sigma-Aldrich). The lysates were kept on ice for 30 min and then centrifuged for 20 min at 12,000 rpm and +4°C for removal of cell debris. The supernatant from the cell lysates was collected for further Western blot analysis.
For inflammasome activation experiments, macrophages were first primed with Toll-like receptor 2 (TLR2) ligand (S)-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser(S)Lys4-OH trihydrochloride (Pam3Cys; 1 μg/ml), which was purchased from EMC Microcollections (Tubingen, Germany) for 4 h. After priming with Pam3Cys, the cells were either stimulated with 100 μg/ml monosodium urate (MSU; Enzo Life Sciences) for 3 h or infected with HSV-1 KOS at an MOI of 1 for 18 h. Cell culture supernatants were collected and pooled for enzyme-linked immunosorbent assay (ELISA) analysis, and total cell lysates were prepared as described above.
iTRAQ labeling, mass spectrometry, and protein identification and quantification.
The proteins were reduced, alkylated, and digested using trypsin (Promega), and the resulting peptides were labeled with fourplex isobaric tags for relative and absolute quantitation (iTRAQs) from a reagents multiplex kit (Applied Biosystems) according to the manufacturer's instructions. After labeling, the samples were pooled and dried, and the peptides were fractionated with strong cation-exchange chromatography (SCX) using a PolySULPHOETHYL A column (200 by 2.1 mm; 202SE0502; PolyLC Inc.) connected to an ÄKTA high-pressure liquid chromatography system (Amersham Biosciences) as previously described (16). Each SCX fraction containing iTRAQ-labeled peptides (approximately 14 fractions) was analyzed twice using an Ultimate 3000 nano-liquid chromatograph (nano-LC; Dionex) coupled to a QSTAR Elite hybrid quadrupole time of flight mass spectrometer (AB Sciex) with nanoelectrospray ionization (16, 25).
Protein identification and relative quantification were performed using ProteinPilot (version 2.0.1) software (Applied Biosystems/MDS Sciex). Data files from both technical replicates of an iTRAQ sample set were processed together. The Universal Protein Resource (UniProt) human protein sequences in the version from January 2009, with close to 34,000 human protein sequences (46), and all UniProt protein sequences in the version from December 2009, with 513,877 protein sequence entries, were used as the search databases. The search criteria were cysteine alkylation with methyl methanethiosulfonate and trypsin digestion, with biological modifications allowed and with a thorough search and a detected protein threshold of 95% confidence (unused ProtScore > 1.3) used. ProteinPilot identification and quantification results were also manually checked: for each identified protein, at least two unique peptides with good-quality tandem mass spectrometry (MS/MS) data were required, and MS/MS spectra with all reporter ion peak heights below 10 counts were manually removed from the quantification results. False discovery rates (FDRs) were calculated using a concatenated normal and reversed sequence database and a previously reported method (7). The calculated FDRs were under 4% in both sample sets.
Bioinformatic analyses.
The identified and quantified human proteins were classified on the basis of their biological functions and cellular compartment using the Gene Ontology (GO) classification (1) with GeneTrail software (2), UniProt as the data source, and Homo sapiens as the organism. GeneTrail was also used to recognize functional Kyoto Encyclopedia of Genes and Genomes (KEGG) (11) pathway categories that were over- or underrepresented in the data. GeneTrail analysis was done using all genes from H. sapiens as a reference set, a significance threshold of 0.05, an FDR-adjusted P value, and 2 as the minimum number of genes per category. The ExoCarta (version 3.1) database was used to determine the proteins that are secreted in exosomes (20). Classically secreted proteins were predicted using the SignalP (version 4.0) program (29).
Western blotting, Luminex assay, and IL-1β ELISA.
Protein concentrations of total cell lysates were measured with the DC protein assay (Bio-Rad) according to the manufacturer's instructions. An equal amount of protein from total cell lysates ranging from 10 to 20 μg or, alternatively, a protein amount that was equal to 10% of the concentrated cell culture supernatants was loaded onto an SDS-polyacrylamide gel. Proteins were transferred from the gel to an Immobilon-P transfer membrane (Millipore), and the membranes were blocked with 5% nonfat milk powder in 0.05% Tris-buffered saline–Tween 20. Incubations with primary antibodies were done overnight at +4°C. Incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (Dako) was done for 1 h at room temperature. Proteins were visualized by a Western Lightning Plus ECL system (Perkin Elmer). The following primary antibodies were used: galectin 3 (9C4) and HSP27 (M20) (both from Santa Cruz Biotechnology), HSP90 (Cell Signaling Technology), HSV-1 gD (DL6; Santa Cruz Biotechnology), interferon-induced tetratricopeptide protein 2 (IFIT2; Sigma-Aldrich), IFIT3 (BD Biosciences), S100-A9 (47-8D3), and signal transducer and activator of transcription 1 (STAT1) p84/p91 (E-23; Santa Cruz Biotechnology). Myxovirus resistance protein A (MxA) (37) and IL-1β (31) antibodies have been previously described.
The proteins CCL2, CCL3, CCL4, and CCL5 were measured in the cell culture supernatants by Luminex assay for with a Luminex system Bio-Plex 200 apparatus (Bio-Rad), using a custom-made Bio-Plex Pro assay (Bio-Rad). The assay was performed according to the manufacturer's instructions. IL-1β concentrations from cell culture supernatants were determined using a human IL-1β Eli-pair ELISA kit (Diaclone) according to the manufacturer's instructions.
RESULTS
IFN-β priming is required for efficient protein secretion from HSV-1-infected human primary macrophages.
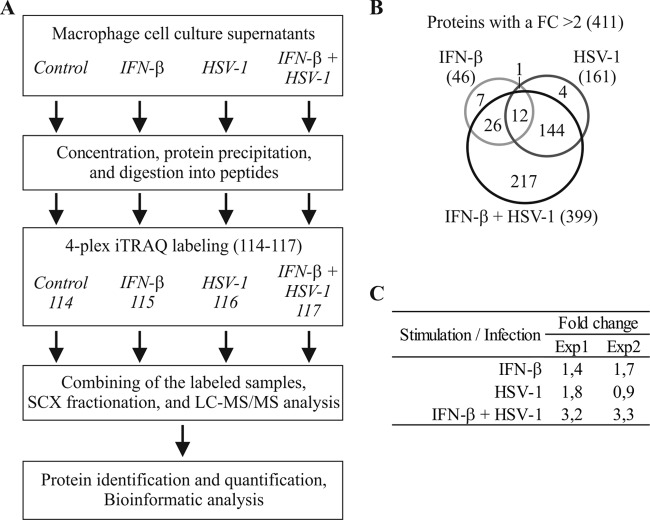
The complex set of proteins secreted from living cells at a given time and under defined conditions is generally referred to as the secretome. In the present study, we have characterized the secretome of IFN-β-primed and HSV-1-infected human primary macrophages using quantitative MS-based proteomics. The secreted proteins were identified and quantified using 4-plex iTRAQ labeling combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the resulting peptides (Fig. 1A). We analyzed two biological replicates, and both sample sets were analyzed twice with LC-MS/MS to improve the quality of the data for protein identification and quantification. Based on the LC-MS/MS data, we identified 1,094 human proteins, and from these we had good-quality quantification information from 516 distinct proteins (see Table S1 in the supplemental material). The secretion of 46, 161, and 399 human proteins increased more than 2-fold upon IFN-β priming, HSV-1 infection, and IFN-β priming and HSV-1 infection, respectively (Fig. 1B). The total amount of iTRAQ-labeled peptides in these secretomes showed that there is an approximately 3-fold increase in the total amount of protein being secreted upon IFN-β priming and HSV-1 infection compared to that for the control sample, whereas in the IFN-β-primed or HSV-1-infected macrophage secretomes, only a modest increase in total protein secretion could be detected (Fig. 1C).
Fig 1.
IFN-β priming increases protein secretion from HSV-1-infected human primary macrophages. (A) Work flow of the experiment. (B) Venn diagram for the human proteins with an FC of > 2 in the secretome upon IFN-β priming or/and HSV-1 infection. (C) Relative amounts of iTRAQ-labeled peptides in the secretomes of IFN-β-stimulated and/or HSV-1-infected macrophages compared to control macrophages.
Our secretome analysis also revealed robust secretion of HSV-1 proteins after HSV-1 infection alone and in combination with IFN-β priming. Based on the LC-MS/MS data, we identified 20 herpesvirus proteins, and from these we had good-quality quantification information from 10 distinct proteins (Fig. 2A; see Table S1 in the supplemental material). To measure HSV-1 replication kinetics in our cell model, we performed Western blot analysis for HSV-1 gD protein from IFN-β-primed and HSV-1-infected macrophage cell lysates at different time points ranging from 3 to 18 hpi (Fig. 2B). Already at 6 hpi, some HSV-1 replication was detected in the cell lysate, and it strengthened until 14 hpi, reaching a plateau and staying at a similar level after 18 hpi (Fig. 2B).
Fig 2.
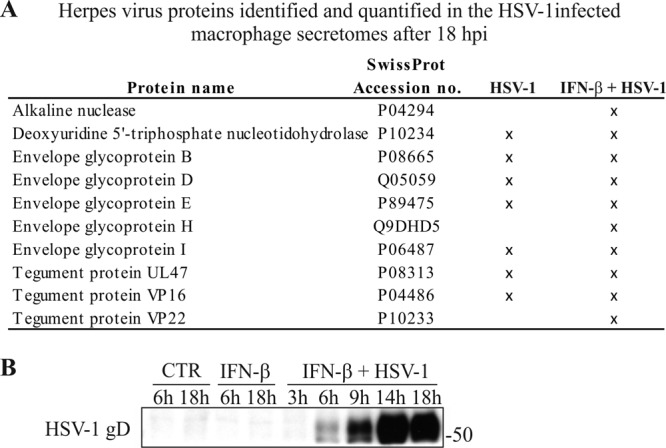
HSV-1 replication in human primary macrophages. (A) Herpesvirus proteins identified and quantified in the HSV-1-infected macrophage secretomes after 18 hpi. (B) HSV-1 replication kinetics in human primary macrophages measured by HSV-1 gD Western blotting from total cell lysates at different time points: 6 and 18 h for control (CTR) and IFN-β-primed macrophages and 3, 6, 9, 14, and 18 hpi for IFN-β-primed and HSV-1-infected macrophages.
HSV-1 infection activates unconventional protein secretion from human primary macrophages.
The human proteins whose secretion increased more than 2-fold upon IFN-β priming and/or HSV-1 infection were classified using the Gene Ontology (GO) classification based on the cellular compartment, which showed that the majority of the secreted proteins were from intracellular compartments (Fig. 3A). A more detailed classification based on the cellular organelle of the secretome data showed that IFN-β priming and HSV-1 infection do not induce protein secretion from any specific cell organelle but, rather, increase the overall secretion of proteins from all of the main organelles (Fig. 3B). To confirm that the increased intracellular protein secretion was not due to cell death, we determined the amount of apoptotic cells in the macrophage cell culture using an APOPercentage apoptosis assay. Upon IFN-β priming or HSV-1 infection alone, no apoptotic cells were seen, and after IFN-β priming and HSV-1-infection, less than 10% of the cells were apoptotic (see Fig. S1 in the supplemental material), suggesting that partial apoptosis may occur in HSV-1-infected macrophages.
Fig 3.
The majority of the human proteins in the HSV-1-induced secretome have an intracellular localization and are known exosomal proteins. (A) Classification of the secretome proteins on the basis of the GO annotations for their cellular compartment. (B) Classification of the intracellular proteins into different cellular organelles in the IFN-β-primed, HSV-1-infected, and IFN-β-primed plus HSV-1-infected macrophage secretomes. ER, endoplasmic reticulum. (C) Classification of all the identified proteins and proteins with an FC of >2 in the secretome with SignalP and ExoCarta to determine classically and exosomally secreted proteins, respectively. GO annotations were done using GeneTrail, classically secreted proteins were predicted using SignalP (version 4.0), and exosomal proteins were identified using the ExoCarta (version 3.1) database.
The proteins identified from the macrophage secretome were next analyzed with SignalP to determine the presence of a signal peptide needed for classical protein secretion. From all the proteins identified in our data set, 24% were predicted to be classically secreted, whereas only 14% of the proteins whose secretion increased more than 2-fold upon IFN-β priming and/or HSV-1 infection were predicted to be classically secreted (Fig. 3C). Unconventional protein secretion includes exosomal release of proteins. Exosomes are membranous vesicles that are actively released to the extracellular space by most mammalian cells (33). From all of the proteins identified, approximately 60% (634 proteins) were found from the exosomal protein database ExoCarta (20), and from the 411 proteins whose secretion increased more than 2-fold upon IFN-β priming and/or HSV-1 infection, almost 80% (314 proteins) were found from ExoCarta (Fig. 3C; see Table S2 in the supplemental material). Taken together, our results strongly suggest that HSV-1 infection induces nonclassical pathways of vesicle-mediated secretion from human primary macrophages.
Proteins related to immune and inflammatory responses and ISGs are efficiently secreted from macrophages upon IFN-β priming and HSV-1 infection.
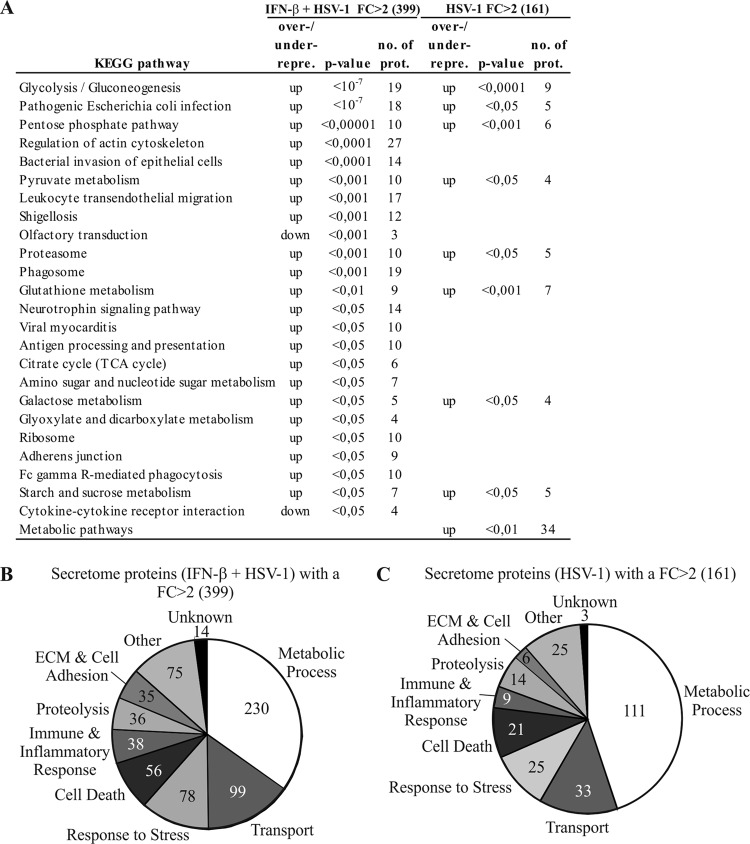
The KEGG database was used for pathway analysis of the human proteins in the secretome with a fold change (FC) of >2 in the HSV-1-infected and the IFN-β-primed plus HSV-1-infected macrophages. This showed 24 pathways as being significantly (P < 0.05) over- or underrepresented after IFN-β priming and HSV-1 infection (Fig. 4A) and 9 pathways as being over- or underrepresented after HSV-1 infection alone (Fig. 4A). The most significant overrepresented KEGG pathways after IFN-β priming and HSV-1 infection included glycolysis/gluconeogenesis, pathogenic Escherichia coli infection, the pentose phosphate pathway, regulation of the actin cytoskeleton, and bacterial invasion of epithelial cells (Fig. 4A). GO classification based on biological functions showed that proteins related to metabolic processes, transport, response to stress, cell death, proteolysis, extracellular matrix (ECM) and cell adhesion, and immune and inflammatory responses were among the most abundant protein groups identified from the secretome after IFN-β priming and HSV-1 infection (Fig. 4B) and after HSV-1 infection alone (Fig. 4C).
Fig 4.
Functional classification of the secreted proteins after HSV-1 infection with and without IFN-β priming. (A) KEGG pathway analysis for macrophage secretome proteins with an FC of >2 after HSV-1 infection with or without IFN-β priming. (B and C) GO classification on the basis of the biological functions for the proteins with an FC of >2 in the secretome upon IFN-β priming and HSV-1 infection (B) and HSV-1 infection alone (C). Most of the proteins belong to more than one GO class. KEGG pathway analysis and GO annotations were done using GeneTrail.
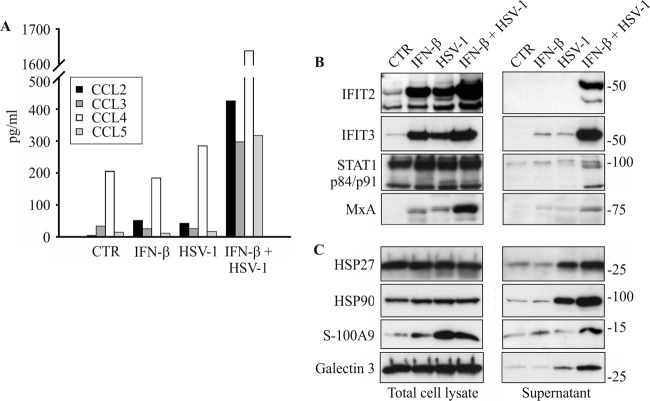
HSV-1 infection is known to activate an immune response in macrophages. We identified altogether 38 immune and inflammatory response proteins that had an FC of >2 in the secretomes of IFN-β-primed and HSV-1-infected macrophages (Fig. 4B), including CXCL10, sialoadhesin, and complement factor 3. Cytokines and chemokines are small signaling proteins that are involved in the activation of the inflammatory response. To complement the MS-based quantification results, we used the fluorescent bead-based immunoassay Luminex to quantify the secreted chemokines: all the quantified chemokines (CCL2-5) were clearly more efficiently secreted after IFN-β priming and HSV-1 infection than control cells, whereas IFN-β priming and HSV-1 stimulation alone had only a modest effect on chemokine secretion (Fig. 5A).
Fig 5.
The secretion of chemokines, interferon-stimulated proteins, and endogenous danger signal proteins is increased by IFN-β priming from HSV-1-infected macrophages. Macrophages were left untreated (control [CTR]) or primed with 200 IU/ml of IFN-β for 4 h, after which they were infected with HSV-1 (MOI, 1) for 18 h. Cell supernatants were collected, and cell lysates were prepared at 18 hpi. (A) Luminex analysis of unconcentrated cell supernatants for selected chemokines from control (0), IFN-β-primed, HSV-1-infected, and IFN-β-primed plus HSV-1-infected macrophages. The assay was done for the following chemokines: CCL2, CCL3, CCL4, and CCL5. (B) Western blot analysis from macrophage total cell lysates and concentrated cell supernatants for interferon-stimulated proteins IFIT2, IFIT3, STAT1 p84/p91, and MxA. (C) Western blot analysis from macrophage total cell lysates and concentrated cell supernatants for endogenous danger signal proteins HSP27, HSP90, S100-A9, and galectin-3.
Type I IFN stimulation and viral infection are both known to induce expression of ISGs (4). We next compared the proteins in our data set to those in the Interferome database, which consists of ISG and protein data collected from public databases (38). We identified and quantified altogether 133 known interferon-induced proteins from the macrophage secretome after IFN-β priming and/or HSV-1 infection (see Table S3 in the supplemental material). From these proteins, 24 had an FC of >2 and have been reported to be regulated by IFNs in at least two previous studies (Table 1). To characterize the intracellular levels and secretion patterns of interferon-induced proteins in more detail, we performed Western blot analysis for selected proteins, namely, IFIT2, IFIT3, MxA, and STAT1. In total cellular lysates, IFIT2, IFIT3, and MxA were clearly induced by IFN-β and HSV-1 stimulation alone, and their levels were the highest in the IFN-β-primed and HSV-1-infected macrophages, whereas activated STAT1 was detected in all samples with similar intensity (Fig. 5B). In contrast, the secretion of ISGs was seen only after IFN-β priming and HSV-1 infection, and IFN-β stimulation or HSV-1 infection alone had only a modest effect on the secretion of ISGs (Fig. 5B).
Table 1.
Interferon-induced proteins with an FC of >2 in IFN-β-primed and/or HSV-1-infected cellsa
| Protein name | SwissProt accession no | ExoCarta | SignalP |
|---|---|---|---|
| Annexin A1 | Q5TZZ9 | x | |
| Complement factor B | Q5JP67 | x | x |
| Cytosol aminopeptidase | P28838 | x | |
| Elongation factor 1-α | Q6IPT9 | x | |
| Fatty acid synthase | Q4LE83 | x | |
| Galectin-3-binding protein precursor | Q08380 | x | x |
| Glyceraldehyde-3-phosphate dehydrogenase | Q53X65 | x | |
| IFIT1 | Q5T7J1 | ||
| IFIT2 | Q8IZ03 | ||
| IFIT3 | Q5T765 | ||
| Intercellular adhesion molecule 1 | Q5NKV8 | x | x |
| ISG15 ubiquitin-like modifier | Q5SVA4 | ||
| Microtubule-associated protein RP/EB family member 1 | Q15691 | ||
| MxA | P20591 | x | |
| Plasma protease C1 inhibitor precursor | P05155 | x | x |
| Proteasome subunit alpha type 3 | P25788 | x | |
| Proteasome subunit beta type 8 | Q5QNR8 | x | |
| Proteasome subunit beta type 9 | Q5JNW4 | x | |
| Protein-l-isoaspartate (d-aspartate) O-methyltransferase | P22061 | ||
| STAT1 | Q68D00 | x | |
| Talin-1 | Q9Y490 | x | |
| Threonyl-tRNA synthetase variant | Q53GX7 | x | |
| Tryptophanyl-tRNA synthetase, cytoplasmic | Q53XB6 | x | |
| Zyxin | Q15942 |
The Interferome database was used. Proteins were shown to be regulated by type I IFN in at least two previous publications.
HSV-1 infection stimulates the secretion of endogenous danger signal proteins.
Cells under stress are known to actively secrete or passively release endogenous danger signal molecules which include proteins and other endogenous molecules, such as ATP and uric acid (3, 14). We identified in total 29 proteins that have been shown or have been suggested to have a role as endogenous danger signals (3, 9) from the secretomes of IFN-β-primed and HSV-1-infected macrophages (Table 2). The majority of these proteins belong to the GO class of biological function response to stress, including heat shock proteins (HSPs), annexins, S100 proteins, thioredoxin superfamily members, and galectins. We next performed Western blot analysis for a selected set of proteins, namely, galectin-3, HSP27, HSP90, and S100-A9 to analyze both their intracellular levels and secretion. The intracellular levels of galectin-3, HSP27, and HSP90 were not markedly affected by IFN-β priming and/or HSV-1 stimulation, whereas in total cellular lysates, S100-A9 was clearly upregulated after HSV-1 infection alone and in combination with IFN-β priming and HSV-1 infection (Fig. 5C). In the secretomes, all of the studied danger signal proteins were clearly more abundant after HSV-1 infection alone and after IFN-β priming and HSV-1 infection than control or IFN-β-primed cells (Fig. 5C).
Table 2.
Endogenous danger signal proteins with an FC of >2 in IFN-β-primed and HSV-1-infected cells
| Protein group and name | SwissProt accession no. | ExoCarta | SignalP |
|---|---|---|---|
| Heat shock proteins | |||
| 60-kDa heat shock protein, mitochondrial precursor | P10809 | x | |
| 78-kDa glucose-regulated protein precursor | P11021 | x | x |
| Heat shock 70-kDa protein 1A variant | Q59EJ3 | x | |
| Heat shock 70-kDa protein 4 | Q2TAL4 | x | |
| Heat shock cognate 71-kDa protein | P11142 | x | |
| Heat shock protein, 105 kDa | Q92598 | ||
| Heat shock protein beta-1 (HSP27) | P04792 | x | |
| HSP90-α | P07900 | x | |
| HSP90-β | P08238 | x | |
| HSPE1 protein | Q53X54 | x | |
| Tumor rejection antigen (Gp96) 1 | Q5CAQ5 | x | x |
| Annexins | |||
| Annexin A1 | Q5TZZ9 | x | |
| Annexin A2 | Q8TBV2 | x | |
| Annexin A4 | Q6P452 | x | |
| Annexin A5 | Q6FHB3 | x | |
| Annexin A6 | P08133 | x | |
| Annexin A11 | Q6ICS0 | x | |
| S100-A proteins | |||
| Protein S100-A4 | P26447 | x | |
| Protein S100-A6 | P06703 | x | |
| Protein S100-A8 | P05109 | x | |
| Protein S100-A9 | P06702 | x | |
| Protein S100-A11 | P31949 | x | |
| Galectins | |||
| Galectin-1 | P09382 | x | |
| Galectin-3 | Q6IBA7 | x | |
| Thioredoxin superfamily members | |||
| Peroxiredoxin-1 | Q06830 | x | |
| Thioredoxin | Q53X69 | x | |
| Thioredoxin reductase 1, cytoplasmic | Q16881 | ||
| Other | |||
| Nucleophosmin | P06748 | x | |
| Peptidyl-proly1 cis-trans isomerase A, cyclophilin A | P62937 | x |
HSV-1 infection of human macrophages does not result in inflammasome activation.
Previous studies have suggested that inflammasomes and active caspase-1 mediate activation of unconventional protein secretion (8, 12). Inflammasome activation is associated with the secretion of proinflammatory cytokines IL-1 and IL-18 as well as its central components apoptosis-associated speck-like protein containing the caspase activation and recruitment domain and caspase-1. Here, we identified only apoptosis-associated speck-like protein containing CARD and not IL-1β, IL-18, or caspase-1 from the secretomes of IFN-β-primed and HSV-1-infected human macrophages. A lack of IL-1β and IL-18 secretion was verified with ELISA (data not shown). We have seen in previous studies that the production of IL-1β in influenza A virus-infected human macrophages is dependent on TLR signaling (35). For this reason, we pretreated macrophages with the TLR2 ligand Pam3Cys for 4 h, after which we infected macrophages with HSV-1 for 18 h or activated the cells with MSU, a known activator of the NOD-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome (19), for 3 h. After this, the cell culture supernatants were collected and total cell lysates were prepared. A robust secretion of IL-1β was seen in Pam3Cys-treated and MSU-stimulated macrophage cell culture supernatants (Fig. 6A). A modest increase in IL-1β secretion was also seen in Pam3Cys-primed and HSV-1-infected human macrophages (Fig. 6A). However, Western blot analysis of cell culture supernatants revealed that the biologically active form of IL-1β is not secreted by Pam3Cys-primed and HSV-1-infected macrophages, demonstrating that the inflammasome is not activated in response to HSV-1 infection (Fig. 6B). In contrast, some pro-IL-1β was seen in cell culture supernatants of Pam3Cys-primed and HSV-1-infected macrophages. The secretion of the biologically active 17-kDa form of IL-1β was readily seen in Pam3Cys-treated and MSU-stimulated macrophage cell culture supernatants (Fig. 6B). Western blot analysis of cell lysates revealed that HSV-1 infection of human macrophages does not result in production of pro-IL-1β, which was readily detectable in Pam3Cys-treated macrophages. In conclusion, our results demonstrate that HSV-1 infection of human macrophages can activate robust unconventional protein secretion in the absence of inflammasome activation.
Fig 6.

IL-1β is not secreted from HSV-1-infected human primary macrophages. Macrophages were first treated with 1 μg/ml of Pam3Cys for 4 h, after which they were either stimulated with MSU (100 μg/ml) for 3 h or infected with HSV-1 (MOI, 1) for 18 h. Cell supernatants were collected, and cell lysates were prepared. (A) IL-1β was analyzed from cell culture supernatants with ELISA. (B) Expression of 31-kDa pro-IL-1b, and 17-kDa mature IL-1b was analyzed by Western blotting from macrophage total cell lysates and concentrated cell culture supernatants.
DISCUSSION
In the present study, we have used an unbiased high-throughput quantitative proteomics approach to characterize the secretome of human primary macrophages infected with HSV-1. Our results show that IFN-β priming increases the total protein secretion from macrophages after HSV-1 infection, leading to the considerably increased secretion of hundreds of proteins. Active protein secretion includes classically secreted proteins which contain a signal peptide that directs their transport to the plasma membrane through the endoplasmic reticulum-Golgi apparatus pathway. In addition, activated immune cells also secrete proteins through unconventional secretory pathways, including vesicle-mediated protein secretion (23). In our data, only 14% of the proteins whose secretion increased more than 2-fold upon IFN-β and/or HSV-1 infection were predicted to be classically secreted, whereas almost 80% of the proteins that were more secreted by IFN-β-primed and/or HSV-1-infected macrophages were found in the ExoCarta database. This demonstrates that exosomal protein release is a major mechanism by which HSV-1-infected macrophages communicate with other cells. We have seen similar exosome-mediated release of proteins in influenza A virus-infected human macrophages (16), underlining the importance of exosomes in cell-to-cell communication during viral infections.
Macrophages play a pivotal role in this response by detecting the viruses through PRRs, which leads to macrophage activation. IFNs and ISGs have a central role in antiviral defense during viral infection (41). HSV-1 infection of macrophages activated robust secretion of several ISGs in IFN-β-primed cells. Interestingly, it has previously been shown that type I signaling is essential for HSV-1-induced NF-κB activation (6), which may explain the enhancing effect of IFN-β priming on ISG secretion seen in our experiments. The secretion of these proteins was not increased by IFN-β priming or HSV-1 infection alone. Secretion of ISGs during viral infection is a novel finding. It was recently suggested that 2′-5′ oligoadenylate synthetases have an extracellular function by stimulating RNase L-independent antiviral activity (15). Our findings suggest an extracellular function for many other ISGs. Especially interesting ISGs found in our data are IFIT2 and IFIT3, which were detected at high levels in the secretomes of IFN-β-primed and HSV-1-infected macrophages. Previous studies have shown that IFIT2 and IFIT3 are involved in the intracellular antiviral response. Mouse IFIT2 has been shown to bind eukaryotic initiation factor 3c, thereby inhibiting translation initiation and protein synthesis (43, 44). IFIT3 has antiviral activity but a yet unidentified mechanism (40). Interestingly, IFIT1 has recently been shown to bind viral RNA, and all the IFITs contain nucleotide binding regions (30). It is tempting to speculate that, similar to IFIT1, IFIT2 and IFIT3 are involved in the extracellular binding of viral nucleotides, DNA, and/or RNA, thereby enabling the recognition of these pathogen-associated molecular patterns. In conclusion, our results suggest a novel extracellular function for many ISGs, including IFIT2 and IFIT3 proteins.
Danger signal proteins are nuclear or cytosolic proteins with defined intracellular functions, and they are usually released through an unconventional protein secretion pathway during the stress response (3). We detected the secretion of many danger signal proteins, including HSPs, annexins, S100 proteins, thioredoxin superfamily members, and galectins, from IFN-β-primed and HSV-1-infeceted macrophages. We have previously shown that these danger signal proteins are also secreted during influenza A virus infection of human macrophages (16). This suggests that these proteins have a general function in the activation of the inflammatory response during viral infections. Many of the identified HSPs have been shown to mediate macrophage activation (10) and induce the secretion of different cytokines (26). Similar to HSPs, galectins 1 and 3, annexins, several S100-A proteins, and thioredoxin all have inflammatory functions in the extracellular space. Galectins have been shown to have a role in the innate immune response against bacterial and fungal infections (39). Our finding that HSV-1 infection also seems to promote secretion of galectins 1 and 3 from macrophages suggests that these proteins also have a role in the host response to viral infection. HSV-1 uses its membrane glycoproteins to gain access to the host cell. It is possible that galectins elicit their antiviral function by binding to these viral glycoproteins and thereby inhibit viral entry to the host cell.
Previous studies have suggested that the secretion of danger signal proteins is mediated by inflammasomes and active caspase-1 (12). However, we did not detect inflammasome activation in HSV-1-infected macrophages. TLR-activated macrophages expressed pro-IL-1β at a high level, but the formation of the biologically active form of IL-1β was not seen in HSV-1-infected human macrophages. These results suggest that HSV-1 infection can activate robust unconventional protein secretion in the absence of inflammasome activation. Possible molecular platforms that could activate caspase-1 and pro-IL-1β processing in human macrophages during HSV-1 infection include absent in melanoma 2 (Aim-2), gamma interferon-inducible protein 16 (IFI16), and NLRP3 inflammasomes. Aim-2 is known to be activated by bacterial and viral DNA, and IFI16 has been reported to recognize the genome of Kaposi's sarcoma-associated herpesvirus (32). NLRP3 in turn is activated by several pathogens of bacterial and viral origin, including influenza A virus (32). Human macrophages express Aim-2, IFI16, and NLRP3 proteins (data not shown), but still, these PRRs fail to detect HSV-1, which readily replicates in macrophages and activates the IFN-mediated antiviral response in these cells. Therefore, HSV-1 must have developed mechanisms by which it successfully evades inflammasome activation. Future studies are needed to elucidate the viral counterstrategies that block inflammasome activation during HSV-1 infection.
In conclusion, we provide the first comprehensive characterization of the secretome of HSV-1-infected macrophages, revealing host factors possibly having a role in antiviral defense. We show that HSV-1 infection can activate robust unconventional protein secretion in the absence of inflammasome activation. A large majority of the identified secretome proteins were known to be exosomal proteins, highlighting the role of unconventional protein secretion as a mechanism by which macrophages communicate with their environment during HSV-1 infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elina Välimäki for the help with the Luminex assay and Jesper Melchjorsen (Århus, Denmark) for providing us with the wild-type KOS strain of HSV-1.
This work was supported by Academy of Finland grants 114437, 135628, and 140950, the Sigrid Juselius Foundation, and the Helsinki Biomedical Graduate Program.
Footnotes
Published ahead of print 12 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ashburner M, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Backes C, et al. 2007. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 35:W186–W192 doi:10.1093/nar/gkm323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianchi ME. 2007. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 81:1–5 [DOI] [PubMed] [Google Scholar]
- 4. Borden EC, et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciborowski P, et al. 2007. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology 363:198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cotter CR, et al. 2011. The virion host shutoff protein of herpes simplex virus 1 blocks the replication-independent activation of NF-kappaB in dendritic cells in the absence of type I interferon signaling. J. Virol. 85:12662–12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4:207–214 [DOI] [PubMed] [Google Scholar]
- 8. Feldmeyer L, et al. 2007. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr. Biol. 17:1140–1145 [DOI] [PubMed] [Google Scholar]
- 9. Gallucci S, Matzinger P. 2001. Danger signals: SOS to the immune system. Curr. Opin. Immunol. 13:114–119 [DOI] [PubMed] [Google Scholar]
- 10. Henderson B, Henderson S. 2009. Unfolding the relationship between secreted molecular chaperones and macrophage activation states. Cell Stress Chaperones 14:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280 doi:10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keller M, Ruegg A, Werner S, Beer HD. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132:818–831 [DOI] [PubMed] [Google Scholar]
- 13. Kodukula P, Liu T, Rooijen NV, Jager MJ, Hendricks RL. 1999. Macrophage control of herpes simplex virus type 1 replication in the peripheral nervous system. J. Immunol. 162:2895–2905 [PubMed] [Google Scholar]
- 14. Kono H, Rock KL. 2008. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kristiansen H, et al. 2010. Extracellular 2′-5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J. Virol. 84:11898–11904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lietzen N, et al. 2011. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 7:e1001340 doi:10.1371/journal.ppat.1001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luber CA, et al. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32:279–289 [DOI] [PubMed] [Google Scholar]
- 18. Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10:417–426 [DOI] [PubMed] [Google Scholar]
- 19. Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440:237–241 [DOI] [PubMed] [Google Scholar]
- 20. Mathivanan S, Simpson RJ. 2009. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9:4997–5000 [DOI] [PubMed] [Google Scholar]
- 21. Melchjorsen J, et al. 2010. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 84:11350–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melchjorsen J, Siren J, Julkunen I, Paludan SR, Matikainen S. 2006. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-kappaB and IRF-3. J. Gen. Virol. 87:1099–1108 [DOI] [PubMed] [Google Scholar]
- 23. Nickel W, Rabouille C. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10:148–155 [DOI] [PubMed] [Google Scholar]
- 24. Nickel W, Seedorf M. 2008. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu. Rev. Cell Dev. Biol. 24:287–308 [DOI] [PubMed] [Google Scholar]
- 25. Öhman T, et al. 2010. Cytosolic RNA recognition pathway activates 14-3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J. Proteome Res. 9:1549–1564 [DOI] [PubMed] [Google Scholar]
- 26. Osterloh A, Breloer M. 2008. Heat shock proteins: linking danger and pathogen recognition. Med. Microbiol. Immunol. 197:1–8 [DOI] [PubMed] [Google Scholar]
- 27. Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. 2011. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 11:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pavlou MP, Diamandis EP. 2010. The cancer cell secretome: a good source for discovering biomarkers? J. Proteomics 73:1896–1906 [DOI] [PubMed] [Google Scholar]
- 29. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 30. Pichlmair A, et al. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12:624–630 [DOI] [PubMed] [Google Scholar]
- 31. Pirhonen J, Sareneva T, Kurimoto M, Julkunen I, Matikainen S. 1999. Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway. J. Immunol. 162:7322–7329 [PubMed] [Google Scholar]
- 32. Rathinam VA, Vanaja SK, Fitzgerald KA. 2012. Regulation of inflammasome signaling. Nat. Immunol. 13:333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Record M, Subra C, Silvente-Poirot S, Poirot M. 2011. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81:1171–1182 [DOI] [PubMed] [Google Scholar]
- 34. Rintahaka J, Lietzen N, Öhman T, Nyman TA, Matikainen S. 2011. Recognition of cytoplasmic RNA results in cathepsin-dependent inflammasome activation and apoptosis in human macrophages. J. Immunol. 186:3085–3092 [DOI] [PubMed] [Google Scholar]
- 35. Rintahaka J, Wiik D, Kovanen PE, Alenius H, Matikainen S. 2008. Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. J. Immunol. 180:1749–1757 [DOI] [PubMed] [Google Scholar]
- 36. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex virus, p 2501–2601 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 37. Ronni T, Melen K, Malygin A, Julkunen I. 1993. Control of IFN-inducible MxA gene expression in human cells. J. Immunol. 150:1715–1726 [PubMed] [Google Scholar]
- 38. Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. 2009. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37:D852–D857 doi:10.1093/nar/gkn732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato S, St-Pierre C, Bhaumik P, Nieminen J. 2009. Galectins in innate immunity: dual functions of host soluble beta-galactoside-binding lectins as damage-associated molecular patterns (DAMPs) and as receptors for pathogen-associated molecular patterns (PAMPs). Immunol. Rev. 230:172–187 [DOI] [PubMed] [Google Scholar]
- 40. Schmeisser H, et al. 2010. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J. Virol. 84:10671–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith JS, Robinson NJ. 2002. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J. Infect. Dis. 186(Suppl 1):S3–S28 [DOI] [PubMed] [Google Scholar]
- 43. Terenzi F, Hui DJ, Merrick WC, Sen GC. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064–34071 [DOI] [PubMed] [Google Scholar]
- 44. Terenzi F, Pal S, Sen GC. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340:116–124 [DOI] [PubMed] [Google Scholar]
- 45. Trost M, et al. 2009. The phagosomal proteome in interferon-gamma-activated macrophages. Immunity 30:143–154 [DOI] [PubMed] [Google Scholar]
- 46. UniProt Consortium 2009. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 37:D169–D174 doi:10.1093/nar/gkn664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whitley RJ, Roizman B. 2001. Herpes simplex virus infections. Lancet 357:1513–1518 [DOI] [PubMed] [Google Scholar]
- 48. Zhang L, Zhang ZP, Zhang XE, Lin FS, Ge F. 2010. Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication. J. Virol. 84:6050–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.