Abstract
Microbial systems are being increasingly developed as production hosts for a wide variety of chemical compounds. Broader adoption of microbial synthesis is hampered by a limited number of high-yielding natural pathways for molecules with the desired physical properties, as well as the difficulty in functionally assembling complex biosynthetic pathways in heterologous hosts. Here, we address both of these challenges by reporting the adaptation of the butanol biosynthetic pathway for the synthesis of odd-chain molecules and the development of a complementary modular toolkit to facilitate pathway construction, characterization, and optimization in engineered Escherichia coli. The modular feature of our pathway enables multientry and multiexit biosynthesis of various odd-chain compounds at high efficiency. By varying combinations of the pathway and toolkit enzymes, we demonstrate controlled production of propionate, trans-2-pentenoate, valerate, and pentanol, compounds with applications that include biofuels, antibiotics, biopolymers, and aroma chemicals. Importantly, and in contrast to a previously used method to identify limitations in heterologous amorphadiene production, our bypass strategy was effective even without the presence of freely membrane-diffusible substrates. This approach should prove useful for optimization of other pathways that use CoA-derivatized intermediates, including fatty acid β-oxidation and the mevalonate pathway for isoprenoid synthesis.
Keywords: biochemicals, metabolic engineering, synthetic biology
The interest in microbial synthesis of fuels and chemicals has increased substantially as efforts to transition toward a “bio-based” economy have gained momentum; however, the ability to more broadly use biological systems for chemical production is somewhat limited by the natural repertoire of biosynthetic pathways. To expand upon these options, it is useful to consider the means by which a limited number of functional conversions can lead to a broad array of structures. In many natural pathways, a carbon skeleton is initially formed from which a handful of compounds with diverse chemical architecture are generated. These compounds can be further modified by several decorating enzymes to yield an enormous array of products that exert physiological functions. For example, a few prenyl diphosphates produced from two simple isoprene units through the central isoprenoid pathway can be modified into over 50,000 functionally diverse isoprenoids by terpene-modifying enzymes (1, 2) (Fig. 1A). Other examples include polyketide biosynthesis, where polyketide chains supplied from the iterative carbon elongation pathway can be derivatized into numerous bioactive compounds (3, 4), and the tricarboxylic acid cycle (TCA cycle) that supplies precursors for biosynthesis of amino acids (5) (Fig. 1A).
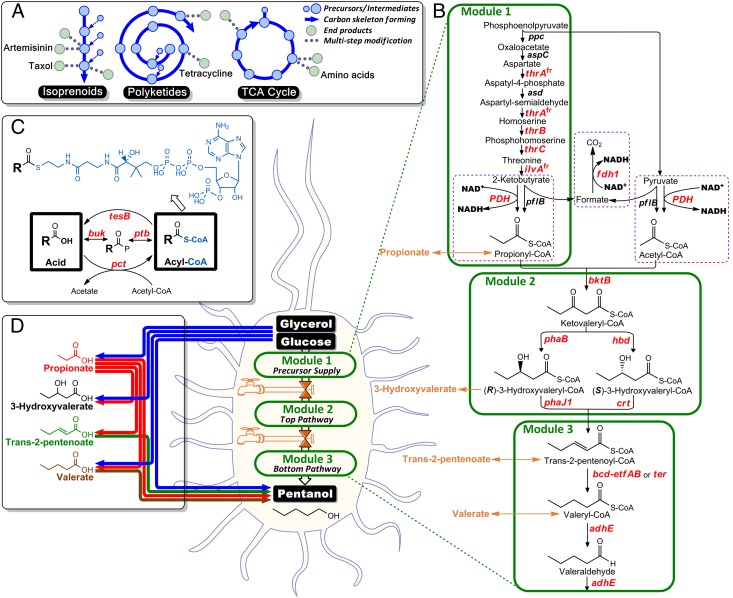
Fig. 1.
Metabolic pathway construction for direct microbial synthesis of pentanol from glucose or glycerol. (A) Nature has evolved an efficient way to supply structurally diverse compounds by initially forming a carbon skeleton from which a handful of bioactive compounds are generated. For example, the highly functionalized antimalarial drug Artemisinin and anticancer drug Taxol can be derived from the isoprenoids pathway; various antibiotics such as Tetracycline can be synthesized from the polyketides pathway; and precursors for synthesis of various amino acids can be supplied from the tricarboxylic acid cycle (TCA). (B) The pentanol biosynthetic pathway consists of three modules, each of which was validated separately and then assembled together. Genes in red and italics were overexpressed from inducible plasmids. (C) An enzyme toolkit was established and used to analyze the pentanol biosynthetic pathway by channeling membrane-impermeable CoA-tagged metabolite intermediates to membrane-permeable free acids, allowing for extracellular detection by HPLC. Similarly, certain free carboxylic acids can be activated with an adduct of a CoA tag using the enzyme toolkit and channeled into the pentanol pathway. (D) Tailor-made synthesis of various odd-chain chemicals can be achieved through particular combinations of the pathway and toolkit enzymes.
In an attempt to mimic nature’s design for supplying structurally diverse compounds, we began with an engineered l-butanol pathway from Clostridium acetobutylicum (6), and incorporated elements from the synthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [poly(3HB-co-3HV)] in Cupriavidus necator (formerly known as Ralstonia eutropha) (7), and threonine in Escherichia coli (8) to create a unique core pathway for the production of industrially relevant chemicals and fuels (Figs. S1 and S2), particularly n-pentanol, a fuel alternative with high energy density and enhanced physical properties that would allow better integration into the current infrastructure (9) (Table S1). Siphoning precursors from threonine enables the in vivo synthesis of odd-chain substrates, expanding upon the natural pool of acids and alcohols that have been produced from simple carbohydrate substrates through fatty acid biosynthesis and β-oxidation (10).
However, successful implementation of a heterologous pathway designed to use nonnatural substrates (in this case to produce pentanol rather than butanol) presents two challenges. First, the extent to which each of the enzymes will accept the five-carbon unnatural substrates is unknown. Second, any limitations in activity and pathway bottlenecks are confounded by the assembly of a unique combination of genes from multiple source organisms into a single heterologous host. To address both of these obstacles, we established a bypass strategy for pathway construction that is conceptually analogous to process control in chemical engineering, where a stream split from the feed to a process unit is combined with the outlet stream from that process, thereby bypassing the unit in question. In our case, the entire pathway was divided into three modules: precursor supply (module 1), top pathway (module 2), and bottom pathway (module 3) (Fig. 1B). Each module was then individually tested with separate inlet and outlet carbon streams to confirm in vivo functionality and establish an operating range for the constituent enzymes. Such a strategy has been used previously to identify limitations in heterologous production systems (11); however, this usually requires freely membrane-diffusible intermediates. In the current pathway, all of the intermediates are present as CoA derivatives and are retained intracellularly. Our bypass approach thus required the development of a CoA-addition/removal enzyme toolkit that would either hydrolyze metabolites within a module to provide a product stream or activate exogenously supplied free acids once inside the cell to provide the desired feed stream (Fig. 1 C and D). Our toolkit consists of the broad-substrate-range enzymes Ptb-Buk (from C. acetobutylicum) (12), TesB (from E. coli) (13), and Pct (from Megasphaera elsdenii) (14). Herein we describe efforts to design, construct, and characterize the multifunctional pathway while exploiting the uniquely designed bypass strategy with the toolkit enzymes.
Results
De Novo Design of a Unique High-Yielding Biosynthetic Pathway.
The carbon skeleton of this unique pathway is established through the action of an acetoacetyl-CoA thiolase, encoded by the bktB gene of C. necator. This enzyme has been previously shown to have highest in vitro enzyme activity toward the C5 substrate (7), so is an ideal candidate enzyme to carry out condensation of acetyl-CoA and propionyl-CoA to form 3-ketovaleryl-CoA (Fig. 1B). The genes used for the conversion of 3-ketovaleryl-CoA to valeryl-CoA include hbd, crt, and bcd-etfAB from C. acetobutylicum, encoding for 3-hydroxybutyryl-CoA dehydrogenase, crotonase, and butyryl-CoA dehydrogenase with its two electron transfer proteins, respectively. Given that Bcd has been shown to be a bottleneck for butanol biosynthesis in engineered E. coli (15, 16), we also used the highly active isozyme, trans-2-enoyl-CoA reductase (Ter). In addition, to explore a broader metabolic space with respect to the intermediates in pentanol biosynthesis, both the hbd and crt genes [involving the (S)-enantiomer] can be replaced with phaB from C. necator H16 and phaJ1 from Pseudomonas aeruginosa [involving the (R)-enantiomer] (17), respectively, to convert ketovaleryl-CoA to trans-2-pentenoyl-CoA. A bifunctional aldehyde/alcohol dehydrogenase, encoded by adhE from C. acetobutylicum, catalyzes the final steps of pentanol synthesis from valeryl-CoA.
Test of Individual Modules for Nonnatural Five-Carbon Substrates.
Because the thiolase reaction is the first committed step and establishes the core five-carbon skeleton, we began with validation of the top pathway (module 2). We previously reported the synthesis of over 2 g/L of (R)- or (S)-3HV from propionate and glucose, with up to 67% of total 3-hydroxyacids as the five-carbon 3HV (18). This finding provided an initial proof-of-concept that high levels of five-carbon intermediates can be obtained. To complete module 2, the conversion of 3HV-CoA to trans-2-pentenoyl-CoA was evaluated. In general, there are two classes of dehydratases: (S)-specific enoyl-CoA hydratase (Crt) and (R)-specific enoyl-CoA hydratase (PhaJ1). Two distinct metabolic routes were examined, including one through (S)-3HV-CoA with the hbd-crt gene pair and the other through (R)-3HV-CoA with the phaB-phaJ1 gene pair (Figs. S3 and S4). We found that either route led to production of trans-2-pentenoate, suggesting all enzymes examined here, Hbd, Crt, PhaB, and PhaJ1, are able to accept five-carbon substrates. The low titers compared with 3HV synthesis (∼55 mg/L versus ∼2 g/L) also identified dehydratase activity as a potential bottleneck in the full pathway, despite previous reports of high enzyme activities measured in vitro in E. coli extracts (19).
To circumvent the need for feeding propionate, we constructed a pathway for propionyl-CoA synthesis from glucose or glycerol through up-regulation of threonine biosynthesis by overexpressing an E. coli thrAfrBC operon along with overexpression of the ilvAfr gene, encoding a feedback-resistant threonine deaminase (18). The resulting 2-ketobutyrate can further be converted to propionyl-CoA by the pyruvate dehydrogenase complex (PDH) enzyme, encoded by the aceEF-lpd operon (Fig. 1B, module 1). Module 1 resulted in propionate production with titers up to 804 mg/L from glycerol when the tesB thiosesterase was overexpressed (Table 1). At this point, both modules 1 and 2 had been validated, and we proceeded to module 3. To examine the bottom half of module 3, valerate was supplemented to cultures with strains expressing the CoA-activators ptb-buk or pct to test whether the alcohol dehydrogenase AdhE could convert valeryl-CoA to pentanol (Fig. S5). Two versions of adhE genes were explored: the original adhE PCR-amplified directly from C. acetobutylicum genomic DNA and a codon-optimized version (denoted as adhEopt) obtained through gene synthesis. We found that pentanol was only detectable in cells containing AdhEopt. This result was corroborated by the appearance of improved protein expression for the codon-optimized variant by SDS/PAGE analysis. Interestingly, the original AdhE gene has been used previously to produce butanol (6, 19, 20). This result might point to a lower activity with the C5 substrates, such that improved expression is now more important. Overall, we succeeded in pentanol production with titers up to 534 mg/L from 20 mM valerate (Fig. S5).
Table 1.
Biosynthesis of various odd-carbon chemicals by different pathway modulation schemes
| Product titers (mg/L) ± SD |
||||||
| Prop |
3HV |
T2P |
Val |
PenOH |
||
| Pathways (substrate*→product) | B | C | D | E | F | Strains† |
| A′→B | 804 ± 20 | ND | ND | ND | ND | Pal25 |
| A→C (S-3HV) | 35 ± 20 | 312 ± 6 | ND | ND | ND | (18) |
| A′→C (R-3HV) | 140 ± 8 | 963 ± 68 | ND | ND | ND | (18) |
| A′→E | 110 ± 10 | ND | ND | 398 ± 15 | ND | Pal24 |
| A′→F | 78 ± 6 | ND | ND | 81 ± 6 | 116 ± 14 | Pal23 |
| B→C (S-3HV)‡ | 2,051 ± 67 | ND | ND | ND | (18) | |
| B→C (R-3HV) ‡ | 1,964 ± 99 | ND | ND | ND | (18) | |
| B→D‡ | 869 ± 54 | 57 ± 2 | ND | ND | Pal3 | |
| B→E‡ | ND | ND | 554 ± 28 | ND | Pal12 | |
| B→F‡ | ND | ND | 360 ± 23 | 358 ± 21 | Pal11 | |
| D→F§ | ND | ND | 799 ± 33 | 1,317 ± 45 | BL12 | |
| E→F§ | ND | ND | ND | 530 ± 18 | BL4 | |
A, glucose; A′, glycerol; B, propionate (Prop); C, 3-hydroxyvalerate (3HV); D, trans-2-pentenoate (T2P); E, valerate (Val); F, pentanol (PenOH); ND, not detected.
*Feed amount: Glucose or glycerol (10 g/L); Prop (20 mM); T2P (2 g/L); Val (20 mM).
†Table S2.
‡Recombinant strains containing synthetic pathways that start with B were grown in media supplemented with 10 g/L of glucose in additional to propionate.
§Nearly all consumed substrate was converted (∼100 mol%) to products listed above.
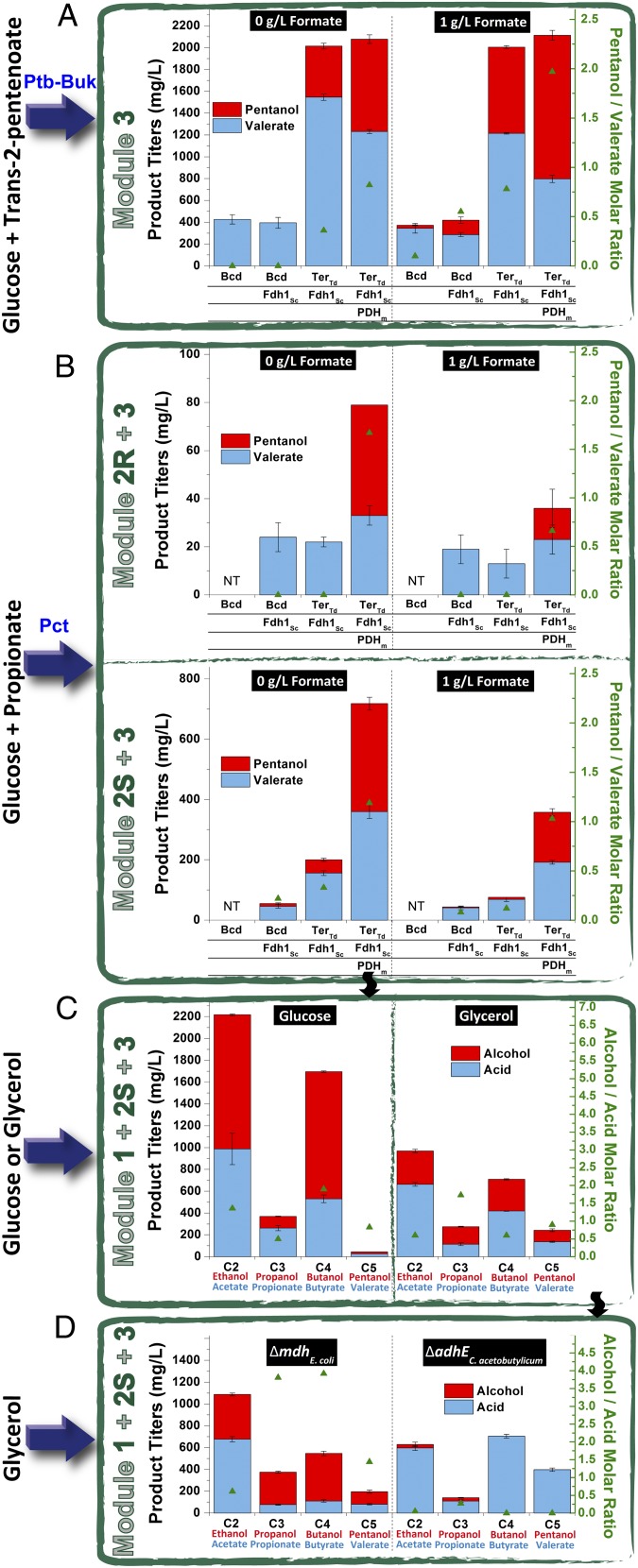
Next, Bcd, the remaining enzyme in module 3, was evaluated. Trans-2-pentenoate was supplemented exogenously and activated to trans-2-pentenoyl-CoA by the CoA-activator Ptb-Buk, followed by sequential conversion to pentanol, catalyzed by Bcd and AdhEopt. Module 3 produced valerate but not pentanol (Fig. 2A and Upper Left plot in Fig. S6). We suspected that low production might be because of insufficient NADH supply. Pentanol synthesis from valeryl-CoA involves two reductions and requires two molecules of NADH, but valerate synthesis from valeryl-CoA, a nonredox reaction, does not require NADH. Additionally, production of valerate through Ptb-Buk yields 1 ATP. Thus, production of valerate would be energetically more favorable than production of pentanol. If this hypothesis is correct, increased NADH availability should facilitate pentanol production.
Fig. 2.
Bottom-up validation of the pentanol biosynthetic pathway using a modular approach. (A) Validation of module 3 was achieved by feeding trans-2-pentenoate and monitoring production of valerate and pentanol. Effects of formate supplementation, the replacement of Bcd with TerTd (trapping carbon flow toward the forward direction of pentanol synthesis), and the overexpression of Fdh1Sc and PDHm (boosting intracellular NADH availability) on pentanol synthesis were examined with respect to product titers and selectivities (pentanol/valerate molar ratios). (B) Two versions of module 2, module 2S with an hbd-crt gene pair and module 2R with a phaB-phaJ1 gene pair, were then stacked onto module 3. The combined modules (module 2S+3 or module 2R+3) were examined across three pathway variants. (C) The propionyl-CoA synthesis module (module 1) was then introduced into the best-performing variant from the previous test with module 2S+3, and the combined pathway (module 1+2S+3) was examined for direct pentanol synthesis from glucose or glycerol. The product profiles, including carboxylic acids and n-alcohols, and product selectivities (alcohol/acid molar ratios) are shown. (D) Effect of gene knockouts, including the mdh gene in the production host genome and the adhEopt gene in the pentanol biosynthetic pathway, on synthesis of carboxylic acids and n-alcohols. NT, not tested.
To address this limitation, we examined three solutions. First, we overexpressed yeast NAD+-dependent formate dehydrogenase, encoded by the fdh1 gene. The native E. coli formate dehydrogenase converts formate to CO2 and H2 without generation of any NADH and the yeast NAD+-dependent FDH1 generates 1 mol of NADH from 1 mol of formate (21). Second, the Bcd enzyme was replaced with the Ter enzyme that directly uses NADH as the electron donor, which was shown to be more efficient (15, 22). As previously reported (23), the Ter enzyme from Treponema denticola accepts C4 (crotonyl-CoA) but not C6 (transhexenoyl-CoA) substrates. On the other hand, Ter from Euglena gracilis works on both C4 and C6 substrates, but its specific activity on crotonyl-CoA is 10-fold lower than Ter from T. denticola. Neither enzyme had been profiled against C5 substrates. Additionally, reactions catalyzed by the two Ter enzymes have been shown to be essentially irreversible with butyryl-CoA as the substrate, but the reaction catalyzed by the Bcd enzyme has an oxidizing activity in the reverse direction (15). Considering the distinct characteristics of the Ter enzymes, both were used in place of Bcd to convert trans-2-pentenoyl-CoA to valeryl-CoA. Under anaerobic growth conditions, 1 mol of glucose yields only 2 mol of NADH because of an inactive PDH enzyme. Because pentanol biosynthesis has an intensive requirement for cofactors, our third solution was to overexpress the PDH mutant lpd101-E354K (PDHm), which functions under anaerobic conditions (24). Using the anaerobically active PDHm was expected to boost the NADH yield on glucose up to four moles of NADH per mole of glucose.
Codon-optimized fdh1 genes from Saccharomyces cerevisiae and Candida boidinii were initially tested. In formate-supplemented cultures, the overexpression of either Fdh1Sc or Fdh1Cb resulted in the synthesis of more reduced products, including pentenol (a monounsaturated five-carbon alcohol) and pentanol (Lower three plots in Fig. S6). The two codon-optimized ter genes from T. denticola and E. gracilis were then compared with respect to enhancement of pentanol synthesis. TerTd was found to have a more profound effect on enhancing pentanol synthesis than TerEg (Fig. S7). Overexpression of Fdh1Sc, TerTd, and PDHm resulted in pentanol titers up to 1,317 mg/L from 2 g/L trans-2-pentenoate in the presence of 1 g/L formate (Fig. 2A). Overall, we succeeded in addressing the NADH deficiency problem and drastically improved pentanol production from an undetectable amount to more than 1 g/L.
Assembly of Individual Modules to Enable Direct Microbial Synthesis of Pentanol from Glucose or Glycerol.
After validating all three modules, we linked them together to produce the C5 alcohol from a single carbon source. Modules 2 and 3 were first assembled, resulting in up to 46 and 358 mg/L of pentanol, respectively, through the phaB-phaJ1 (module 2R+3) or hbd-crt routes (module 2S+3) (Fig. 2B). These titers are between those obtained when only module 2 was used to produce trans-2-pentenoate and when only module 3 was used to produce pentanol. These results validate previous reports that emphasize the importance of considering the complete pathway for optimization of productivity (25), and also suggest that the full synthetic capacity of module 3 is being underused, perhaps because of increased cofactor demands when modules 2 and 3 are combined or because of limited flux through Crt/PhaJ1. Because the hbd-crt route outperformed the phaB-phaJ1 route, the former was chosen for further study. Recombinant strains containing all three modules (module 1+2S+3) were constructed, and the resulting strain synthesized 19 mg/L of pentanol from glucose and 109 mg/L of pentanol from glycerol (Fig. 2C). The lower titers from a sole carbon source are consistent with the results seen when only evaluating module 1 and the top half of module 2 (18), and indicate a need to increase the precursor supply. In general, the use of glycerol provides more reducing equivalents [e.g., NAD(P)H] because of its higher reduction state relative to glucose (26–28). A more reduced substrate favors energetically expensive pentanol synthesis, resulting in enhanced production compared with the use of glucose (Fig. 2C). The use of glycerol is also expected to have less acetyl-CoA overflow as a result of lower glycolytic flux. The reduced supply of the C2 precursor would lead to product redistribution, resulting in decreased production of even-chain alcohols (ethanol and butanol), which is consistent with our observations (Fig. 2C).
Host Engineering to Adjust Product Profiles.
The observation of concomitant production of carboxylic acids in an engineered pentanol-producing E. coli strain that lacks any toolkit enzyme for hydrolysis suggests the existence of an endogenous enzymatic activity for cleaving acyl-CoA and releasing free acids. Given that phosphotransacetylase (encoded by pta), one of the acetate synthesis enzymes, has been deleted in the host strain E. coli Pal(DE3) (Table S2), we suspected that the CoA-cleaving activity came from an E. coli endogenous tesB gene. However, tesB deletion failed to reduce acid synthesis. Guided by our previous observation that increased NADH availability not only enhanced pentanol production but boosted the pentanol/valerate molar ratios (Fig. 2 A and B), we then deleted the mdh gene, encoding malate dehydrogenase that catalyzes the conversion of oxaloacetate to malate using NADH as a coenzyme. Pursuing the mdh deletion was also motivated by our observation of malate and succinate byproduct formation in our recombinant E. coli strains. For physiological reasons, wild-type E. coli produces a large amount of ethanol and lactate as a way to regenerate NAD+ from NADH under anaerobic conditions. Our E. coli host strain, however, is deficient in these reactions because of the adhE and ldhA deletions and, therefore, resorts to malate and succinate synthesis. To preserve NADH for our pentanol pathway as well as reduce formation of malate and succinate byproducts, we deleted the mdh gene. As expected, the mdh deletion led to product redistribution toward alcohols with increased alcohol/acid molar ratios (Fig. 2D), most likely a result of further increased NADH availability. Additionally, we also shifted the product distribution in the opposite direction by removing the adhE gene in the pentanol biosynthetic pathway, resulting in near abolishment of alcohol synthesis (Fig. 2D). Taken together, our results demonstrated a functional and feasible pentanol biosynthetic pathway from a sole carbon source in E. coli.
Rewiring of the Biosynthetic Pathway for Synthesis of Various Fuels and Chemicals.
With a functional pentanol biosynthetic pathway, we then focused on rerouting carbon flow in the pathway for custom synthesis of various odd-carbon numbered chemicals (Fig. 1D). By varying the combination of pathway and toolkit enzymes and supplying various carbon sources, we achieved controlled production of propionate, chiral 3HV, trans-2-pentenoate, and valerate (Table 1): molecules with applications that include biofuels [valerate esters (29, 30) and propane (31)], antibiotics [6-deoxyerythronolide B (3)], biopolymers [poly-3HV (32)], and aroma chemicals [trans-2-pentenoate (33)] (Fig. S2).
Discussion
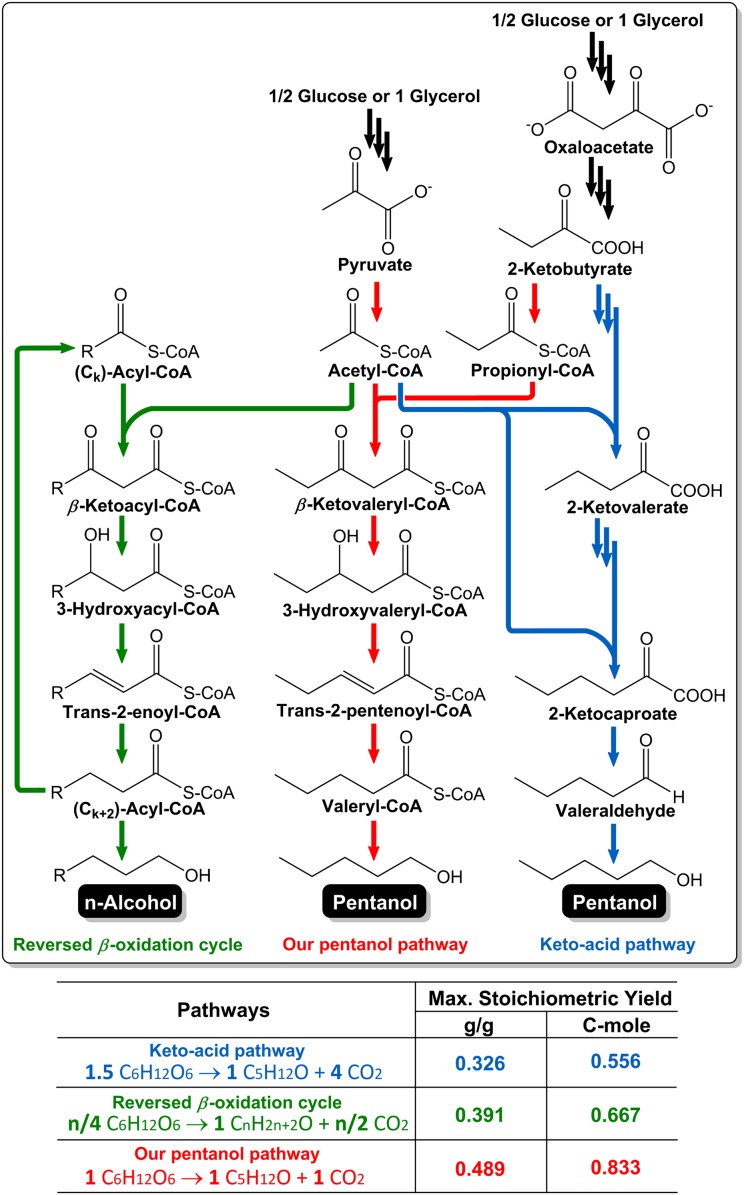
Recently, two alternative alcohol biosynthetic routes, the keto-acid pathway (34, 35) and engineered reversal of the fatty acid β-oxidation cycle (10), have been investigated for pentanol synthesis in engineered E. coli, but pentanol was observed only as a minor component in the product mixture. In the latter example, propionate supplementation was also required to observe pentanol production. Moreover, these two routes are less efficient with respect to the carbon balance than our pathway, a crucial metric in the production of biofuels (36). As illustrated in the calculation of maximum theoretical yields of pentanol on glucose or glycerol (Fig. 3), our pathway has the potential to conserve the most carbon for alcohol synthesis. This high theoretical yield provides a benchmark for future scale-up development. The butanol pathway was also recently extended to produce 1-hexanol (37). The reported titers were low and this pathway, as shown in previous work (10), suffers in being limited to carbon chain elongation by two carbon units.
Fig. 3.
Comparison of theoretical maximum yields of pentanol biosynthesis through various biosynthetic pathways. The detailed reaction steps of each pathway are shown, including the keto-acid pathway (34), reversed β-oxidation cycle (10), and our pentanol pathway.
In this work, we have demonstrated the redesign of a natural pathway combined with inclusion of elements from other pathways to produce a unique route toward a variety of odd-chain chemicals, including pentanol. The bypass strategy we employed enabled the analysis of individual modules to both validate in vivo functionality of a complex multistep pathway and to identify potential bottlenecks in the fully assembled pathway. Importantly, this strategy was used even without the presence of freely membrane-diffusible substrates and should prove useful for optimization of other pathways that use CoA-derivatized intermediates, including fatty acid β-oxidation and the mevalonate pathway for isoprenoid synthesis. In the present pathway, further engineering of the internal redox metabolism, as well as improved substrate specificity and activity of pathway enzymes, should lead to economically viable and precisely controlled production of desired products.
Materials and Methods
Plasmids.
Codon-optimized genes, including adhE (CAP0035) from C. acetobutylicum ATCC 824, fdh1 from S. cerevisiae, fdh1 from C. boidinii, ter from T. denticola, and ter E. gracilis were purchased from Genscript. Genes derived from C. acetobutylicum ATCC 824 (hbd, crt, bcd, etfAB, adhE, and ptb-buk), C. necator (formerly known as R. eutropha) H16 (bktB and phaB), P. aeruginosa (phaJ1), M. elsdenii (pct), Corynebacterium glutamicum ATCC 13032 (ilvAfr), E. coli MG1655 (tesB), E. coli SE2378 (aceEF-lpd101), and E. coli ATCC 21277 (thrAfrBC opeon) were obtained by PCR using genomic DNA (gDNA) templates. All gDNAs were prepared using the Wizard Genomic DNA Purification Kit (Promega). Custom oligonucleotides (primers) were purchased for all PCR amplifications (Sigma-Genosys) (Table S3). In all cases, Phusion High Fidelity DNA polymerase (Finnzymes) was used for DNA amplification. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. Recombinant DNA techniques were performed according to standard procedures (38).
Compatible vectors pETDuet-1, pCDFDuet-1, pACYCDuet-1, and pCOLADuet-1 (Novagen) were used to provide individual expression of each gene under a T7lac promoter and a ribosome binding site. The PCR products were digested with restriction enzymes corresponding to the restriction site incorporated into them by their respective primers and ligated directly into similarly digested Duet vectors. Ligation reactions using pETDuet-1, pACYCDuet-1, or pCOLADuet-1 as the vector were used to transform E. coli DH10B, and ligations using pCDFDuet-1 were used to transform E. coli ElectroTen-Blue. All constructs were confirmed to be correct by restriction enzyme digestion and nucleotide sequencing. Once all plasmids were constructed, they were used to cotransform, as appropriate, E. coli BL21Star(DE3), Pal(DE3), Pal(DE3 ΔtesB), or Pal(DE3 Δmdh) to create production strains.
Bacterial Strains.
E. coli MG1655(Δpta ΔadhE ΔldhA) was donated by Gregory Stephanopoulos of the Department of Chemical Engineering at the Massachusetts Institute of Technology, Cambridge, MA. E. coli Pal(DE3) was then constructed from E. coli MG1655(Δpta ΔadhE ΔldhA) using a λDE3 Lysogenization Kit (Novagen) to allow the expression of genes under the T7lac promoter (16). Deletions of tesB and mdh genes in Pal(DE3) were achieved with P1 transduction using the Keio collection strains as donor cells (39). The kanamycin cassette was removed using plasmid pCP20 as described by Datsenko and Wanner (40) and the successfully constructed mutant strains were verified by colony PCR using appropriate primers (Table S3).
E. coli BL21Star(DE3) (Invitrogen) was used as the host strain for substrate feeding experiments, including pentanol synthesis from valerate (strains BL1–BL5 in Table S2) or trans-2-pentenoate (strains BL6–BL12 in Table S2), and Pal(DE3) was the production host strain used for the rest of experiments, including trans-2-pentenoate synthesis from glucose and propionate (strains Pal1–Pal4 in Table S2), butanol synthesis from glucose (strains Pal5–Pal6 in Table S2), pentanol synthesis from glucose and propionate (strains Pal7–Pal16 in Table S2), pentanol synthesis solely from glucose or glycerol (strains Pal17–Pal24, Palm1, and Palt1 in Table S2), and propionate synthesis solely from glycerol (strain Pal25 in Table S2).
Culture Conditions.
For trans-2-pentenoate synthesis from glucose and propionate, seed cultures of the recombinant strains (strains Pal1–Pal4) were grown in TB medium at 30 °C overnight on a rotary shaker at 250 rpm, and were used to inoculate, at an inoculation volume of 10%, 50 mL TB medium in 250-mL flasks for aerobic growth, and 13 mL TB medium in 15-mL glass tubes (Bellco Glass) stoppered with a butyl rubber septum for microaerobic or anaerobic growth. The septum was pierced with a 26-gauge syringe needle to achieve microaerobic conditions. All cell cultures were supplemented with 10 g/L glucose. Cultures were induced with 0.5 mM IPTG at 2 h postinoculation and incubated for another 24 h.
For the substrate feeding experiments, seed cultures of the recombinant strains (strains BL1–BL12, Table S2) were grown in TB medium at 30 °C overnight on a rotary shaker at 250 rpm, and were used to inoculate 45 mL TB medium supplemented with 10 g/L glucose at an inoculation volume of 10% in 50-mL glass culture tubes. Cultures were induced with 0.5 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) at 2 h postinoculation and incubated for another 72 h. One of the substrates, including neutralized valerate (10 mM or 20 mM) and trans-2-pentenoate (2 g/L), was supplemented to strains BL1–BL5 and BL6–BL12, respectively, at the same time of induction to provide the precursor needed for pentanol synthesis.
For pentanol synthesis, seed cultures of the recombinant E. coli strains (strains Pal7–Pal25, Palm1, and Palt1 in Table S2) were grown in TB medium at 30 °C overnight, and used to inoculate 3 mL TB medium supplemented with 10 g/L glucose or 10 g/L glycerol at an inoculation volume of 10% in 15 mL Falcon tubes. Cultures were induced with 0.5 mM IPTG at 2 h postinoculation and incubated for another 96 h. For strains Pal7–Pal16 (Table S2), 20 mM neutralized propionate was supplemented at the same time of induction. For propionate synthesis, seed cultures of the recombinant strain Pal25 were grown in LB medium at 30 °C overnight and used to inoculate 50 mL LB medium supplemented with 20 g/L glycerol at an inoculation volume of 2% in 250 mL flasks. Cultures were induced with 0.5 mM IPTG at 3 h postinoculation (OD600 reached 0.8∼1.0) and incubated for another 48 h. For profiling experiments with various headspace to culture volume ratios, the seed cultures of the recombinant strains (strains Pal5 and Pal6 in Table S2) were used to inoculate, at an inoculation volume of 10%, 10 mL, 5 mL, 3 mL of TB medium in 15-mL Falcon tubes, 25 mL and 10 mL of TB medium in 250-mL flasks with caps screwed on, and 25 mL of TB medium in 250-mL flask with loose caps. All cultures were supplemented with 10 g/L glucose and then incubated at 30 °C on a rotary shaker. Cultures were induced with 0.5 mM IPTG at 2 h postinoculation and incubated for another 48 h. A headspace-to-culture volume ratio at 4 was found to achieve the highest specific butanol production (Fig. S8), so this culture condition was chosen to conduct any following culture experiments.
In all cases, culture medium was supplemented with 50 mg/L ampicillin, 50 mg/L streptomycin, 34 mg/L chloramphenicol, and 25 mg/L kanamycin, as required. One milliliter of culture was withdrawn at the end of the incubation period for HPLC analysis. In general, experiments were performed in triplicates, and data are presented as the averages and SDs of the results.
Metabolite Analysis.
Culture samples were pelleted by centrifugation and aqueous supernatant collected for HPLC analysis using an Agilent 1200 series instrument with a refractive index detector and a diode array detector at 210 nm. Analytes were separated using an Aminex HPX-87H anion-exchange column (Bio-Rad Laboratories) and a 5 mM H2SO4 mobile phase. Glucose, glycerol, acetate, 3-hydroxybutyrate, 3-hydroxyvalerate, crotonate, trans-2-pentenoate, butyrate, valerate, butanol, pentenol, and pentanol were quantified using commercial standards on a 1200 HPLC series instrument by linear interpolation from calibration of external standards.
Supplementary Material
Acknowledgments
We thank K. T. Shanmugam (Department of Microbiology and Cell Science, University of Florida) for the gift of the Escherichia coli SE2378 strain, and M. J. Sheppard for cloning and characterizing the codon-optimized adhE gene. This work was supported by Shell Global Solutions.
Footnotes
Conflict of interest statement: A patent application based on the work described herein has been filed by the Massachusetts Institute of Technology.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209002109/-/DCSupplemental.
References
- 1.Vranová E, Coman D, Gruissem W. Structure and dynamics of the isoprenoid pathway network. Mol Plant. 2012;5(2):318–333. doi: 10.1093/mp/sss015. [DOI] [PubMed] [Google Scholar]
- 2.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2(12):674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291(5509):1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 4.Boghigian BA, Pfeifer BA. Current status, strategies, and potential for the metabolic engineering of heterologous polyketides in Escherichia coli. Biotechnol Lett. 2008;30(8):1323–1330. doi: 10.1007/s10529-008-9689-2. [DOI] [PubMed] [Google Scholar]
- 5.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen DR, et al. Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng. 2009;11(4–5):262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Slater S, et al. Multiple beta-ketothiolases mediate poly(beta-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180(8):1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, et al. Metabolic engineering of a reduced-genome strain of Escherichia coli for L-threonine production. Microb Cell Fact. 2009;8:2. doi: 10.1186/1475-2859-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoekman SK. Biofuels in the U.S.–Challenges and opportunities. Renew Energy. 2009;34(1):14–22. [Google Scholar]
- 10.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476(7360):355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 11.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 12.Liu SJ, Steinbüchel A. Exploitation of butyrate kinase and phosphotransbutyrylase from Clostridium acetobutylicum for the in vitro biosynthesis of poly(hydroxyalkanoic acid) Appl Microbiol Biotechnol. 2000;53(5):545–552. doi: 10.1007/s002530051655. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Z, et al. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl Environ Microbiol. 2004;70(7):3807–3813. doi: 10.1128/AEM.70.7.3807-3813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi S, et al. A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc Natl Acad Sci USA. 2008;105(45):17323–17327. doi: 10.1073/pnas.0805653105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bond-Watts BB, Bellerose RJ, Chang MC. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7(4):222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 16.Fischer CR, Tseng HC, Tai M, Prather KL, Stephanopoulos G. Assessment of heterologous butyrate and butanol pathway activity by measurement of intracellular pathway intermediates in recombinant Escherichia coli. Appl Microbiol Biotechnol. 2010;88(1):265–275. doi: 10.1007/s00253-010-2749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuge T, et al. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol Lett. 2000;184(2):193–198. doi: 10.1111/j.1574-6968.2000.tb09013.x. [DOI] [PubMed] [Google Scholar]
- 18.Tseng HC, Harwell CL, Martin CH, Prather KL. Biosynthesis of chiral 3-hydroxyvalerate from single propionate-unrelated carbon sources in metabolically engineered E. coli. Microb Cell Fact. 2010;9:96. doi: 10.1186/1475-2859-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inui M, et al. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol. 2008;77(6):1305–1316. doi: 10.1007/s00253-007-1257-5. [DOI] [PubMed] [Google Scholar]
- 20.Atsumi S, et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10(6):305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Berríos-Rivera SJ, Bennett GN, San KY. Metabolic engineering of Escherichia coli: Increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab Eng. 2002;4(3):217–229. doi: 10.1006/mben.2002.0227. [DOI] [PubMed] [Google Scholar]
- 22.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol. 2011;77(9):2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tucci S, Martin W. A novel prokaryotic trans-2-enoyl-CoA reductase from the spirochete Treponema denticola. FEBS Lett. 2007;581(8):1561–1566. doi: 10.1016/j.febslet.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Ingram LO, Shanmugam KT. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J Bacteriol. 2008;190(11):3851–3858. doi: 10.1128/JB.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ajikumar PK, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HC, Kim JS, Jang W, Kim SY. Thymidine production by overexpressing NAD+ kinase in an Escherichia coli recombinant strain. Biotechnol Lett. 2009;31(12):1929–1936. doi: 10.1007/s10529-009-0097-z. [DOI] [PubMed] [Google Scholar]
- 27.Murarka A, Dharmadi Y, Yazdani SS, Gonzalez R. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microbiol. 2008;74(4):1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dharmadi Y, Murarka A, Gonzalez R. Anaerobic fermentation of glycerol by Escherichia coli: A new platform for metabolic engineering. Biotechnol Bioeng. 2006;94(5):821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 29.Lange J-P, et al. Valeric biofuels: A platform of cellulosic transportation fuels. Angew Chem Int Ed Engl. 2010;49(26):4479–4483. doi: 10.1002/anie.201000655. [DOI] [PubMed] [Google Scholar]
- 30.Bozell JJ. Chemistry. Connecting biomass and petroleum processing with a chemical bridge. Science. 2010;329(5991):522–523. doi: 10.1126/science.1191662. [DOI] [PubMed] [Google Scholar]
- 31.Fischer CR, Peterson AA, Tester JW. Production of C3 hydrocarbons from biomass via hydrothermal carboxylate reforming. Ind Eng Chem Res. 2011;50(8):4420–4424. [Google Scholar]
- 32.Steinbüchel A, Debzi E-M, Marchessault RH, Timm A. Synthesis and production of poly(3-hydroxyvaleric acid) homopolyester by Chromobacterium violaceum. Appl Microbiol Biotechnol. 1993;39(4):443–449. [Google Scholar]
- 33.Xie JC, Sun BG, Zhang FP, Luo CS. Study on synthesis of flavor 2-pentenoic acid. Food Science. 2007;28(10):252–254. [Google Scholar]
- 34.Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci USA. 2008;105(52):20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 36.Dugar D, Stephanopoulos G. Relative potential of biosynthetic pathways for biofuels and bio-based products. Nat Biotechnol. 2011;29(12):1074–1078. doi: 10.1038/nbt.2055. [DOI] [PubMed] [Google Scholar]
- 37.Dekishima Y, Lan EI, Shen CR, Cho KM, Liao JC. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J Am Chem Soc. 2011;133(30):11399–11401. doi: 10.1021/ja203814d. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 39.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.