Abstract
Over-expression of AdeABC efflux pump stimulated continuously by the mutated AdeRS two component system has been found to result in antimicrobial resistance, even tigecycline (TGC) resistance, in multidrug-resistant Acinetobacter baumannii (MRAB). Although the insertion sequence, ISAba1, contributes to one of the AdeRS mutations, the detail mechanism remains unclear. In the present study we collected 130 TGC-resistant isolates from 317 carbapenem resistant MRAB (MRAB-C) isolates, and 38 of them were characterized with ISAba1 insertion in the adeS gene. The relationship between the expression of AdeABC efflux pump and TGC resistant was verified indirectly by successfully reducing TGC resistance with NMP, an efflux pump inhibitor. Further analysis showed that the remaining gene following the ISAba1 insertion was still transcribed to generate a truncated AdeS protein by the Pout promoter on ISAba1 instead of frame shift or pre-termination. Through introducing a series of recombinant adeRS constructs into a adeRS knockout strain, we demonstrated the truncated AdeS protein was constitutively produced and stimulating the expression of AdeABC efflux pump via interaction with AdeR. Our findings suggest a mechanism of antimicrobial resistance induced by an aberrant cytoplasmic sensor derived from an insertion element.
Introduction
Acinetobacter baumannii, a Gram-negative coccobacillus usually found in soil and water, has emerged as a highly problematic nosocomial pathogen [1]. A. baumannii constitutes a major public health problem due to its intrinsic resilience to numerous drugs and its ability to readily acquire new resistance determinants [2], [3]. Multidrug-resistant A. baumannii resistant to carbapenem (MRAB-C) has been increasingly reported worldwide, which raises serious concern about the limited antimicrobial treatment options [4].
Multidrug efflux pumps belonging to the resistance-nodulation-cell division (RND) family have been shown to play an important role in the antimicrobial resistance of A. baumannii [3], [5]. To date, three different RND pumps have been identified in A. baumannii, namely AdeABC, AdeIJK and AdeFGH. AdeABC efflux pump is the first characterized RND system in A. baumannii [3]. Overexpression of the AdeABC efflux pump has been shown to pump out aminoglycosides, beta-lactam, fluoroquinolone, tetracycline, macrolide, chloramphenicol, trimethoprim, and even tigecycline (TGC). TGC is the first member of the glycylcycline and is considered a bacteriostatic drug with high efficacy in treating a broad range of antibiotic-resistant bacteria [4], [6]. Previous studies have shown that TGC has poor activities against MRAB-C due to over-expression of the AdeABC efflux pump system [3], [7], [8], [9], [10], [11]. The AdeIJK efflux pump contributes minimally to acquired resistance due to its toxicity for the host [12]. The third efflux pump, AdeFGH, its overexpression confers high-level resistance to most antibiotics except β-lactam and aminoglycosides [13]. Its overexpression is therefore least expected to result in the multidrug resistance in MRAB-C isolates. The expression of AdeABC efflux pump is tightly regulated by the two-component system which contains a sensor kinase (SK) AdeS and a response regulator (RR) AdeR, encoded by the adeRS operon. The adeRS operon is located at the upstream of adeABC operon and is transcribed in the opposite direction [3], [7], [14]. The promoters of the adeABC operon and the adeRS operon were predicted at the region between the two operons [14].
In micro-organisms, the two-component system is a very important signal transduction system for adaptation to drastic and immediate changes in external or internal environmental conditions [15]. This system is usually composed of a RR and a SK. The SK monitor's certain signal is caused by environmental changes. Signal bound to sensor domains of SK normally results in inducting an ATP dependent auto-phosphorylation of the histidine kinase domain in the catalytic core of the enzyme and transmitting the information to RR via a phosphoryl-transfer reaction to aspartate residue in RR. The RR is shifted to its active state by the phosphate group received and is directly bound to the promoter region of its target genes as a transcriptional regulator [16]. The typical type of SK is a homodimeric integral membrane protein in which the sensor domain, direct detection of extracellular signals, is formed by an extracellular loop contained between two membrane-spanning segments. The transmitter domain contains a histidine kinase A (HisKA) domain, and an ATP-binding domain follows the last transmembrane segment at the C-terminus and is localized within the cytoplasm [15]. However, some SKs which deviate from the typical type of SK model with their sensor domains within the membrane or fully cytoplasmic are found in bacteria, fungi, and plant [16]. The cytoplasmic SK is a protein located in the cytoplasm comprising a sensor domain and transmitter domain, without a transmembrane domain [16], [17], [18], [19], [20].
Several families of Insertion sequence (IS) in A. baumannii have been associated with acquired antimicrobial resistance mechanisms [21]. Many IS elements are related to antibiotic resistance through two mechanisms. Some of the IS elements contain promoters which can enhance the expression of their downstream antibiotic resistance genes [22]. ISAba1 has been found in upstream of ampC, bla OXA-23, bla OXA-27, bla OXA-51 and sulII antibiotic resistance genes [22], [23], [24], [25], [26], [27], [28], [29], [30]. Other elements, such as ISAba2, ISAba3, and ISAba4 contain preceding promoters that enhance the expression of carbapenem resistant genes, bla OXA-58 and the bla OXA-23 genes [31], [32], [33]. Moreover, the IS element may be inserted into and may disrupt the coding region of an outer membrane protein gene. The loss of the outer membrane protein results in resistance by blocking the input of antibiotics. Previous research described that an insertion mutation within the carO outer membrane protein gene by the ISAba1, ISAba125, or ISAba825 element was associated with carbapenem resistant [22], [30], [34].
Previous studies found that ISAba1 insertion mutation within the adeS gene can enhance overexpression of the AdeABC efflux pump system and cause TGC resistant [9]. Excessive transcription of adeABC operon was triggered due to a strong promoter in ISAba1 or the disruption of the AdeS protein. However, the detail mechanism regarding how the ISAba1 element controls the AdeABC efflux pump over- expression remains unknown. In the present study, we investigated the prevalence, genotype, AdeABC efflux pump expression and TGC resistance pattern of MRAB-C clinical isolates in which adeRS operon was uniquely destroyed by insertion of an IS element. Further, we identified a truncated cytoplasmic AdeS encoded by an extra operon driven by the promoter within the ISAba1 element. The truncated AdeS protein was then responsible for enhancing AdeABC efflux pump over- expression.
Materials and Methods
Bacterial strains, growth media and plasmids
A total of 317 distinct clinical isolates of MRAB-C were collected in this study at Tri-Service General Hospital from 2008 to 2010. The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown routinely in Luria–Bertani (LB) broth (BD Difco, Franklin Lakes, NJ) at 37°C with shaking. Plasmids were maintained and selected in E. coli and A. baumannii hosts using a medium supplemented with the following antibiotics: ampicillin (Sigma-Aldrich, Poole, UK) (100 mg/L); kanamycin (Sigma-Aldrich) (50 mg/L); and tetracycline (Sigma-Aldrich) (5 mg/L).
Table 1. Bacterial strains and plasmids used in this study.
| Strains or plasmids | Relevant characteristics | Reference | |
| E. coli strains | DH5α | Φ80d, lacZM15, endA1, recA1,hsdR17 (r−, m+), supE44, thi-1, gyrA96, relA1, F– (lacZYA- argF)U169 | Lab stock |
| Acinetobacter baumannii strains | ATCC 15151 | A. baumannii reference strain | ATCC |
| ABmut02 | Derived from BCRC15884. adeRS mutant obtained by plasmid insertion. KanR, AmpR | This study | |
| AB039 | clinical MDR strain, TGC MIC = 2 mg/L, adeRS without IS element insertion | ||
| AB090 | clinical MDR strain, TGC MIC = 1 mg/L, adeS mutant obtained by ISaba1 insertion | This study | |
| AB096 | clinical MDR strain, TGC MIC = 16 mg/L, adeRS without IS element insertion | ||
| AB260 | clinical MDR strain, TGC MIC = 1 mg/L, adeS mutant obtained by ISaba1 insertion | This study | |
| AB293 | clinical MDR strain, TGC MIC = 16 mg/L, adeS mutant obtained by ISaba1 insertion | This study | |
| AB392 | clinical MDR strain, TGC MIC≦ 0.5 mg/L, adeR mutant obtained by ISaba125 insertion | This study | |
| Plasmids | pMU125 | E. coli-Acinetobacter shuttle plasmid with A. baumannii replication origin | [37] |
| pCR2.1-Topo | Suicide plasmid for A. baumannii. KanR, AmpR | Invitrogen | |
| pCR2.1-adeRint | pCR2.1-Topo containing a 513-bp internal fragment of the adeR. KanR, AmpR | This study | |
| pYM102 | mini-CTX::lacIq-PT7-lacZ | [40] | |
| pS01 | E. coli-Acinetobacter shuttle plasmid with A. baumannii replication origin; mini-CTX::lacIq-PT7-lacZ | This study | |
| pRS-adeRS (wt) | Derived from pRS, mini-CTX:: adeRS-Pade-lacZ (adeRS from ATCC 15151) | This study | |
| pRS-adeRS (AB090) | Derived from pRS, mini-CTX:: adeRS-Pade-lacZ (adeRS from AB260) | This study | |
| pRS-adeRS (AB293) | Derived from pRS, mini-CTX:: adeRS-Pade-lacZ (adeRS from AB293) | This study | |
| pRS-adeR (wt) | Derived from pRS, mini-CTX:: adeR-Pade-lacZ (adeR from ATCC 15151) | This study | |
| pRS-adeR (AB260) | Derived from pRS, mini-CTX:: adeR-Pade-lacZ (adeR from AB260) | This study | |
| pRS-adeR (AB293) | Derived from pRS, mini-CTX:: adeR-Pade-lacZ (adeR from AB293) | This study | |
| pRS-Pade-Pout-truncated adeS | Derived from pRS-adeRS (AB293), SphI digested and self-ligatioin. mini-CTX:: truncated adeS-Pout-Pade-lacZ | This study | |
| pRS-Pout-truncated adeS | Derived from pRS, mini-CTX:: truncated adeS -Pout-lacZ (soluble adeS from AB293) | This study | |
| pRS-Pout-truncated adeS (SDM) | Derived from pRS_Pout_truncated adeS, mutated at the start code ATG to AAG by site-directed mutagenesis | This study |
Antimicrobial susceptibility tests
All bacterial cultures were prepared on the day of antimicrobial susceptibility testing. The U.S. Food and Drug Administration (FDA) recommended tigecycline (TGC) susceptibility breakpoints for Enterobacteriaceae (susceptible MIC, ≦2 g/L; intermediate MIC, >2 and <8 g/L; resistant MIC, ≧8 g/L) were used as MIC interpretation criteria. TGC (Wyeth, Madison, NJ) powder was obtained from commercial sources and was prepared in a solution with sterilized water, and the solution was then frozen in aliquots at −80°C. TGC MICs were initially determined by the E-test (AB Biodisk, Solna, Sweden) and confirmed by the agar dilution test using Mueller-Hinton II agar (BD Difco). Susceptibility tests of other antimicrobials were performed by the VITEK-2 system (bioMérieux, Marcy l'Etoile, France). All strains were tested for susceptibility to the following antibiotics: ampicillin, cefazolin, gentamicin, amikacin, ampicillin/sulbactam, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, ceftazidime, ceftriaxone, imipenem, ciprofloxacin, cefepime, nitrofurantoin, tigecycline and levofloxacin. To verify the over- expression of efflux pump, we prepared Mueller–Hinton (M-H) agar plates with and without adding the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine (NMP) (Sigma-Aldrich). The final concentration of NMP in the M-H agar was 64 mg/L. The MICs of TGC in the presence or absence of NMP were determined by the E-test described above.
DNA manipulations
Genomic DNA was isolated using the protocol suggested in the literature [35]. Plasmid DNA was isolated by the Viogene plasmid DNA extraction miniprep system (Viogene biotek corporation, Taiwan). DNA concentration measurements were performed with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA). Standard PCR amplifications were performed with the DyNAzyme DNA polymerase (Finnzymes, Espoo, Finland). When necessary, high fidelity and blunt-ended PCR products were amplified with Phusion DNA Polymerase (Finnzymes). PCR products were purified with Virogene Gel and PCR Clean-Up kit (Viogene). Oligonucleotide was synthesized by Genomics BioScience and Technologies, Inc. (Taiwan). All oligonucleotides used in the study are listed in Table 2. Thermal cycle reactions were performed using the 9700 GeneAmp thermocycler (Applied Biosystems, Foster City, CA). Nucleotide sequencing was performed with BigDye Terminator Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Capillary electrophoresis was performed with a 3730XL Genetic Analyzer (Applied Biosystems) and then analyzed by BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Restriction enzymes were purchased from New England Biolabs (New England Biolabs, Beverly, MA) and used according to the manufacturer's recommendation. Fast-Link DNA Ligation Kit was purchased from Epicentre (Epicentre, Madison, Wisconsin) and used according to the manufacturer's recommendation. Competent cells were prepared and transformed by electroporation as previously described [36]. The sequence was analyzed with the NCBI BlastX bioinformatic tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the NCBI open reading frame (ORF) finder (http://www.ncbi.nlm.nih.gov/projects/gorf/).
Table 2. Oligonucleotides used in this study.
| Primer name | Sequence | Features/purpose | Source |
| adeRS_1f | ATGTTTGATCATTCTTTTTCTTTTG | Detection the IS element insertion within the adeRS operon and Sequencing of adeRS operon | [41] |
| adeRS_1849r | TTAGTTATTCATAGAAATTTTTATG | ||
| adeRS_689r | TTAATTAACATTTGAAATATG | ||
| adeRS_776f | ATGAAAAGTAAGTTAGGAATTAGTAAG | ||
| ISAba1-F | CATTGGCATTAAACTGAGGAGAAA | with ade RS_1849r primer to confirme the nsertion of ISaba1 within the adeS operon | This study |
| ISAba1-R | TTGGAAATGGGGAAAACGAA | with ade RS_1f primer to confirme the insertion of ISaba1 within the adeS operon | This study |
| pWH1266_ori_PstI_F | GCCTGCAGGATCGTAGAAATATCTATGA | Cloning of A. baumannii replication origin from pMU125 with engineered PstI site | This study |
| pWH1266_ori_PstI_SmaI_R | AACTGCAGACCCGGGGGATTTTAACATTTTGCGTTG | ||
| AdeA_promoter_f_BamHI | CACGGATCCAACCTAGTGAGTTTTTGATGTTCG | Cloning adeRS operon and promoter ade with engineered SmaI and BamHI site into pRS to form pRS- adeRS | This study |
| AdeS_r_SmaI | CGCCCCGGGTTAGTTATTCATAGAAATTTTTATG | ||
| AdeR_r_SmaI | CACCCCGGGTATTTAGGCGTCATCTTTTACAGC | With AdeA_promoter_f BamHI primer to clone adeR operon and promoter ade with engineered SmaI and BamHI site into pRS to form pRS- adeR | This study |
| ISaba1Pout_f_BamHI | CACGGATCCAAGCATGATGAGCGCAAAG | With AdeS_r SmaI primer to clone soluble adeS operon and ISaba1 Pout with engineered SmaI and BamHI site into pRS to form pRS-Pout-soluble adeS | This study |
| adeR_6f | TGATCATTCTTTTTCTTTTGATTGCCAAGA | Cloning the partial sequence of adeR into pCR2.1- Topo to form pTOPOadeR | This study |
| adeR_514r | CAGTCAGCGTCAGATTAAGCA | ||
| adeA_F | TTGATCGTGCTTCTATTCCTCAAG | RT- PCR of adeA gene | |
| adeA_R | GGCTCGCCACTGATATTACGTT | ||
| adeB_F | GGATTATGGCGACAGAAGGA | RT- PCR of adeB gene | |
| adeB_R | AATACTGCCGCCAATACCAG | ||
| QPCR_adeRS1661_f | ACCGAGTTCCAAGACGAT | RT- PCR of adeS gene | This study |
| QPCR_adeRS1782_r | CCTTTCAGTGCCACAATA | ||
| QPCR_adeR96_f | AAAACGTGAAGGCATGAGTG | RT- PCR of adeR gene | This study |
| QPCR_adeR220_r | CTTCCCAACCGTTTAATTCG | ||
| rpoB_f | TCCGCACGTAAAGTAGGAAC | RT- PCR of rpoB gene | |
| rpoB_r | ATGCCGCCTGAAAAAGTAAC | ||
| TruncatedAdeS_SDMut_sense | ATAATTTTAATGATAAGGCTCAAAAGC | change the start code ATG to AAG in soluble adeS by site-directed mutagenesis at pRS-Pout-soluble adeS | This study |
Quantitative real-time PCR
To detect the expression of adeR, adeS, adeA and adeB gene, quantitative real-time RT-PCR was performed individually for each. Primers for the quantitative real-time were designed using Primer3 software. The efficiency of each primer pair in quantitative real-time PCR has been determined by a 10-fold serial dilution of the template. By plotting the log values of template concentrations on the x-axis and the Ct values on the y-axis, standard linear lines with slopes (m) ranging from −3.58 to −3.27 were obtained. We obtained 95.1∼105.0% amplification efficiencies (EAmp) for the primer pairs based on the calculation equation, EAmp = 10−1/m−1. A. baumannii cells were grown aerobically in LB broth until reaching the mid-log phase. DNaseI-treated RNA templates were prepared using an RNeasy kit (QIAGEN Sciences, MD). The concentrations of the RNA were quantified with a spectrophotometer. Reverse transcription was performed using a high-capacity cDNA archive kit (Applied Biosystems) with 200 ng of RNA in a 20 µL reaction mixture containing 1× (each) reverse transcription buffer, deoxynucleoside triphosphate mix, and random primers as well as 2.5 U/ml multiscribe reverse transcriptase. Negative-control reactions included equal concentrations of RNA and all reagents while omitting reverse transcriptase. Incubation for 10 min at 25°C followed by 2 h at 37°C was carried out with a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems). Real time PCR was performed with the DyNAmo™ Flash SYBR Green qPCR Kit (Finnzymes) and a 1∶10 final dilution of the cDNA product on the ABI PRISM 7500 Real-Time PCR system (Applied Biosystems). Each PCR analysis contained one primer pair. Cycling conditions were as follows: denature at 95°C for 10 min, followed with 40 cycles of denature at 95°C for 15 s and annealing/extension at 56°C for 30 s, and finalized with 1 cycle at 95°C for 30 s and 28°C for 30 s. Following PCR cycling, melting point data were collected and the dissociation curve was examined for each well. The critical threshold cycle (Ct) numbers were determined by Sequence Detection Systems version 2.2.2 (Applied Biosystems). In order to omit efficiency variation among primer pairs, ΔCt and ΔΔCt were applied for comparing the gene expression level. The ΔCt for gene expression was calculated against that for the rpoB housekeeping gene, and the ΔΔCt was calculated against that for the TGC-susceptible control strain (ATCC 15151, wt) as suggested in the literature.
Knockout the adeRS operon by homologous recombination
Plasmid inserted with the adeRS operon of A. baumannii was carried out as previously described [36]. Briefly, kanamycin and ampicillin resistant plasmid pCR2.1-Topo, unable to replicate in A. baumannii, was used as a suicide vector. An internal fragment (513 bp) of the adeR gene was amplified by PCR with AdeR_6f and AdeR_514r primers (Table 2) from a template of A. baumannii ATCC 15151 genomic DNA. The PCR product was cloned into the pCR2.1-Topo vector and electroporated into E. coli to yield the pCR2.1-adeRint plasmid (Table 1). Recombinant plasmid (100 ng) was then introduced in the kanamycin and ampicillin susceptible A. baumannii ATCC 15151 by electroporation [36]. Mutants were selected on LB agar plates containing kanamycin and ampicillin. Inactivation of the adeRS operon by insertion of the plasmid via a single crossover recombination was confirmed by sequencing the amplified PCR products with the AdeA_promoter_f_BamHI and AdeRS_1849r primer pairs (Table 2).
Construction of the E. coli-Acinetobacter shuttle vector pS01
The DNA fragment of A. baumannii replication origin was obtained by PCR using primers pWH1266_ori_PstI_F and pWH1266_ori_PstI_SmaI_R (Table 2) in a reaction mixture (50 µL) containing 1 U of Phusion DNA Polymerase, 0.2 mM of each deoxynucleoside triphosphate, 1× PCR buffer, 5 pmol of each primer, 2 mM MgSO4 and 10 ng of pMU125 DNA as the template [37]. For PCR, the reaction mixture was subjected to an initial 1 min denaturation step at 98°C, followed by 35 cycles of 10 s at 98°C, 10 s at 55°C, and 30 s at 72°C and a 10 min final elongation at 72°C. The purified PCR product was then cloned into plasmid pYM102 through the Pst I site. The resulting construct, pS01, was sequenced to verify the absence of PCR-introduced mutations.
Construction of pRS series for expressing the adeRS, adeR and Pout-truncated adeS genes
The plasmid pS01 was digested with SmaI and BamHI for 2 h at 37°C (or 25°C in the case of SmaI) to delete its lacI q gene and T7 early gene promoter. The digested plasmids were ligated with the respective gene fragments to generate a series of recombinant pRS clones, including pRS-adeRS (wt), pRS-adeRS (AB090), pRS-adeRS (AB293), pRS-adeR (wt), pRS-adeR (AB260), pRS-adeR (AB293) and pRS-Pout-truncated_adeS. The adeRS and adeR gene fragments of ATCC 15151 and AB260 were amplified by PCR using common forward primer AdeA_promoter_f_BamHI with reverse primers AdeS_r_SmaI and AdeR_r_SmaI, respectively (Table 2). The Pout-truncated adeS gene fragment of AB293 was amplified by PCR using primers ISAba1Pout_f_BamHI and AdeS_r_SmaI. Another plasmid pRS-Pade-Pout-truncated adeS was obtained by splicing out the DNA fragment between Pade and Pout of ISAbaI in pRS-adeRS (AB293) with SphI digestion and re-ligation.
Site Directed Mutagenesis
The site-directed mutagenesis (SDM) protocol was modified from the previous study [38]. We used plasmid pRS-Pout-truncated adeS as the template for SDM to disrupt start codon (ATG to AAG). The plasmid was amplified using a mutation bearing primer TruncatedAdeS_SDMut_sense in PCR reaction mixture (25 µL) containing 1 µg template, 1 U of Phusion DNA Polymerase, 0.2 mM of each deoxynucleoside triphosphate, 1× PCR buffer, 5 pmol of SDM primer, 2 mM MgSO4 and 10% (vol/vol) dimethyl sulfoxide (DMSO). For PCR, the reaction mixture was subjected to an initial 30 sec denaturation step at 98°C, followed by 28 cycles of 10 s at 98°C, 30 s at 50°C, and 4 min at 72°C and a 10-min final elongation at 72°C. The PCR product was further treated with 5 U of DpnI at 37°C for 1 hr. The DpnI-treated PCR product was directly transformed into DH5α competent cells. Transformants were incubated in 5 ml LB medium containing 5 mg/L tetracycline overnight. Tetracycline resistant colonies were isolated and extracted for plasmid content. Mutant start codon in the successfully transformed plasmid was confirmed by DNA sequencing.
Pulsed-field gel electrophoresis
All clinical A. baumannii isolates with the IS element in their adeRS operon were genotyped by the pulsed-field gel electrophoresis (PFGE). Total DNA was prepared and PFGE was performed as described previously [39]. Endonuclease ApaI was used to digest the bacterial DNA, and then restriction fragments were analyzed by PFGE in 1% SeaKem Gold agarose gels (Cambrex Bio Science, Rockland, ME, USA) in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA, pH 8.0) using a Bio-Rad CHEF-Mapper apparatus (Bio-Rad Laboratories, Hercules, CA). Gels were stained with ethidium bromide and photographed under ultraviolet light. Dendrograms showing percentage similarity were prepared with the Molecular Analyst Fingerprinting, Fingerprinting Plus and Fingerprinting DST Software (Bio-Rad Laboratories) and were compared using the UPGMA clustering method as proposed in previous studies [10], [39]. Isolates with similarity over 90% were considered to be identical; isolates with below 70% similarity (>3 band difference) were considered to be non-clonally related, using previously described criteria [39].
Measurement of β-galactosidase activity
β-galactosidase activity assay was modified from the method described by Miller (1992) [40]. Briefly, 0.5 mL bacterial suspension of 37°C overnight culture was placed on ice for 20 min, and then harvested by centrifugation at 8000×g for 5 min at 4°C. The pellets were re-suspended in 0.5 mL of Z buffer (60 mMNa2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, 50 mM β-mercaptoethanol, pH 7.0) and the OD600 of the resuspended cells was measured (blank against Z buffer). To lysis the cells, 0.1 mL of the cell suspension was mixed with 0.4 mL of Z buffer, 50 µL of chloroform, and 25 µL of 0.1% Sodium dodecyl sulfate, vortexed for 10 sec, and incubated at 28°C. At a time point defined as zero, 0.1 mL of ONPG (o-nitrophenyl-β-D-galactopyranoside) (Sigma-Aldrich) (4 g/L) was added to the lysed cell suspension. The reaction was stopped by adding 0.25 mL of 1 MNa2CO3 at a time defined as T and the OD was then measured at 420 nm and 550 nm using a Synergy HT multi-mode microplate reader (BioTek Instruments, Winooski, VT). The activity of β-galactosidase was calculated using the following equation: 1000×[OD420−(1.75×OD550)]/[(T (min)×0.2 (mL)×OD600)], expressed in Miller units.
Nucleotide sequence accession number
The 3038 -bp DNA sequences of the adeRS operon of AB260 and AB293 corresponding to the ISAba1-interrupted adeS gene were submitted to the GenBank database as accession number JQ690823 and Q690824, respectively. The 2941-bp DNA sequence of the adeRS operon of AB392 corresponding to the ISAba125-interrupted adeR gene was submitted to the GenBank database as accession number JQ690825.
Results
Compatible antibiotics susceptibility with PFGE genotyping results in MRAB-C isolates
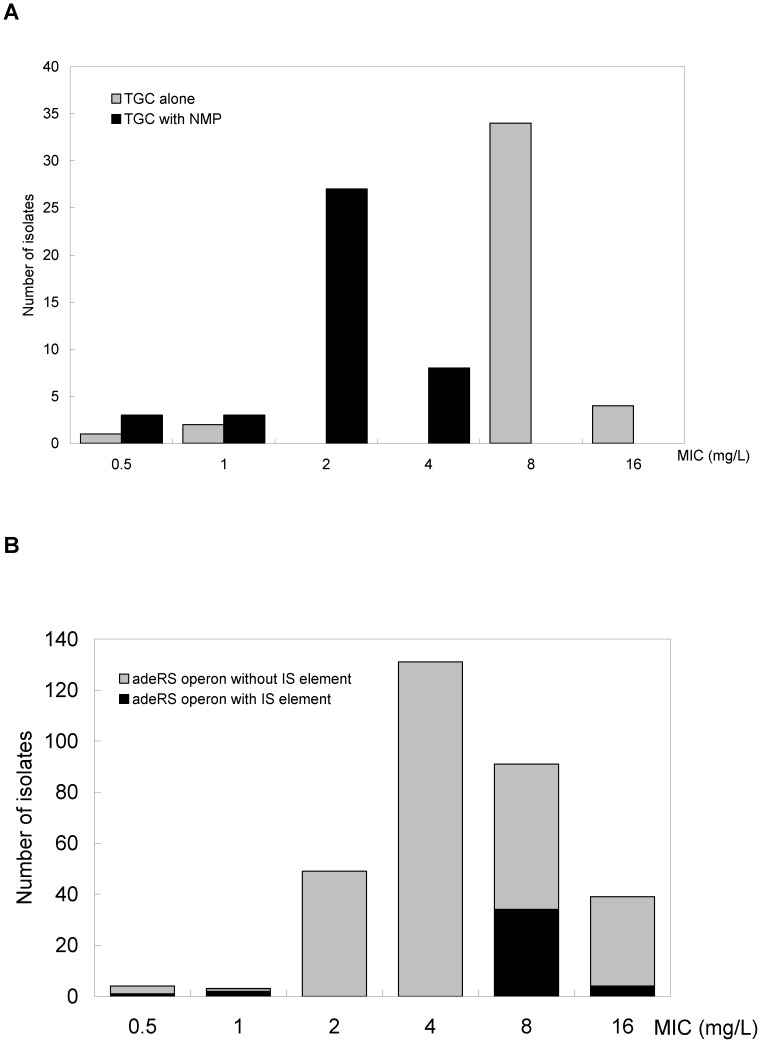
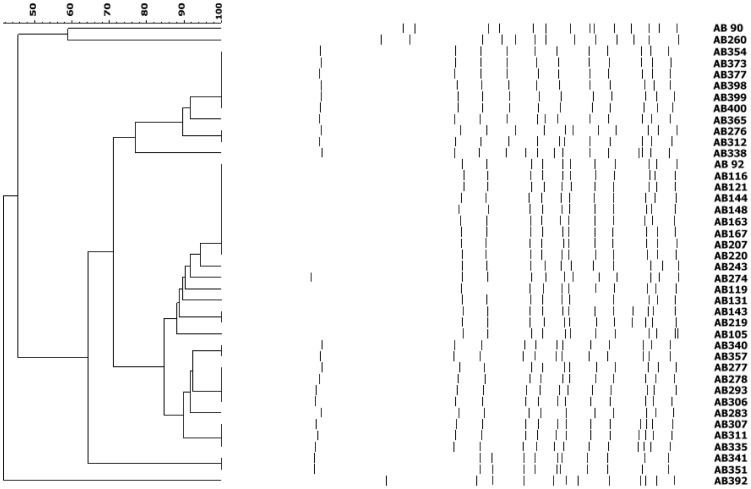
Antimicrobial susceptibility testing revealed that the 317 clinical MRAB-C isolates were resistant to all antibiotics tested in the Vitek-2 system. Using the FDA criteria for the agar dilution method, 130 (41%), 131 (41.3%), and 56 (17.7%) isolates were resistant, intermediate, and susceptible to TGC, respectively (Figure 1A). The full length of adeRS operon in each MRAB-C isolate was expected to be consistent in size (1.8 kb) after PCR amplification. However, the PCR products from 41 isolates were about 3.0 kb, which indicates additional DNA sequences within the adeRS operon. Among the 41 isolates, 38 (92.7%) isolates were resistant and 3 (7.3%) were susceptible to TGC. In a previous study, we demonstrated the expression of AdeABC efflux pump is significantly correlated with MIC of TGC in clinical isolates during the period [10]. To verify the effect of AdeABC efflux pump on TGC resistance, all MRAB-C isolates were tested for their MIC of TGC in the presence of NMP, an efflux pump inhibitor (Figure 1B). On plates with NMP, their E-test MIC (0.5–4 mg/L) of TGC showed 2–8 folds lower values than those without NMP (8–16 mg/L). TGC resistance was therefore related to AdeABC efflux pump overexpression in these isolates. After comparing the genotype patterns by pulsed field gel electrophoresis (PFGE) with ApaI-digestion, the 41 isolates were classified into 3 clusters and 3 sporadic clones based on PFGE criteria (Figure 2). The genotypic grouping result was compatible with phenotypes of TGC susceptibility. Two major clusters (AB354-AB338 and AB92-AB335 in Figure 2) and one minor cluster (AB341 and AB351) contained 38 (92.7%, 38/41) of all the MRAB-C isolates that were all TGC resistant. The remaining 3 sporadic clones (AB90, AB260 and AB392) were TGC susceptible.
Figure 1. Tigecycline MIC profile of the clinical A. baumannii isolates.
(A) Distribution of tigecycline MIC of the clinical A. baumannii isolates with (black bar) and without (grey bar) insertion of additional DNA in their adeRS operon (B) MIC to tigecycline alone (grey bar) and to tigecycline/NMP combination (black bar) for the 41 clinical A. baumannii isolates with insertion of additional DNA in their adeRS operon.
Figure 2. Pulsed-field gel electrophoresis (PFGE) for ApaI of 41 A. baumannii isolates with insertion of additional DNA in their adeRS operon.
Dendrogram was generated by UPGMA clustering.
IS elements within the adeRS operon found by sequencing analysis
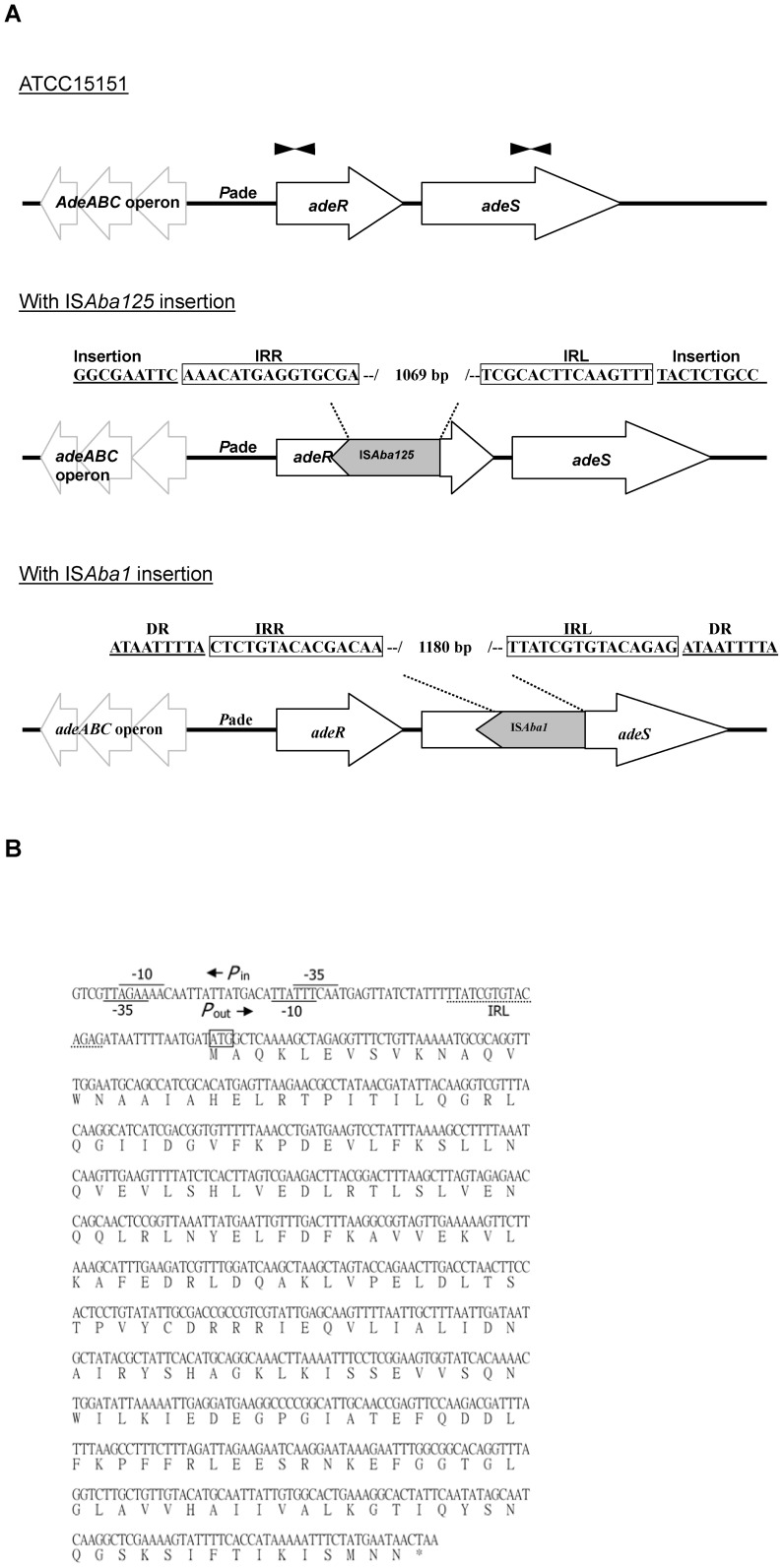
To determine the additional DNA insertion in adeRS operon, direct sequencing of adeRS operon PCR products showed the presence of the ISAba125 element within the adeR gene in AB392 isolates and the ISAba1 element within the adeS gene in the remaining 40 strains (Figure 3A). The ISAba125 element was 1,087-bp in length. It contained a 969-bp open reading frame (ORF) encoding a putative transposase, and was bounded by 15-bp inverted repeat sequences which are common to members of the IS30 group. Two 9-bp individual sequences (GGCGAATTC and TACTCTGCC) flanked the ISAba125 at the predicted insertion site in adeR gene of AB392. The presence of ISAba125 mediated disruptive events on the adeR gene in AB392, resulting in the loss function of adeR, and maybe affecting the expression of the AdeABC efflux pump system. The ISAba1 element was 1180-bp in length, containing two consecutive and overlapping ORFs, 567-bp ORF1 and 535-bp ORF2. It was therefore likely that a frame-shift is necessary to generate a unique protein as functional transposase. The ISAba1 was bounded by 15-bp inverted repeat sequences which are common to members of the IS4 family (Figure 3A). A 9-bp duplication (ATAATTTTA) flanked ISAba1 at the predicted insertion site in adeS gene of the other isolates. The presence of ISAba1 mediated disruptive events on the adeS gene, resulting in the loss function of adeS gene, and maybe affecting the expression of the AdeABC efflux pump system. However, we noted the most striking difference in the distribution of TGC MIC in the isolates. Only 2 strains (AB090 and AB260) were TGC susceptible and the remaining 38 isolates were TGC resistant. Hence, it was necessary to investigate whether the phenomenon was caused by DNA mutation in the promoter region or in adeRS operon. Nucleotide sequences of the promoter region and the adeRS operon with ISAba1 insertion were analyzed by multiple sequence alignment. DNA sequences of the promoter region and the destroyed adeS gene were identical among the 38 TGC resistant isolates. However, two point mutations (Met197Ile) and (Ser200Cys) in adeR gene were identified in all isolates, except for the two TGC sensitive isolates.
Figure 3. Alteration of the adeRS operons by insertion of ISAba125 or ISAba1 in multidrug resistant A. baumannii isolates.
(A) Genetic scheme of wide type adeRS operon in ATCC 15151 and two insertion mutations. ISAba125 disrupts the adeR gene and ISAba1 disrupts the adeS gene in clinical multidrug resistant A. baumannii isolates. Open arrows indicate coding sequences and direction of transcription. Pade is the promoter of the adeABC operon. Closed arrowheads indicate the position and orientation of the primers used in quantitative adeR and adeS real-time PCR (all positions are according to accession no. JQ690823-5 in GenBank). IRR, inverted repeat-right; IRL, inverted repeats-left; DR, direct repeat. (B) Presumed promoters in ISAba1 and downstream coding sequence of the truncated AdeS. The deduced amino-acid sequence is designated in a single letter code below the nucleotide sequence and the star sign indicates stop codon. The inverted repeat sequences (IRL) of ISAba1 are dash-underlined. The −35 and −10 motifs of promoters are indicated as numbers. Pin is the promoter for transposase gene in ISAba1, and Pout is the promoter for the truncated AdeS derived from ISAba1 insertion. The truncated AdeS start codon is boxed.
Transcript analysis of adeABC operon and adeRS operon
As the synergistic effect of NMP on TGC susceptibility described above, the TGC resistance may be associated with up-regulation of the AdeABC efflux pump in those isolates with their adeS gene destroyed by ISAba1. We investigated the transcription activity of adeRS operon and adeABC operon in representative isolates with various TGC MICs using quantitative real-time RT-PCR (Table 3). The two isolates (AB039 and AB096) were discussed in our previous research [10], [41], [42]. They both have higher MIC of TGC than strain ATCC 15151. The MICs of TGC were 2 mg/L and 16 mg/L for AB39 and AB96, respectively. Although the detail mechanism was not explored, we have demonstrated the higher MIC is proportionally correlated with the adeB expression level. We used these two strains simply to show the consistency of our experiment. We were still not able to identify the exact cause for their difference. In this manuscript, nonetheless, we focused on the group of isolates with an insertion element.
Table 3. Relationship between tigecycline MICs and expression of adeRS and adeABC operons in clinical strains of A. baumannii.
| Differential quantification of gene expression (fold) | ||||||
| Strains | Strains | TGC MIC (mg/L) | adeR | adeS | adeA | adeB |
| Without insertion | ATCC 15151 | ≦0.5 | 1.00 | 1.00 | 1.00 | 1.00 |
| AB39 | 2 | 1.50±0.73 | 0.65±0.2 | 24.15±5.95 | 42.64±22.49 | |
| AB96 | 16 | 1.55±0.15 | 0.67±0.05 | 285.75±106.12 | 622.03±189.32 | |
| With insertion | AB392 (ISAba125) | ≦0.5 | 1.21±0.25 | 0.08±0.02 | 0.55±0.33 | 1.05±0.64 |
| AB260 (ISAba1) | 1 | 1.87±0.14 | 2.82±0.88 | 1.02±0.54 | 1.71±0.59 | |
| AB293 (ISAba1) | 16 | 2.45±0.98 | 2.12±0.35 | 51.32±9.21 | 81.75±68.78 | |
To quantify the adeRS operon expression in strains with various IS element insertion, the first set of PCR primers for adeR gene was targeting the region between its promoter and the ISAba125 insertion site in AB392 (Figure 3A). The second set of PCR primers was targeting the terminal region of adeS gene after the ISAba1 insertion site in AB293 (Figure 3A). We first compared the adeB gene expression in TGC resistant isolates (AB096 and AB293) with the proposed basal expression in control strain ATCC 15151. The expression of adeB gene increased 622 times and 82 times in AB096 and AB293, respectively. In contrast, only a 42-fold, 1.1-fold and 1.7-fold increase in the adeB gene expression was observed in TGC susceptible clinical strains, AB039, AB392 and AB260, respectively. The adeA gene showed the same increasing expression pattern in TGC resistant clinical isolates as the adeB gene. All the TGC susceptible isolates (ATCC15151, AB260 and AB392) had less transcription of adeA and adeB consistently. In contrast, the transcription levels of the adeA and adeB genes in TGC resistant isolates were significantly higher than those in TGC susceptibility isolates (Mann-Whitney Test, P value <0.05). We tried to correlate the overexpression of adeA and adeB genes with their regulatory compartment by quantifying the transcripts of adeR and adeS genes in the TGC resistant isolates. The level of transcription of adeR in AB260 (1.87±0.14) and AB293 (2.45±0.98) was not significantly different (Mann-Whitney Test; P value = 0.38). However, the adeS gene transcription in AB392 was 12.5-fold lower than in the ATCC 15151 control strain. Lower adeS transcription might result from the polar effect after an ISAba125 insertion in AB392. The ISAba1 insertion also resulted in the same polar effect after an ISAba1 insertion in AB260, but did not cause the same polar effect to reduce the adeS gene transcription in AB293. Our findings suggested that an extra promoter within the ISaba1 element of AB293 constitutively drives the transcription for the remaining adeS gene fragment.
A truncated AdeS predicted by DNA sequencing
To determine the structure and function domain of AdeS, we analyzed the full adeS gene sequence in ATCC 15151 by the NCBI tool and the transmembrane domain prediction tool. The N-terminus of the AdeS contained two transmembrane segments (amino acid positions 9–36 and 58–85), which suggests that AdeS is a membrane anchored protein. The C-terminus of AdeS after the second transmembrane segment was the transmitter compartment containing a phosphorylatable HisKA domain and an ATP -binding domain. The insertion site of ISAba1 was located between the second transmembrane segment and the transmitter compartment in AB293. Also, a Pout promoter overlapping with Pin promoter was found near left inverted repeat sequence (IRL) within the ISAba1 element (Figure 3B). By analyzing the downstream sequences of the Pout promoter, we predicted an ORF encoding a protein with 229 amino acids by ORF finder. The amino acid sequence of the predicted protein was the same as the carboxyl-terminal transmitter domain of AdeS protein. We deduced a truncated AdeS driven by the Pout promoter in the ISAba1 element (Figure 3B). The predicted protein contained neither transmembrane helices nor a signal peptide. It was suggested to be a truncated protein within the cytoplasm. The truncated AdeS shared 100% amino acid identity with the kinase sensor protein annotated as a hypothetical protein of the MRAB-C strain 1656-2 (GenBank accession number ADX03920). We further analyzed the biological function of the truncated AdeS protein by investigating its possible relationship with two point mutations of AdeR in determining TGC resistance.
Acquired TGC resistance induced by coexistence of the truncated AdeS and mutated AdeR
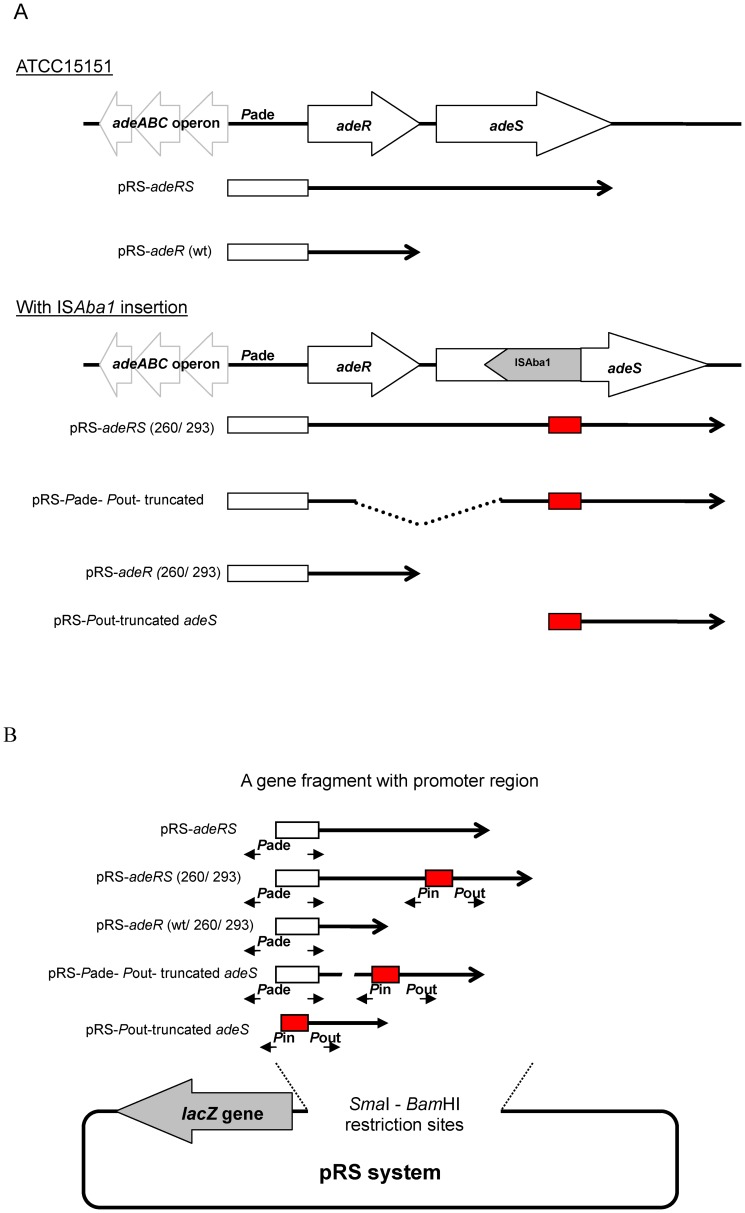
To further assess the relationship between the truncated AdeS and mutated AdeR in TGC resistant isolates, DNA fragments of the full length adeRS operon and the adeR gene from different strains (wt, AB260 and AB293) were cloned into the pRS system to construct pRS-adeRS (wt, AB260, AB293) and pRS-adeR (wt, AB260, AB293), respectively (Figure 4A). The predicted truncated adeS operon of AB293 was also cloned into pRS to generate pRS-Pout-truncated adeS. The pRS-adeRS (AB293) was digested with SphI and was self-ligated to generate pRS-Pade-Pout-truncated adeS. The adeRS opreon of ATCC 15151 was destroyed to generate a mutant bacterial strain (ABmut02) by homologous recombination with a suicide plasmid. These recombinants and the ABmut02 mutant strain were used to assess the regulation of AdeABC efflux pump expression. The recombinant plasmids were transformed respectively into ATCC 15151 and ABmut02 to obtain the transformed strains. The transformed strains were then tested for their antimicrobial susceptibilities. Results of the antibiotic susceptibility assays were summarized in Table 4. Assays on ATCC15151 wild type strain showed an 8-fold TGC MIC increase after transforming ATCC 15151 with pRS-adeRS (AB260), pRS-adeRS (AB293), pRS-Pade-Pout-truncated adeS or pRS-Pout-truncated adeS. The AdeRS-destroyed mutant ABmut02 shared the same TGC MIC with wild type ATCC 15151 after transforming with the pRS-adeRS (AB260) or pRS-adeRS (AB293). However, contrary to expectation, transformation with pRS-Pade-Pout-truncated_adeS or pRS-Pout-truncated_adeS did not show higher TGC MIC in ABmut02. We demonstrated that the truncated AdeS must cooperate with AdeR to induce TGC resistance. And the two point mutations in AdeR were proven to have no significant effect on its interaction with the truncated AdeS. To determine whether the multidrug resistance was directly induced by the truncated AdeS in the pRS system, we disrupted the start codon of the truncated AdeS by site directed mutagenesis. We found 16-fold lower TGC MIC and multidrug MIC reduction in ATCC 15151 transformed with the start codon disrupted mutant, pRS-Pout-truncated_adeS (SDM). Accordingly, we suggest that truncated AdeS is associated with acquired TGC and multidrug resistance.
Figure 4. Construction of the pRS recombinant series.
(A) Strategies for obtaining a series of DNA fragments containing promoter region of adeABC operon and a series of adeRS operon variants from wild type (wt) ATCC 15151 and drug resistant strains (260, 293) by PCR. Open arrows denote coding sequences and indicate the direction of transcription in the bacterial genome. The white rectangle box denotes the region of Pade promoter (Pade and Pre). The red rectangle box denotes the promoter region in ISAba1 (Pin and Pout). Arrows following the rectangle boxes together indicate the PCR fragments used to generate pRS recombinant series. (B) Construction map of the promoter-less lacZ gene of pRS system. The pRS system was derived from pS01, which remove lacIq-PT7 promoter by engineered SmaI and BamHI cutting. The pRS system was obtained and used for cloning PCR fragments into upstream of the promoter-less lacZ gene of pRS system between SmaI and BamHI restriction sites. The rectangle box indicates the region of promoters. The region of Pade promoter region contains two overlapped promoters in opposite direction (Pade and Pre). Two overlapped promoters, Pin and Pout, were in the region of promoter region in ISAba1. The expression level of lacZ gene represented for the activity of promoter of adeABC operon or Pin promoter of ISAba1.
Table 4. Antimicrobial susceptibilities of the various A. baumannii transformants.
| MIC (mg/liter) of | ||||||
| Strains | with plasmid | TGC | TGC+ NMP | FEP | CIP | LEV |
| ATCC 15151 (wt adeRS) | no plasmid | 0.25 | 0.25 | 2 | ≦0.25 | ≦0.12 |
| pS01 | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeRS (wt) | 0.25 | 0.125 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeRS (AB260) | 2 | 0.25 | 16 | 1 | 0.25 | |
| pRS-adeRS (AB293) | 2 | 0.25 | 16 | 1 | 0.25 | |
| pRS-adeR (wt) | 0.25 | 0.125 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeR (AB260) | 0.25 | 0.125 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeR (AB293) | 0.25 | 0.125 | 2 | ≦0.25 | ≦0.12 | |
| pRS-Pade-Pout-truncated adeS | 2 | 0.25 | 16 | 0.5 | 0.25 | |
| pRS-Pout-truncated adeS | 2 | 0.25 | 16 | 0.5 | 0.25 | |
| pRS-Pout-truncated adeS (SDM) | 0.25 | 0.125 | 2 | ≦0.25 | ≦0.12 | |
| ABmut02 (adeRS destruct) | no plasmid | 0.25 | 0.25 | 2 | ≦0.25 | ≦0.12 |
| pS01 | 0.25 | 0.25 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeRS (wt) | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeRS (AB260) | 2 | 0.25 | 16 | 0.5 | 0.25 | |
| pRS-adeRS (AB293) | 2 | 0.25 | 32 | 1 | 0.25 | |
| pRS-adeR (wt) | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeR (AB260) | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-adeR (AB293) | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-Pade-Pout-truncated adeS | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-Pout-truncated adeS | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
| pRS-Pout-truncated adeS (SDM) | 0.125 | 0.0625 | 2 | ≦0.25 | ≦0.12 | |
Quantifying the Pade promoter activity by the β-galactosidase assay
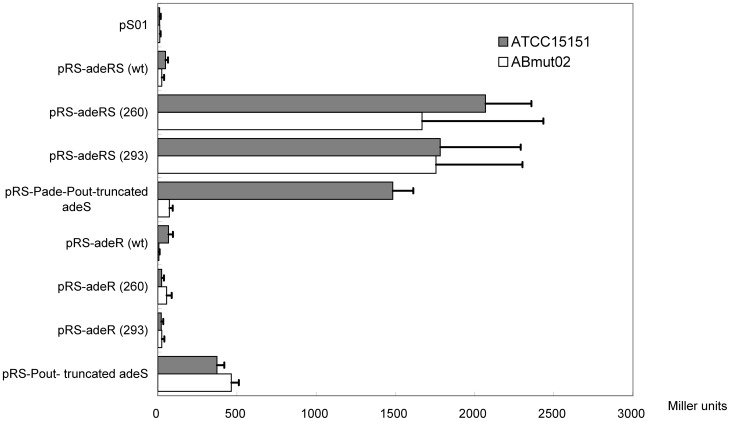
To further differentiate the promoter effect from either Pade of adeABC operon or Pin of ISaba1, we quantified adeA and adeB gene expression in all the transformants listed in Table 4. We constructed a lacZ gene reporter system instead of laborious quantification RT-PCR of adeA and adeB. The lacZ gene was cloned as a substitute for the adeABC gene in the pRS series (Figure 4B). Then we were able to assay the promoter effect by directly quantifying β-galactosidase activity. The β-galactosidase activities in ATCC 15151 strain with pS01 (13±7 units) and in ABmut02 strain with pS01 (12±6 units) were measured as background. In ATCC 15151, β-galactosidase activities increased about 172-fold (2068±290 units), 149-fold (1783±508 units) and 123-fold after transformation with pRS-adeRS (260), pRS-adeRS (293), and pRS-Pade-Pout-truncated adeS, respectively (Mann-Whitney Test; P value <0.05). In ABmut02, β-galactosidase activities also increased about 128-fold (1669±766 units) and 135-fold (1757±546 units) after transformation with pRS-adeRS (260) and pRS-adeRS (293), respectively (Mann-Whitney Test; P value <0.05) (Figure 5). Transformation of pRS-Pade-Pout-truncated adeS can only increase β-galactosidase activitiy by 20 fold while AdeR is lacking in ABmut02 strain (Mann-Whitney Test; P value <0.05). The β-galactosidase activity did not increase while being transformed with the recombinant plasmids expressing adeR only (wt, 260, 293) into either ATCC 15151 or ABmut02 strains. These results suggested that the expression of the adeABC efflux pump was directly controlled by the two-component system, truncated AdeS and AdeR. The expression of β-galactosidase activity was significantly higher in ATCC15151 with pRS-Pade-Pout-truncated adeS than with pRS-Pout-truncated adeS (Mann-Whitney Test; P value <0.05). And the lower expression of β-galactosidase activity was observed in ABmut02 with pRS-Pade-Pout-truncated adeS than with pRS-Pout-truncated adeS (Mann-Whitney Test; P value <0.05). These results suggested that AdeR protein is necessary for stimulating Pade promoter and enhancing the expression of the adeABC efflux pump. In order to rule out the possibility of Pin promoter of ISAba1 on enhancing the expression of adeABC operon, we only increased the β-galactosidase activity of ATCC 15151 and ABmut02 transformed with the Pin containing plasmid pRS-Pout-truncated_adeS by 31-fold (374±46 units) and 36-fold (465±48 units), respectively. The β-galactosidase assay successfully quantified the significantly higher lacZ activity in the strain transformed with pRS-adeRS compared with pRS-Pout-truncated adeS. The biological function of truncated adeS was further demonstrated besides converting the MIC presentation, and the truncated adeS with coexistence of adeR can constitutively stimulate the promoter Pade to express the lacZ gene.
Figure 5. β-galactosidase activities of transformed strains with various pRS recombinants.
ATCC 15151 and ABmut02 cells transformed with various pRS recombinants were grown in LB medium at 37°C overnight. The measurement of β-galactosidase activity was repeated at least three times, and the mean values of one representative experiment performed in triplicate are reported (error bars indicate the standard deviation). The measurement of β-galactosidase activity represents for the activity driven by either Pade of adeABC operon or Pin of ISAba1.
Discussion
Antibiotic resistance is a major challenge in treating infections caused by A. baumannii [1]. The adeABC operon is located in chromosomal DNA and it endorses the intrinsic low-level expression in A. baumannii. [3], [7]. The expression of AdeABC efflux pump is tightly regulated by the two-component regulatory system AdeR-AdeS, encoded by the adeRS operon that is located upstream of adeABC operon and transcribed in the opposite direction [3], [7], [14].. The overexpression of the AdeABC efflux pump, responsible for reducing accumulation of many antibiotics, is an efficient mechanism for multidrug resistance in A. baumannii [43]. AdeS is a membrane-integrated sensor protein with a histidine kinase domain which converts and transfers environmental stimuli to a response regulator AdeR for regulating the AdeABC gene expression [3], [7]. On the basis of our current data, we proposed an N-terminal truncated AdeS generated by the Pout promoter within the ISAba1 insertion of adeS gene. The truncated AdeS protein was able to activate AdeR and then enhance the adeABC gene expression.
Ruzin et al. [9] reported they found the disruption of AdeS by the insertion sequence ISAba1 in two TGC-resistant isolates of Acinetobacter calcoaceticus–A. baumannii complex. The AdeABC pumps were overexpressed in these strains and associated with TGC resistant. However, the prevalence, genotype and mechanism of the strain with an ISAba1 insertion in adeS gene had not been well investigated. In this study, we found 41 of the 317 isolates contained DNA insertions in their adeRS operon. Besides the ISAba1 insertion of adeS gene in 40 isolates as presumed, the remaining one isolate was found with ISAba125 insertion in its adeR gene. Compared with the TGC susceptibility phenotype, we found that 38 of the above 40 isolates accounted for 29.2% (38/130) of the TGC resistant isolates, and they demonstrated differences in the PFGE patterns. This result may indicate a common ancestor shared by these isolates with ISAba1 insertion in adeS gene.
Previous studies including ours have indicated that the sequence of AdeR and AdeS are widely variable in clinical isolates, but several mutations are still recommended for causing overexpression of the AdeABC efflux pump [10], [41], [42]. Most of the publications describe point mutations in the sensor domain (Asp30Gly), in the linker domain (Aln94Val) between the recognition domain and the histidine box, and in the histidine box (Tro153Met) of the AdeS [14], [44]. A mutation (Pro116Leu) in AdeR is also reported to cause adeABC operon overexpression likely due to altering the structural conformation of the regulator [14]. In addition, AdeR mutation (Aln91Val) and AdeS mutation (Gly103Asp) occur concurrently in a susceptibility conversion strain after exposure to TGC. However, the detail interaction among these mutations has not been ascertained [42]. Although we found two new mutations (Met197Ile) and (Ser200Cys) on the DNA binding domain of AdeR protein of all TGC resistant strains included in this study, strain AB260 with the same mutations showed lower expression level of the adeABC operon and was susceptible to TGC. However, the transformed strains with pRS-adeRS(AB293) and with pRS-adeRS(AB260) were presented with the presumed higher MIC to TGC. We therefore suggested that the adeABC operon expression in the susceptible strain AB260 might be inhibited by some unknown mechanism. Future studies will focus on analyzing the inhibition mechanism of adeABC operon expression in the TGC susceptibility strain.
ISAba1 copies were found in the chromosome and plasmids, which have been known to be related to antibiotic resistance only in Acinetobacter spp [27]. Previous genome-wide studies on evaluating the drug resistance mechanism in clinical A. baumannii isolates found multidrug resistant isolates carry more ISAba1 elements copies (9 to 428) than clinical susceptible strains and laboratory strains [21], [45], [46], [47]. The ISAba1 element flanks an antibiotic resistant gene and is involved in the overexpression of the genes from an outward-facing promoter, Pout in the IS element. Another example, disruption of an antibiotics influx pump protein, designated CarO, has also been reported by the ISAba1 element insertional inactivation [34]. Other ISAba1 insertions are polymorphic and are thus unlikely to be directly related to drug resistance in these strains [48]. Some ISAba1 insertion locations are also found in or adjacent to hypothetical protein genes. Previous genome-wide studies have identified an ISAba1 insertion in the adeS gene and even a presumed sensor kinase protein [21], [45]. In this study, a similar ISAba1 insertion into adeS was demonstrated to generate a truncated AdeS which shared the same amino acid sequence as the predicted sensor kinase protein of the genome-wide study [45]. We therefore suggested that the A. baumannii strain with a truncated adeS gene through ISAba1 insertion may have a worldwide distribution and require further investigation.
Instead of laboriously comparing the possible genomic difference among A. baumannii strains, we converted the reference strain ATCC 15151 to an AdeRS-knockout laboratory strain, ABmut02. A series of pRS recombinants carrying different adeRS fragments and the lacZ reporter gene were then able to be analyzed for their regulatory effect in the mutant strain. By directly quantifying the expression of the lacZ gene, we can compare the triggering effect of various adeRS operons on the adeABC promoter. Because adeR and adeS operon array sequentially after the same promoter, we did not obtain a laboratory strain solely with adeR gene disruption. The MIC of TGC can be as high as 16 mg/L for clinical resistant isolates. However, MICs of TGC were only mildly elevated (MIC = 2 mg/L, 8-folds) after transforming the truncated AdeS into ATCC 15151 and its adeRS-knockout derivative. The partially resistance-converting result suggests there are other mechanisms affecting the AdeABC efflux system. We presume some inhibitors offset the truncated AdeS effect in ATCC 15151 and AB260, but not in AB293. This inhibitor proposal may explain why isolate AB260 only demonstrates moderate MIC to TGC (1 mg/L) instead of MIC 16 mg/L in AB293. In addition, boosting AdeS by transforming a plasmid encoded the truncated adeS operon into ATCC 15151 did convert its susceptibility to TGC, while the same plasmid alone was unable to overcome the proposed inhibitor in ATCC 15151 mutant (ABmut02) with the destructed adeRS. The observed alteration of MIC to TGC for those artificial transformants supports our proposal indirectly. However, we need more direct evidence to prove this proposal. Moreover, an extraordinary efflux pump or other resistance mechanism may coexist to result in the aberrantly high MIC to TGC in AB293.
In this report, we have shown the distribution of multidrug resistant A. baumannii with the ISAba1 insertion in adeS gene in Taiwan. We have also demonstrated the existence of a truncated AdeS and its biological function. The truncated AdeS is able to induce multidrug resistance by constitutively stimulating the expression of AdeABC efflux pump through motivating the AdeR to interact with and activate the adeABC promoter. Our findings not only confirm the previously presumed additional adeS gene after ISAba1 insertion, but also further characterize it as another mechanism for ISAba1-related antibiotic resistance. As the two-component system is a vital structure of the bacteria and is used to adapt to changing environmental conditions, insight and applications resulting from studies on A. baumannii will probably be broadly applicable to other bacteria.
Acknowledgments
We thank Luis Actis for providing plasmid pMU125. We are grateful to Yoshiaki Kawamura for providing plasmid pYM102.
Funding Statement
This study was supported by grants TSGH-C101-128 from the Tri-Service General Hospital, Taiwan, Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Peleg AY, Seifert H, Paterson DL (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21: 538–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5: 939–951. [DOI] [PubMed] [Google Scholar]
- 3. Coyne S, Courvalin P, Perichon B (2011) Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55: 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gordon NC, Wareham DW (2009) A review of clinical and microbiological outcomes following treatment of infections involving multidrug-resistant Acinetobacter baumannii with tigecycline. J Antimicrob Chemother 63: 775–780. [DOI] [PubMed] [Google Scholar]
- 5. Cortez-Cordova J, Kumar A (2011) Activity of the efflux pump inhibitor phenylalanine-arginine beta-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int J Antimicrob Agents 37: 420–424. [DOI] [PubMed] [Google Scholar]
- 6. Yahav D, Lador A, Paul M, Leibovici L (2011) Efficacy and safety of tigecycline: a systematic review and meta-analysis. J Antimicrob Chemother 66: 1963–1971. [DOI] [PubMed] [Google Scholar]
- 7. Wieczorek P, Sacha P, Hauschild T, Zorawski M, Krawczyk M, et al. (2008) Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol 46: 257–267. [DOI] [PubMed] [Google Scholar]
- 8. Vila J, Marti S, Sanchez-Cespedes J (2007) Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother 59: 1210–1215. [DOI] [PubMed] [Google Scholar]
- 9. Ruzin A, Keeney D, Bradford PA (2007) AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother 59: 1001–1004. [DOI] [PubMed] [Google Scholar]
- 10. Sun JR, Chan MC, Chang TY, Wang WY, Chiueh TS (2010) Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob Agents Chemother 54: 4934–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruzin A, Immermann FW, Bradford PA (2010) RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Microb Drug Resist 16: 87–89. [DOI] [PubMed] [Google Scholar]
- 12. Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P (2008) AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B (2010) Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54: 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchand I, Damier-Piolle L, Courvalin P, Lambert T (2004) Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48: 3298–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szurmant H, White RA, Hoch JA (2007) Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol 17: 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheung J, Hendrickson WA (2010) Sensor domains of two-component regulatory systems. Curr Opin Microbiol 13: 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Essen LO, Mailliet J, Hughes J (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci U S A 105: 14709–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X, Kuk J, Moffat K (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci U S A 105: 14715–14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mern DS, Ha SW, Khodaverdi V, Gliese N, Gorisch H (2010) A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: indication of four levels of sensor kinases and response regulators. Microbiology 156: 1505–1516. [DOI] [PubMed] [Google Scholar]
- 20. Gorisch H (2003) The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. Biochim Biophys Acta 1647: 98–102. [DOI] [PubMed] [Google Scholar]
- 21. Adams MD, Chan ER, Molyneaux ND, Bonomo RA (2010) Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 54: 3569–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poirel L, Nordmann P (2006) Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12: 826–836. [DOI] [PubMed] [Google Scholar]
- 23. Hu WS, Yao SM, Fung CP, Hsieh YP, Liu CP, et al. (2007) An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob Agents Chemother 51: 3844–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, et al. (2003) AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J Antimicrob Chemother 52: 629–635. [DOI] [PubMed] [Google Scholar]
- 25. Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, et al. (2006) The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258: 72–77. [DOI] [PubMed] [Google Scholar]
- 26. Segal H, Nelson EC, Elisha BG (2004) Genetic environment and transcription of ampC in an Acinetobacter baumannii clinical isolate. Antimicrob Agents Chemother 48: 612–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Segal H, Garny S, Elisha BG (2005) Is IS(ABA-1) customized for Acinetobacter? FEMS Microbiol Lett 243: 425–429. [DOI] [PubMed] [Google Scholar]
- 28. Segal H, Jacobson RK, Garny S, Bamford CM, Elisha BG (2007) Extended −10 promoter in ISAba-1 upstream of blaOXA-23 from Acinetobacter baumannii. Antimicrob Agents Chemother 51: 3040–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, et al. (2007) Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1 blaOXA-23 genes in a Chinese hospital. J Med Microbiol 56: 1076–1080. [DOI] [PubMed] [Google Scholar]
- 30. Lee Y, Kim CK, Lee H, Jeong SH, Yong D, et al. (2011) A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the bla(OXA-23) gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob Agents Chemother 55: 361–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen TL, Wu RC, Shaio MF, Fung CP, Cho WL (2008) Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob Agents Chemother 52: 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P (2008) Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother 52: 1252–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poirel L, Nordmann P (2006) Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob Agents Chemother 50: 1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mussi MA, Limansky AS, Viale AM (2005) Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob Agents Chemother 49: 1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Syn CK, Swarup S (2000) A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal Biochem 278: 86–90. [DOI] [PubMed] [Google Scholar]
- 36. Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, et al. (2010) A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dorsey CW, Tomaras AP, Actis LA (2002) Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl Environ Microbiol 68: 6353–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, et al. (2005) Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43: 4328–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morita Y, Narita S, Tomida J, Tokuda H, Kawamura Y (2010) Application of an inducible system to engineer unmarked conditional mutants of essential genes of Pseudomonas aeruginosa. J Microbiol Methods 82: 205–213. [DOI] [PubMed] [Google Scholar]
- 41. Peleg AY, Adams J, Paterson DL (2007) Tigecycline Efflux as a Mechanism for Nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 51: 2065–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, et al. (2010) AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65: 1589–1593. [DOI] [PubMed] [Google Scholar]
- 43. Magnet S, Courvalin P, Lambert T (2001) Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coyne S, Guigon G, Courvalin P, Perichon B (2010) Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother 54: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park JY, Kim S, Kim SM, Cha SH, Lim SK, et al. (2011) Complete genome sequence of multidrug-resistant Acinetobacter baumannii strain 1656-2, which forms sturdy biofilm. J Bacteriol 193: 6393–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, et al. (2008) Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190: 8053–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, et al. (2008) Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3: e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murat D, Bance P, Callebaut I, Dassa E (2006) ATP hydrolysis is essential for the function of the Uup ATP-binding cassette ATPase in precise excision of transposons. J Biol Chem 281: 6850–6859. [DOI] [PubMed] [Google Scholar]