Summary
Through characterising the geographic and functional spectrum of human genetic variation, the 1000 Genomes Project aims to build a resource to help understand the genetic contribution to disease. We describe the genomes of 1,092 individuals from 14 populations, constructed using a combination of low-coverage whole-genome and exome sequencing. By developing methodologies to integrate information across multiple algorithms and diverse data sources we provide a validated haplotype map of 38 million SNPs, 1.4 million indels and over 14 thousand larger deletions. We show that individuals from different populations carry different profiles of rare and common variants and that low-frequency variants show substantial geographic differentiation, which is further increased by the action of purifying selection. We show that evolutionary conservation and coding consequence are key determinants of the strength of purifying selection, that rare-variant load varies substantially across biological pathways and that each individual harbours hundreds of rare non-coding variants at conserved sites, such as transcription-factor-motif disrupting changes. This resource, which captures up to 98% of accessible SNPs at a frequency of 1% in populations of medical genetics focus, enables analysis of common and low-frequency variants in individuals from diverse, including admixed, populations.
Recent efforts to map human genetic variation through sequencing exomes1 and whole genomes2-4 have characterised the vast majority of common SNPs and many structural variants across the genome. However, while over 95% of common (>5% frequency) variants were discovered in the Pilot Phase of the 1000 Genomes Project, lower-frequency variants, particularly outside the coding exome, remain poorly characterised. Low-frequency variants are enriched for potentially functional mutations, for example protein-changing variants, under weak purifying selection1,5,6. Furthermore, low-frequency variants, because they tend to be recent in origin, exhibit increased levels of population differentiation6-8. Characterising such variants, for both point mutations and structural changes, across a range of populations is thus likely to identify many variants of functional significance and is critical in interpreting individual genome sequences; for example to help separate shared variants from those private to families.
We now report on the genomes of 1,092 individuals sampled from 14 populations drawn from Europe, East Asia, sub-Saharan Africa and the Americas (Figs. S1,S2), analysed through a combination of low-coverage (2-6x) whole-genome sequence (WGS) data, targeted deep exome sequence data (50-100x) and dense SNP genotype data (Tables 1, S1-S3). This design was shown by the Pilot Phase2 to be powerful and cost-effective in discovering and genotyping all but the rarest SNP and short insertion and deletion (indel) variants. Here, the approach was augmented with statistical methods for selecting higher quality variant calls from candidates obtained using multiple algorithms and to integrate SNP, indel and larger structural variants (SVs) within a single framework (see Box and Fig. S1). Because of the challenges of identifying large and complex structural variants and shorter indels in regions of low complexity, we focused on conservative but high quality subsets: biallelic indels and large deletions.
Table 1.
Summary of 1000 Genomes Phase 1 data
| Autosomes | Chromosome X |
GENCODE regionsa |
|
|---|---|---|---|
| Samples | 1092 | 1092 | 1092 |
| Total raw bases (Gb) | 19,049 | 804 | 327 |
| Mean mapped depth (x) | 5.1 | 3.9 | 80.3 |
| SNPs | |||
| No. sites overall | 36.7 M | 1.3 M | 498 K |
| Novelty rateb | 58% | 77% | 50% |
| No. Syn / NonSyn / Nonsense |
NA | 4.7 / 6.5 / 0.097 K |
199 / 293 / 6.3 K |
| Avg. no. SNPs per sample | 3.60 M | 105 K | 24.0 K |
| Indels | |||
| No. sites overall | 1.38 M | 59 K | 1,867 |
| Novelty rateb | 62% | 73% | 54% |
| No. in-frame / frameshift | NA | 19 / 14 | 719 / 1,066 |
| Avg. no. indels per sample |
344 K | 13 K | 440 |
| Genotyped large deletions | |||
| No. sites overall | 13.8 K | 432 | 847 |
| Novelty rateb | 54% | 54% | 50% |
| Avg. no. variants per sample |
717 | 26 | 39 |
Autosomal genes only.
Compared to dbSNP release 135 (Oct 2011) excluding contribution from Phase 1 1000 Genomes (or equivalent data for large deletions).
Overall, we discovered and genotyped 38 million SNPs, 1.4 million bi-allelic indels and 14 thousand large deletions (Table 1). Multiple technologies were used to validate a frequency-matched set of sites to assess and control the false discovery rate (FDR) for all variant types. Where results were clear, 3/185 exome sites (1.6%), 5/281 low-coverage sites (1.8%) and 72/3415 (2.1%) large deletions could not be validated (Supplementary Information and Tables S4-S9). The initial indel call-set was found to have a high FDR (27/76), which led to the application of additional filters, leaving an implied FDR of 5.4% (Table S6; Supplementary Information). Moreover, for 2.1% of low-coverage SNP and 18% of indel sites we found inconsistent or ambiguous results indicating the substantial challenges remaining in characterising variation in low-complexity genomic regions. We previously described the “accessible genome”: the fraction of the reference genome where short-read data can lead to reliable variant discovery. Through longer read-lengths the fraction accessible has increased from 85% in the Pilot to 94% (available as a genome annotation; see Supplementary Information) and 1.7 million low-quality SNPs from the Pilot Phase have been eliminated.
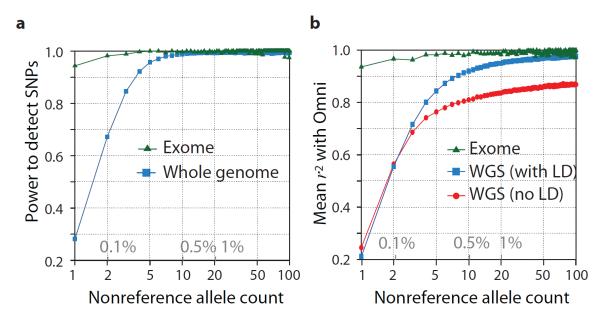
By comparison to external SNP and high-depth sequencing data, we estimate the power to detect SNPs present at a frequency of 1% in the study samples is 99.3% across the genome and 99.8% in the consensus exome target (Fig. 1a). Moreover, the power to detect SNPs at 0.1% frequency in the study is over 90% in the exome and nearly 70% across the genome. The accuracy of individual genotype calls at heterozygous sites is over 99% for common SNPs and 95% for SNPs at frequency of 0.5% (Fig. 1b). By integrating LD information, genotypes from low-coverage data are as accurate as those from high depth exome data for SNPs with frequency >1%. For very rare SNPs (≤0.1%, therefore present in 1 or 2 copies), there is no gain in genotype accuracy from incorporating LD information and accuracy is lower. Variation among samples in genotype accuracy is primarily driven by sequencing depth (Fig. S3) and technical issues such as sequencing platform and version (detectable by PCA; Fig. S4) rather than population-level characteristics. The accuracy of inferred haplotypes at common SNPs was estimated by comparison to SNP data collected on mother-father-offspring trios for a subset of the samples. This indicates that a phasing (switch) error is made, on average, every 300-400 kb (Fig. S5).
Figure 1. Power and accuracy.
a, Power to detect SNPs as a function of variant count (and proportion) across the entire set of samples, estimated by comparison to independent SNP array data in the exome (green) and whole genome (blue). b, Genotype accuracy compared to the same SNP array data as a function of variant frequency summarised by the r2 between true and inferred genotype (coded as 0, 1 and 2) within the exome (green), whole genome after haplotype integration (blue) and whole genome without haplotype integration (red).
A key goal of the 1000 Genomes Project was to identify over 95% of SNPs at 1% frequency in a broad set of populations. Our current resource includes ~50%, 98% and 99.7% of the SNPs with frequencies of ~0.1%, 1.0% and 5.0% respectively in ~2,500 UK-sampled genomes (the Wellcome Trust-funded UK10K project), thus meeting this goal. However, coverage may be lower for populations not closely related to those studied. For example, our resource includes only 23.7%, 76.9% and 99.3% of the SNPs with frequencies of ~0.1%, 1.0% and 5.0% respectively in ~2,000 genomes sequenced in a study of the isolated population of Sardinia (the SardiNIA study).
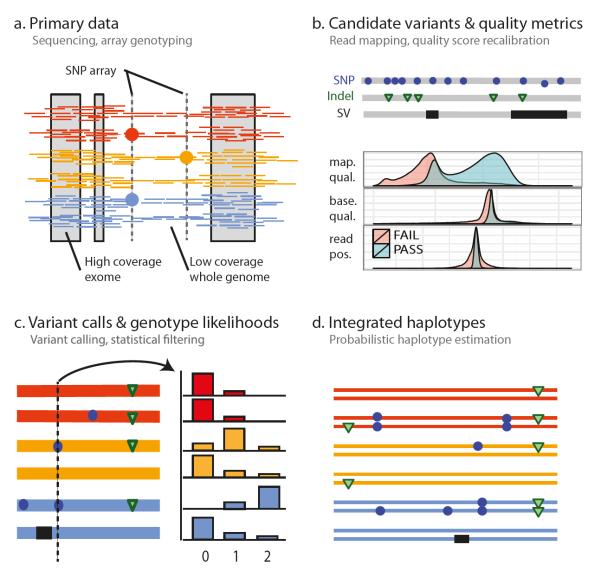
Box: Constructing an integrated map of variation.
The 1,092 haplotype-resolved genomes released as Phase 1 by the 1000 Genomes Project are the result of integrating diverse data from multiple technologies generated by several centres between 2008 and 2010. The figure describes the process leading from primary data production to integrated haplotypes. a. Unrelated individuals (though see Table S10) were sampled in groups of up to 100 from related populations (Wright’s FST typically <1%) within broader geographical or ancestry-based groups2. Primary data generated for each sample consist of low-coverage (average 5x) whole-genome and high-coverage exome (average 80x across a consensus target of 24 Mb spanning over 15,000 genes) sequence data and high density SNP array information. b. Following read-alignment, multiple algorithms were used to identify candidate variants. For each variant, quality metrics were obtained, including information about uniqueness of the surrounding sequence (e.g., mapping quality), the quality of evidence supporting the variant (e.g., the position of variant bases within reads), and the distribution of variant calls in the population (e.g,. inbreeding coefficient). Machine-learning approaches using this multidimensional information were trained on sets of high-quality known variants (e.g., the high-density SNP array data), allowing variants sites to be ranked in confidence and subsequently thresholded to ensure low FDR. c. Genotype likelihoods were used to summarise the evidence for each genotype at bi-allelic sites (0, 1 or 2 copies of the variant) in each sample at every site. d, As the evidence for a single genotype is typically weak in the low-coverage data, and can be highly variable in the exome data, statistical methods were used to leverage information from patterns of linkage disequilibrium, allowing haplotypes (and genotypes) to be inferred.
The distribution of genetic variation within and between populations
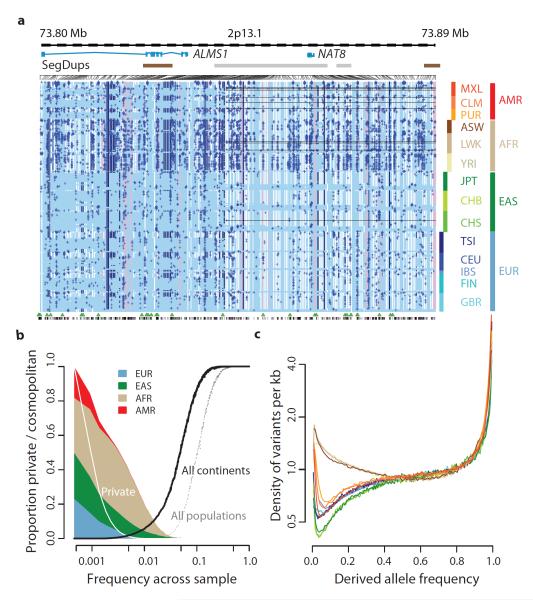
The integrated data set provides a detailed view of variation across multiple populations (illustrated in Fig. 2a). Most common variants (94% of variants with frequency ≥5% in the figure) were known prior to the current phase of the project and had their haplotype structure mapped through earlier projects2,9. In contrast, only 62% of variants in the range 0.5-5% and 13% of variants with frequency ≤ 0.5% had been described previously. For analysis, populations are grouped by the predominant component of ancestry: Europe (CEU, TSI, GBR, FIN, IBS), Africa (YRI, LWK, ASW), East Asia (CHB, JPT, CHS) and the Americas (MXL, CLM, PUR). Variants present at 10% and above across the entire sample are almost all found in all populations studied. In contrast, 17% of low-frequency variants in the range 0.5-5% were observed in a single ancestry group and 53% of rare variants at 0.5% were observed in a single population (Fig. 2b). Within ancestry groups, common variants are weakly differentiated (most within-group estimates of FST are < 1%; Table S11), although below 0.5% frequency variants are up to twice as likely to be found within the same population compared to random sample from the ancestry group (Fig. S6a). The degree of rare-variant differentiation varies between populations. For example, within Europe, the IBS and FIN populations carry excesses of rare variants (Fig. S6b), which can arise through events such as recent bottlenecks10, ‘clan’ breeding structures11 and admixture with diverged populations12.
Figure 2. The distribution of rare and common variants.
a, Summary of inferred haplotypes across a 100 kb region of chromosome 2 spanning the genes ALMS1 and NAT8, variation in which has been associated with kidney disease45. Each row represents an estimated haplotype, with the population of origin indicated on the right. Reference alleles are indicated by the light blue background. Variants (non-reference alleles) above 0.5% frequency are indicated by pink (typed on the high density SNP array), white (previously known) and dark blue (not previously known). Low frequency variants (<0.5%) are indicated by blue crosses. Indels are indicated by green triangles and novel variants by dashes below. A large, low-frequency deletion (black line) spanning NAT8 is present in some populations. Multiple structural haplotypes mediated by segmental duplications are present at this locus, including copy number gains, which were not genotyped for this study. Within each population haplotypes are ordered by total variant count across the region. b, The fraction of variants identified across the project that are found in only one population (white line), are restricted to a single ancestry-based group (defined as in part A, solid colour), are found in all groups (solid black line) and are found in all populations (dotted black line). c, The density of the expected number of variants per kb carried by a genome drawn from each population, as a function of variant frequency (see Supplementary Information). Colours as for part a. Under a model of constant population size, the expected density is constant across the frequency spectrum.
Some common variants show strong differentiation between populations within ancestry-based groups (Table S12), many of which are likely to have been driven by local adaptation either directly or through hitch-hiking. For example, the strongest differentiation between AFR populations is in NRSF transcription-factor peak (PANC1-cell-line)13 upstream of ST8SIA1 (difference in derived allele frequency LWK-YRI of 0.475 at rs7960970), whose product is involved in ganglioside generation14. Overall, we find a range of 17-343 SNPs (fewest = CEU-GBR, most = FIN-TSI) showing a difference in frequency of at least 0.25 between pairs of populations within an ancestry-group.
The derived allele frequency distribution shows substantial divergence between populations below a frequency of 40% (Fig. 2c), such that individuals from populations with substantial African ancestry (YRI, LWK, ASW) carry up to three times as many low-frequency variants (0.5-5% frequency) as those of European or East Asian origin, reflecting ancestral bottlenecks in non-African populations15. However, individuals from all populations show an enrichment of rare (<0.5%) variants, reflecting recent explosive increases in population size and the effects of geographic differentiation6,16. Compared to the expectations from a model of constant population size, individuals from all populations show a substantial excess of high-frequency derived variants (>80% frequency).
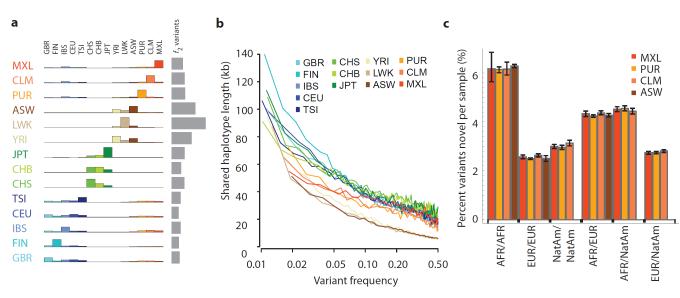
Because rare variants are typically recent, their patterns of sharing can reveal aspects of population history. Variants present twice across the entire sample (referred to as f2 variants), typically the most recent of informative mutations, are found within the same population in 53% of cases (Fig. 3a). However, between-population sharing identifies recent historical connections. For example, where one of the individuals carrying an f2 variant is from the Spanish population (IBS) and the other is not (referred to as IBS-X), the other individual is more likely to come from the AMR populations (48%, correcting for sample size) than elsewhere in Europe (41%). Within the East Asian populations, CHS and CHB show stronger f2 sharing to each other (58% and 53% of CHS-X and CHB-X variants respectively) than either does to JPT, but JPT is closer to CHB than to CHS (44% versus 35% of JPT-X variants). Within African-ancestry populations, the ASW are closer to the YRI (42% of ASW-X f2 variants) compared to the LWK (28%), in line with historical information17 and genetic evidence based on common SNPs18. Some sharing patterns are surprising; for example, 2.5% of the f2 FIN-X variants are shared with YRI or LWK populations.
Figure 3. Allele sharing within and between populations.
a, Sharing of f2 variants, those found exactly twice across the entire sample, within and between populations. Each row represents the distribution across populations for the origin of samples sharing an f2 variant with the target population (indicated by the left-hand side). The grey bar represents the average number of f2 variants carried by a randomly-chosen genome in each population. b, Median length of haplotype identity (excluding cryptically-related samples and singleton variants and allowing for up to two genotype errors) between two chromosomes that share variants of a given frequency in each population. Estimates are from 200 randomly-sampled regions of 1 Mb each and up to 15 pairs of individuals for each variant. c, The average proportion of variants that are novel (compared to the pilot phase of the project) among those found in regions inferred to have different ancestries within ASW, PUR, CLM and MXL. Error bars represent 95% bootstrap confidence intervals.
Independent evidence about variant age comes from the length of the shared haplotypes on which they are found. We find, as expected, a negative correlation between variant frequency and the median length of shared haplotypes, such that chromosomes carrying variants at 1% frequency share haplotypes of 100-150 kb (typically 0.08-0.13 cM; Figs. 3b and S7a), although the distribution is highly skewed and 2-5% of haplotypes around the rarest SNPs extend over 1 Mb (Figs. S7b,c). Haplotype phasing and genotype calling errors will limit the ability to detect long shared haplotypes and the observed lengths are a factor of 2-3 shorter than predicted by models that allow for recent explosive growth6 (Fig. S7a). Nevertheless, the haplotype length for variants shared within and between populations is informative about relative allele age. Within populations and between populations where there is recent shared ancestry (e.g., through admixture and within continents) f2 variants typically lie on long shared haplotypes (median within ancestry group 103 kb, Fig. S8). In contrast, between populations with no recent shared ancestry, f2 variants are present on very short haplotypes, for example, an average of 11 kb for FIN-YRI f2 variants (median between ancestry groups excluding admixture is 15 kb), and are therefore likely to reflect recurrent mutations and chance ancient coalescent events.
To analyse populations with substantial historical admixture, statistical methods were applied to each individual to infer regions of the genome with different ancestries. Populations and individuals vary substantially in admixture proportions. For example, the MXL population contains the greatest proportion of Native American ancestry (47% on average compared to 24% in CLM and 13% in PUR), but the proportion varies from 3% to 92% between individuals (Fig. S9a). Rates of variant discovery, the ratio of nonsynonymous to synonymous variation and the proportion of variants that are novel vary systematically between regions with different ancestries. Regions of Native American ancestry show less variation, but a higher fraction of the variants discovered are novel (3.0% of variants per sample, Fig. 3c) compared to regions of European ancestry (2.6%). Regions of African ancestry show the highest rates of novelty (6.2%) and heterozygosity (Fig. S9b,c).
The functional spectrum of human variation
The Phase 1 data enable us to compare, for different genomic features and variant types, the effects of purifying selection on evolutionary conservation19, the allele frequency distribution and the level of differentiation between populations. At the most highly conserved coding sites, 85% of nonsynonymous (NonSyn) variants and over 90% of STOP gain and splice-disrupting variants are below 0.5% in frequency , compared to 65% of synonymous (Syn) variants (Fig. 4a). In general, the rare variant excess tracks the level of evolutionary conservation for variants of most functional consequence, but varies systematically between types (e.g., for a given level of conservation enhancer variants have a higher rare variant excess than variants in transcription factor motifs). However, STOP gains and, to a lesser-extent, splice-site disrupting changes, show elevated rare-variant excess whatever the conservation of the base in which they occur, as such mutations can be highly deleterious whatever the level of sequence conservation. Interestingly, the least conserved splice-disrupting variants show rare-variant load similar to synonymous and non-coding regions suggesting that these alternative transcripts are under very weak selective constraint. Sites at which variants are observed are typically less conserved than average (for example, sites with NonSyn variants are, on average, as conserved as third codon positions, Fig S10).
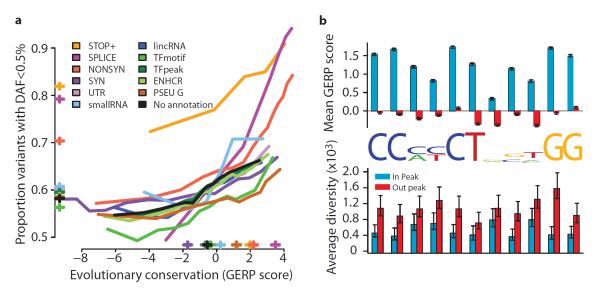
Figure 4. Purifying selection within and between populations.
a, The relationship between evolutionary conservation (measured by GERP score19) and rare variant proportion (fraction of all variants with derived allele frequency < 0.5%) for variants occurring in different functional elements and with different coding consequences. Crosses indicate the average GERP score at variant sites (x-axis) and proportion of rare variants (y-axis) in each category. b, Levels of evolutionary conservation (mean GERP score, top) and genetic diversity (per nucleotide pairwise differences, bottom) for sequences matching the CTCF-binding motif within CTCF-binding peaks as experimentally identified by ChIP-Seq in the ENCODE project13 (blue) and in a matched set of motifs outside peaks (red). The logo plot shows the distribution of identified motifs within peaks. Error bars represent ± 2 s.e.m.
A simple way of estimating the segregating load arising from rare, deleterious mutations across a set of genes comes from comparing the ratios of NonSyn to Syn variants in different frequency ranges. The NonSyn to Syn ratio among rare (<0.5%) variants is typically in the range 1-2 and among common variants in the range 0.5-1.5, suggesting that 25-50% of rare NonSyn variants are deleterious. However, the segregating rare load among gene groups in KEGG pathways20 varies substantially (Fig. S11a; Table S13). Certain groups (e.g., ECM-receptor interaction, DNA replication and pentose phosphate pathway) show a substantial excess of rare coding mutations, which is only weakly correlated with the average degree of evolutionary conservation. Pathways and processes showing an excess of rare functional variants vary between continents (Fig. S11b). Moreover, the excess of rare NonSyn variants is typically higher in populations of European and East Asian ancestry (for example, the ECM-receptor interaction pathway load is strongest in EUR). Other groups of genes (for example, those associated with allograft rejection) actually have a high NonSyn:Syn ratio in common variants, potentially indicating the effects of positive selection.
Genome-wide data provide important insights into the rates of functional polymorphism in the non-coding genome. For example, we consider motifs matching the consensus for transcriptional repressor CTCF, which has a well-characterised and highly conserved binding motif21. Within CTCF-binding peaks experimentally defined by chromatin-immunoprecipitation sequencing (ChIP-seq), average levels of conservation within the motif are comparable to third codon positions, while outside peaks there is no conservation (Fig. 4c). Within peaks levels of genetic diversity are typically reduced 25-75%, depending on the position in the motif (Fig. 4c). Unexpectedly, the reduction in diversity at some degenerate positions, for example position 8 in the motif, is as great as that at nondegenerate positions, suggesting that motif degeneracy may not have a simple relationship with functional importance. Variants within peaks show a weak but consistent excess of rare variation (proportion with frequency <0.5% is 61% within peaks compared to 58% outside peaks, Fig. S12) supporting the hypothesis that regulatory sequences harbour substantial amounts of weakly deleterious variation.
Purifying selection can also affect population differentiation if its strength and efficacy vary among populations. Although the magnitude of the effect is weak, nonsynonymous variants consistently show greater levels of population differentiation than synonymous variants, for variants of frequency less than 10% (Fig. S13).
Uses of 1000 Genomes Project data in medical genetics
Data from the 1000 Genomes Project are widely used to screen variants discovered in exome data from individuals with genetic disorders22 and in cancer genome projects23. The enhanced catalogue presented here improves the power of such screening. Moreover, it provides a ‘null expectation’ for the number of rare, low-frequency and common variants with different functional consequences typically found in randomly-sampled individuals from different populations.
Estimates of the overall numbers of variants with different sequence consequences are comparable to previous values 1,20-22 (Table S14). However, only a fraction of these are likely to be functionally-relevant. A more accurate picture of the number of functional variants is given by the number of variants segregating either at conserved positions (here defined as sites with a GERP19 conservation score of >2), or where the function (e.g., STOP gain) is strong and independent of conservation (Table 2). We find that individuals typically carry over 2,500 nonsynonymous variants at conserved positions, 20-40 variants identified as damaging24 at conserved sites and about 150 loss-of-function variants (LOF: STOP gains, frameshift indels in coding sequence and disruptions to essential splice-sites). However, most of these are common (>5%) or low-frequency (0.5-5%) such that the numbers of rare (<0.5%) variants in these categories (which might be considered as pathological candidates) are much lower; 130-400 nonsynonymous variants per individual, 10-20 LOF variants, 2-5 damaging mutations and 1-2 variants identified previously from cancer genome sequencing25. By comparison to synonymous variants, we can estimate the excess of rare variants; those mutations that are sufficiently deleterious that they will never reach high frequency. We estimate that individuals carry an excess of 76-190 rare deleterious nonsynonymous variants and up to 20 LOF and disease-associated variants. Interestingly, the overall excess of low-frequency variants is similar to that of rare variants (Table 2). Because many variants contributing to disease risk are likely to be segregating at low frequency, we recommend that variant frequency be considered when using the resource to identify pathological candidates.
Table 2.
Per individual variant load at conserved sites
| Variant type | Number of derived variant sites per individual |
Excess rare deleterious |
Excess low- frequency deleterious |
||
|---|---|---|---|---|---|
| Derived allele frequency across sample | |||||
| <0.5% | 0.5%-5% | >l5% | |||
| All sites | 30K-150K | 120K- 680K |
3.6M- 3.9M |
- | - |
| Synonymous a | 29-120 | 82-420 | 1.3K-1.4K | - | - |
| Nonsynonymous a | 130-400 | 240-910 | 2.3K-2.7K | 76-190b | 77-130b |
| Stop-gain a | 3.9-10 | 5.3-19 | 24-28 | 3.4-7.5b | 3.8-11b |
| Stop-loss | 1.0-1.2 | 1.0-1.9 | 2.1-2.8 | 0.81-1.1b | 0.80-1.0b |
| HGMD-DM a | 2.5-5.1 | 4.8-17 | 11-18 | 1.6-4.7b | 3.8-12b |
| COSMIC a | 1.3-2.0 | 1.8-5.1 | 5.2-10 | 0.93-1.6b | 1.3-2.0b |
| Indel-frameshift | 1.0-1.3 | 11-24 | 60-66 | -d | 3.2-11b |
| Indel-non-frameshift | 2.1-2.3 | 9.5-24 | 67-71 | -d | 0-0.73b |
| Splice site donor | 1.7-3.6 | 2.4-7.2 | 2.6-5.2 | 1.6-3.3b | 3.1-6.2b |
| Splice site acceptor | 1.5-2.9 | 1.5-4.0 | 2.1-4.6 | 1.4-2.6b | 1.2-3.3b |
| UTR a | 120-430 | 300-1.4K | 3.5K-4.0K | 0-350c | 0-1.2Kc |
| Non-coding RNA a | 3.9-17 | 14-70 | 180 -200 | 0.62-2.6c | 3.4-13c |
|
Motif gain in TF
peak a |
4.7-14 | 23-59 | 170-180 | 0-2.6c | 3.8-15c |
|
Motif loss in TF
peak a |
18-69 | 71-300 | 580-650 | 7.7-22c | 37-110c |
| Other conserved a | 2.0K-9.9K | 7.1K-39K | 120K-130K | - | |
| Total conserved | 2.3K-11K | 7.7K-42K | 130K-150K | 150-510 | 250-1.3K |
Only sites where ancestral state can be assigned with high confidence reported.
Ranges reported are across populations.
Sites with GERP>2
Using Synonymous sites as base-line
Using ‘Other conserved’ as base-line
Rare indels were filtered in Phase 1
The combination of variation data with information about regulatory function13 can potentially improve the power to detect pathological non-coding variants. We find that individuals typically harbour several thousands of variants (and several hundred rare variants) in conserved (GERP conservation score >2) UTRs, non-coding RNAs and transcription-factor binding motifs (Table 2). Within experimentally-defined transcription factor binding sites, individuals carry 700-900 conserved motif losses (for the transcription factors analysed, see Supplementary Information), of which 18-69 are rare (<0.5%) and which show strong evidence for being selected against. Motif gains are rarer (~200 per individual at conserved sites) but they also show evidence for an excess of rare variants compared to conserved sites with no functional annotation (Table 2). Many of these changes are likely to have weak, slightly deleterious effects on gene regulation and function.
A second major use of the 1000 Genomes Project data in medical genetics is imputing genotypes in existing genome-wide association studies (GWAS)26. For common variants, the accuracy of using the Phase 1 data to impute genotypes at sites not on the original GWAS chip is typically 90-95% in non-African and approximately 90% in African-ancestry genomes (Figs. 5a, S14a), which is comparable to the accuracy achieved with high quality benchmark haplotypes (Fig. S14b). Imputation accuracy is similar for intergenic SNPs, exome SNPs, indels and large deletions (see also Fig. S14c), despite the different amounts of information about such variants and accuracy of genotypes. For low-frequency variants (1-5%), imputed genotypes have between 60% and 90% accuracy in all populations, including those with admixed ancestry (also comparable to the accuracy from trio-phased haplotypes; Fig. S14b).
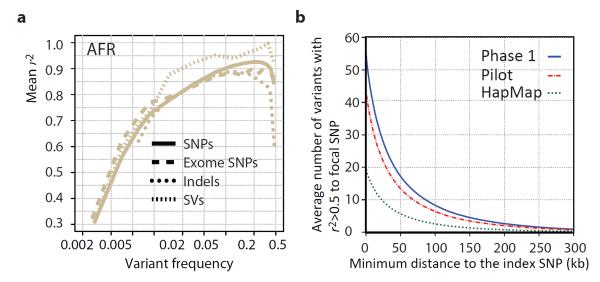
Figure 5. Implications of Phase 1 1000 Genomes data for GWAS.
a, Accuracy of imputation of genome-wide SNPs, exome SNPs and indels (using sites on the Illumina 1M array) into 10 individuals of African ancestry (3 LWK, 4 Masaai from Kenya - MKK, 2 YRI) sequenced to high coverage by an independent technology3. Only indels in regions of high sequence complexity with frequency >1% are analysed. Deletion imputation accuracy estimated by comparison to array data46 (note this is for a different set of individuals though with a similar ancestry, but included on the same plot for clarity). Accuracy measured by squared Pearson correlation coefficient between imputed and true dosage across all sites in a frequency range estimated from the 1000 Genomes data. Lines represent whole genome SNPs (solid), exome SNPs (long dashes), short indels (dotted) and large deletions (short dashes). b, The average number of variants in linkage disequilibrium (r2>0.5 among EUR) to focal SNPs identified in GWAS47 as a function of distance from the index SNP. Lines indicate the number of HapMap, Pilot and Phase 1 variants.
Imputation has two primary uses: fine-mapping existing association signals and detecting novel associations. GWAS have had only a few examples of successful fine-mapping to single causal variants27,28, often because of extensive haplotype structure within regions of association29,30. We find that, in Europeans, each previously reported GWAS signal31 is, on average, in linkage disequilibrium (r2 ≥ 0.5) with 56 variants: 51.5 SNPs and 4.5 indels. In 19% of cases at least one of these variants changes the coding sequence of a nearby gene (compared to 12% in control variants matched for frequency, distance to nearest gene and ascertainment in GWAS arrays) and in 65% of cases at least one of these is at a site with GERP>2 (68% in matched controls). The size of the associated region is typically <200 kb in length (Figure 5b). Our observations suggest that trans-ethnic fine-mapping experiments are likely to be especially valuable: among the 56 variants that are in strong linkage disequilibrium with a typical GWAS signal, ~15 show strong disequilibrium across our four continental groupings (Table S15). Compared to earlier catalogs, our current resource increases the number of variants in linkage disequilibrium with each GWAS signal by 25% compared to the Pilot phase of the project and by greater than 2-fold compared to the HapMap resource.
Discussion
The success of exome sequencing in Mendelian disease genetics32 and the discovery of rare and low-frequency disease-associated variants in genes associated with complex diseases27,33,34 strongly support the hypothesis that, in addition to factors such as epistasis35,36 and gene-environment interactions37, many additional genetic risk factors of substantial effect size remain to be discovered through studies of rare variation. The data generated by the 1000 Genomes Project not only aid the interpretation of all genetic association studies, but also provide lessons on how best to design and analyse sequencing-based studies of disease.
The utility and cost-effectiveness of collecting multiple data types (low-coverage whole genome sequence, targeted exome data, SNP genotype data) for finding variants and reconstructing haplotypes are demonstrated here. Exome capture provides private and rare variants that are missed by low-coverage data (approximately 60% of the singleton variants in the sample were detected only from exome data compared to 5% only detected from low-coverage data, Fig. S15). However, whole-genome data enable characterisation of functional non-coding variation and accurate haplotype estimation, which are essential for the analysis of cis-effects around genes, for example those arising from variation in upstream regulatory regions38. There are also benefits from integrating SNP array data, for example to improve genotype estimation39 and to aid haplotype estimation where array data have been collected on additional family members. In principle, any sources of genotype information (e.g., from array CGH) could be integrated using the statistical methods developed here.
Major methodological advances in Phase 1, including improved methods for detecting and genotyping variants40, statistical and machine-learning methods for evaluating the quality of candidate variant calls, modelling of genotype likelihoods and performing statistical haplotype integration41, have generated a high-quality resource. However, regions of low sequence complexity, satellite regions, large repeats and many large-scale structural variants, including copy-number polymorphisms, segmental duplications and inversions (which constitute most of the “inaccessible genome”), continue to present a major challenge for short-read technologies. Some issues are likely to be improved by methodological developments such as better modelling of read-level errors, integrating de novo assembly42,43 and combining multiple sources of information to aid genotyping of structurally-diverse regions40,44. Importantly, even subtle differences in data type, data processing or algorithms may lead to systematic differences in false-positive and false negative error modes between samples. Such differences complicate efforts to compare genotypes between sequencing studies. Moreover, analyses that naively combine variant calls and genotypes across heterogeneous data sets are vulnerable to artifact. Analyses across multiple data sets must therefore either process them in standard ways or use meta-analysis approaches that combine association statistics (but not raw data) across studies.
Finally, the analysis of low-frequency variation demonstrates both the pervasive effects of purifying selection at functionally-relevant sites in the genome and how this can interact with population history to lead to substantial local differentiation, even when standard metrics of structure such as FST are very small. The effect arises primarily because rare variants tend to be recent and thus tend to be geographically restricted6-8. The implication is that the interpretation of rare variants in individuals with a particular disease should be within the context of the local (either geographic or ancestry-based) genetic background. Moreover, it argues for the value of continuing to sequence individuals from diverse populations to characterise the spectrum of human genetic variation and support disease studies across diverse groups. A further 1500 individuals from 11 new populations, including at least 15 high-depth trios, will form the final phase of this project.
Methods summary
All details concerning sample collection, data generation, processing and analysis can be found in the Supplementary Information. Fig. S1 summarises the process and indicates where relevant details can be found.
Supplementary Material
Figure 6.
Acknowledgements
We thank many people who contributed to this project: A. Naranjo, M.V. Parra, and C. Duque for help in the collection of the Colombian samples; N. Kälin and F. Laplace for valuable discussions; A. Schlattl and T. Zichner for assistance in managing data sets; E. Appelbaum, H. Arbery, E. Birney, S. Bumpstead, J. Camarata, J. Carey, G. Cochrane, M. DaSilva, S. Dökel, E. Drury, C. Duque, K. Gyaltsen, P. Jokinen, B. Lenz, S. Lewis, D. Lu, A. Naranjo, S. Ott, I. Padioleau, M.V. Parra, N. Patterson, A. Price, L. Sadzewicz, S. Schrinner, N. Sengamalay, J. Sullivan, F. Ta, Y. Vaydylevich, O. Venn, K. Watkins, A. Yurovsky.
We thank the people who generously contributed their samples, from these populations: Yoruba in Ibadan, Nigeria; the Han Chinese in Beijing, China; the Japanese in Tokyo, Japan; the Utah CEPH community; the Luhya in Webuye, Kenya; people with African Ancestry in the Southwest United States; the Toscani in Italia; people with Mexican Ancestry in Los Angeles, California; the Southern Han Chinese in China; the British in England and Scotland; the Finnish in Finland; the Iberian Populations in Spain; the Colombians in Medellin, Colombia; and the Puerto Ricans in Puerto Rico.
This research was supported in part by Wellcome Trust grants WT098051 to R.D., M.E.H., C.T.S.; WT090532/Z/09/Z, WT085475/Z/08/Z, and WT095552/Z/11/Z to P.D.; WT086084/Z/08/Z and WT090532/Z/09/Z to G.A.M.; WT089250/Z/09/Z to I.M.; WT085532AIA to P.F.; Medical Research Council grant G0900747(91070) to G.A.M.; British Heart Foundation grant RG/09/12/28096 to C.A.A.; the National Basic Research Program of China (973 program no. 2011CB809201, 2011CB809202, 2011CB809203); the Chinese 863 program (2012AA02A201); the National Natural Science Foundation of China (30890032,31161130357); the Shenzhen Key Laboratory of Transomics Biotechnologies (CXB201108250096A); the Shenzhen Municipal Government of China (grants ZYC200903240080A, ZYC201105170397A); Guangdong Innovative Research Team Program (NO. 2009010016); BMBF grant 01GS08201 to H.L.; BMBF Grant 0315428A to R.H.; the Max Planck Society; Swiss National Science Foundation 31003A_130342 to E.T.D.; Swiss National Science Foundation NCCR “Frontiers in Genetics” to E.T.D.; Louis Jeantet Foundation grant to E.T.D.; Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I021213/1 to A.R-L; German Research Foundation (Emmy Noether Fellowship KO 4037/1-1) to J.O.K.; Netherlands Organization for Scientific Research VENI grant 639.021.125 to K.Y.; Beatriu de Pinos Program grants 2006BP-A 10144 and 2009BP-B 00274 to M.V.; Israeli Science Foundation grant 04514831 to E.H.; Genome Québec and the Ministry of Economic Development, Innovation and Trade grant PSR-SIIRI-195 to P.A.; NIH grants UO1HG5214, RC2HG5581, and RO1MH84698 to G.R.A.; R01HG4719 and R01HG3698 to G.T.M; RC2HG5552 and UO1HG6513 to G.R.A. and G.T.M; R01HG4960 and R01HG5701 to B.L.B.; U01HG5715 to C.D.B. and A.G.C.; T32GM8283 to D.C.; U01HG5208 to M.J.D.; U01HG6569 to M.A.D.; R01HG2898 and R01CA166661 to S.E.D.; UO1HG5209, UO1HG5725, P41HG4221 to C.L.; P01HG4120 to E.E.E.; U01HG5728 to Y.F.; U54HG3273 and U01HG5211 to R.A.G.; R01HL95045 to S.G.; U41HG4568 to S.J.K.; P41HG2371 to W.J.K; ES015794, AI077439, HL088133, and HL078885 to E.G.B.; RC2HL102925 to S.G. and D.M.A.; R01GM59290 to L.B.J. and M.A.B.; U01HG5715 to A.K.; U54HG3067 to E.S.L. and S.G.; T15LM7033 to B.K.M.; T32HL94284 to J.L.R-F.; DP2OD6514 and BAA-NIAID-DAIT-NIHAI2009061 to P.C.S.; T32GM7748 to X.S.; U54HG3079 to R.K.W.; UL1RR024131 to R.D.H.; HHSN268201100040C to the Coriell Institute for Medical Research; a Sandler Foundation award and an American Asthma Foundation award to E.G.B.; an IBM Open Collaborative Research Program award to Y.B.; an A.G. Leventis Foundation scholarship to D.K.X.; a Wolfson Royal Society Merit Award to P.D.; a Howard Hughes Medical Institute International Fellowship award to P.H.S.; a grant from T. and V. Stanley to S.C.Y.; and a Mary Beryl Patch Turnbull Scholar Program award to K.C.B. E.H. is a faculty fellow of the Edmond J. Safra Bioinformatics program at Tel-Aviv University. E.E.E. and D.H. are investigators of the Howard Hughes Medical Institute. M.V.G. is a long-term fellow of EMBO.
Footnotes
Author information All primary data, alignments, individual call sets, consensus call sets, integrated haplotypes with genotype likelihoods and supporting data including details of validation is available from the project web-site http://www.1000genomes.org. Variant and haplotypes for specific genomic regions and specific samples can be viewed and downloaded through the project browser at http://browser.1000genomes.org/. Common project variants with no known medical impact have been compiled by dbSNP for filtering; see http://www.ncbi.nlm.nih.gov/variation/docs/human_variation_vcf/. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare the following financial interests: P.A. is an advisor for Illumina and Ancestry.com; E.T.D. is an advisor for DNAnexus; A.C. is on the scientific advisory board for Affymetrix; C.D.B. is on the scientific advisory boards for Personalis, Inc., Ancestry.com, Locus Development, and the 23 and Me.com project “Roots into the future”; D.H. is on the scientific advisory board for Pacific Biosciences; E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., SynapDx Corp, and DNAnexus, Inc.; P.F. is on the scientific advisory board for Omicia, Inc.; C.L. is on the scientific advisory board for BioNano Genomics and is a senior scientific advisor for Samsung; E.R.M. holds shares in Life Technologies and serves on Illumina’s Speaker’s Bureau; R.A.G. and D.M. hold a co-investment with Life Technologies; J.K.B., C.J.D., J.G., J.P.S.,T.W., B.W., and Y.Z. work at Affymetrix; J.K.B. works at Ancestry.com; N.H. works at Life Technologies; F.M.D. used to work and hold shares at Life Technologies; W.J.K. works at Kent Informatics; B.B., M.B., D.R.B., R.K.C.,T.C., M.E., S.H., S.K., L.M., J.P., and R.S. work at Illumina.
Participant list
The 1000 Genomes Consortium (Participants are arranged by project role, then by institution alphabetically, and finally alphabetically within institutions except for Principal Investigators and Project Leaders, as indicated.)
Corresponding Author: Gil A. McVean (mcvean@well.ox.ac.uk)1,2
Steering Committee: David M. Altshuler3-5 (Co-Chair), Richard M. Durbin6 (Co-Chair), Gonçalo R. Abecasis7, David R. Bentley8, Aravinda Chakravarti9, Andrew G. Clark10, Peter Donnelly1,2, Evan E. Eichler11, Paul Flicek12, Stacey B. Gabriel3, Richard A. Gibbs13, Eric D. Green14, Matthew E. Hurles6, Bartha M. Knoppers15, Jan O. Korbel16, Eric S. Lander3, Charles Lee17, Hans Lehrach18,27, Elaine R. Mardis19, Gabor T. Marth20, Gil A. McVean1,2, Deborah A. Nickerson21, Jeanette P. Schmidt22, Stephen T. Sherry23, Jun Wang24, Richard K. Wilson19
Production Group: Baylor College of Medicine Richard A. Gibbs (Principal Investigator)13, Huyen Dinh13, Christie Kovar13, Sandra Lee13, Lora Lewis13, Donna Muzny13, Jeff Reid13, Min Wang13, BGI-Shenzhen Jun Wang (Principal Investigator)24-26, Xiaodong Fang24, Xiaosen Guo24, Min Jian24, Hui Jiang24, Xin Jin24, Guoqing Li24, Jingxiang Li24, Yingrui Li24, Zhuo Li24, Xiao Liu24, Yao Lu24, Xuedi Ma24, Zhe Su24, Shuaishuai Tai24, Meifang Tang24, Bo Wang24, Guangbiao Wang24, Honglong Wu24, Renhua Wu24, Ye Yin24, Wenwei Zhang24, Jiao Zhao24, Meiru Zhao24, Xiaole Zheng24, Yan Zhou24, Broad Institute of MIT and Harvard Eric S. Lander (Principal Investigator)3, David M. Altshuler3-5, Stacey B. Gabriel (Co-Chair)3, Namrata Gupta3, European Bioinformatics Institute Paul Flicek (Principal Investigator)12, Laura Clarke12, Rasko Leinonen12, Richard E. Smith12, Xiangqun Zheng-Bradley12, Illumina David R. Bentley (Principal Investigator)8, Russell Grocock8, Sean Humphray8, Terena James8, Zoya Kingsbury8, Max Planck Institute for Molecular Genetics Hans Lehrach (Principal Investigator)18,27, Ralf Sudbrak (Project Leader)18, Marcus W. Albrecht28, Vyacheslav S. Amstislavskiy18, Tatiana A. Borodina28, Matthias Lienhard18, Florian Mertes18, Marc Sultan18, Bernd Timmermann18, Marie-Laure Yaspo18, US National Institutes of Health Stephen T. Sherry (Principal Investigator)23, University of Oxford Gil A. McVean (Principal Investigator)1,2, Washington University in St. Louis Elaine R. Mardis (Co-Principal Investigator) (Co-Chair)19, Richard K. Wilson (Co-Principal Investigator)19, Lucinda Fulton19, Robert Fulton19, George M. Weinstock19, Wellcome Trust Sanger Institute Richard M. Durbin (Principal Investigator)6, Senduran Balasubramaniam6, John Burton6, Petr Danecek6, Thomas M. Keane6, Anja Kolb-Kokocinski6, Shane McCarthy6, James Stalker6, Michael Quail6
Analysis Group: Affymetrix Jeanette P. Schmidt (Principal Investigator)22, Christopher J. Davies22, Jeremy Gollub22, Teresa Webster22, Brant Wong22, Yiping Zhan22, Albert Einstein College of Medicine: Adam Auton (Principal Investigator)29, Baylor College of Medicine Richard A. Gibbs (Principal Investigator)13, Fuli Yu (Project Leader)13, Matthew Bainbridge13, Danny Challis13, Uday S. Evani13, James Lu13, Donna Muzny13, Uma Nagaswamy13, Jeff Reid13, Aniko Sabo13, Yi Wang13, Jin Yu13, BGI-Shenzhen Jun Wang (Principal Investigator)24-26, Lachlan J.M. Coin24, Lin Fang24, Xiaosen Guo24, Xin Jin24, Guoqing Li24, Qibin Li24, Yingrui Li24, Zhenyu Li24, Haoxiang Lin24, Binghang Liu24, Ruibang Luo24, Nan Qin24, Haojing Shao24, Bingqiang Wang24, Yinlong Xie24, Chen Ye24, Chang Yu24, Fan Zhang24, Hancheng Zheng24, Hongmei Zhu24, Boston College Gabor T. Marth (Principal Investigator)20, Erik P. Garrison20, Deniz Kural20, Wan-Ping Lee20, Wen Fung Leong20, Alistair N. Ward20, Jiantao Wu20, Mengyao Zhang20, Brigham and Women’s Hospital Charles Lee (Principal Investigator)17, Lauren Griffin17, Chih-Heng Hsieh17, Ryan E. Mills17,41, Xinghua Shi17, Marcin von Grotthuss17, Chengsheng Zhang17, Broad Institute of MIT and Harvard Mark J. Daly (Principal Investigator)3, Mark A. DePristo (Project Leader)3, David M. Altshuler3-5, Eric Banks3, Gaurav Bhatia3, Mauricio O. Carneiro3, Guillermo del Angel3, Stacey B. Gabriel3, Giulio Genovese3, Namrata Gupta3, Robert E. Handsaker3,5, Chris Hartl3, Eric S. Lander3, Steven A. McCarroll3, James C. Nemesh3, Ryan E. Poplin3, Stephen F. Schaffner3, Khalid Shakir3, Cold Spring Harbor Laboratory Seungtai C. Yoon (Principal Investigator)30, Jayon Lihm30, Vladimir Makarov31, Dankook University Hanjun Jin (Principal Investigator)32, Wook Kim33, Ki Cheol Kim33, European Molecular Biology Laboratory Jan O. Korbel (Principal Investigator)16, Tobias Rausch16, European Bioinformatics Institute Paul Flicek (Principal Investigator)12, Kathryn Beal12, Laura Clarke12, Fiona Cunningham12, Javier Herrero12, William M. McLaren12, Graham R.S. Ritchie12, Richard E. Smith12, Xiangqun Zheng-Bradley12, Cornell University Andrew G. Clark (Principal Investigator)10, Srikanth Gottipati34, Alon Keinan10, Juan L. Rodriguez-Flores10, Harvard University Pardis C. Sabeti (Principal Investigator)3,35, Sharon R. Grossman3,35, Shervin Tabrizi3,35, Ridhi Tariyal3,35, Human Gene Mutation Database David N. Cooper (Principal Investigator)36, Edward V. Ball36, Peter D. Stenson36, Illumina David R. Bentley (Principal Investigator)8, Bret Barnes37, Markus Bauer8, R. Keira Cheetham8, Tony Cox8, Michael Eberle8, Sean Humphray8, Scott Kahn37, Lisa Murray8, John Peden8, Richard Shaw8, Leiden University Medical Center Kai Ye (Principal Investigator)38, Louisiana State University Mark A. Batzer (Principal Investigator)39, Miriam K. Konkel39, Jerilyn A. Walker39, Massachusetts General Hospital Daniel G. MacArthur (Principal Investigator)40, Monkol Lek40, Max Planck Institute for Molecular Genetics Ralf Sudbrak (Project Leader)18, Vyacheslav S. Amstislavskiy18, Ralf Herwig18, Pennsylvania State University Mark D. Shriver (Principal Investigator)42, Stanford University Carlos D. Bustamante (Principal Investigator)43, Jake K. Byrnes44, Francisco M. De La Vega10, Simon Gravel43, Eimear E. Kenny43, Jeffrey M. Kidd43, Phil Lacroute43, Brian K. Maples43, Andres Moreno-Estrada43, Fouad Zakharia43, Tel-Aviv University Eran Halperin (Principal Investigator)45-47, Yael Baran45, Translational Genomics Research Institute David W. Craig (Principal Investigator)48, Alexis Christoforides48, Nils Homer110, Tyler Izatt48, Ahmet A. Kurdoglu48, Shripad A. Sinari48, Kevin Squire49, US National Institutes of Health Stephen T. Sherry (Principal Investigator)23, Chunlin Xiao23, University of California, San Diego Jonathan Sebat (Principal Investigator)50,51, Vineet Bafna52, Kenny Ye53, University of California, San Francisco Esteban G. Burchard (Principal Investigator)54, Ryan D. Hernandez (Principal Investigator)54, Christopher R. Gignoux54, University of California, Santa Cruz David Haussler (Principal Investigator)55,111, Sol J. Katzman55, W. James Kent55, University of Chicago Bryan Howie56, University College London Andres Ruiz-Linares (Principal Investigator)57, University of Geneva Emmanouil T. Dermitzakis (Principal Investigator)58,59,104, Tuuli Lappalainen58,59,104, University of Maryland School of Medicine Scott E. Devine (Principal Investigator)60, Xinyue Liu60, Ankit Maroo60, Luke J. Tallon60, University of Medicine and Dentistry of New Jersey Jeffrey A. Rosenfeld (Principal Investigator)61,62, Leslie P. Michelson61, University of Michigan Gonçalo R. Abecasis (Principal Investigator) (Co-Chair)7, Hyun Min Kang (Project Leader)7, Paul Anderson7, Andrea Angius106, Abigail Bigham63, Tom Blackwell7, Fabio Busonero7,105,106, Francesco Cucca105,106, Christian Fuchsberger7, Chris Jones107, Goo Jun7, Yun Li64, Robert Lyons108, Andrea Maschio7,105,106, Eleonora Porcu7,105,106, Fred Reinier107, Serena Sanna106, David Schlessinger109, Carlo Sidore7,105,106, Adrian Tan7, Mary Kate Trost7, University of Montréal Philip Awadalla (Principal Investigator)65, Alan Hodgkinson65, University of Oxford Gerton Lunter (Principal Investigator)1, Gil A. McVean (Principal Investigator) (Co-Chair)1,2, Jonathan L. Marchini (Principal Investigator)1,2, Simon Myers (Principal Investigator)1,2, Claire Churchhouse2, Olivier Delaneau2, Anjali Gupta-Hinch1, Zamin Iqbal1, Iain Mathieson1, Andy Rimmer1, Dionysia K. Xifara1,2, University of Puerto Rico Taras K. Oleksyk (Principal Investigator)66, University of Texas Health Sciences Center at Houston Yunxin Fu (Principal Investigator)67, Xiaoming Liu67, Momiao Xiong67, University of Utah Lynn Jorde (Principal Investigator)68, David Witherspoon68, Jinchuan Xing69, University of Washington Evan E. Eichler (Principal Investigator)11, Brian L. Browning (Principal Investigator)70, Can Alkan21,71, Iman Hajirasouliha102, Fereydoun Hormozdiari21, Arthur Ko21, Peter H. Sudmant21 Washington University in St. Louis Elaine R. Mardis (Co-Principal Investigator)19, Ken Chen103, Asif Chinwalla19, Li Ding19, David Dooling19, Daniel C. Koboldt19, Michael D. McLellan19, John W. Wallis19, Michael C. Wendl19, Qunyuan Zhang19, Wellcome Trust Sanger Institute Richard M. Durbin (Principal Investigator)6, Matthew E. Hurles (Principal Investigator)6, Chris Tyler-Smith (Principal Investigator)6, Cornelis A. Albers72, Qasim Ayub6, Senduran Balasubramaniam6, Yuan Chen6, Alison J. Coffey6, Vincenza Colonna6,73, Petr Danecek6, Ni Huang6, Luke Jostins6, Thomas M. Keane6, Heng Li3,6, Shane McCarthy6, Aylwyn Scally6, James Stalker6, Klaudia Walter6, Yali Xue6, Yujun Zhang6, Yale University Mark B. Gerstein (Principal Investigator)74-76, Alexej Abyzov74, 76, Suganthi Balasubramanian76, Jieming Chen74, Declan Clarke77, Yao Fu74, Lukas Habegger74, Arif O. Harmanci74, Mike Jin76, Ekta Khurana76, Xinmeng Jasmine Mu74, Cristina Sisu74
Structural Variation Group: BGI-Shenzhen Yingrui Li24, Ruibang Luo24, Hongmei Zhu24, Brigham and Women’s Hospital Charles Lee (Principal Investigator) (Co-Chair)17, Lauren Griffin17, Chih-Heng Hsieh17, Ryan E. Mills17,41, Xinghua Shi17, Marcin von Grotthuss17, Chengsheng Zhang17, Boston College Gabor T. Marth (Principal Investigator)20, Erik P. Garrison20, Deniz Kural20, Wan-Ping Lee20, Alistair N. Ward20, Jiantao Wu20, Mengyao Zhang20, Broad Institute of MIT and Harvard Steven A. McCarroll (Project Lead)3, David M. Altshuler3-5, Eric Banks3, Guillermo del Angel3, Giulio Genovese3, Robert E. Handsaker3,5, Chris Hartl3, James C. Nemesh3, Khalid Shakir3, Cold Spring Harbor Laboratory Seungtai C. Yoon (Principal Investigator)30, Jayon Lihm30, Vladimir Makarov31, Cornell University Jeremiah Degenhardt10, European Bioinformatics Institute Paul Flicek (Principal Investigator)12, Laura Clarke12, Richard E. Smith12, Xiangqun Zheng-Bradley12, European Molecular Biology Laboratory Jan O. Korbel (Principal Investigator) (Co-Chair)16, Tobias Rausch16, Adrian M. Stütz16, Illumina David R. Bentley (Principal Investigator)8, Bret Barnes37, R. Keira Cheetham8, Michael Eberle8, Sean Humphray8, Scott Kahn37, Lisa Murray8, Richard Shaw8, Leiden University Medical Center Kai Ye (Principal Investigator)38, Louisiana State University Mark A. Batzer (Principal Investigator)39, Miriam K. Konkel39, Jerilyn A. Walker39, Stanford University Phil Lacroute43, Translational Genomics Research Institute David W. Craig (Principal Investigator)48, Nils Homer110, US National Institutes of Health Deanna Church23, Chunlin Xiao23, University of California, San Diego Jonathan Sebat (Principal Investigator)50,51, Vineet Bafna52, Jacob J. Michaelson79, Kenny Ye53, University of Maryland School of Medicine Scott E. Devine (Principal Investigator)60, Xinyue Liu60, Ankit Maroo60, Luke J. Tallon60, University of Oxford Gerton Lunter (Principal Investigator)1, Gil A. McVean (Principal Investigator)1,2, Zamin Iqbal1, University of Utah David Witherspoon68, Jinchuan Xing69, University of Washington Evan E. Eichler (Principal Investigator) (Co-Chair)11, Can Alkan21,71, Iman Hajirasouliha102, Fereydoun Hormozdiari21, Arthur Ko21, Peter H. Sudmant21, Washington University in St. Louis Ken Chen103, Asif Chinwalla19, Li Ding19, Michael D. McLellan19, John W. Wallis19, Wellcome Trust Sanger Institute Matthew E. Hurles (Principal Investigator) (Co-Chair)6, Ben Blackburne6, Heng Li6, Sarah J. Lindsay6, Zemin Ning6, Aylwyn Scally6, Klaudia Walter6, Yujun Zhang6, Yale University Mark B. Gerstein (Principal Investigator)74-76, Alexej Abyzov74,76, Jieming Chen74, Declan Clarke77, Ekta Khurana76, Xinmeng Jasmine Mu74, Cristina Sisu74
Exome Group: Baylor College of Medicine Richard A. Gibbs (Principal Investigator) (Co-Chair)13, Fuli Yu (Project Leader)13, Matthew Bainbridge13, Danny Challis13, Uday S. Evani13, Christie Kovar13, Lora Lewis13, James Lu13, Donna Muzny13, Uma Nagaswamy13, Jeff Reid13, Aniko Sabo13, Jin Yu13, BGI-Shenzhen Xiaosen Guo24, Yingrui Li24, Renhua Wu24, Boston College Gabor T. Marth (Principal Investigator) (Co-Chair)20, Erik P. Garrison20, Wen Fung Leong20, Alistair N. Ward20, Broad Institute of MIT and Harvard Guillermo del Angel3, Mark A. DePristo3, Stacey B. Gabriel3, Namrata Gupta3, Chris Hartl3, Ryan E. Poplin3, Cornell University Andrew G. Clark (Principal Investigator)10, Juan L. Rodriguez-Flores10, European Bioinformatics Institute Paul Flicek (Principal Investigator)12, Laura Clarke12, Richard E. Smith12, Xiangqun Zheng-Bradley12, Massachusetts General Hospital Daniel G. MacArthur (Principal Investigator)40, Stanford University Carlos D. Bustamante (Principal Investigator)43, Simon Gravel43, Translational Genomics Research Institute David W. Craig (Principal Investigator)48, Alexis Christoforides48, Nils Homer110, Tyler Izatt48, US National Institutes of Health Stephen T. Sherry (Principal Investigator)23, Chunlin Xiao23, University of Geneva Emmanouil T. Dermitzakis (Principal Investigator)58,59,104, University of Michigan Gonçalo R. Abecasis (Principal Investigator)7, Hyun Min Kang7, University of Oxford Gil A. McVean (Principal Investigator)1,2, Washington University in St. Louis Elaine R. Mardis (Principal Investigator)19, David Dooling19, Lucinda Fulton19, Robert Fulton19, Daniel C. Koboldt19, Wellcome Trust Sanger Institute Richard M. Durbin (Principal Investigator)6, Senduran Balasubramaniam6, Thomas M. Keane6, Shane McCarthy6, James Stalker6, Yale University Mark B. Gerstein (Principal Investigator)74-76, Suganthi Balasubramanian76, Lukas Habegger74
Functional Interpretation Group: Boston College Erik P. Garrison20, Baylor College of Medicine Richard A. Gibbs (Principal Investigator) 13, Matthew Bainbridge13, Donna Muzny13, Fuli Yu13, Jin Yu13, Broad Institute of MIT and Harvard Guillermo del Angel3, Robert E. Handsaker3,5, Cold Spring Harbor Laboratory Vladimir Makarov31, Cornell University Juan L. Rodriguez-Flores10, Dankook University Hanjun Jin (Principal Investigator)32, Wook Kim33, Ki Cheol Kim33, European Bioinformatics Institute Paul Flicek (Principal Investigator)12, Kathryn Beal12, Laura Clarke12, Fiona Cunningham12, Javier Herrero12, William M. McLaren12, Graham R.S. Ritchie12, Xiangqun Zheng-Bradley12, Harvard University Shervin Tabrizi3,35, Massachusetts General Hospital Daniel G. MacArthur (Principal Investigator)40, Monkol Lek40, Stanford University Carlos D. Bustamante (Principal Investigator)43, Francisco M. De La Vega10, Translational Genomics Research Institute David W. Craig (Principal Investigator)48, Ahmet A. Kurdoglu48, University of Geneva Tuuli Lappalainen58,59,104, University of Medicine and Dentistry of New Jersey Jeffrey A. Rosenfeld (Principal Investigator)61,62, Leslie P. Michelson61,62, University of Montréal Philip Awadalla (Principal Investigator)65, Alan Hodgkinson65, University of Oxford Gil A. McVean (Principal Investigator)1,2, Washington University in St. Louis Ken Chen103, Wellcome Trust Sanger Institute Chris Tyler-Smith (Principal Investigator) (Co-Chair)6, Yuan Chen6, Vincenza Colonna6,73, Adam Frankish6, Jennifer Harrow6, Yali Xue6, Yale University Mark B. Gerstein (Principal Investigator) (Co-Chair)74-76, Alexej Abyzov74,76, Suganthi Balasubramanian76, Jieming Chen74, Declan Clarke77, Yao Fu74, Arif O. Harmanci74, Mike Jin76, Ekta Khurana76, Xinmeng Jasmine Mu74, Cristina Sisu74
Data Coordination Center Group: Baylor College of Medicine Richard A. Gibbs (Principal Investigator)13, Christie Kovar13, Divya Kalra13, Walker Hale13, Gerald Fowler13, Donna Muzny13, Jeff Reid13, BGI-Shenzhen Jun Wang (Principal Investigator)24,26, Xiaosen Guo24, Guoqing Li24, Yingrui Li24, Xiaole Zheng24, Broad Institute of MIT and Harvard David M. Altshuler3-5, European Bioinformatics Institute Paul Flicek (Principal Investigator) (Co-Chair)12, Laura Clarke (Project Lead)12, Jonathan Barker12, Gavin Kelman12, Eugene Kulesha12, Rasko Leinonen12, William M. McLaren12, Rajesh Radhakrishnan12, Asier Roa12, Dmitriy Smirnov12, Richard E. Smith12, Ian Streeter12, Iliana Toneva12, Brendan Vaughan12, Xiangqun Zheng-Bradley12, Illumina David R. Bentley (Principal Investigator)8, Tony Cox8, Sean Humphray8, Scott Kahn37, Max Planck Institute for Molecular Genetics Ralf Sudbrak (Project Lead)18, Marcus W. Albrecht28, Matthias Lienhard18, Translational Genomics Research Institute David W. Craig (Principal Investigator)48, Tyler Izatt48, Ahmet A. Kurdoglu48, US National Institutes of Health Stephen T. Sherry (Principal Investigator) (Co-Chair)23, Victor Ananiev23, Zinaida Belaia23, Dimitriy Beloslyudtsev23, Nathan Bouk23, Chao Chen23, Deanna Church23, Robert Cohen23, Charles Cook23, John Garner23, Timothy Hefferon23, Mikhail Kimelman23, Chunlei Liu23, John Lopez23, Peter Meric23, Chris O’Sullivan80, Yuri Ostapchuk23, Lon Phan23, Sergiy Ponomarov23, Valerie Schneider23, Eugene Shekhtman23, Karl Sirotkin23, Douglas Slotta23, Chunlin Xiao23, Hua Zhang23, University of California, Santa Cruz David Haussler (Principal Investigator)55,111, University of Michigan Gonçalo R. Abecasis (Principal Investigator)7, University of Oxford Gil A. McVean (Principal Investigator)1,2, University of Washington Can Alkan21,71, Arthur Ko21, Washington University in St. Louis David Dooling19, Wellcome Trust Sanger Institute Richard M. Durbin (Principal Investigator)6, Senduran Balasubramaniam6, Thomas M. Keane6, Shane McCarthy6, James Stalker6
Samples and ELSI Group: Aravinda Chakravarti (Co-Chair)9, Bartha M. Knoppers (Co-Chair)15, Gonçalo R. Abecasis7, Kathleen C. Barnes81, Christine Beiswanger82, Esteban Burchard54, Carlos D. Bustamante43, Hongyu Cai24, Hongzhi Cao24, Richard M. Durbin6, Neda Gharani82, Richard A. Gibbs13, Christopher R. Gignoux54, Simon Gravel43, Brenna Henn43, Danielle Jones34, Lynn Jorde68, Jane S. Kaye83, Alon Keinan10, Alastair Kent84, Angeliki Kerasidou1, Yingrui Li24, Rasika Mathias85, Gil McVean1,2, Andres Moreno-Estrada43, Pilar N. Ossorio86,87, Michael Parker88, David Reich5, Charles N. Rotimi89, Charmaine D. Royal90, Karla Sandoval43, Yeyang Su24, Ralf Sudbrak18, Zhongming Tian24, Bernd Timmermann18, Sarah Tishkoff91, Lorraine H. Toji82, Chris Tyler-Smith6, Marc Via92, Yuhong Wang24, Huanming Yang24, Ling Yang24, Jiayong Zhu24
Sample Collection: British from England and Scotland (GBR) Walter Bodmer93, Colombians in Medellín, Colombia (CLM) Gabriel Bedoya94, Andres Ruiz-Linares57, Han Chinese South (CHS) Cai Zhi Ming24, Gao Yang95, Chu Jia You96, Finnish in Finland (FIN) Leena Peltonen‡, Iberian Populations in Spain (IBS) Andres Garcia-Montero97, Alberto Orfao98, Puerto Ricans in Puerto Rico (PUR) Julie Dutil99, Juan C. Martinez-Cruzado66, Taras K. Oleksyk66
Scientific Management: Lisa D. Brooks100, Adam L. Felsenfeld100, Jean E. McEwen100, Nicholas C. Clemm100, Audrey Duncanson101, Michael Dunn101, Eric D. Green14, Mark S. Guyer100, Jane L. Peterson100
Writing Group: Goncalo R. Abecasis7, Adam Auton29, Lisa D. Brooks100, Mark A. DePristo3, Richard M. Durbin6, Robert E. Handsaker3,5, Hyun Min Kang7, Gabor T. Marth20, Gil A. McVean1,2
Wellcome Trust Centre for Human Genetics, Oxford University, Oxford OX3 7BN, UK.
Dept of Statistics, Oxford University, Oxford OX1 3TG, UK.
The Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, Massachusetts 02142, USA.
Center for Human Genetic Research, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Dept of Genetics, Harvard Medical School, Cambridge, Massachusetts 02142, USA.
Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, CB10 1SA, UK.
Center for Statistical Genetics, Biostatistics, University of Michigan, Ann Arbor, Michigan 48109, USA.
Illumina United Kingdom, Chesterford Research Park, Little Chesterford, Nr Saffron Walden, Essex CB10 1XL, UK.
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Center for Comparative and Population Genomics, Cornell University, Ithaca, New York 14850, USA.
Dept of Genome Sciences, University of Washington School of Medicine and Howard Hughes Medical Institute, Seattle, Washington 98195, USA.
European Bioinformatics Institute, Wellcome Trust Genome Campus, Cambridge, CB10 1SD, UK.
Baylor College of Medicine, Human Genome Sequencing Center, Houston, Texas 77030, USA.
US National Institutes of Health, National Human Genome Research Institute, 31 Center Drive, Bethesda, Maryland 20892, USA.
Centre of Genomics and Policy, McGill University, Montréal, Québec H3A 1A4, Canada.
European Molecular Biology Laboratory, Genome Biology Research Unit, Meyerhofstraße 1, 69117 Heidelberg, Germany.
Dept of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts 02115, USA.
Max Planck Institute for Molecular Genetics, Ihnestraße 63-73, 14195 Berlin, Germany.
The Genome Center, Washington University School of Medicine, St Louis, Missouri 63108, USA.
Dept of Biology, Boston College, Chestnut Hill, Massachusetts 02467, USA.
Dept of Genome Sciences, University of Washington School of Medicine, Seattle, Washington 98195, USA.
Affymetrix, Inc., Santa Clara, California 95051, USA.
US National Institutes of Health, National Center for Biotechnology Information, 45 Center Drive, Bethesda, Maryland 20892, USA.
BGI-Shenzhen, Shenzhen 518083, China.
The Novo Nordisk Foundation Center for Basic Metabolic Research, University of Copenhagen, DK-2200 Copenhagen, Denmark.
Dept of Biology, University of Copenhagen, DK-2100 Copenhagen, Denmark.
Dahlem Centre for Genome Research and Medical Systems Biology, D-14195 Berlin-Dahlem, Germany.
Alacris Theranostics GmbH, D-14195 Berlin-Dahlem, Germany.
Dept of Genetics, Albert Einstein College of Medicine, Bronx, New York 10461, USA.
Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA.
Seaver Autism Center and Dept of Psychiatry, Mount Sinai School of Medicine, New York, New York 10029, USA.
Department of Nanobiomedical Science, Dankook University, Cheonan 330-714, Korea.
Department of Biological Sciences, Dankook University, Cheonan 330-714, Korea.
Dept of Biological Statistics and Computational Biology, Cornell University, Ithaca, New York 14853, USA.
Center for Systems Biology and Dept Organismic and Evolutionary Biology, Harvard University, Cambridge, Massachusetts 02138, USA.
Institute of Medical Genetics, School of Medicine, Cardiff University, Heath Park, Cardiff CF14 4XN, UK.
Illumina, Inc., San Diego, California 92122, USA.
Molecular Epidemiology Section, Dept of Medical Statistics and Bioinformatics, Leiden University Medical Center 2333 ZA, The Netherlands.
Dept of Biological Sciences, Louisiana State University, Baton Rouge, Louisiana 70803, USA.
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Dept of Computational Medicine and Bioinfomatics, University of Michigan, Ann Arbor, Michigan 48109, USA.
Dept of Anthropology, Penn State University, University Park, Pennsylvania 16802, USA.
Dept of Genetics, Stanford University, Stanford, California 94305, USA.
Ancestry.com, San Francisco, California 94107, USA.
Blavatnik School of Computer Science, Tel-Aviv University, 69978 Tel Aviv, Israel.
Dept of Microbiology, Tel-Aviv University, 69978 Tel Aviv, Israel.
International Computer Science Institute, Berkeley, California 94704, USA.
The Translational Genomics Research Institute, Phoenix, Arizona 85004, USA.
Dept of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, California 90024, USA.
Dept of Psychiatry, University of California, San Diego, La Jolla, California 92093, USA.
Dept of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, California 92093, USA.
Dept of Computer Science, University of California, San Diego, La Jolla, California 92093, USA.
Dept of Epidemiology and Population Health, Albert Einstein College of Medicine, Bronx, New York 10461, USA.
Dept of Bioengineering and Therapeutic Sciences and Medicine, University of California, San Francisco, California 94158, USA.
Center for Biomolecular Science and Engineering, University of California, Santa Cruz, California 95064, USA.
Dept of Human Genetics, University of Chicago, Chicago, Illinois 60637, USA.
Dept of Genetics, Evolution and Environment, University College London, London WC1E 6BT, UK.
Dept of Genetic Medicine and Development, University of Geneva Medical School, Geneva, 1211 Switzerland.
Institute for Genetics and Genomics in Geneva (iGE3), University of Geneva, 1211 Geneva, Switzerland.
Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA.
IST/High Performance and Research Computing, University of Medicine and Dentistry of New Jersey, Newark, New Jersey 07107, USA.
Department of Invertebrate Zoology, American Museum of Natural History, New York, New York 10024, USA.
Dept of Anthropology, University of Michigan, Ann Arbor, Michigan 48109, USA.
Dept of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Dept of Pediatrics, University of Montréal, Ste. Justine Hospital Research Centre, Montréal, Québec H3T 1C5,Canada.
Dept of Biology, University of Puerto Rico, Mayagüez, Puerto Rico 00680, USA.
The University of Texas Health Science Center at Houston, Houston, Texas 77030, USA.
Eccles Institute of Human Genetics, University of Utah School of Medicine, Salt Lake City, Utah 84112, USA.
Dept of Genetics, Rutgers University,The State University of New Jersey, Piscataway, New Jersey 08854, USA.
Dept of Medicine, Division of Medical Genetics, University of Washington, Seattle, Washington 98195, USA.
Dept of Computer Engineering, Bilkent University, TR-06800 Bilkent, Ankara, Turkey.
Dept of Haematology, University of Cambridge and National Health Service Blood and Transplant, Cambridge CB2 1TN, UK.
Institute of Genetics and Biophysics, National Research Council (CNR), 80125 Naples, Italy.
Program in Computational Biology and Bioinformatics, Yale University, New Haven, Connecticut 06520, USA.
Dept of Computer Science, Yale University, New Haven, Connecticut 06520, USA.
Dept of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA.
Dept of Chemistry, Yale University, New Haven, Connecticut 06520, USA.
Dept of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, USA.
Beyster Center for Genomics of Psychiatric Diseases, University of California, San Diego, La Jolla, California 92093, USA.
US National Institutes of Health, National Human Genome Research Institute, 50 South Drive, Bethesda, Maryland 20892, USA.
Division of Allergy and Clinical Immunology, School of Medicine, Johns Hopkins University, Baltimore, Maryland 21205, USA.
Coriell Institute for Medical Research, Camden, New Jersey 08103, USA.
Centre for Health, Law and Emerging Technologies, University of Oxford, Oxford OX3 7LF, UK.
Genetic Alliance, London, N1 3QP, UK.
Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Dept of Medical History and Bioethics, Morgridge Institute for Research, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA.
University of Wisconsin Law School, Madison, Wisconsin 53706, USA.
The Ethox Centre, Department of Public Health, University of Oxford, Old Road Campus, Oxford, OX3 7LF, UK.
US National Institutes of Health, Center for Research on Genomics and Global Health, National Human Genome Research Institute, 12 South Drive, Bethesda, Maryland 20892, USA.
Institute for Genome Sciences and Policy, Duke University, Durham, North Carolina 27708, USA.
Dept of Genetics, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, USA.
Dept of Animal Biology, Unit of Anthropology, University of Barcelona, 08028 Barcelona, Spain.
Cancer and Immunogenetics Laboratory, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Laboratory of Molecular Genetics, Institute of Biology, University of Antioquia, Medellín, Colombia.
Peking University Shenzhen Hospital, Shenzhen, 518036, China.
Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Kunming 650118, China.
Instituto de Biologia Molecular y Celular del Cancer, Centro de Investigacion del Cancer/IBMCC (CSIC-USAL), Institute of Biomedical Research of Salamanca (IBSAL) & Banco Nacional de ADN Carlos III, University of Salamanca, 37007 Salamanca, Spain.
Instituto de Biologia Molecular y Celular del Cancer, Centro de Investigacion del Cancer/IBMCC (CSIC-USAL), Institute of Biomedical Research of Salamanca (IBSAL) & Cytometry Service and Dept of Medicine, 37007 University of Salamanca, Salamanca, Spain.
Ponce School of Medicine and Health Sciences, Ponce, Puerto Rico 00716, USA.
US National Institutes of Health, National Human Genome Research Institute, 5635 Fishers Lane, Bethesda, Maryland 20892, USA.
Wellcome Trust, Gibbs Building, 215 Euston Road, London NW1 2BE, UK.
Dept of Computer Science, Simon Fraser University, Burnaby, British Columbia V5A 1S6, Canada.
Dept of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77230, USA.
Swiss Institute of Bioinformatics, 1211 Geneva, Switzerland.
Dipartimento di Scienze Biomediche, Università delgi Studi di Sassari, 07100 Sassari, Italy.
Istituto di Ricerca Genetica e Biomedica, CNR, Monserrato, 09042 Cagliari, Italy.
Center for Advanced Studies, Research, and Development in Sardinia (CRS4), AGCT Program, Parco Scientifico e tecnologico della Sardegna, 09010 Pula, Italy.
University of Michigan Sequencing Core, University of Michigan, Ann Arbor, Michigan 48109, USA.
National Institute on Aging, Laboratory of Genetics, Baltimore, Maryland 21224,USA.
Life Technologies, Beverly, Massachusetts 01915, USA.
Howard Hughes Medical Institute, Santa Cruz, California 95064, USA.
Deceased
References
- 1.Tennessen JA, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012 doi: 10.1126/science.1219240. doi:10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. doi:10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 4.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. doi:10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marth GT, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. doi:10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. doi:10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–246. doi: 10.1038/ng.1074. doi:10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. doi:10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmela E, et al. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS ONE. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. doi:10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. doi:10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. doi:10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. doi:10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki K, et al. Expression cloning of a novel Gal beta (1-3/1-4) GlcNAc alpha 2,3-sialyltransferase using lectin resistance selection. J Biol Chem. 1993;268:22782–22787. [PubMed] [Google Scholar]
- 15.Marth G, et al. Sequence variations in the public human genome data reflect a bottlenecked population history. Proc Natl Acad Sci U S A. 2003;100:376–381. doi: 10.1073/pnas.222673099. doi:10.1073/pnas.222673099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. doi:10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall GM. Slavery and African Ethnicities in the Americas: Restoring the Links. Univ North Carolina Press; 2005. [Google Scholar]
- 18.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. doi:10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. doi:10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. doi:10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. doi:10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. doi:10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Altas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. doi:10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome medicine. 2009;1:13. doi: 10.1186/gm13. doi:10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. doi:10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. doi:10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanna S, et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. doi: 10.1371/journal.pgen.1002198. doi:10.1371/journal.pgen.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory AP, Dendrou CA, Bell J, McVean G, Fugger L. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassanein MT, et al. Fine mapping of the association with obesity at the FTO locus in African-derived populations. Hum Mol Genet. 2010;19:2907–2916. doi: 10.1093/hmg/ddq178. doi:10.1093/hmg/ddq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maller J, The Wellcome Trust Case Control Consortium Fine mapping of 14 loci identified through genome-wide association analyses. Nat Genet. 2012 In press. [Google Scholar]
- 31.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamshad MJ, et al. The Centers for Mendelian Genomics: A new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012 doi: 10.1002/ajmg.a.35470. doi:10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43–47. doi: 10.1038/ng.733. doi:10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 34.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. doi:10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. doi:10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. doi:10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. doi:10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. doi:10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flannick J, et al. Efficiency and power as a function of sequence coverage, SNP array density, and imputation. PLoS Comput Biol. 2012;8:e1002604. doi: 10.1371/journal.pcbi.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–276. doi: 10.1038/ng.768. doi:10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. doi:10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet. 2012;44:226–232. doi: 10.1038/ng.1028. doi:10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson JT, Durbin R. Efficient construction of an assembly string graph using the FM-index. Bioinformatics. 2010;26:i367–373. doi: 10.1093/bioinformatics/btq217. doi:10.1093/bioinformatics/btq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. doi:10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers JC, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42:373–375. doi: 10.1038/ng.566. doi:10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hindorff LA, et al. A Catalog of Published Genome-Wide Association Studies. 2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.