Abstract
Background
The trnH–psbA intergenic spacer region has been used in many DNA barcoding studies. However, a comprehensive evaluation with rigorous sequence preprocessing and statistical testing on the utility of trnH–psbA and its combinations as DNA barcodes is lacking.
Methodology/Principal Findings
Sequences were searched from GenBank for a meta-analysis on the usefulness of trnH–psbA and its combinations as DNA barcodes. After preprocessing, we constructed full and matching data sets that contained 17 983 trnH–psbA sequences and 2190 sets of trnH–psbA, matK, rbcL, and ITS2 sequences from the same sample, repectively. These datasets were used to analyze the ability of trnH–psbA and its combinations to discriminate species by the BLAST and BLAST+P methods. The Fisher's exact test was used to evaluate the significance of performance differences. For the full data set, the identification success rates of trnH–psbA exceeded 70% in 18 families and 12 genera, respectively. For the matching data set, the identification rates of trnH–psbA were significantly higher than those of the other loci in two families and four genera. Similarly, the identification rates of trnH–psbA+ITS2 were significantly higher than those of matK+rbcL in 18 families and 21 genera.
Conclusion/Significane
This study provides valuable information on the higher utility of trnH–psbA and its combinations. We found that trnH–psbA+ITS2 combination performs better or equally well compared with other combinations in most taxonomic groups investigated. This information will guide the optimal usage of trnH–psbA and its combinations for species identification.
Introduction
Accurate species identification is a prerequisite for conducting numerous basic and applied studies on monitoring and conserving natural resources, blocking the traffic of endangered and invasive species, as well as controlling the quality of pharmaceutical and food products. Species identification is primarily based on morphology [1]. The classic method of species determination frequently also warrants expert interpretation. However, most species are studied by only a few specialists, and even specialists frequently encounter specimens that cannot be reliably identified. Thus, a rapid, subjective, and accurate method for species identification is urgently needed.
DNA barcoding is based on sequence diversity within a short and standardized gene region for species discrimination. This method can identify known species and discover novel ones [2], [3]. In animals, the CO1 gene is widely used as the DNA barcode for efficient and accurate species determination [2], [4]–[9]. In plants, the situation is much more complicated. A variety of DNA regions have been proposed as barcodes for plant identification [10]–[20], but no region has been found to have characteristics for DNA barcoding as favorable as those of CO1. The Plant Working Group of the Consortium for the Barcode of Life [21] examined the suitability of seven leading candidate markers and proposed the two-locus combination of matK+rbcL as the core plant barcode. However, the lack of discriminatory power of this proposed barcode in some plant groups and the absence of primer universality for matK render the barcode questionable [22]. Further evaluation of other noncoding markers, particularly trnH–psbA and the internal transcribed spacers of nuclear ribosomal DNA (nrITS/nrITS2), is necessary before a universal plant barcode can be designated [22]. Recently, the usefulness of the ITS region as a DNA barcode has been further assessed by the China Plant BOL Group, who proposed that ITS should be used as the core plant barcode [23]. Although the trnH–psbA region has been evaluated in such studies in comparison with other markers alone or in combination, several shortcomings exist. First, most studies focus only on particular groups of plants. Second, the sequences are not rigorously processed. For example, intraspecific inversions and rps19 insertions, which are both well known to be present in the trnH–psbA region, are not explicitly examined and processed, which may negatively affect the species identification success rate. Third, the trnH–psbA+ITS2 combination is not evaluated at the family and genus levels. Most importantly, the statistical tests for the differences in identification success rates among various single markers or combinations are not performed.
In the present study, we performed a meta-analysis based on multiple sources of data available in public databases to evaluate the utility of the trnH–psbA region and its combinations with other markers as plant DNA barcodes.
Methods
Search Strategy, Inclusion and Exclusion Criteria, and Data Extraction
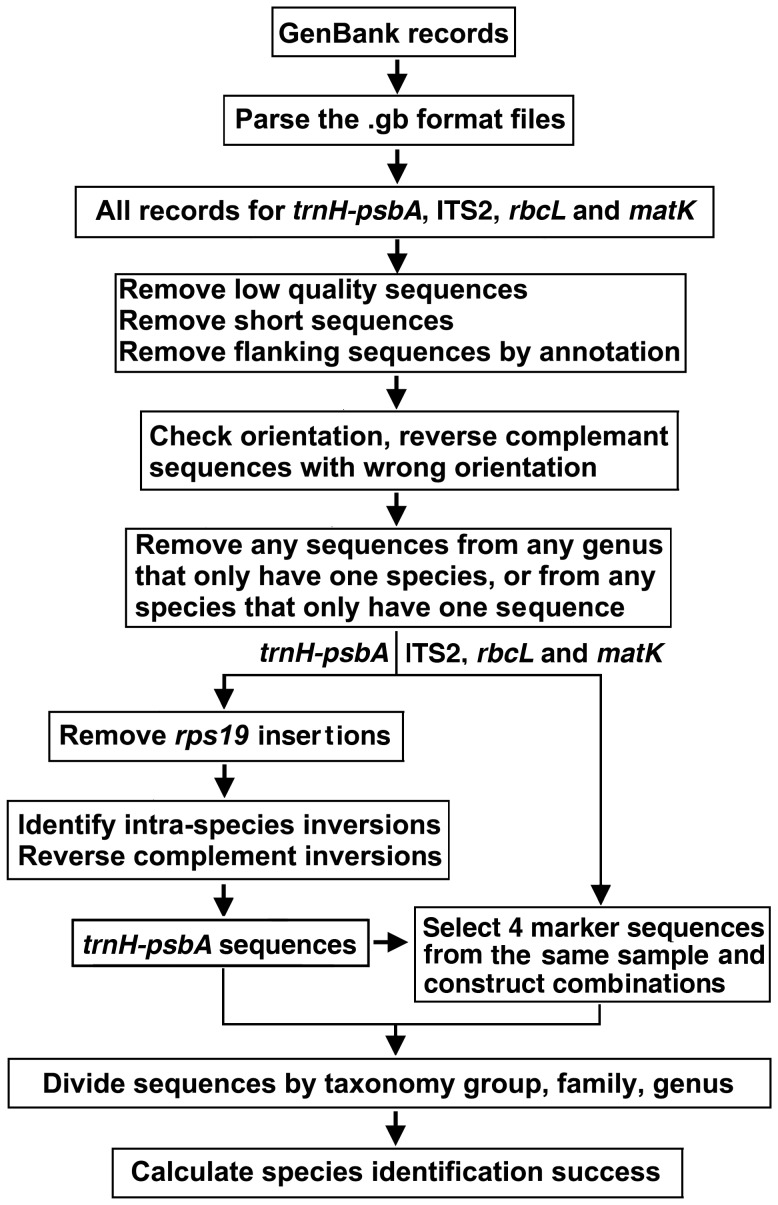
The sequences of the trnH–psbA intergenic region as well as its psbA and trnH flanking regions from eudicotyledons, monocotyledons, gymnosperms, mosses, and ferns were extracted from GenBank (at end of July 2012) using the keywords “psbA” and “trnH” in the annotations. The trnH–psbA intergenic regions were further annotated using either BLAST or a hidden Markov model program to identify and trim the flanking regions. The plant sequences of matK, rbcL, and ITS2 were extracted from GenBank using the keywords “matK”, “rbcL”, and “Internal Transcribed Spacer 2”, respectively. Sequences from public databases such as GenBank/EMBL are generally not assessed following the standards of barcoding with respect to access to vouchers and sequence quality, among others. Thus, all sequences were subjected to rigorous processing to reduce dimensions of errors further. Low-quality sequences with more than 1% nucleotides being “Ns” were removed. The longest and shortest 1% sequences were treated as outliers and excluded. The orientation of each sequence was checked, and sequences in the psbA–trnH orientation were reverse-complemented to the trnH–psbA orientation. All sequences were searched using BLAST against a local rps19 reference database that contained all sequences downloaded from GenBank. An E value less than 1×10−5 was used as a cutoff to determine if a sequence had an rps19 insertion. The species with putative rps19 sequences are listed in Table S1. To minimize the inflation of sequence divergence by intraspecific inversion, we developed custom Perl scripts to identify inversions, select a reference orientation, and reverse-complement all inversions that were in opposite orientation to the reference. The species with sequence inversions are shown in Table S2. Sequences belonging to any genus or family that contained only one species and those belonging to species for which only one sequence was available were also removed. The data processing pipeline is shown in Figure 1. The PRISMA checklist and flow diagram are available in the Supporting Information (PRISMA Checklist S1 and PRISMA Flow Diagram S1).
Figure 1. Workflow diagram for data processing and analysis.
After processing, the remaining sequences were used to construct data sets. The full data set (Table S3) contained a total of 17 983 trnH–psbA sequences from 3495 species representing 498 genera in 149 families, which were used to analyze the ability of trnH–psbA to identify species in various taxonomic groups. We also constructed a matching data set that consisted of trnH–psbA, matK, rbcL, and ITS2 sequences with the same voucher number (same sample). This data set contained 2190 sets of trnH–psbA, matK, rbcL, and ITS2 sequences from 586 species belonging to 71 genera and 47 families (Table S4). The data set was used to assess the relative species identification performance of trnH–psbA and its combinations compared with other suggested single markers and matK+rbcL.
Data Analysis
Sequence lengths were calculated for all trnH–psbA sequences of eudicotyledons, monocotyledons, gymnosperms, mosses, and ferns. The preprocessed sequences were aligned using ClustalW, followed by manual correction of the alignment, and Kimura's two-parameter distances were calculated using DNADIST from the EMBOSS package (Version 6.2). Three parameters were used to evaluate interspecific differences: (i) average interspecific distance between all species in each genus with more than one species (all interspecific distances), (ii) mean pairwise distance within each genus with at least two species (theta prime), and (iii) smallest interspecific distance within each genus with more than one species (minimum interspecific distance). Another set of three parameters was used to evaluate intraspecific differences: (i) average intraspecific distance between all samples within each species with at least two representatives (all intraspecific differences), (ii) mean pairwise distance within each species with more than one individual (theta), and (iii) maximum intraspecific distance within each species with more than one representative (coalescent depth). The intraspecific differences and interspecific variations of congeneric species of trnH–psbA in the five major plant taxonomic groups were calculated using a custom Perl script as previously described [17], [24]–[26].
BLAST and BLAST+P distance were performed using custom-written Perl scripts. Reference databases containing all sequences from the matching data set or the full data set were constructed. Using BLAST, we searched the reference database using the query sequence and inferred its identity based on the top hit, as previously described [27]. For the BLAST+P method, we first performed the BLAST search to select the significant (E value<1×10−5) and top (maximum number, 100) sequences. Multiple sequence alignment, distance calculation, and identity determination were then performed for these sequences. The identity of a query was inferred based on those of its neighbors with the shortest distance. The significant differences between the identification success rates of markers were tested by the Fisher's exact test implemented using JMP software (SAS Corp, NC, USA).
Results
Universality of the Intraspecific Inversion and rps19 Insertions in trnH–psbA
Two characteristics of trnH–psbA in some taxa reportedly complicate the use of the region as a DNA barcode. These characteristics are intraspecific inversions and rps19 insertions, which inflate intraspecific variation. In this study, we calculated the percentages of species with inversions and sequences with rps19 insertions in different families and genera to evaluate the universality of the two characteristics. Generally, sequences from 57 families and 111 genera contained inversions, accounting for 38.3% and 22.3% of all 149 families and 498 genera, respectively (Tables S5 and S6). On the other hand, sequences from 41 families and 135 genera had rps19 insertions, comprising 27.5% and 27.1% of the total of 149 families and 498 genera, respectively (Tables S7 and S8). The percentages of species with inversions in 14 of the 57 families exceeded 30% (Table S5), and the percentages of sequences with rps19 insertions were higher than 50% in 24 of the 41 families containing rps19 insertions and 100% in 14 (Table S7). Among the 111 genera with inversions, the percentages of species with inversions were more than 30% for 58 genera and 100% for 5 (Table S6). In the 135 genera with rps19 insertions, the percentages of sequences with rps19 insertions exceeded 50% in 108 genera and 100% in 97 (Table S8). We included steps to identify the inversions and rps19 insertions as well as perform the necessary corrections (such as the reorientation of the inversions and removal of the rps19 insertions) in our data processing pipeline. All data reported hereafter were subjected to these corrections.
Sequence Length of trnH–psbA
The sequence length of a marker affects the efficiency of PCR amplification. Consequently, we examined the length distribution of the trnH–psbA sequences. As shown in Figure S1, the lengths of trnH–psbA sequences in eudicotyledons, monocotyledons, gymnosperms, ferns, and mosses ranged from 152 bp to 851 bp, from 151 bp to 905 bp, from 283 bp to 1006 bp, from 167 bp to 547 bp, and from 103 bp to 265 bp, respectively. The corresponding average lengths obtained were 357, 357, 573, 429, and 141 bp, respectively.
Genetic Divergences of trnH–psbA
For ferns, the maximal intraspecific distance of trnH–psbA was generally smaller than the corresponding smallest interspecific divergence between congeneric species (Table 1). For the other four groups, the intraspecific variation and interspecific distance did not show clear separation (Table S9). trnH–psbA demonstrated a clear DNA barcoding gap only in ferns (Figure S2).
Table 1. Identification success rates of trnH–psbA in the five major plant taxonomic groups using BLAST and BLAST+P distance.
| Taxa | No. of families | No. of genera | No. of species | No. of samples | BLAST | BLAST+P distance |
| Success (%) | Success (%) | |||||
| Eudicotyledons | 92 | 332 | 2437 | 13727 | 51.1 | 64.5 |
| Monocotyledons | 32 | 126 | 782 | 3054 | 45.7 | 54.7 |
| Gymnosperms | 7 | 12 | 126 | 633 | 35.5 | 37.0 |
| Mosses | 9 | 14 | 52 | 277 | 72.2 | 78.3 |
| Ferns | 9 | 14 | 98 | 292 | 75.0 | 75.3 |
Ability of trnH–psbA to Identify Species in Various Taxonomic Groups
The BLAST and BLAST+P distance methods were used to evaluate the performance of trnH–psbA in species identification. Using BLAST, the trnH–psbA region correctly identified 51.1%, 45.7%, 35.5%, 72.2%, and 75.0% of 13 727 eudicotyledon, 3054 monocotyledon, 633 gymnosperm, 277 moss, and 292 fern sequences at the species level, respectively (Table 1). The success rates of trnH–psbA identification in the five groups were higher using the BLAST+P distance method than using BLAST. When the BLAST+P distance method was used, the corresponding species identification rates of trnH–psbA for these groups were 64.5%, 54.7%, 37.0%, 78.3%, and 75.3%, respectively (Table 1).
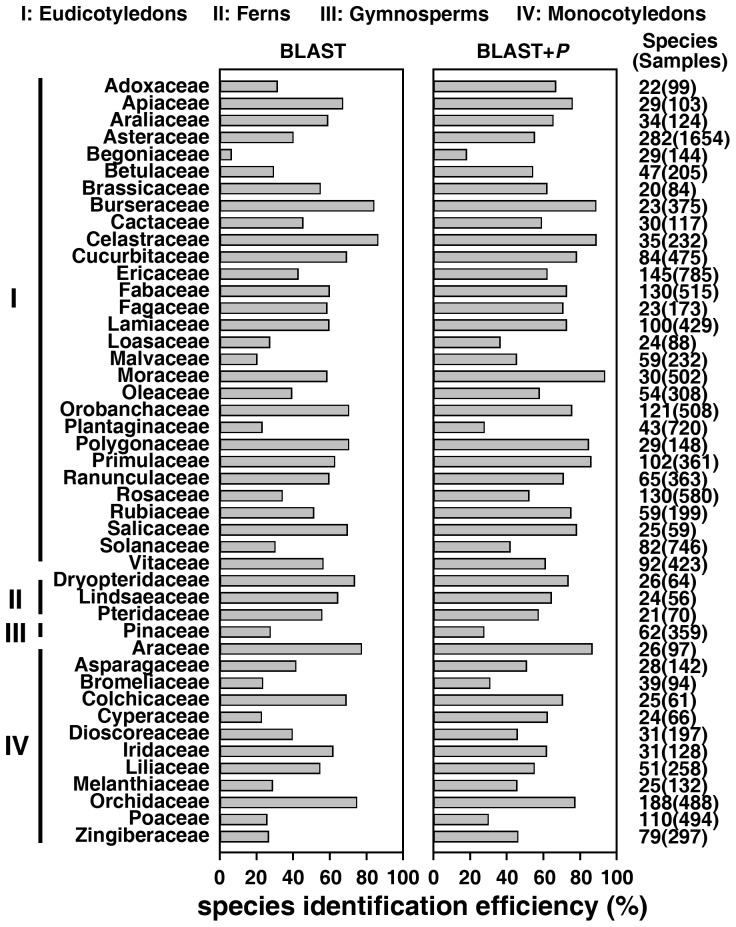
We also analyzed the species identification ability of trnH–psbA in different plant families. Among the 45 families with at least 20 species, the rates of successful identification of 7 families were higher than 70% using BLAST (Figure 2). The success rate of using trnH–psbA in discriminating members of family Begoniaceae was the lowest at 6.3%. On the other hand, the success rate of using trnH–psbA to differentiate among family Celastraceae was the highest at 86.2% (Figure 2). Using the BLAST+P distance method, identification success rates exceeded 70% in 18 families (Figure 2). The species identification success rate for Begoniaceae was the lowest at 18.1%, whereas the rate for Moraceae was the highest at 93.4% (Figure 2). The success rates of using trnH–psbA to identify families with fewer than 20 species are listed in Table S10.
Figure 2. Success rates of trnH–psbA in discriminating closely related species in families with at least 20 species using BLAST (left) and BLAST+P distance (right).
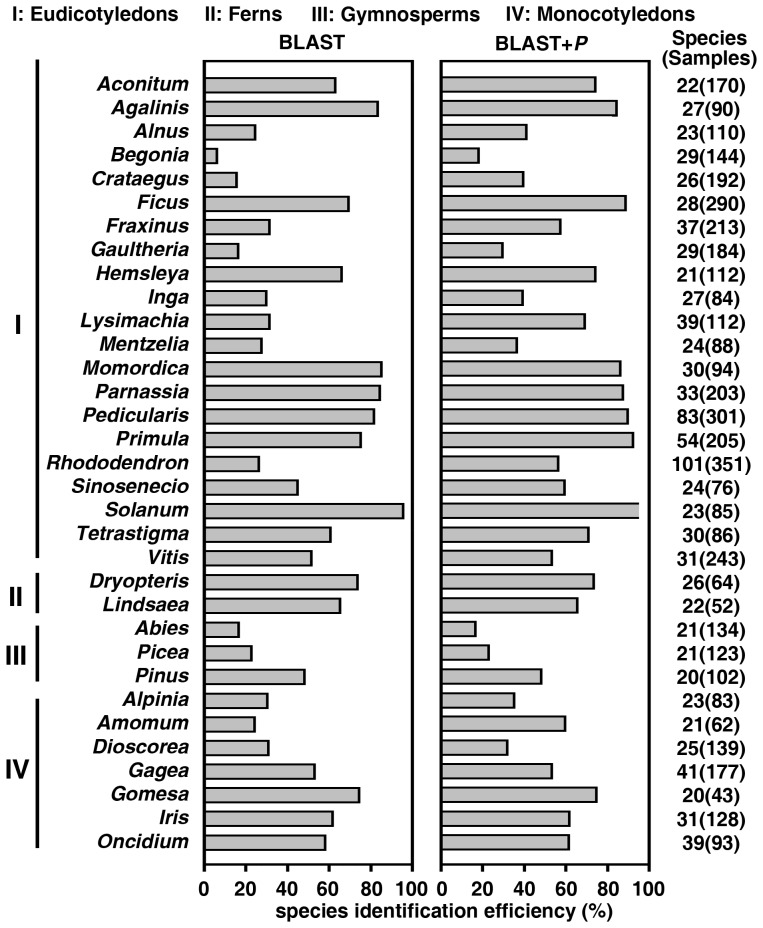
We also analyzed the species identification ability of trnH–psbA in different genera. Among the 33 genera with at least 20 species, identification success rates exceeded 70% for 8 genera using BLAST (Figure 3). When the BLAST+P distance method was used, the success rates of trnH–psbA identification were higher than 70% in 12 genera. The success rates for discriminating among the Abies, Begonia, and Picea genera were the lowest at 16.4%, 18.1%, and 22.8%, respectively (Figure 3). The success rates for the identification of Ficus, Pedicularis, Primula, and Solanum species were the highest at 88.6%, 89.7%, 92.2%, and 96.5%, respectively (Figure 3). For 12 species-rich, taxonomically complex genera (Vitis, Gagea, Rhododendron, Fraxinus, Oncidium, Iris, Lysimachia, Tetrastigma, Momordica, Parnassia, Pedicularis, and Primula), with 86–351 samples from 30–101 species in this study, trnH–psbA exhibited successful identification rates within the range of 53.1%–92.2% (Figure 3). The success rates for the identification of genera with fewer than 20 species are presented in Table S11.
Figure 3. Identification success rates of trnH–psbA in genera with at least 20 species using BLAST (left) and BLAST+P distance (right).
To determine easily the family- or genus-specific discriminatory power of trnH–psbA, we added a Check Performance module in the trnH–psbA Web server (http://psba-trnh-plantidit.dnsalias.org). Potential users of DNA barcoding technology can check the discriminatory power of trnH–psbA in various taxonomic groups and determine whether trnH–psbA works well for a given group.
Relative Performance of trnH–psbA and Its Combinations in Species Identification Compared with Other Suggested Single Markers and the Core Barcode matK+rbcL
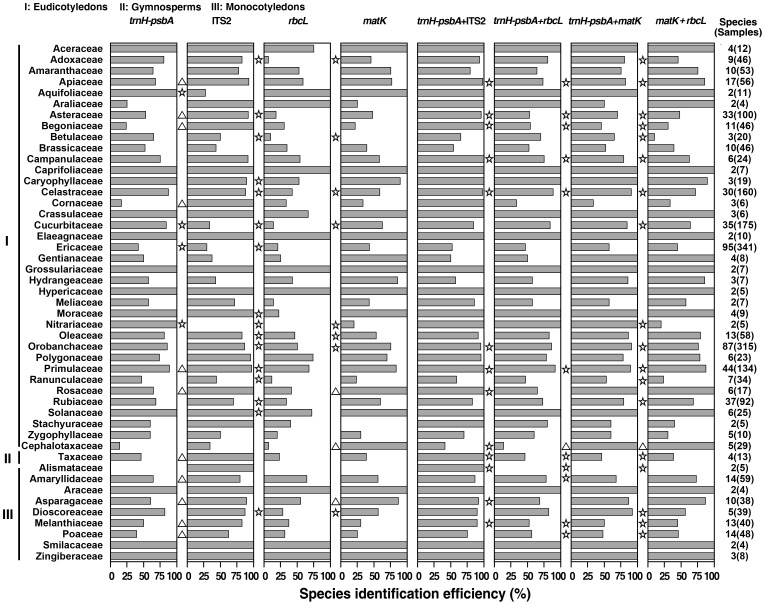
To further evaluate the discriminatory ability of the trnH–psbA region, we compared its identification rates with those of the other three most recommended markers (matK, rbcL, and ITS2) based on the matching data set using the BLAST+P distance method. For the 47 families tested, trnH–psbA exhibited the highest identification efficiency among the four DNA barcodes in 19 families (Figure 4). Using the Fisher's exact test, we found that trnH–psbA had a significantly higher discrimination rate than the other three DNA barcodes in Cucurbitaceae and Nitrariaceae (Figure 4). Detailed information on the identification success rate of single markers at the family level can be found in Table S12. Among the 71 genera tested, the discrimination rates of ITS2, trnH–psbA, matK, and rbcL were higher than 80% in 45, 34, 30, and 19 genera, respectively (Figure 5). The trnH–psbA region had the highest identification rate among the four DNA barcodes in 36 genera (Figure 5). Fisher's exact test demonstrated that the identification success rates of trnH–psbA were significantly higher than those of the other three DNA barcodes in Nitraria, Thladiantha, Hemsleya, and Rhododendron (Figure 5). ITS2 and trnH–psbA showed superior identification ability in Primula (97.0% with ITS2, 88.8% with trnH–psbA), Pedicularis (87.9% with ITS2, 89.6% with trnH–psbA), and Parnassia (88.1% with ITS2, 87.5% with trnH–psbA), which had at least 30 species each. No significant difference was found between the identification rates of the two markers in Pedicularis and Parnassia (Figure 5). The four DNA barcodes showed low discrimination rates in Rhododendron (24.2% with ITS2, 47.4% with trnH–psbA, 20.4% with rbcL, 36.0% with matK) (Figure 5). Detailed information on the identification success rate of single markers at the genus level can be found in Table S13.
Figure 4. Comparison of identification success rates for markers in different families and the corresponding statistical test results.
The statistical tests were carried out between trnH–psbA and the other three single markers, or trnH–psbA+ITS2 and the other three marker combinations, respectively. The significant differences are indicated to the left of the column for the corresponding marker or marker combination. “☆” indicates that the identification success rates for trnH–psbA or trnH–psbA+ITS2 are significantly higher than those of the corresponding marker or marker combination. “▵” indicates that the identification success rates for trnH–psbA or trnH–psbA+ITS2 are significantly lower than those of the corresponding marker or marker combination.
Figure 5. Comparison of the identification success rates for markers in different genera and the corresponding statistical test results.
The statistical tests were carried out between trnH–psbA and the other three single markers, or trnH–psbA+ITS2 and three other marker combinations, respectively. The significant differences are indicated to the left of the column for the corresponding marker or marker combination. “☆” indicates that the identification success rates for trnH–psbA or trnH–psbA+ITS2 are significantly higher than those of the corresponding marker or marker combination. “▵” indicates that the identification success rates for trnH–psbA or trnH–psbA+ITS2 are significantly lower than those of the corresponding marker or marker combination.
In addition to comparing the performance of individual markers, we also combined trnH–psbA with the above-described three loci and compared the various combinations with matK+rbcL. The trnH–psbA+ITS2 combination showed the highest discrimination rate among the four two-locus combinations in 41 of the 47 families (Figure 4). Fisher's exact test showed that trnH–psbA+ITS2 had significantly higher identification efficiency than the other three combinations in nine families (Begoniaceae, Apiaceae, Asteraceae, Celastraceae, Primulaceae, Campanulaceae, Alismataceae, Taxaceae, and Melanthiaceae) (Figure 4). The trnH–psbA+ITS2 combination had significantly higher identification efficiency than matK+rbcL in 18 families, whereas matK+rbcL had significantly higher identification efficiency than trnH–psbA+ITS2 in one family (Figure 4). Detailed information on the identification success rate of two-locus combinations at the family level can be found in Table S14. Among the 71 genera tested, trnH–psbA+ITS2 provided the highest discrimination rate among the four combinations for 61 genera (Figure 5). Fisher's exact test showed that the discrimination rates of trnH–psbA+ITS2 were significantly higher than those of the other three combinations in nine genera (Begonia, Parnassia, Ligularia, Alisma, Cyananthus, Amentotaxus, Primula, Paris, and Sinosenecio) (Figure 5). In particular, trnH–psbA+ITS2 had significantly higher identification efficiency than matK+rbcL in 21 genera, whereas the discrimination success rates of matK+rbcL were significantly higher than those of trnH–psbA+ITS2 for one genus (Figure 5). trnH–psbA+ITS2 performed well in discriminating among the species-rich genera Primula, Parnassia, Pedicularis, and Rhododendron, for which the discrimination success rates were 100.0%, 98.1%, 97.0%, and 52.6%, respectively. The identification rates of trnH–psbA+matK, trnH–psbA+rbcL, and matK+rbcL were as follows: 89.6%, 91.8%, and 87.3% for Primula; 90.6%, 88.8%, and 71.9% for Parnassia; 94.0%, 90.3%, and 77.9% for Pedicularis; and 57.3%, 50.7%, and 36.5% for Rhododendron, respectively. Detailed information on the identification success rate of two-locus combinations at the genus level can be found in Table S15.
Discussion
trnH–psbA is widely accepted as a supplementary DNA barcode. A number of studies have also examined the relative performance of trnH–psbA in a taxonomic setting [28], [29]. However, these studies have focused on particular taxon settings using different data, different analytical methods, and inconsistent parameters to evaluate the performance of this spacer region. Consequently, the results of studies on different taxa, even on the same taxon, are difficult to compare and generalize. A systematic evaluation is needed to estimate uniformly the efficacy of using trnH–psbA in species determination across various groups. The present study aimed to perform such comprehensive evaluation in a systematic and standard way.
For the full data set constructed in this study, the trnH–psbA region demonstrated different discrimination abilities in various plant taxonomic groups. For example, under the BLAST+P distance method, the identification success rates for 13 727 eudicotyledons, 3054 monocotyledons, 633 gymnosperms, 277 mosses, and 292 ferns were 64.5%, 54.7%, 37.0%, 78.3%, and 75.3% at the species level, respectively. Among the 45 families with at least 20 species tested, trnH–psbA provided a high discrimination rate (>70%) in 18 families, with Moraceae being the highest at 93.4%. Among 33 genera with at least 20 species, the success rates of trnH–psbA identification were higher than 70% in 12 genera, with Solanum being the highest at 96.5%. For the matching data set, trnH–psbA had a significantly higher identification rate than the other three suggested loci in Cucurbitaceae, Nitrariaceae, Nitraria, Thladiantha, Hemsleya, and Rhododendron. To the best of our knowledge, this study is by far the most comprehensive and systematic analysis of trnH–psbA as a DNA barcode. The results guide the optimal use of trnH–psbA for species identification.
Three problems that may generate spurious results are inherent in the current study design. First, the sequences from GenBank/EMBL may not be assessed following the standards of barcoding. Our solution was to subject the sequence data to a very rigorous preparation process. We initially downloaded all sequences from GenBank. Some species had only one representative sequence, and these sequences may have contained more errors. Intraspecific variation also cannot be calculated for these species; similarly, some genera have only one species. Thus, congeneric, interspecific distances cannot be calculated for these genera. Consequently, sequences for single-sequence species or single-species genera were filtered, and only those belonging to genera with at least two species and species with at least two sequences were retained.
Second, the trnH–psbA intergenic spacer is highly problematic [30]–[32] because of very frequent intrapopulation inversions that can lead to an overestimation of species diversity when directly correlating sequence information to taxonomic entities. A fragment of rps19 was frequently found as an insertion in some groups, which also led to an overestimation of the species diversity. Our study also systematically evaluated the prevalence of intraspecific inversions and rps19 insertions in trnH–psbA sequences. Among the total of 149 families and 498 genera tested, 57 families and 111 genera contained species with inversions and 41 families and 135 genera included sequences with rps19 insertions.
To solve this problem, we developed a pipeline that can perform the following steps: (1) identify and mask out the rps19 fragment, and (2) identify intraspecific inversions, select a reference orientation, and reverse-complement inversions in the wrong orientations. These steps led to an improved alignment of trnH–psbA sequences and will be described in detail in a separate paper. We also tested novel methods based on hybrid local alignment, global alignment, and alignment-free methods to compare the query sequence and the database, which significantly improved the discriminatory power of trnH–psbA (http://psba-trnh-plantidit.dnsalias.org).
Lastly, this study was designed as a meta-analysis. Accordingly, we adopted a heterogeneous sampling strategy instead of other strategies in a particular geographical (all species in an area) or taxon (all species in a genus or family) setting. To ensure the reliability of the results, we carried out a stringent data preprocessing step. Such a systematic evaluation enabled this study to provide information on the prevalence of intraspecific inversions and rps19 insertions, and offers a species identification performance metric for trnH–psbA in various taxon settings. Such information cannot be obtained using sequences from particular geographical or taxonomic settings.
The essence of DNA barcoding technology is the use of a universal barcode. However, in reality, “regardless of the region(s) ultimately adopted for plant barcoding, there will always be some species that would be better resolved by some other region” [12]. For example, research has shown that trnH–psbA exhibits better discrimination ability than matK+rbcL in angiosperm genera, such as Ficus [33] and Alnus [34]. The matK+rbcL barcode is even more problematic for non-angiosperms, particularly ferns, because of the difficulty in generating full-length matK sequences [35], [36] or the insufficient discriminatory capacity of rbcL [37]–[39]. In the present study, we compared the combinations of trnH–psbA with three other popular markers (ITS2, rbcL, and matK) to the core DNA barcode matK+rbcL using the matching data set. The trnH–psbA+ITS2 combination performed the best among all four two-locus combinations in the majority of taxa examined at the family or genus level. Among the 47 families and 71 genera tested, trnH–psbA+ITS2 had significantly higher identification efficiency than matK+rbcL in 18 families and 21 genera, wherease the discrimination success rates of matK+rbcL were significantly higher than those of trnH–psbA+ITS2 only for one family and one genus. These findings indicated the need to describe supplementary barcodes to complete the core barcode matK+rbcL. This study showed that the trnH–psbA+ITS2 combination may even be a more effective plant DNA barcode than the core barcode.
Supporting Information
List of sequences containing rps19 .
(PDF)
List of species with inversions in their sequences.
(PDF)
List of trnH – psbA samples used in this study. The names of the corresponding group, family, genus, and species, as well as the GenBank accession number for each sample are shown.
(PDF)
List of trnH – psbA , ITS2, rbcL , and matK samples with the same voucher numbers. The names of the corresponding group, family, genus, and species, the voucher number, as well as the GenBank accession number for each sample are shown.
(PDF)
Percentages of species with inversions in different families.
(PDF)
Percentages of species with inversions in different genera.
(PDF)
Percentages of sequences with rps19 insertions in different families.
(PDF)
Percentages of sequences with rps19 insertions in different genera.
(PDF)
Intra- and interspecific distances of congeneric species in the five major plant taxonomic groups.
(PDF)
Identification success rates of trnH – psbA using BLAST and BLAST+P distance in selected families with fewer than 20 species.
(PDF)
Identification success rates of trnH – psbA using BLAST and BLAST+P distance in genera with fewer than 20 species.
(PDF)
Detailed information on the identification success rate of single markers at the family level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of single markers at the genus level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of two-locus combinations at the family level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of two-locus combinations at the genus level and the corresponding statistical test results.
(PDF)
Box plots of the lengths of trnH – psbA sequences in the five major plant taxonomic groups. In each box plot, the box shows the interquartile range of the data, which is defined as the difference between the 75th and 25th percentiles. The continuous and dotted lines across the box represent the median and average values, respectively.
(TIF)
Inter- and intraspecific divergences of trnH – psbA sequences in the five major plant taxonomic groups. Sequence divergence across all species for which sequences of multiple individuals are presented is illustrated. Divergence is shown as a scatter plot between the maximal intraspecific and minimal interspecific distances. A black line is drawn where the two distances are equal.
(TIF)
(DOC)
(DOC)
Funding Statement
This work was supported by a start fund from the Chinese Academy of Medical Science (No. 431118) granted to C. Liu, Basic Scientific Research Operation Grants for State-Level Public Welfare Scientific Research Initiatives (No. YZ-12-08 granted to X. Pang and No. YZ-12-04 granted to C. Liu), Beijing Municipal Science and Technology Foundation (No. D08080203640901) granted to S. Chen, Research grant for returned Overseas Chinese Scholars, Ministry of Human Resources (No. 431207) granted to C. Liu, and Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (No. IRT1150). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Heinrich M (2007) The identification of medicinal plants. J Ethnopharmacol 111: 440. [Google Scholar]
- 2. Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitchell A (2008) DNA barcoding demystified. Aust J Entomol 47: 169–173. [Google Scholar]
- 4. Evans KM, Wortley AH, Mann DG (2007) An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158: 349–364. [DOI] [PubMed] [Google Scholar]
- 5. Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci USA 103: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA (2007) DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 23: 167–172. [DOI] [PubMed] [Google Scholar]
- 7. Hogg ID, Hebert PDN (2004) Biological identification of springtails (Hexapoda: Collembola) from the Canadian Arctic, using mitochondrial DNA barcodes. Can J Zool 82: 749–754. [Google Scholar]
- 8. Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R (2005) Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Phil Trans R Soc B 360: 1805–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia's fish species. Phil Trans R Soc B 360: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, et al. (2005) Land plants and DNA barcodes: short-term and longterm goals. Phil Trans R Soc B 360: 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56: 295–299. [Google Scholar]
- 12. Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, et al. (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3: e2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2: e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kress WJ, Erickson DL (2008) DNA barcodes: genes, genomics, and bioinformatics. Proc Natl Acad Sci USA 105: 2761–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar S, Hahn FM, McMahan CM, Cornish K, Whalen MC (2009) Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol 9: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105: 2923–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newmaster SG, Fazekas AJ, Ragupathy S (2006) DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot 84: 335–341. [Google Scholar]
- 19. Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008) Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour 8: 480–490. [DOI] [PubMed] [Google Scholar]
- 20. Pennisi E (2007) Taxonomy. Wanted: a barcode for plants. Science 318: 190. [DOI] [PubMed] [Google Scholar]
- 21. CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6: e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. China Plant BOL Group (2011) Comparative analysis of a large data set indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108: 19641–19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen SL, Yao H, Han JP, Liu C, Song JY, et al. (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5: e8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meier R, Zhang GY, Ali F (2008) The use of mean instead of smallest interspecific distances exaggerates the size of the “Barcoding gap” and leads to misidentification. Syst Biol 57: 809–813. [DOI] [PubMed] [Google Scholar]
- 26. Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3: 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ross HA, Murugan S, Li WLS (2008) Testing the reliability of genetic methods of species identification via simulation. Syst Biol 57: 216–230. [DOI] [PubMed] [Google Scholar]
- 28. Seberg O, Petersen G (2009) How many loci does it take to DNA barcode a crocus? PLoS ONE 4: e4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korotkova N, Borsch T, Quandt D, Taylor NP, Müller KF, et al. (2011) What does it take to resolve relationships and to identify species with molecular markers? An example from the epiphytic Rhipsalideae (Cactaceae). Am J Bot 98: 1549–1572. [DOI] [PubMed] [Google Scholar]
- 30. Štorchová H, Olson MS (2007) The architecture of the chloroplast psbA-trnH non-coding region in angiosperms. Plant Syst Evol 268: 235–256. [Google Scholar]
- 31. Borsch T, Quandt D (2009) Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Syst Evol 282: 169–199. [Google Scholar]
- 32. Whitlock BA, Hale AM, Groff PA (2010) Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS ONE 5: e11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, et al. (2010) Universal plant DNA barcode loci may not work in complex groups: A case study with Indian Berberis species. PloS ONE 5: e13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ren BQ, Xiang XG, Chen ZD (2010) Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Mol Ecol Resour 10: 594–605. [DOI] [PubMed] [Google Scholar]
- 35. de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, et al. (2011) Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PloS ONE 6: e16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuo LY, Li FW, Chiou WL, Wang CN (2011) First insights into fern matK phylogeny. Mol Phylogenet Evol 59: 556–566. [DOI] [PubMed] [Google Scholar]
- 37. Jansen T, Schneider H (2005) Exploring the evolution of humus collecting leaves in drynarioid ferns (Polypodiaceae, Polypodiidae) based on phylogenetic evidence. Plant Syst Evol 252: 175–197. [Google Scholar]
- 38. Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, et al. (2004) Phylogeny and evolution of ferns (Monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot 91: 1582–1598. [DOI] [PubMed] [Google Scholar]
- 39. Schneider H, Russell SJ, Cox CJ, Bakker F, Henderson S, et al. (2004) Chloroplast phylogeny of asplenioid ferns based on rbcL and trnL-F spacer sequences (Polypodiidae, Aspleniaceae) and its implications for biogeography. Syst Bot 29: 260–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of sequences containing rps19 .
(PDF)
List of species with inversions in their sequences.
(PDF)
List of trnH – psbA samples used in this study. The names of the corresponding group, family, genus, and species, as well as the GenBank accession number for each sample are shown.
(PDF)
List of trnH – psbA , ITS2, rbcL , and matK samples with the same voucher numbers. The names of the corresponding group, family, genus, and species, the voucher number, as well as the GenBank accession number for each sample are shown.
(PDF)
Percentages of species with inversions in different families.
(PDF)
Percentages of species with inversions in different genera.
(PDF)
Percentages of sequences with rps19 insertions in different families.
(PDF)
Percentages of sequences with rps19 insertions in different genera.
(PDF)
Intra- and interspecific distances of congeneric species in the five major plant taxonomic groups.
(PDF)
Identification success rates of trnH – psbA using BLAST and BLAST+P distance in selected families with fewer than 20 species.
(PDF)
Identification success rates of trnH – psbA using BLAST and BLAST+P distance in genera with fewer than 20 species.
(PDF)
Detailed information on the identification success rate of single markers at the family level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of single markers at the genus level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of two-locus combinations at the family level and the corresponding statistical test results.
(PDF)
Detailed information on the identification success rate of two-locus combinations at the genus level and the corresponding statistical test results.
(PDF)
Box plots of the lengths of trnH – psbA sequences in the five major plant taxonomic groups. In each box plot, the box shows the interquartile range of the data, which is defined as the difference between the 75th and 25th percentiles. The continuous and dotted lines across the box represent the median and average values, respectively.
(TIF)
Inter- and intraspecific divergences of trnH – psbA sequences in the five major plant taxonomic groups. Sequence divergence across all species for which sequences of multiple individuals are presented is illustrated. Divergence is shown as a scatter plot between the maximal intraspecific and minimal interspecific distances. A black line is drawn where the two distances are equal.
(TIF)
(DOC)
(DOC)