Abstract
We describe a solid phase microextraction (SPME), multistep elution, transient isotachophoresis (tITP) CE-MS/MS procedure which employs a high sensitivity porous ESI sprayer for the proteomic analysis of a moderately complex protein mixture. In order to improve comprehensiveness and sensitivity over a previously reported proteomic application of the ESI sprayer, we evaluated preconcentration with SPME and multistep elution prior to tITP stacking and CE separation. To maximize separation efficiency we primarily employed electrokinetic methods for elution and separation after loading the sample by application of pressure. Conditions were developed for optimum simultaneous electrokinetic elution and sample stacking using a tryptic digest of 16 proteins to maximize peptide identifications and minimize band broadening. We performed comparative proteomic analysis of a dilution series using CE and nanoflow liquid chromatography (nLC). We found complementary peptide and protein identifications with larger quantities (100 ng) of a Pyrococcus furiosus tryptic digest, but with mass-limited amounts (5 ng) CE was 3 times more effective at identifying proteins. We attribute these gains in sensitivity to lower noise levels with the porous CE sprayer, illustrated by better signal-to-noise ratios of peptide precursor ions and associated higher XCorr values of identified peptides when compared directly to nLC. From comparative analysis of SPME-tITP-CE with direct injection CE, the SPME-tITP process improved comprehensiveness and sensitivity.
Keywords: solid phase microextraction, capillary electrophoresis, sheathless electrospray ionization, peptides, proteomics
Proteomic methods are defined by a series of analytical technologies, typically sample clean-up and preparation, liquid chromatography, electrospray ionization, and tandem mass spectrometry.1 For complex samples, multidimensional liquid chromatography with electrospray ionization has become an essential step within this series of technologies for maximizing proteomic coverage.2, 3 In particular, nanoflow liquid chromatography (nLC) has provided a suitable balance of robust multidimensional separation capacity and electrospray ionization (ESI) efficiency. Efforts to minimize the flow rate of nLC separations have illustrated the gains in sensitivity for proteomic analyses.4-7 Although nLC separations at low nL/min flow rates are achievable, the flow rate generated from electroosmotic flow in capillary electrophoresis (CE) is inherently well matched for this high sensitivity ESI regime.
The development of various electrospray interfaces for CE-ESI has facilitated numerous successful applications in proteomics.8, 9 Electrospray potential is commonly applied using a sheath liquid format. Recently, two low-flow sheath liquid sprayers have been designed independently by the Chen and Dovichi Labs to minimize band broadening and dilution of analytes.10-12 The Dovichi sprayer was shown to be as sensitive as UPLC-MS/MS for peptide analysis, also providing complementary peptide identifications. Other efforts have focused on development of sheathless CE-ESI interfaces which completely eliminate the sheath liquid to maximize electrospray sensitivity. A subset of these interfaces use pulled capillaries coated with various conductive materials.13-17 These sprayers are highly functional yet generally suffer from clogging of the tip or evaporation and flaking of the coating, minimizing their lifetime. Another major subset is the porous junction interface which applies the electrospray voltage through a conductive region of the capillary. They generally require the use of hydrofluoric acid to etch a region of the capillary until porous. Thus, the fabrication and usage of sheathless CE-ESI setups with porous junctions has been primarily confined to labs with this specific expertise.18-21 The recent optimizations and evaluations of a prototype sheathless CE platform for peptide analysis illustrated the potential for widespread usage.22-25 The demonstrations by the Mayboroda Lab utilized transient isotachophoresis (tITP) to address the mass-limited injections associated with capillary electrophoresis. In the evaluation by the Lindner Lab, CE-MS/MS was 10-fold more sensitive than nLC-MS/MS for analysis of histone H1, even without the use of tITP. Despite the proven operation and gain in sensitivity, these demonstrations only allowed for the analysis of a small aliquot volumes (~5 – 250 nL) from simple proteomic samples. Most proteomic preparations result in volumes at least 1000-fold greater than the single injection possible with tITP-CE-MS/MS.26 Additionally, for analysis of more complex protein mixtures an orthogonal microfluidic separation dimension is necessary to improve comprehensiveness. Solid-phase microextraction (SPME) has been previously shown to allow for both preconcentration of peptides and sequential elution of peptide fractions for improved proteomic coverage.27-31 When used in conjunction with tITP, further sensitivity gains have been shown.32, 33 In the work described here, a SPME cartridge was employed to both concentrate samples and perform sequential elution to improve comprehensiveness prior to tITP-CE-MS/MS using the high sensitivity porous sprayer. We optimized critical SPME elution conditions and evaluated the sensitivity of the sheathless SPME-tITP-CE-MS/MS platform for analysis of a moderately complex proteomic mixture by comparison to both conventional nLC-MS/MS results over the same analysis time and CE without SPME-tITP using direct sample injections.
METHODS AND MATERIALS
Reagents and Chemicals
Unless otherwise stated, chemicals were purchased from Thermo Fisher Scientific (Waltham, MA). Deionized water (18.2 MΩ, Barnstead, Dubuque, IA), hydrochloric acid (Mallinckrodt Chemicals), ammonium acetate (Spectrum Chemical MFG Corp), ammonium hydroxide (J.T. Baker), methanol (J.T. Baker), acetonitrile (J.T. Baker), and isopropanol (J.T. Baker) were used for preparations.
Protein Digestion
A 16 protein mixture (Beta-galactosidase (E.Coli), Glycogen phosphorylase, muscle form (Rabbit), Serum albumin precursor (Bovine), Catalase(Bovine), Ovalbumin (Chicken), Serotransferrin precursor (Bovine), Alpha-actin 1 (Bovine), Alcohol dehydrogenase I (yeast), Glyceraldehyde-3-phosphate dehydrogenase (Rabbit), Carbonic anhydrase II (Bovine), Alpha-S2 casein precursor (Bovine), Beta casein precursor (Bovine), Myoglobin (Horse), Alpha-lactalbumin precursor (Bovine), Lysozyme C precursor (Chicken)) and a complex protein standard from Pyrococcus furiosus (Pfu, Cat # 400510, Agilent Technologies, Santa Clara, CA) were digested by first denaturing and reducing in 8 M urea, 100 mM Tris(hydroxyethylamine) pH 8.5, and 5 mM tris(2-carboxyethyl)phosphine for 30 min. Cysteine residues were acetylated with 10 mM iodoacetamide for 15 min in the dark. The sample was diluted to 2 M urea with 100 mM Tris(hydroxyethylamine) pH 8.5. Trypsin was added at a 1:50 protease:protein ratio for an overnight digestion at 37°C. Peptide samples were then acidified to 5% formic acid and spun at 18,000 × g.
SPME-CE-MS/MS
Prototype fused-silica CE capillaries (90 cm length, 30 μm ID, 150 μm OD) with a 3 cm porous tip for electrospray were obtained from Beckman Coulter, Inc (Brea, CA). The CE capillary was positively coated using polyethylenimine (PEI) as described in U.S. Patent 6923895. A C8 SPME column was prepared by creating a Kasil frit at one end of an 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc, Phoenix, AZ), then packed with 1 mm reverse phase particles (Zorbax C8, 5 μm dia., 300Å pores, Agilent Technologies, Santa Clara, CA). The Kasil frit was prepared by polymerizing a mixture of 300 μL Kasil 1624 (PQ Corporation, Malvern, PA) and 100 μL formamide at 100°C for 4 hrs. The Kasil frit was cut to 1 mm. The other end of the C8 trap column was then cut to 3 cm long and connected to the 30 μm ID inlet capillary using a microfilter union (M-520, IDEX Health & Science LLC, Lake Forest, IL). The Kasil frit end was then connected to the CE capillary by a zero dead volume union (P-720, IDEX Health & Science LLC). A schematic of the SPME-CE-MS/MS configuration is shown in Supporting Figure 1.
CE separations were performed on a PA 800 plus Pharmaceutical Analysis System (Beckman Coulter, Inc.). The CE capillary with a 3 cm long porous segment at one end was inserted into a sprayer interface and enabled the electric contact through a conductive liquid delivered by a 2nd capillary. 95mM HOAc - 5% MeOH (pH 3.1) was used as the background electrolyte (BGE) buffer as well as the conductive liquid. The peptide samples were first loaded onto the SPME column using 75 psi pressure, then flushed with BGE for 10 min at 75 psi. The elution buffer was injected either hydrodynamically or electrokinetically. The separation was then conducted by applying 30 kV to the injection end and 1.5 kV at the electrospray tip. All CE-MS/MS experiments were conducted on a LTQ XL instrument (Thermo Fisher Scientific). The MS method was set up as data dependent acquisition with 1 full MS followed by 5 MS/MS scans. MS scan was set from 400-1800 m/z. Data-dependent acquisition of MS/MS spectra were performed with the following settings: inlet capillary temperature 200 °C, full scan automatic gain control target 30K ions, MSn scan automatic gain control target 10K ions, ESI voltage 1.5 kV, maximum injection time 10 ms for full MS and 100 ms for MS2, dynamic exclusion time 60 sec, and 5 MS/MS per MS on the most intense precursor ions.
Direct CE-MS/MS analysis
CE–MS/MS with direct injection was conducted on PEI coated prototype fused silica capillary (90 cm length, 30 μm ID/150 μm OD). 95 mM HOAc - 5% MeOH (pH 3.1) was used as BGE. The peptide samples were injected at 5 psi for 30 sec followed by an injection of a small BGE plug (5 psi for 5 sec), then subjected to CE separation at 30 kV with 5 psi vacuum. The MS acquisition was set up the same as for SPME-tITP-CE-MS/MS analysis.
nLC-MS/MS analysis
Capillary columns were prepared in-house for nLC-MS/MS analysis from particle slurries in methanol. An analytical RPLC column was generated by pulling a 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc, Phoenix, AZ) to 5 μm ID tip. Reverse phase particles (Aqua C18, 5 μm dia., 125 Å pores, Phenomenex, Torrance, CA) were packed directly into the pulled column at 800 psi until 15 cm long. The column was further packed, washed, and equilibrated at 100 bar with LC buffer A. nLC-MS/MS analysis was performed on an Agilent 1100 HPLC pump and Thermo LTQ XL using an in-house built electrospray stage. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 μm ID, P-775-01, IDEX Health &Sciences LLC) directly downstream of a 1:1000 split flow used to reduce the flow rate to 350 nL/min through the analytical column. 3 hr gradients were from 100 % buffer A to 15% buffer B in 10 min, then to 50% buffer B over 140min, up to 100% B over 15 min, held at 100% B for 10 min, then back to 100% A for a 10 min column re-equilibration. Buffer A was 5% acetonitrile 0.1% formic acid, B was 80% acetonitrile 0.1% formic acid. The MS acquisition was set up the same as CE-MS/MS except 2.5 kV was used for the ESI voltage.
Database Searching
Tandem mass spectra were extracted from raw files using RawExtract 1.9.9 and processed with Integrated Proteomics Pipeline (IP2, http://www.integratedproteomics.com/). Spectra were searched using ProLuCID34 against either a yeast database with the 16 standard proteins added or a Pfu database, both with their reverse sequences appended. The search space included all fully- and semi-tryptic peptide candidates with a minimum sequence length of six amino acid residues. Carbamidomethylation of cysteine was considered as a static modification. DTASelect was used to filter peptide candidates to a 0.5% false discovery rate based on the number of reverse sequences identified. In-house developed scripts were used to extract precursor intensities, estimate noise, and calculate the S/N in the full MS scan for each identified precursor ion.35 The background noise was defined as the average intensity of the lowest 10% of peaks from the defined precursor MS scan and S/N was calculated by dividing the peptide precursor intensities by the background noise.
RESULTS AND DISCUSSION
One of the major advantages of CE over LC is the remarkable separation efficiencies from the absence of pressure-driven flow. Previous demonstrations of SPME-CE and similar methodologies, however, indicated band broadening and loss of separation efficiency from preconcentration and elution may be a significant aspect to consider.33, 36, 37 Thus many of our experiments focused on minimizing the band broadening processes associated with the addition of SPME while maximizing peptide and protein identifications.
Effective capillary length with SPME elutions
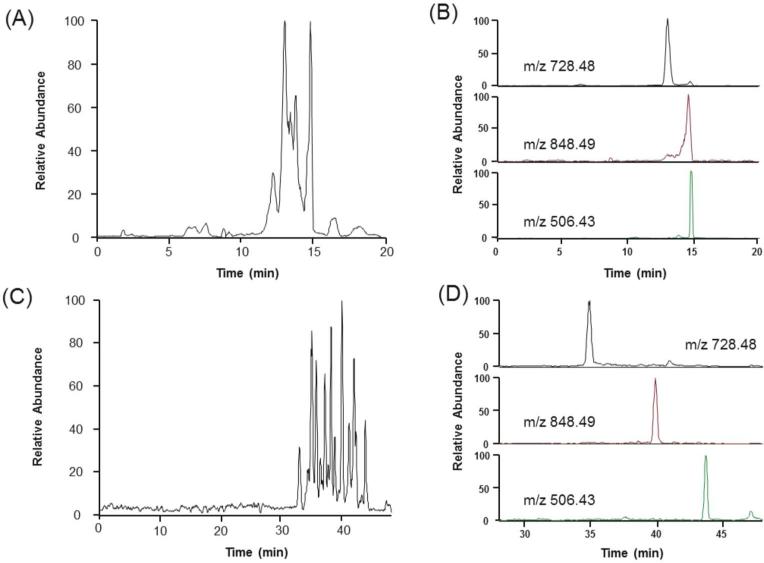
We first tested whether we could achieve comparable separation efficiencies using SPME with a single elution to those recently published with tITP. The initial separations we performed by eluting peptides from the SPME cartridge had poor separation resolution (Figure 1A). This phenomenon was expected since the migration of peptides have been slowed previously by lowering the separation voltage30 or applying a vacuum.22 However, a reduction in separation voltage or application of a small vacuum did not remedy this inefficiency (data not shown). When we extracted ion chromatograms for a few representative peptides (Figure 1B) we noticed the limitation was poor separation efficiency and not premature ESI. The main changes to our setup in comparison to the vacuum-assisted setup by Busnel et al. were capillaries and connections for insertion of the SPME cartridge (Supporting Figure 1), the lack of vacuum, and the introduction of an organic elution plug. The organic elution plug and SPME cartridge decrease the effective length of the capillary and the electric field over the remaining section of the capillary.36, 38 This problem was likely accentuated by the reduction in capillary inner diameter from the common 50 μm to 30 μm. From the inner diameter reduction, the capillary volume was reduced almost 3-fold, also reducing potential elution volumes 3-fold. Similar problems have been encountered with membrane preconcentration prior to CE-MS with small ID capillaries.39 Longer capillaries have been implicated in ameliorating this problem by increasing the effective length of the capillary for large elution volumes.36 To test this for our small ID (30 μm) and volume (~600 nL) capillary, we doubled the capillary length. Indeed, this modification improved the resolution (Figure 1C) and efficiency (Figure 1D) of peptide peaks. To ensure the effects were from the relative decrease in elution volume and not a reduction in EOF velocity from decreasing the electric field, we performed separations with the shorter capillary with similar electric fields (Supporting Figure 2). The distribution, resolution, and bandwidth of the peptide peaks were more similar between the same capillary lengths (90 cm) than the same electric fields (~160 V/cm). These results indicate that the reduction in the relative elution plug length was indeed the major cause of the separation improvement and not reduction in EOF.
Figure 1.
Effect of capillary length on SPME-CE-MS/MS separation. The electropherograms of one step elution and CE separation of 16 protein mixture tryptic digests using (A) 90 cm and (C) 190 cm. The extracted ion electropherograms of 3 representative ions are shown from both (B) 90 cm and (D) 190 cm capillary lengths. Run conditions: 95 mM HOAc - 5% MeOH (pH 3.1) was used as the background electrolyte. Peptides were eluted with 100 nL 95% MeOH - 50 mM NH4OAc, pH 4.5. 30 kV was applied at the inlet of both 90 cm and 190 cm capillaries. MS/MS acquisition began upon application of separation voltage.
Peptide recovery from SPME
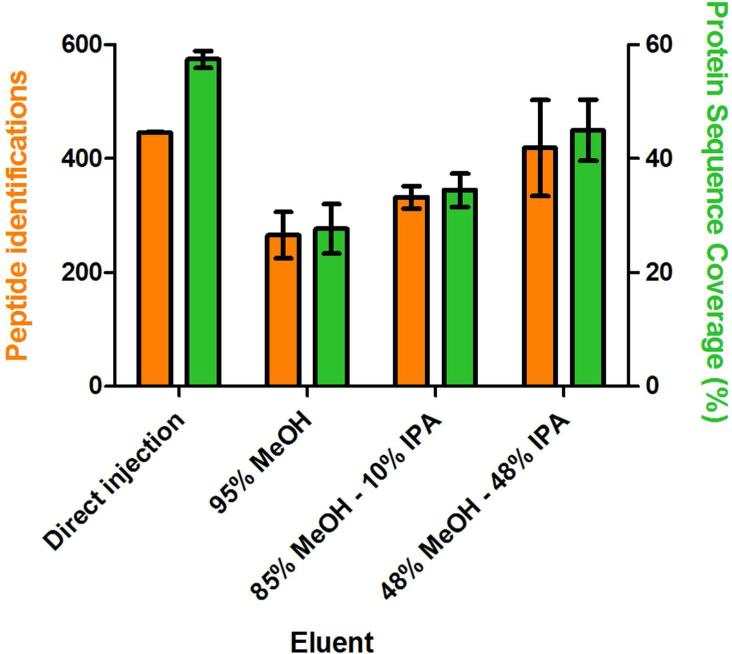
With an appropriate balance between BGE and elution plug volume within the smaller capillary, we sought to maximize the recovery of peptides from SPME using different elution conditions. We had also found the required elution volumes for efficient elution of peptides from a 1 mm SPME cartridge containing C18 resin were suboptimum even with the capillary volume doubled (data not shown). To remedy this problem, we instead employed a 1 mm SPME cartridge using C8 resin which both adequately captured peptides and allowed for reasonable elution volumes. We also tested different elution solvents to maximize peptide identifications. We initially tested methanol alone to elute the peptides but got very limited peptide identifications and low protein sequence coverages. Isopropanol is a more nonpolar modifier and has been used to elute more hydrophobic peptides.40, 41 Thus, we investigated different combinations of methanol and isopropanol. These results are shown in Figure 2. The 16 protein mixture tryptic digests were first loaded onto the SPME column and hydrodynamically eluted with 100 nL of the three different elution buffers (95% MeOH, 85% MeOH - 10% IPA, and 48% MeOH - 48% IPA. Peptide identifications and protein sequence coverages were used to assess the effectiveness of the elution in comparison to sample directly injected without the use of SPME. We found that an elution buffer with equal methanol and isopropanol compositions maximized recovery based on peptide identifications, having 93% of the peptide identifications from direct injection. Using only isopropanol gave similar peptide identifications compared to methanol alone, but band broadening was higher due to a higher viscosity and back pressure (data not shown).
Figure 2.
Effect of elution buffer on the elution efficiency, peptide identifications, and protein sequence coverages. The 16 protein mixture tryptic digests were eluted hydrodynamically with 100 nL elution volumes of 3 different buffers (95% MeOH, 85% MeOH - 10% isopropanol (IPA) and 48% MeOH - 48% IPA), followed by CE-MS/MS. An equal amount of protein was directly injected onto the capillary without SPME to simulate an elution efficiency of 100% for comparison. Peptide identifications and the average protein sequence coverage from triplicate analyses are plotted. 82×74mm (300 × 300 DPI)
Electrokinetic elution optimization with SPME
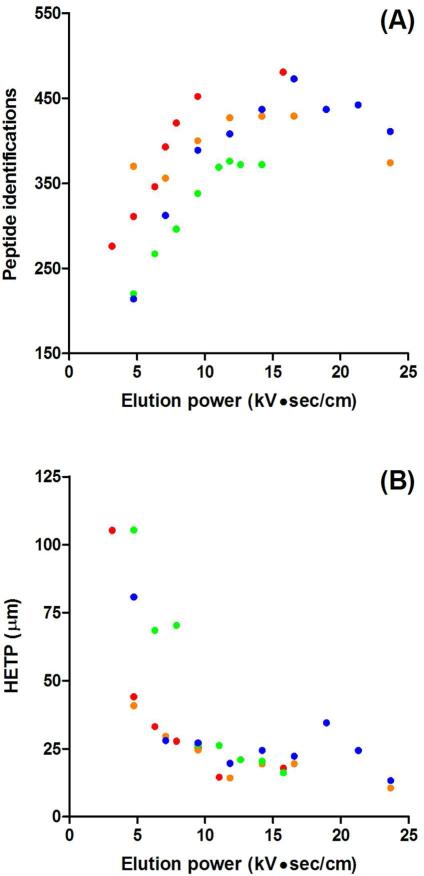
The effect of elution volume on peptide separation efficiency has been previously characterized for SPME-tITP-CE with UV detection and pressure-driven elution.37 In addition to monitoring the effects of elution volume with MS, we also used electrokinetic elutions to further minimize band broadening from pressure-driven flow. Electrokinetic elutions use EOF to drive the elution solvent through the SPME cartridge and can even allow for staggered, overlapping elutions to reduce analysis time.31 We tested different volumes using electrokinetic elutions to maximize peptide identifications. The results of this consideration are in Figure 3A where we plotted the number of peptide identifications as a function of the elution power, the product of the electric field and the elution time (Et). The elution power is proportional to the elution volume and was plotted as such for potential generalization to other SPME-CE configurations where different capillary coatings or solvents would change EOF velocity and thus plug length. As expected, as the elution volume was increased we generally identified more peptides. With increased elution volume, more peptide mass was eluted, resulting in higher peptide identifications. However, we did observe a plateauing of the identified peptides at median elution powers and even a decrease at higher powers. Possible explanations for a plateau in peptide identifications are saturation of our MS detector signal or sampling speed. Thus an elution power between 10 and 15 kV●sec/cm appeared well matched to the mass spectrometer employed. When peptide identifications were compared based on elution buffer ionic strength, a set of elutions performed at 10 kV with 50 mM (green points) and 95 mM (red points) acetic acid showed a significant difference. However, the same comparison at 15 kV – 50 mM (blue points) and 100 mM (orange points) acetic acid – nearly overlapped each other. A higher ionic strength of the BGE would be expected to suppress EOF. Thus, both apparent systematic and random error within the experiments made it difficult to conclude if the ionic strength changed EOF enough to affect peptide identifications. We also considered the bandwidth of peptides, represented as the height equivalent of theoretical plates (HETP) in Figure 3B. We found under most elution conditions, we were near optimum separation efficiency. The 10 – 15 kV●sec/cm range which maximized peptide identifications also had the best separation efficiency. In fact considering all elution powers, an inverse trend between peptide identifications and separation efficiency based on elution volume was found. This would indicate that optimizing elution volumes from the SPME cartridge was important to maximizing peptide identifications. Injection volume has been shown to affect peak width under different FESS conditions42 and may be the source of reduced efficiency in our study. With these experiments we were able to find a range of optimum elution electric fields and times to maximize peptide identifications and minimize band broadening. An understanding of the two key parameters for optimization, electric field and elution time, will make future optimizations more straightforward with other solvent systems and capillary coatings. We continued to use the optimized elution conditions,10 kV●sec/cm from 2.5 min – 15 kV elutions and 95 mM acetic acid BGE, for further analyses.
Figure 3.
Parameter optimization for electrokinetic elution. (A) Peptide identifications based on elution power (electrokinetic elution electric field × elution time). The 16 protein mixture tryptic digests were electrokinetically eluted with 50 mM NH4OAc - 48% MeOH - 48% IPA at 10 kV (red and green dots) or 15 kV (orange and blue) for increasing times (1 – 5 min in 0.5 min increments) followed by CE-MS/MS. Separations were performed with both 50 mM (blue and green) and 95 mM (red and orange) HOAc - 5% MeOH background electrolyte. (B) HETP plotted based on elution power. The average HETP was calculated using variance and migration time from the same three representative peptides (481.01, 636.61, and 729.71 m/z) among all separations with different elution powers. 82×152mm (300 × 300 DPI)
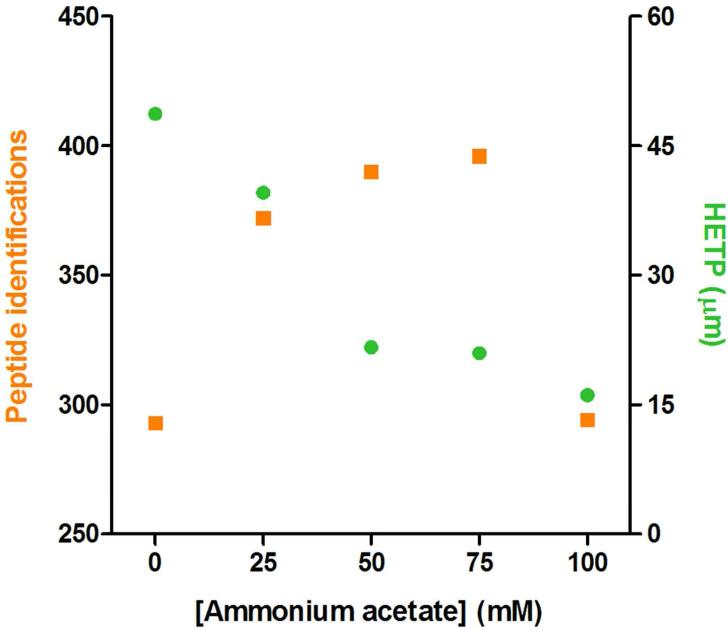
Using the elution solvents and electrokinetic elutions which maximized peptide recovery and identifications while minimizing band broadening, we further tested elution buffers to optimize tITP during the elution process. For its compatibility with ESI-MS, we also employed ammonium acetate as the leading electrolyte (LE) as in the Busnel et al study.22 Since we used organic elution instead of direct injection, the tITP process is slightly more complicated. Briefly, with the application of high voltage, the LE ions migrate faster than the peptide ions, allowing for the peptide ions to stack behind the LE with constant velocity based on their mobilities. This process may take place in both the elution plug and the separation buffer, but a pseudo-tITP effect can also be obtained through the application of organic solvent.47 Thus, the peptides are primarily stacked by the elution plug, then mobility-ordered between the ammonium acetate LE plug and the elution plug, acting as a trailing electrolyte (TE). The LE zone is gradually disrupted as higher mobility protons migrate through the zone and the tITP process is stopped. The peptides then begin to separate in the normal CZE mode based on differential electrophoretic mobility. To test the effects of tITP, we varied the concentration of ammonium acetate in the elution buffer and monitored the resulting peptide identifications and bandwidth (Figure 4). We found that increasing concentrations of ammonium ions (50 mM and 75 mM) indeed improved peptide identifications by ~33% over analysis without ammonium acetate and tITP. The improved identifications correlated with peptide stacking, reducing the average HETP from 49 μm to 21 μm. We found that the best separation efficiency did not give the maximum peptide identifications and attribute this to an appropriate match between mass spectrometer sampling speed and peptide peak width. In this study, a Thermo LTQ instrument was used which has an acquisition rate of 3-5 MS/MS per second. The use of a newer instrument with the capabilities to perform ~20 MS/MS per second43, 44 may yield higher identifications at higher ammonium acetate concentrations. Thus we employed 50 mM ammonium acetate in the elution buffer for subsequent experiments which maximized peptide identifications with our setup.
Figure 4.
tITP effects on SPME-CE separation efficiency and peptide identifications. The 16 protein mixture tryptic digests were electrokinetically eluted at 15 kV for 2.5 min with 48% MeOH - 48% IPA and the addition of various concentration of ammonium acetate (0 – 100 in 25 mM increments) pH 4.5 followed by CE-MS/MS. Peptide identifications are plotted on the left axis and the average HETP for 3 representative peptides on the right axis were calculated as in Figure 3. 82×72mm (300 × 300 DPI)
Protein identification sensitivity comparison to nLC-MS/MS
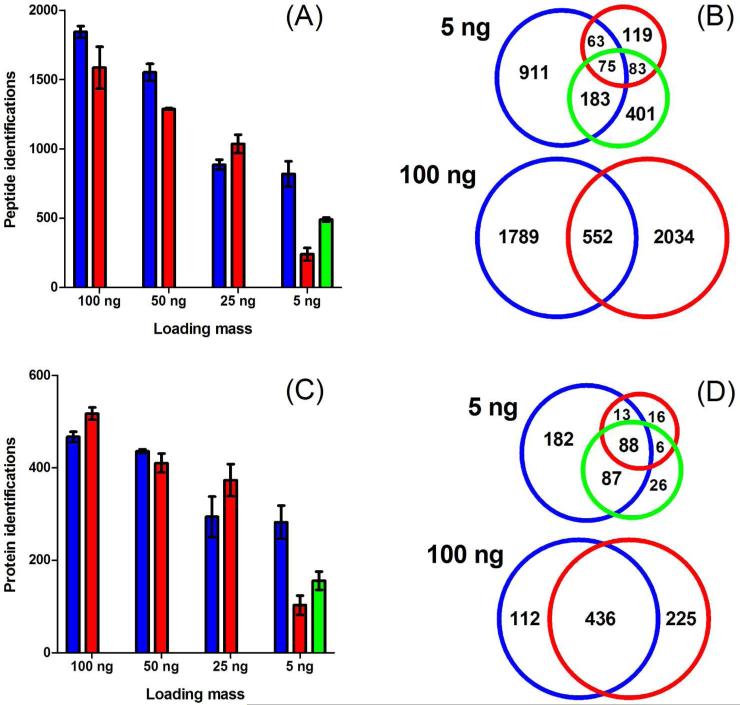
With optimized elution conditions from analysis of protein standard digests with SPME-tITP-CE-MS/MS, we next conducted multistep peptide elution and analysis on Pfu tryptic digests. Pfu has a moderately complex proteome in comparison to yeast or mammalian cells with 2,125 predicted proteins. Of the predicted proteins, 900 have been identified previously by shotgun proteomics.45 Thus, the Pfu proteome was suitably complex for comparison of CE and nLC methods. We compared the protein identification sensitivity for a dilution series of Pfu tryptic digests with results obtained by nLC-MS/MS over the same analysis time. To perform multistep SPME-tITP-CE-MS/MS, the Pfu tryptic digests were first hydrodynamically loaded onto the SPME column. The first step elution buffer (10%-10% MeOH-IPA - 50 mM NH4OAc, pH 4.5) was then electrokinetically driven at 15 kV for 2.5 min and the eluted peptides were subjected to a 35 min tITP-CE separation. The remaining multistep elutions (20%-20%, 30%-30%, 40%-40%, and 48%-48% MeOH-IPA – 50 mM NH4OAc, pH 4.5) and separations were repeated consecutively in the same fashion until the last organic elution and separation were complete. For nLC-MS/MS analysis, the tryptic digests were pressure loaded onto the reverse phase C18 analytical column followed by 3 hr gradient LC-MS/MS analysis. A representative electropherogram and chromatogram are shown in Supporting Figures 3 and 4, respectively. The results of the comparison are shown in Figure 5. Generally, both methods had comparable identifications and reproducibility for peptides (Figure 5A) and proteins (Figure 5C), particularly from 25 – 100 ng of Pfu tryptic digest starting mass. However, the sensitivity of SPME-tITP-CE-MS/MS was dramatically better than nLC-MS/MS when only 5 ng of material was analyzed. Notably, from a Venn comparison of the 5 ng starting mass data, 86% of the proteins identified by nLC were also identified by CE (Figure 5D). We attribute these identification sensitivity gains to the combination of greater signal–to-noise (S/N) ratio possible with the low-flow sheathless porous sprayer,22, 24, 25 the improved separation efficiency with tITP,22, 25 and the improved separation capacity with multistep elution SPME.30, 31 This concept is illustrated and discussed further in the next section. We also analyzed 5 ng of Pfu digest using a direct injection with CE-MS/MS. Using direct CE injection without SPME and tITP, twice as many proteins and peptides could be identified in comparison to nLC in 20% of the analysis time. These results further validate previous CE analysis with the high sensitivity porous sprayer which did not perform MS/MS22 or were performed on less complex samples.24 The addition of SPME-tITP doubled these identification results. Peptides were more adequately fractionated by hydrophobicity with SPME multistep elution into consecutive CE separations, further increasing peptide and protein identifications. Thus, comparison of peptide and protein identification results of 5 ng Pfu digest among direct injection CE, SPME-tITP-CE, and nLC begins to validate the comprehensiveness gains from SPME multistep elution and the sensitivity gains with porous CE sprayer. Further comparison is also described in the next section.
Figure 5.
Peptide (A) and protein (C) identification sensitivity comparison of 5-step SPME-CE-MS/MS (blue), direct injection CE-MS/MS (green), and nLC-MS/MS (red) of Pfu tryptic digests run in duplicate. Venn comparisons of identified peptides (B) and proteins (D) from combined duplicate runs of 5 ng and 100 ng Pfu digests using SPME-tITP-CE and nLC. 209×204mm (300 × 300 DPI)
Venn comparison of the identified peptides and proteins at 100 ng starting mass showed that the SPME-tITP-CE and nLC methods are complementary when not sensitivity-limited (Figures 5B and 5D, 100 ng comparisons). A comparison of peptides identified from all starting masses analyzed illustrates better peptide complementarity with the two methods than at the protein level (Supporting Figure 5). Similar peptide identification complementary was found in the recent CE and nLC comparison study by the Dovichi Lab.46 They found that CE biased towards identification of basic, hydrophilic peptides of low molecular mass. Additionally, the Faserl study found that CE identified smaller peptides that were unretained by C18 and thus unidentified by nLC.24 We found the same trends through our comparison of CE and nLC analysis of a more complex mixture of peptides (Supporting Figure 6). These trends can also be primarily explained by the difference in electrospray conditions between methods. That is, nLC has an increasing organic composition of the solvent throughout the run known to effect ESI response of different peptide hydrophobicities,47 while CE ESI conditions are essentially constant.
Proteomic metrics in comparison to nLC-MS/MS
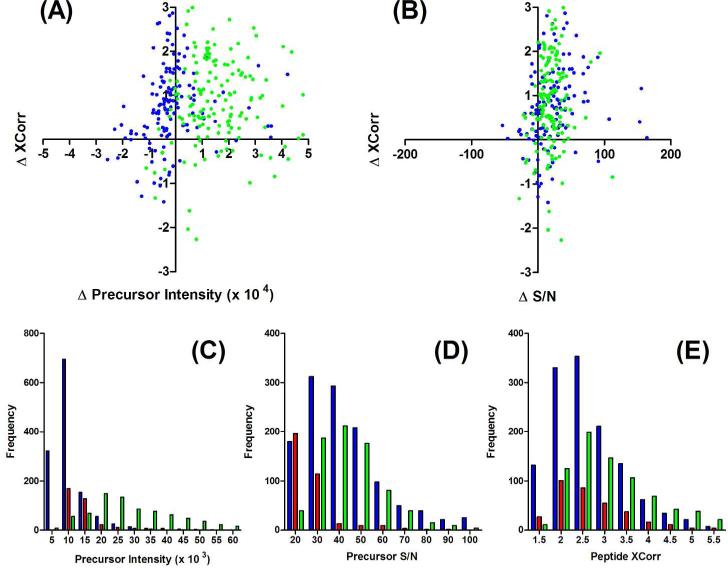
To better understand the sensitivity gains achieved over nLC-MS/MS using CE-MS/MS with and without SPME-tITP, we compared proteomic metrics between these three methods. Two important metrics that typically define proteomic performance are peptide precursor intensity and peptide XCorr values.48 Higher peptide precursor intensities often lead to better MS/MS sampling and higher XCorr values. We directly compared the difference in precursor intensity and XCorr values for peptides identified by nLC-MS/MS and CE-MS/MS with and without SPME-tITP when 5 ng Pfu lysate were analyzed (Figure 6A). Notably, nearly all peptides identified by either of the CE-based methods had higher XCorr values. Thus, there is generally higher confidence in the peptides identified by CE when mass-limited. Surprisingly, the precursor intensities for CE with SPME-tITP were lower than without the SPME and tITP concentration steps. This may be due to differences in flow rate which were difficult to match with and without a SPME cartridge that adds backpressure to the system. However, when we instead compared S/N ratios, both CE-based methods were better than nLC and had similar trends (Figure 6B). The higher S/N can thus be at least partially attributed to the lower flow rate associated with CE and the porous sprayer. These trends were also observed with the SPME-tITP-CE and nLC comparison performed with 100 ng Pfu lysate (Supporting Figures 7A & B). Additionally, since SPME-tITP had similar S/N to direct injection, but lower precursor intensities, it must also have lower noise levels from the sample cleanup and concentration steps than a direct injection. Indeed the average noise levels for SPME-tITP (204 ± 111) were lower than direct CE analysis (715 ± 376). Somewhat surprisingly the direct CE analysis noise was higher than nLC (518 ± 120) and could be from the lack of a solid phase within the setup, common to the SPME-tITP-CE and nLC setups, which facilitates sample cleanup. The Faserl et al. study illustrated the remarkable sensitivity and S/N for angiotensin in base peak electropherograms, extracted ion electropherograms, and MS/MS spectra.24 Our results clearly illustrate the benefits of the high sensitivity sprayer on a mixture of peptides where matrix and sample complexity can adversely affect S/N ratios.
Figure 6.
Comparison of peptide precursor intensity, signal-to-noise ratio (S/N), and XCorr from duplicate analysis of 5 ng of Pfu tryptic digests using 5-step SPME-tITP-CE-MS/MS (blue), direct injection CE-MS/MS (green), and nLC-MS/MS (red). Correlation plots of changes (CE values – nLC values) in (A) precursor intensity and (B) S/N to changes in XCorr values of peptides identified in both CE and nLC experiments. Distributions of peptide (C) precursor intensities, (D) S/N ratios and (E) XCorr values were plotted for SPME-tITP-CE (blue), direct CE (green), and nLC (red) experiments. Noise was calculated by averaging the intensities of the lowest 10% peaks from the precursor MS scan. S/N ratios were calculated by dividing peptide precursor intensities by the noise within each scan. 209×165mm (300 × 300 DPI)
When we compared the distributions of all peptides identified by both CE and nLC methods, the improved sensitivity from CE was further illustrated. Similar to trends with common peptides identified between methods, direct CE analysis had the highest precursor intensities (Figure 6C), but S/N were again comparable with SPME-tITP and both were considerably better than nLC (Figure 6D). The SPME-tITP method had higher frequencies over all S/N ranges between the two CE methods. These results would indicate that the SPME fractionation improved S/N for all peptide precursor intensities. The gains from these S/N improvements with CE again appeared to improve identifications over nLC through higher XCorr values (Figure 6E). Comparing SPME-tITP with direct CE XCorr values, the SPME-tITP method identified more peptides with lower XCorr (< 4). Thus, both the S/N and XCorr distribution results would indicate that the multistep elutions with SPME-tITP increased the separation and ion trap space to improve dynamic range. Peptides with lower S/N could then be sampled and identified with SPME-tITP that weren't possible with a direct CE injection. Comparison of peptide metrics between SPME-tITP-CE and nLC analysis when not mass-limited at 100 μg shows similar performance (Supporting Figure 7). Thus, SPME-tITP-CE-MS/MS exploits the high S/N possible with CE analysis using the high sensitivity porous sprayer with the comprehensiveness through multistep elution SPME-tITP.
CONCLUSIONS
We have demonstrated the feasibility and sensitivity gains achieved through coupling solid-phase microextraction and transient isotachophoresis to CE-MS/MS with a 30 μm ID porous sprayer capillary. Improvements to prefractionation using SPME should improve comprehensiveness and sensitivity further. In light of this and other recent CE-MS studies with new electrospray interfaces and mass spectrometers, CE is reemerging as a viable and sensitive method for proteomic analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Hans A. Dewald, Thomas E. Horton, Jean-Marc Busnel, Jeff D. Chapman, Chitra Ratnayake, and Lena Sripitisawad from Beckman Coulter, Inc. for providing the sheathless porous sprayer capillaries and electrospray interface. The authors would like to acknowledge funding to J.R.Y from the National Institute of Health grants R01DK074798, U19AI063603, RFP-NHLBI-HV-10-5, and P41RR011823.
Footnotes
Supporting Information Available: This material is available free of charge at http://pubs.acs.org.
Raw data files can be found at http://fields.scripps.edu/published/SPME-CE2012/.
REFERENCES
- 1.Ning Z, Zhou H, Wang F, Abu-Farha M, Figeys D. Analytical aspects of proteomics: 2009-2010. Anal Chem. 2011;83(12):4407–26. doi: 10.1021/ac200857t. [DOI] [PubMed] [Google Scholar]
- 2.Mohammed S, Heck A., Jr. Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr Opin Biotechnol. 2011;22(1):9–16. doi: 10.1016/j.copbio.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Motoyama A, Yates JR., 3rd Multidimensional LC separations in shotgun proteomics. Anal Chem. 2008;80(19):7187–93. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74(16):4235–49. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 5.Marginean I, Kelly RT, Prior DC, LaMarche BL, Tang K, Smith RD. Analytical characterization of the electrospray ion source in the nanoflow regime. Anal Chem. 2008;80(17):6573–9. doi: 10.1021/ac800683s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly RT, Page JS, Tang K, Smith RD. Array of chemically etched fused-silica emitters for improving the sensitivity and quantitation of electrospray ionization mass spectrometry. Anal Chem. 2007;79(11):4192–8. doi: 10.1021/ac062417e. [DOI] [PubMed] [Google Scholar]
- 7.Luo Q, Tang K, Yang F, Elias A, Shen Y, Moore RJ, Zhao R, Hixson KK, Rossie SS, Smith RD. More sensitive and quantitative proteomic measurements using very low flow rate porous silica monolithic LC columns with electrospray ionization-mass spectrometry. J Proteome Res. 2006;5(5):1091–7. doi: 10.1021/pr050424y. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell EJ, Chen DD. Twenty years of interface development for capillary electrophoresis-electrospray ionization-mass spectrometry. Anal Chim Acta. 2008;627(1):25–33. doi: 10.1016/j.aca.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 9.Fonslow BR, Yates JR., 3rd Capillary electrophoresis applied to proteomic analysis. J Sep Sci. 2009;32(8):1175–88. doi: 10.1002/jssc.200800592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxwell EJ, Zhong X, Chen DD. Asymmetrical emitter geometries for increased range of stable electrospray flow rates. Anal Chem. 2010;82(20):8377–81. doi: 10.1021/ac1017953. [DOI] [PubMed] [Google Scholar]
- 11.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun Mass Spectrom. 2010;24(17):2554–60. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 12.Zhong X, Maxwell EJ, Chen DD. Mass transport in a micro flow-through vial of a junction-at-the-tip capillary electrophoresis-mass spectrometry interface. Anal Chem. 2011;83(12):4916–23. doi: 10.1021/ac200636y. [DOI] [PubMed] [Google Scholar]
- 13.Bendahl L, Hansen SH, Olsen J. A new sheathless electrospray interface for coupling of capillary electrophoresis to ion-trap mass spectrometry. Rapid Commun Mass Spectrom. 2002;16(24):2333–40. doi: 10.1002/rcm.865. [DOI] [PubMed] [Google Scholar]
- 14.Chang YZ, Her GR. Sheathless capillary electrophoresis/electrospray mass spectrometry using a carbon-coated fused-silica capillary. Anal Chem. 2000;72(3):626–30. doi: 10.1021/ac990535e. [DOI] [PubMed] [Google Scholar]
- 15.Chen YR, Her GR. A simple method for fabrication of silver-coated sheathless electrospray emitters. Rapid Commun Mass Spectrom. 2003;17(5):437–41. doi: 10.1002/rcm.940. [DOI] [PubMed] [Google Scholar]
- 16.Ramsey RS, Mcluckey SA. Capillary Electrophoresis Electrospray-Ionization Ion-Trap Mass-Spectrometry Using a Sheathless Interface. Journal of Microcolumn Separations. 1995;7(5):461–469. [Google Scholar]
- 17.Zamfir AD, Dinca N, Sisu E, Peter-Katalinic J. Copper-coated microsprayer interface for on-line sheathless capillary electrophoresis electrospray mass spectrometry of carbohydrates. J Sep Sci. 2006;29(3):414–22. doi: 10.1002/jssc.200500374. [DOI] [PubMed] [Google Scholar]
- 18.Fanali S, D'Orazio G, Foret F, Kleparnik K, Aturki Z. On-line CE-MS using pressurized liquid junction nanoflow electrospray interface and surface-coated capillaries. Electrophoresis. 2006;27(23):4666–73. doi: 10.1002/elps.200600322. [DOI] [PubMed] [Google Scholar]
- 19.Moini M. Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal Chem. 2007;79(11):4241–6. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 20.Moini M, Huang H. Application of capillary electrophoresis/ electrospray ionization-mass spectrometry to subcellular proteomics of Escherichia coli ribosomal proteins. Electrophoresis. 2004;25(13):1981–7. doi: 10.1002/elps.200305906. [DOI] [PubMed] [Google Scholar]
- 21.Whitt JT, Moini M. Capillary electrophoresis to mass spectrometry interface using a porous junction. Anal Chem. 2003;75(9):2188–91. doi: 10.1021/ac026380j. [DOI] [PubMed] [Google Scholar]
- 22.Busnel JM, Schoenmaker B, Ramautar R, Carrasco-Pancorbo A, Ratnayake C, Feitelson JS, Chapman JD, Deelder AM, Mayboroda OA. High capacity capillary electrophoresis-electrospray ionization mass spectrometry: coupling a porous sheathless interface with transient-isotachophoresis. Anal Chem. 2010;82(22):9476–83. doi: 10.1021/ac102159d. [DOI] [PubMed] [Google Scholar]
- 23.Ramautar R, Busnel JM, Deelder AM, Mayboroda OA. Enhancing the coverage of the urinary metabolome by sheathless capillary electrophoresis-mass spectrometry. Anal Chem. 2011 doi: 10.1021/ac202407v. [DOI] [PubMed] [Google Scholar]
- 24.Faserl K, Sarg B, Kremser L, Lindner H. Optimization and evaluation of a sheathless capillary electrophoresis-electrospray ionization mass spectrometry platform for peptide analysis: comparison to liquid chromatography-electrospray ionization mass spectrometry. Anal Chem. 2011;83(19):7297–305. doi: 10.1021/ac2010372. [DOI] [PubMed] [Google Scholar]
- 25.Heemskerk AA, Busnel JM, Schoenmaker B, Derks RJ, Klychnikov O, Hensbergen PJ, Deelder AM, Mayboroda OA. Ultra-low flow electrospray ionization-mass spectrometry for improved ionization efficiency in phosphoproteomics. Anal Chem. 2012;84(10):4552–9. doi: 10.1021/ac300641x. [DOI] [PubMed] [Google Scholar]
- 26.Fonslow BR, Yates JR., 3rd . Proteolytic digestion approaches for shotgun proteomics. In: Pawliszyn J, editor. Comprehensive Sampling and Sample Preparation: Analytical Techniques for Scientists. Elsevier Inc; USA: 2012. [a]-[b] [Google Scholar]
- 27.Figeys D, Corthals GL, Gallis B, Goodlett DR, Ducret A, Corson MA, Aebersold R. Data-dependent modulation of solid-phase extraction capillary electrophoresis for the analysis of complex peptide and phosphopeptide mixtures by tandem mass spectrometry: application to endothelial nitric oxide synthase. Anal Chem. 1999;71(13):2279–87. doi: 10.1021/ac9813991. [DOI] [PubMed] [Google Scholar]
- 28.Figeys D, Ducret A, Aebersold R. Identification of proteins by capillary electrophoresis-tandem mass spectrometry. Evaluation of an on-line solid-phase extraction device. J Chromatogr A. 1997;763(162):295–306. doi: 10.1016/s0021-9673(96)00847-3. [DOI] [PubMed] [Google Scholar]
- 29.Figeys D, Ducret A, Yates JR, 3rd, Aebersold R. Protein identification by solid phase microextraction-capillary zone electrophoresis-microelectrospray-tandem mass spectrometry. Nat Biotechnol. 1996;14(11):1579–83. doi: 10.1038/nbt1196-1579. [DOI] [PubMed] [Google Scholar]
- 30.Tong W, Link A, Eng JK, Yates JR., 3rd Identification of proteins in complexes by solid-phase microextraction/multistep elution/capillary electrophoresis/tandem mass spectrometry. Anal Chem. 1999;71(13):2270–8. doi: 10.1021/ac9901182. [DOI] [PubMed] [Google Scholar]
- 31.Lee WH, Wang CW, Her GR. Staggered multistep elution solid-phase extraction capillary electrophoresis/tandem mass spectrometry: a high-throughput approach in protein analysis. Rapid Commun Mass Spectrom. 2011;25(15):2124–30. doi: 10.1002/rcm.5091. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Tomlinson AJ, Naylor S. Peer Reviewed: Membrane Preconcentration CE. Anal Chem. 1999;71(5):183A–9A. doi: 10.1021/ac990246q. [DOI] [PubMed] [Google Scholar]
- 33.Medina-Casanellas S, Benavente F, Barbosa J, Sanz-Nebot V. Transient isotachophoresis in on-line solid phase extraction capillary electrophoresis time-of-flight-mass spectrometry for peptide analysis in human plasma. Electrophoresis. 2011;32(13):1750–9. doi: 10.1002/elps.201100017. [DOI] [PubMed] [Google Scholar]
- 34.Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR., 3rd ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5(10):S174. [Google Scholar]
- 35.Wong CC, Cociorva D, Venable JD, Xu T, Yates JR., 3rd Comparison of different signal thresholds on data dependent sampling in Orbitrap and LTQ mass spectrometry for the identification of peptides and proteins in complex mixtures. J Am Soc Mass Spectrom. 2009;20(8):1405–14. doi: 10.1016/j.jasms.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strausbauch MA, Madden BJ, Wettstein PJ, Landers JP. Sensitivity enhancement and second-dimensional information from solid phase extraction-capillary electrophoresis of entire high-performance liquid chromatography fractions. Electrophoresis. 1995;16(4):541–8. doi: 10.1002/elps.1150160189. [DOI] [PubMed] [Google Scholar]
- 37.Strausbauch MA, Landers JP, Wettstein PJ. Mechanism of peptide separations by solid phase extraction capillary electrophoresis at low pH. Anal Chem. 1996;68(2):306–14. doi: 10.1021/ac9503217. [DOI] [PubMed] [Google Scholar]
- 38.Chien RL, Burgi DS. Sample Stacking of an Extremely Large Injection Volume in High-Performance Capillary Electrophoresis. Anal Chem. 1992;64(9):1046–1050. [Google Scholar]
- 39.Tomlinson AJ, Benson LM, Jameson S, Naylor S. Rapid loading of large sample volumes, analyte cleanup, and modified moving boundary transient isotachophoresis conditions for membrane preconcentration-capillary electrophoresis in small diameter capillaries. Electrophoresis. 1996;17(12):1801–7. doi: 10.1002/elps.1150171203. [DOI] [PubMed] [Google Scholar]
- 40.Bollhagen R, Schmiedberger M, Grell E. High-performance liquid chromatographic purification of extremely hydrophobic peptides: transmembrane segments. J Chromatogr A. 1995;711(1):181–6. doi: 10.1016/0021-9673(95)00056-s. [DOI] [PubMed] [Google Scholar]
- 41.Hynek R, Svensson B, Jensen ON, Barkholt V, Finnie C. Enrichment and identification of integral membrane proteins from barley aleurone layers by reversed-phase chromatography, SDS-PAGE, and LC-MS/MS. J Proteome Res. 2006;5(11):3105–13. doi: 10.1021/pr0602850. [DOI] [PubMed] [Google Scholar]
- 42.Burgi DS, Chien RL. Optimization in Sample Stacking for High-Performance Capillary Electrophoresis. Anal Chem. 1991;63(18):2042–2047. [Google Scholar]
- 43.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal Chem. 2009;81(18):7757–65. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wojcik R, Li Y, Maccoss MJ, Dovichi NJ. Capillary electrophoresis with Orbitrap-Velos mass spectrometry detection. Talanta. 2012;88:324–9. doi: 10.1016/j.talanta.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee AM, Sevinsky JR, Bundy JL, Grunden AM, Stephenson JL., Jr. Proteomics of Pyrococcus furiosus, a hyperthermophilic archaeon refractory to traditional methods. J Proteome Res. 2009;8(8):3844–51. doi: 10.1021/pr801119h. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Champion MM, Sun L, Champion PA, Wojcik R, Dovichi NJ. Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry as an alternative proteomics platform to ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry for samples of intermediate complexity. Anal Chem. 2012;84(3):1617–22. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cech NB, Krone JR, Enke CG. Predicting electrospray response from chromatographic retention time. Anal Chem. 2001;73(2):208–13. doi: 10.1021/ac0006019. [DOI] [PubMed] [Google Scholar]
- 48.Fonslow BR, Carvalho PC, Academia K, Freeby S, Xu T, Nakorchevsky A, Paulus A, Yates JR., 3rd Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J Proteome Res. 2011;10(8):3690–700. doi: 10.1021/pr200304u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.