Abstract
The high noise level found in single-particle electron cryo-microscopy (cryo-EM) image data presents a special challenge for three-dimensional (3D) reconstruction of the imaged molecules. The spectral signal-to-noise ratio (SSNR) and related Fourier shell correlation (FSC) functions are commonly used to assess and mitigate the noise-generated error in the reconstruction. Calculation of the SSNR and FSC usually includes the noise in the solvent region surrounding the particle and therefore does not accurately reflect the signal in the particle density itself. Here we show that the SSNR in a reconstructed 3D particle map is linearly proportional to the fractional volume occupied by the particle. Using this relationship, we devise a novel filter (the “single-particle Wiener filter”) to minimize the error in a reconstructed particle map, if the particle volume is known. Moreover, we show how to approximate this filter even when the volume of the particle is not known, by optimizing the signal within a representative interior region of the particle. We show that the new filter improves on previously proposed error-reduction schemes, including the conventional Wiener filter as well as figure-of-merit weighting, and quantify the relationship between all of these methods by theoretical analysis as well as numeric evaluation of both simulated and experimentally collected data. The single-particle Wiener filter is applicable across a broad range of existing 3D reconstruction techniques, but is particularly well suited to the Fourier inversion method, leading to an efficient and accurate implementation.
Keywords: Electron microscopy, wiener filter, single particle, protein structure, SNR, SSNR, spectral signal-to-noise ratio
INTRODUCTION
In electron cryo-microscopy (cryo-EM), as in X-ray crystallography, an important goal of the data processing is to minimize the effects of noise in a density map. In recent years, cryo-EM has matured into a tool capable of providing near-atomic-resolution reconstructions of non-crystalline (single particle) biomolecules (Grigorieff and Harrison, 2011), thus bypassing certain limitations of X-ray crystallography (for example, the requirement that the target molecule be grown into a crystal) and NMR spectroscopy (which is limited to highly-concentrated, relatively low-molecular mass samples). Key advances that led to this breakthrough include the development of better electron optical systems, as well as improvements in image processing methodology for three-dimensional (3D) reconstructions of the resulting electron micrographs.
In a high-resolution cryo-EM experiment there will typically be ~104 - 106 images of the target molecule, each of which suffers from high noise levels, and is corrupted by a contrast transfer function (CTF) of the microscope. After determining the orientations and positions of each molecule in the images, a reconstruction algorithm merges the images into a 3D density representing the molecule. A large body of literature exists on various aspects of the reconstruction step (Penczek, 2010), but due to its importance it remains the subject of ongoing investigation.
In this work we address the reconstruction step; specifically, we seek a method to estimate a so-called ‘optimal’ map, where the mean-squared error compared to the ideal, unknown noise-free reference volume is minimized. Several studies have addressed this problem using different formalisms. At least two studies have reported implementations of the Wiener filter applied to the problem of 3D reconstruction of single-particle cryo-EM data (Zhang et al., 2008a; Scheres, 2012). The underlying assumption (either implicit or explicit) in these studies was that this filter should minimize the mean-squared error in the resulting 3D map, with respect to the signal present in the image data. Similarly, a so-called ‘figure-of-merit’ (FOM) filtering scheme was proposed as a post-processing step intended to generate a ‘best map’ (i.e., lowest mean-squared error) given the data (Rosenthal and Henderson, 2003). The error remaining in a map when subjected to such filter schemes has not been carefully scrutinized in these reports, thus leaving the essential premise of the filters (error reduction) untested. Moreover, we have recently demonstrated that, in order to minimize error in averages of aligned two-dimensional (2D) images, the bulk solvent surrounding the particle must be adequately accounted for through the addition of a scale factor. This resulted in a modification to the Wiener filter which we called the ‘single-particle Wiener filter’ (SPW filter).
Here we extend our previous results with the SPW filter to the more involved problem of 3D reconstruction. We test the various assumptions of our theory by applying the resulting SPW filter to synthetic and experimentally acquired test data sets. We find that the resulting algorithm is generally applicable to reconstruction problems with single particles, and quantitatively minimizes the error within the particle density map in cases where neither the conventional Wiener filter nor the FOM filter is as effective. Our algorithm is the first adaptation of the Wiener filter to specifically address problems caused by the presence of bulk solvent surrounding the particle. We demonstrate that this approach leads to better real-space and Fourier space fidelity for reconstructed maps using a highly efficient Fourier inversion framework. The SPW filter described here has been implemented in the single particle software FREALIGN (Grigorieff, 2007) starting with version 8.10.
THEORY
The Wiener filter (Wiener, 1949) has been applied to both 2D and 3D cryo-EM image processing problems, with the goal of optimally combining noisy images into a “best” representation of the noise-free object being imaged (Saxton, 1978; Ludtke et al., 2001; Zhang et al., 2008a; Scheres, 2012). If one can obtain an estimate of the signal-to-noise ratio (SNR) of the Fourier-space representation of the data, the Wiener filter will suppress the noise in poorly measured parts of the Fourier space in order to obtain better agreement with the noise-free signal. However, the utility of the Wiener filter is compromised in single-particle imaging applications by an ambiguity in the definition of the SNR: as noted in Sindelar and Grigorieff (2011), the SNR of a particular imaged particle can be made arbitrarily low just by increasing the field of view to include more noise in the surrounding solvent area. Thus, the behavior of the Wiener filter depends on the selected image size, for a given particle, and in general tends to give over-filtered results for images of single particles (Sindelar and Grigorieff, 2011).
The above deficiency can be linked to the observation that the Wiener filter is only guaranteed to be optimal for stationary processes, where the expected mean and variance of the target function does not vary under translation (Van Trees, 2001). In fact, the target function in the currently considered case, a 3D density map of a single particle, is highly non-stationary: the mean and variance of the density inside a particle will always be substantially different than the mean and variance in the solvent region. We therefore seek a modified filter that better captures the properties of single particles.
Deriving a 3D “single-particle” Wiener filter
To a first approximation, a large number of randomly oriented images will contribute a variable number of Fourier space measurements (i=1, 2, ... nhkl) to each discrete point shkl in the 3D discrete Fourier transform, or DFT, of the particle map ρ(r) (see Appendix A). Here, shkl represents a discrete grid point in the 3D DFT having integer indices hkl, nhkl is the number of measurements for shkl contributing to this grid point. Here and in the following, bold symbols are vectors and italicized non-bold symbols refer to the length of the corresponding vectors. In particular, shkl will be the radial spatial frequency corresponding to grid point shkl. If the SNR of the measurements is available as a function of shkl, then the Wiener filter supplies a set of linear coefficients that minimize the average error in the resulting DFT. By Parseval's theorem, the error is also minimized in the corresponding real-space 3D map ρW(r) obtained by Fourier inversion. The Wiener expression (Saxton, 1978) generalized for 3D is (see Appendix A):
| (1) |
where CTFi,hkl are the previously estimated CTF values of the microscope for the given Fourier space measurement, accounting for the image defocus level, astigmatism, etc.
We now derive a modification to Eq. (1) that addresses the special properties of single particles. Following our approach for the case of aligned 2D images (Sindelar and Grigorieff, 2011), we define a 3D binary enveloping function, env3D(r), outside of which the target particle density is known to be zero. We then seek the new set of linear coefficients to the measurements that yield a real-space map where the error is specifically minimized inside the envelope. Applying a set of assumptions that are expected to be reasonable for single-particle cryo-EM data sets (for example, that the data set is sufficiently large to yield a well-localized particle map), it is straightforward to adapt the previously presented 2D SPW filter to its 3D analog (Appendix A). After including a “gridding” formalism to account for the fact that, in the 3D case, most Fourier space measurements do not fall exactly on the discrete grid points shkl (see Appendix B) we arrive at the following expressions for the 3D SPW filter:
| (2) |
with
| (3) |
and
| (4) |
(note that fparticle refers to a fraction of a 3D volume whereas in Sindelar and Grigorieff (2011) it referred to a fraction of a 2D image). Here and in the following, PSSNR and SSNR are functions of radial spatial frequency s. They approximate SNR values found at grid points shkl by averaging over all values in a resolution shell. It is important to note here that Eqs. (2) - (4), as is the case with the equivalent 2D SPW expressions, can be applied in the absence of any specific knowledge about the shape of the envelope function. Instead, all that is required is the mean squared value of the envelope function, fparticle, which is equal to the fractional volume occupied by the envelope within the boundary of the reconstructed box. Eqs. (2) - (4) will then minimize the reconstruction error of the particle density inside the envelope. Below we will describe how to find a “best” value for fparticle that optimizes the map within the particle itself.
Accurate estimation of the image SSNR by masking
In order to implement Eq. (2) it becomes necessary to obtain an accurate estimate of the SSNR of the raw data images. The SSNR can be most accurately obtained from a ‘masked’ FSC calculated from two volumes each containing half the data (Harauz and Van Heel, 1986) where solvent noise surrounding the particle is suppressed with a soft-edged mask function envmask. The term envmask differs from env3D as it usually has a simpler shape such as a sphere and therefore contains substantially more volume than the actual particle volume. As shown by Sindelar and Grigorieff (2011), for a set of aligned 2D images,
| (5) |
where the FRC is the 2D analog of the FSC, formed by comparing two independently averaged image data sets (Harauz and Van Heel, 1986), S(s) is a resolution shell centered around radial spatial frequency s, and nS is the number of Fourier space pixels contained within S.
For a 3D data set, the corresponding result is (see Appendix C):
| (6) |
where is the mean-squared value of the soft-edged mask function evaluated over the 3D (real-space) reconstruction volume. This expression estimates the SSNR found in the raw data images, including the noise found in the solvent region, and thus may be combined with Eq. (3) to obtain the PSSNR (assuming knowledge of fparticle; see below). Here and in the following we make the assumption that the SSNR does not vary significantly between images and therefore, an average SSNR for the entire data set can be assumed. In the Discussion, we will consider the case of variable SSNR in a data set.
Derivation of a related post-processing SPW filter
The PSSNR term in the denominator in Eq. (2) systematically down-weights structure factors FSPW where the number of measurements is not sufficient to overcome the measurement noise. FSPW thus represents an optimal estimate of the true structure factors (in the least squares sense and ignoring gridding-related artifacts), and its calculation requires incorporation of the SSNR found in the 2D image data during calculation of the final reconstruction. An alternative scheme has been described that uses a filter based on an FOM (Rosenthal and Henderson, 2003). Unlike the Wiener filter and its SPW derivative described above, the FOM filter is not incorporated directly into the 3D reconstruction step, and is instead applied in a post-processing step after the reconstruction has been calculated. To compare these filtering methods, we relate FSPW to unfiltered gridded reconstruction using Eqs. (2) and (A2.6):
| (7) |
where we have left out the small ε term from Eq. (A2.6), which is expected to have a negligible effect on this expression. The above expression represents a voxel-by-voxel correction to the unfiltered reconstruction FLSQ. We now substitute our estimate of the SSNR as a function of the masked FSC given in Eq. (6):
| (8) |
The above expression still requires knowledge of the individual CTF terms in the 2D image data. To further simplify this expression, we now assume that the filter is approximately constant within a given resolution shell. This requires that the sum of squared CTF values (this can be considered as the effective number of Fourier-space measurements) is similar for all structure factors within the resolution shell. This condition will be met when (1) a sufficiently large number of images have been collected, such that every point in Fourier space is measured many times by a spread of defocus values, and (2) there are no strongly preferred orientations in the data set (note that the presence of astigmatism in the images would not affect our analysis, under the above conditions). The expected value of this filter is then
| (9) |
where the brackets denote the average value for all possible instances of the noise in resolution shell S(shkl). The above expression is expected, upon application to a non-filtered 3D reconstruction, to optimally filter the density map to reduce noise.
Eq. (9) describes how to obtain an approximation to the SPW algorithm (Eq. (2)), by defining a post-processing filter to be applied to the unfiltered reconstruction (FLSQ). This result may be compared with the FOM filter described by Rosenthal and Henderson (2003), which is written in our terminology as:
| (10) |
In contrast, we see that in the limit of fparticle = fmask Eq. (9) reduces to:
| (11) |
Note that while Rosenthal and Henderson applied masks to their reconstructed volumes prior to calculating the FSC, they did not explicitly consider the effects of masking in their expressions for Cref.
De novo estimation of fparticle from FSC half volumes
The above results indicate that successful application of the SPW filter requires an accurate estimate for fparticle. However, fparticle is defined by the shape of the solvent envelope of the particle, which is frequently challenging to obtain in experimental applications. Here we present a strategy for estimating fparticle using only information available from the input images. We begin with the property that the SPW filter minimizes the expected error within the particle region, compared with a noise-free reference volume. We further note that the SPW filter minimizes the reconstruction error everywhere in the particle simultaneously. In other words, if a chosen value of fparticle minimizes the error in any given region within the solvent envelope, the error should also be minimized at all other regions within the envelope as well, assuming equal quality of the map in all regions. Thus, one may restrict the above error evaluation to a small mask located within a “core” region of the particle, which is straightforward to establish even when the solvent boundary is indistinct (see below). If the noise-free reference volume is available, it is therefore possible to estimate fparticle by systematically varying this quantity during application of the SPW filter. The best estimate of fparticle will be the value that minimizes the error in the “core” region of the filtered reconstruction, with respect to the reference volume. In this way, fparticle can be estimated without knowledge of the precise shape of the particle envelope.
In experimental studies, the noise-free reference volume remains unknown, requiring further modification to the above strategy. It is straightforward to show, however, that because the SPW filter minimizes the error with respect to the noise-free reference volume, this filter also minimizes the error with respect to a noisy reference volume (so long as the added noise is random, and the reference is otherwise unfiltered). For any given experimental data set, moreover, a noisy reference volume is readily obtained by gridded Fourier inversion (Eq. (A2.6)). This observation implies that we may use an experimentally obtained reference volume in the above estimation procedure for fparticle, rather than using a noise-free reference, and still obtain the same result.
We thus arrive at the following scheme for estimating fparticle: From one half of the data we calculate an unfiltered, noisy estimate, FLSQ, of the reconstruction (Eq. (A2.6)). The second half of the data is used to calculate a filtered estimate, FSPW in which we now allow fparticle to vary (Eq. (2)). We subsequently perform a series of reconstructions using values of fparticle that range from 0 to 1, comparing the real-space cross-correlation coefficient (CCC) between core regions of the filtered half-data density map and the unfiltered, noisier half-data map. We then choose the value of fparticle that optimizes the core region CCC. This value of fparticle is thus expected to give a SPW filter whose output minimizes the error in the particle region. Moreover, this value of fparticle is expected to correspond (at least approximately) to the fraction of the reconstructed volume that is occupied by particle density (this property will be tested below). Thus, the procedure just described will yield an approximation of the SPW filter using only the images and Euler angles that are standard input in any 3D reconstruction algorithm.
FLOW CHART: Integrated SPW filter
- Insert projections into Fourier volume via box convolution (equivalent to nearest-neighbor interpolation if box dimension is 1 × 1 × 1 in voxel units):
- Calculate sum in numerator of Eq. (2), stored on a per-voxel basis.
- Calculate sum in denominator of Eq. (2), also on a per-voxel basis (this and the preceding step are identical to the previously published FREALIGN implementation).
- Gather separate numerator, denominator tallies for two half-data-set reconstructions, for FSC computation.
Perform Fourier inversion (Eq. (A2.6)) to obtain both half-data-set reconstructions, and compute the FSC between the two maps (using a smoothed mask where fmask is conservatively chosen to significantly exceed the volume of the particle) to obtain a lower bound on the reconstruction resolution.
Estimate the whole-image SSNR from the masked FSC, by Eq. (6).
Select a ‘core region’ of the density by low-pass-filtering the reconstruction several times lower than the resolution lower bound computed in the last step, and defining the binary envelope to enclose a small fraction (i.e. ~10%) of the filtered reconstruction density.
- Perform a series of reconstructions using the second half data set, according to the formula:
where f, representing the unknown quantity fparticle, is varied between 0 and 1. Estimate fparticle as the value of f that maximizes the real-space CCC between core regions of the first (unmodified) half-data set reconstruction and the filtered reconstructions generated in step E.
FLOW CHART: Post-processing SPW filter
If the SPW filter is implemented with a post-processing filter rather than as an integrated reconstruction algorithm, any reconstruction algorithm may be used and fewer steps are necessary:
Obtain unfiltered half-data-set reconstructions and compute the masked FSC and full-data-set reconstructions.
Define a ‘core region’ of the density, as described in step D in the integrated SPW procedure.
- Apply a series of filters to the second half-data-set reconstruction, using the following form of the SPW post-processing filter:
where f, representing the unknown quantity fparticle, is varied between 0 and 1. Note that this post-processing filter has been modified from Eq. (9) in order to take into account the reduced signal-to-noise ratio found in a reconstruction made with half the data, compared with a full-data-set reconstruction. Estimate fparticle as the value of f that maximizes the real-space CCC between core regions of the first half-data set reconstruction and the filtered reconstructions generated in step C.
Apply the post-processing SPW filter (Eq. (9)), using the value for fparticle estimated in part D, to the unfiltered full-data-set reconstruction step A.
RESULTS
Normalized SSNR estimation via the masked FSC
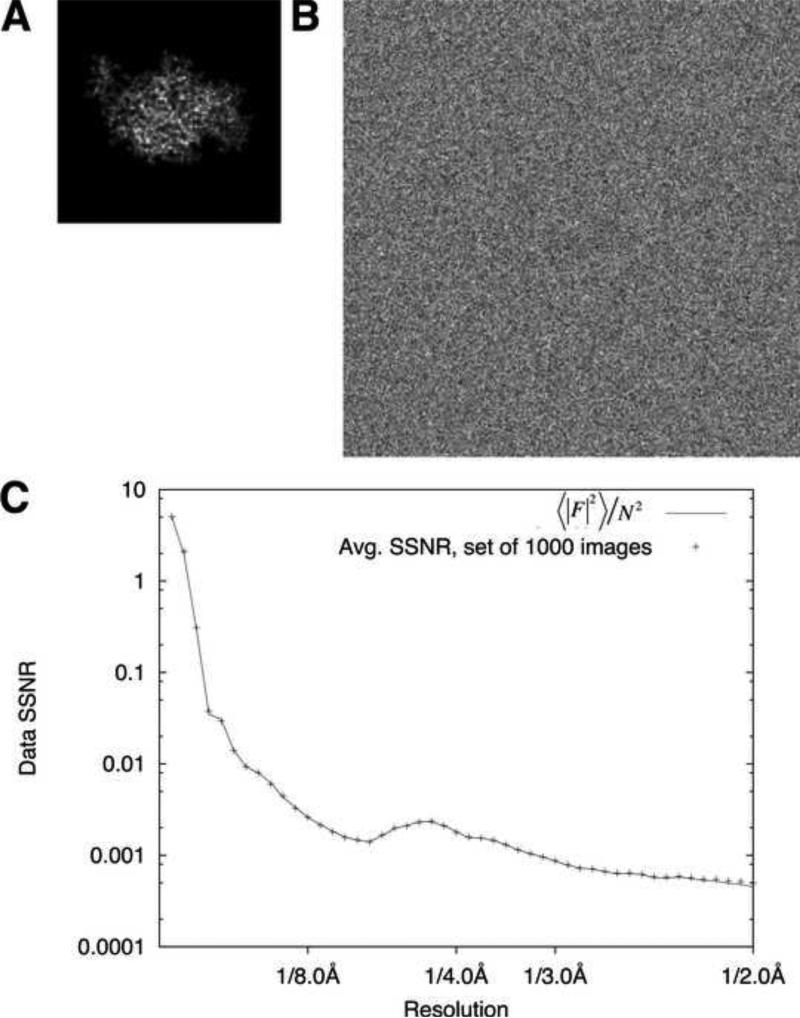
Eq. (6) predicts that using the FSC to estimate the SSNR for a reconstructed particle map will yield a result that is inversely proportional to the fraction of solvent that is included in the FSC comparison. We tested this prediction using a synthetic data set composed of noisy projection images of a small (~35kD) protein molecule (crystal structure of the kinesin motor domain, PDB ID 1MKJ), from randomly sampled viewing orientations (Fig. 1A, B). Special care was taken to avoid interpolation artifacts during the projection process (see Methods), thus allowing the SSNR characteristic of the projection images to be precisely established a priori (Fig. 1C). Images were divided into two equal sets and subjected to gridded Fourier inversion (Eq. (A2.6)) using the exact (known) Euler angles of the projections in order to compute a pair of 3D reconstructions from each set, and a third reconstruction for the combined full image set. We then performed FSC comparisons of the resulting reconstructions, after multiplying the maps with a solvent mask. Three different mask sizes were used: a tight binary mask (Fig. 2E), generated from the reconstruction itself by the method of Wang (Wang, 1985) with parameters chosen such that the mask volume was ~2x the particle volume (see Methods); a looser mask (Fig. 2F), generated from the former mask by applying a cosine edge smoothing function (mask volume was ~5x the particle volume); and a smoothed spherical mask (Fig. 2G) where the radius matched the maximum linear dimension of the particle map (net mask volume was ~10x the particle volume).
Figure 1. Volume-normalized estimation of the SSNR.
A. Noise-free, interpolation-free projection of kinesin monomer crystal structure (PDB ID 1MKJ), at 1 Å/pixel in a 96 × 96 pixel image.
B. Noisy, CTF-modulated image derived from (A) with SNR = 0.002. The SNR value is computed from the entire image. Image size is enlarged to 256 × 256 pixels in order to retain information delocalized by the CTF.
C. Composite SSNR behavior for 1000 synthetic images generated as in panel B (crosses), compared to the idealized SSNR computed by dividing the rotationally averaged structure factors of the noise-free volume ( by the mean squared amplitude N2 for the white noise added to the images.
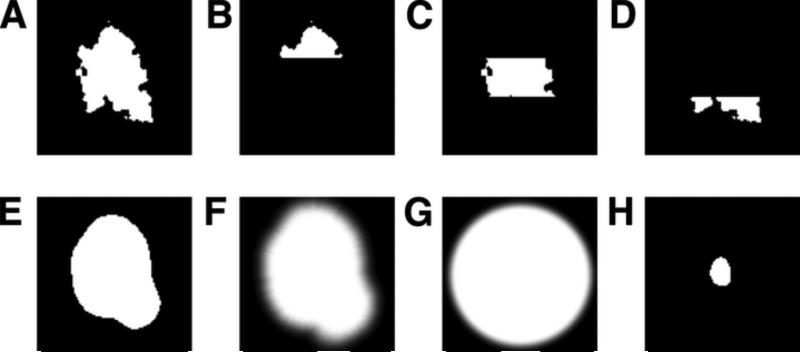
Figure 2. Masks used for real-space and/or Fourier-space cross-correlation calculations.
Representative z slices are shown.
A. Molecular surface mask (fmask = 0.0236).
B-D. Subdivided pieces of the molecular surface mask in A.
E. Binary envelope mask derived from a reconstruction low-pass filtered to 30 Å resolution (fmask = 0.051).
F. Smoothed mask derived from E by applying a cosine edge filter (fmask = 0.101).
G. Smoothed spherical mask (fmask = 0.223).
H. Core mask (fmask = 0.0059).
The results of these FSC calculations (Fig. 3; note that FSC values are scaled into estimates of Cref using Eq. (9)) illustrate that the application of masking yields substantially different results, due to the varying amount of solvent noise eliminated by the masking. However, we can resolve this discrepancy by defining a quantity PSSNRfinal that places the SSNR of the reconstruction onto an absolute scale, applying the same logic that was used to derive Eq. (6):
| (12) |
Following application of Eq. (12), the scaled Cref estimates converge to approximately the same value throughout most of spatial frequency range (Fig. 3A, B), indicating that Eq. (12) yields a consistent resolution measure.
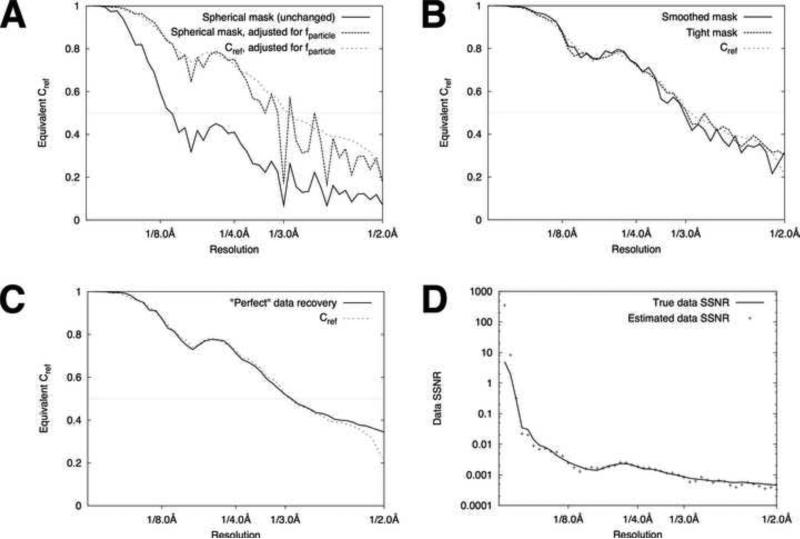
Figure 3. FSC/SSNR analysis of reconstructed volumes.
A. Applying a generous mask for FSC calculations underestimates the reconstruction quality. A spherical mask (Fig. 2G) was applied to half-data set reconstructions (Fourier inversion, no filter applied) and the FSC was computed, for the synthetic data set described in Fig. 1. For comparison purposes, FSC values were then transformed to the equivalent Cref values using Eq. 12 (solid curve). The middle curve (dashed) shows the same FSC values after being transformed to account for the particle volume via Eq. 13, using 0.023 as the estimated value for fparticle (see text). The lighter dashed curve indicates the ‘true’ Cref values obtained by masked comparison between the full-data-set reconstruction and the noise-free reference.
B. Decreasing the mask size leads to more accurate estimates for Cref . The ‘true’ Cref values are carried over from panel A. The heavy solid curve shows the estimates for Cref obtained by performing FSC calculations using the smoothed mask from Fig. 2F; the heavy dashed curve shows Cref values estimated by FSC using the binary mask from Fig. 2E.
C. Measured SSNR indicates near-optimal reconstruction algorithm. FSCref (identical to panels A, B) is compared with the “ideal” case where all measurements contribute “perfectly” to signal recovery (see text). The horizontal line at FSCref = 0.5 indicates the nominal resolution of the reconstruction as given by Rosenthal and Henderson (2003).
D. Recovering the data SSNR from masked FSC calculations. The composite SSNR of the raw data images was estimated from masked FSC calculations via Eq. (6), and compared with the known SSNR characteristic of the synthetic data set (Fig. 1C).
We cross-validated these estimates by separately computing the Fourier shell correlation between the noise-free reference volume and a masked full-dataset reconstruction (we refer to this latter function as Cref, following the convention of Rosenthal and Henderson (2003). The resulting Cref curve was scaled to form an estimate of PSSNRfinal by combining Eqs. (6), (9) and (12), and is also shown in Fig. 3A-C. The estimates for PSSNRfinal generated from this latter approach are in excellent agreement with the FSC-generated estimates. As with the FSC calculations, the Cref calculations showed smaller fluctuations (indicating higher fidelity) as tighter masking was applied (results not shown). Thus, while tight masking is desirable to reduce the random error in the PSSNRfinal estimates, our results demonstrate that the mask size may be expanded as necessary (for example, to avoid mask-related artifacts in the FSC computation; see Discussion) without introducing systematic under-estimation of the reconstruction resolution, so long as the values are adjusted by Eq. (6).
Using the known SSNR characteristic of the synthetic images, we then derived an upper bound for the expected value of PSSNRfinal for an idealized Fourier inversion algorithm (assuming no reconstruction artifacts):
| (13) |
This limiting function is defined purely by the signal and noise characteristics of the data images, together with the number of images taken, imaging geometry, CTF conditions, and microscope parameters; all of these values are precisely known for the synthetic data set used here. As shown in Fig. 3C, the values for PSSNRideal are in excellent agreement with the Cref function derived from the conventional Fourier inversion reconstruction. The estimated SSNR showed higher fluctuations about the known value in the lowest-resolution shells (corresponding to resolutions lower than 10 Å), due to the combination of poor statistics (fewer voxels per shell) and small CTF values at these spatial frequencies, leading to higher noise variance. These errors, however, did not strongly affect the performance of the SPW filter (see below) because of the high overall SSNR of the final reconstruction at low resolution. The estimated PSSNR also showed a tendency to under-estimate the known SSNR values at resolutions higher than 3 Å, likely due to incomplete sampling of Fourier transform by the data. Again, however, these errors did not significantly affect the performance of the SPW filter because these errors occurred at spatial frequencies beyond the nominal resolution of the reconstruction (Cref < 0.5, Fig. 3C). Thus, the agreement between these three different SSNR estimation methods (FSC-derived, Cref-derived, and ‘ideal’) indicate that our expressions are self-consistent and quantitative, under the given (simulated) imaging conditions.
Estimating the SSNR of the data
We estimated the SSNR in our data set by applying Eq. (6), using the soft-edged mask in Fig. 2F (fmask = 0.101); we then back-calculated an estimate of PSSNR for the original data images by applying Eq. (3). We note that this back-calculation formula is based on the assumption of a perfect, artifact-free Fourier inversion algorithm, which our tests indicated was approximately valid (see above). As shown in Fig. 3D, the resulting estimates for the image SSNR were in excellent agreement with the known, pre-defined SSNR characteristic of the synthetic images used in these tests, although minor deviations below the known value are visible at the highest spatial frequencies.
Evaluation of the conventional Wiener filter in a 3D Fourier inversion reconstruction algorithm
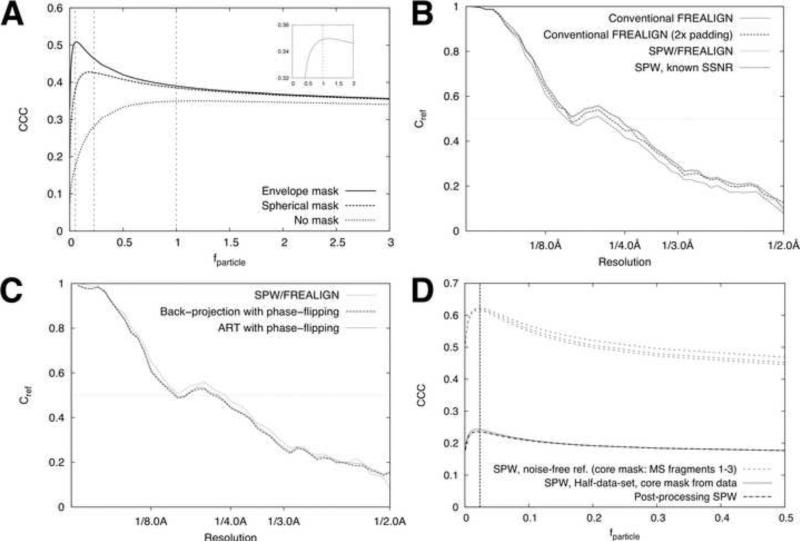
To test the validity of the Wiener filter when applied within a 3D Fourier inversion scheme, we performed a series of 3D reconstructions using the synthetic data images from Fig. 1 as inputs, and employing the known SSNR characteristic of the images for the Wiener filter. As shown in Fig. 4A-B, applying the Wiener filter within a Fourier inversion scheme filters away high-resolution noise from the resulting 3D reconstruction, improving the real-space agreement with the noise-free 3D reference map. To further test the validity of the Wiener filter within the approximations inherent in our gridded reconstruction algorithm, we systematically perturbed the SSNR term in the denominator of Eq. (1) above and below its true value in order to test whether the mean-squared error was properly minimized with respect to the reference volume. This test is mathematically equivalent to applying Eqs. (2) - (4) using values of fparticle scaled above and below 1, which is how the results are presented here (Fig. 5A, inset). These calculations show that, as expected, the error is minimized near fparticle=1, although the peak is relatively broad. This perturbation experiment thus indicates that incorporating the Wiener filter into a Fourier inversion reconstruction scheme approximately minimizes the mean-squared error of the full 3D reconstruction volume with respect to the filtering parameters.
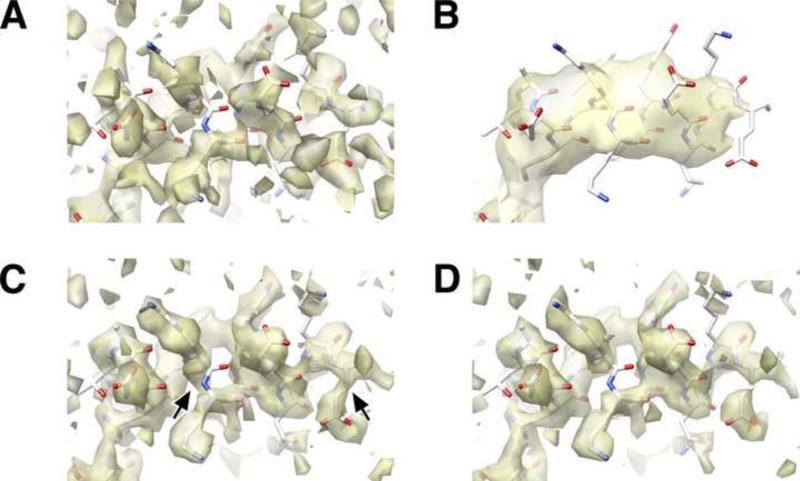
Figure 4. Performance of various filters, as indicated by features found in a representative region of the reconstructed maps.
Density maps are represented as isosurfaces by UCSF Chimera (Pettersen et al., 2004).
A. Reconstruction of the dataset in panel C generated by the published Fourier inversion scheme of FREALIGN. Reconstruction is post-filtered by a 3 Å resolution low-pass filter, corresponding to the nominal resolution as indicated by Fig. 3C.
B. Reconstruction as in panel A, but using a conventional Wiener filter incorporated into the Fourier inversion scheme. Severe over-filtering is apparent, relative to panel A.
C. Density map generated by the integrated SPW method.
D. Result of applying the post-processing filter (Eq. (10)) to an unfiltered Fourier inversion reconstruction (Eq. A2.6). To facilitate comparison, the threshhold level in this density map was chosen such that the isosurface contains the identical volume as the map in panel C.
Figure 5. Validating the SPW filter with masked CCC calculations.
A. Real-space cross correlation coefficients between reconstructions and the noise-free reference volume are shown for the case of no masking, as well as a spherical mask (Fig. 2G) and a smoothed molecular envelope mask (Fig. 2F). The vertical dashed lines indicate the computed values of fmask for each case (1, 0.223, and 0.101, respectively). The inset shows a rescaled plot of the ‘no mask’ curve.
B. Estimating fparticle by masked correlation comparison of core regions in the reconstructed maps. The upper curves show the results from comparing full-data-set reconstructions (integrated SPW method) with the noise-free reference volume. The lower curves show the results of equivalent masked CCC calculations that compare half-data-set reconstructions. For the lower curves, results from both integrated and postprocessing forms of the SPW filter are shown. The vertical dash line indicates our estimate of fmask (0.023, see text).
C. Single-particle Wiener filter improves the resolution of reconstructions, as indicated by Cref comparisons. The reconstructed volume was multiplied by the mask in Fig. 2F prior to computing the Cref values. The ‘conventional FREALIGN’ calculations used the Fourier inversion method (Eq. (A2.6)); for the 2x padded reconstruction, images were padded by zeros. SPW reconstructions were also computed with 2x padding, applying Eq. (2) either with estimated SSNR values or the known SSNR values (see Fig. 3C). D. Single-particle Wiener filter outperforms other reconstruction algorithms. Results of two other algorithms are shown (see text). Unlike in Fig. 3, the Cref values here and in panel C are not scaled to account for particle volume.
Fig. 4B also shows that the 3D density map that results from the Wiener filter reconstruction appears to be strongly over-filtered, especially when compared with the output of the SPW reconstruction methods (see below). This over-filtering results from the Wiener filter's sensitivity to the noise in the solvent region, such that the larger the solvent region, the lower the measured SSNR and hence the greater the over-filtering effect ((Sindelar and Grigorieff, 2011); see Eq. (3) above).
Single-particle Wiener filter improvement over the conventional Wiener filter
The above drawback in the Wiener filter can be corrected by re-defining the reconstruction problem to neglect the reconstruction error that occurs within the solvent region, and instead to minimize the error within the particle envelope only. The resulting SPW filter (Eq. (2)) is predicted to minimize the mean-squared error within an arbitrarily shaped enveloping function characterized by a fractional volume fparticle, so long as the envelope fully encloses the particle. We note that the mask function itself is not a required input to the SPW filter; instead, fparticle is the only additional input required (with respect to the conventional Wiener filter).
To test the performance of the SPW filter within the Fourier inversion scheme, we applied both the integrated as well as the post-processing SPW filters to our synthetic image data set. The resulting density maps (Figs. 4C-D) were visibly improved relative to the unfiltered or Wiener filtered maps. We tested the SPW filtered maps by real-space cross-correlation comparison with the noise-free reference volume, confining the comparison within either (1) a relatively tight binary mask (envelope mask in Fig. 2E), generated from a moderately filtered reconstruction (see Methods); or (2) a large spherical binary mask having a diameter slightly larger than the longest particle dimension. We then systematically perturbed fparticle throughout the range from 0 to 3.0 and computed the masked CCC where the comparison was restricted to the defined envelope region. As predicted (Fig. 5A), the SPW filter reduced the error within both envelopes, for values of fparticle close to the exactly computed value for these envelopes. For the tight mask, fparticle was estimated as 0.06 vs. the known value of 0.051; for the spherical mask the estimated value was 0.19, compared to the known value of fparticle = 0.223.
In contrast, the whole-volume CCC for the map produced by the SPW filter was not minimized as a function of the SSNR function, and indeed was substantially lower than the whole-volume CCC yielded by the conventional Wiener filter (data not shown). Thus, CCC comparisons indicate that the SPW filter optimizes the error within the particle envelope, but that this improvement is accomplished at the expense of increased noise in the solvent region. The increased noise in the solvent region, however, is readily removed by multiplying the reconstruction with the binary particle envelope, yielding a highest-quality map where the error has been completely eliminated from the solvent region and minimized within the particle envelope.
These results demonstrate that our modified Wiener filter expression specifically tunes the noise suppression in the particle volume defined by fparticle. It follows that fparticle should be made as small as possible, while still corresponding to an envelope that fully encloses the particle, in order to completely minimize the error within the particle region. Below we evaluate our scheme for empirically determining such a value of fparticle even in the absence of precise description of the particle shape.
SPW filter yields improved FSC values relative to other reconstruction schemes
To assess the Fourier-space signal of the SPW reconstruction scheme compared with other reconstruction methods, we computed masked Fourier shell correlation functions comparing the reconstructions with the noise-free reference map. The resulting Cref curve was increased across the entire spatial frequency range, relative to the corresponding result for the equivalent unfiltered reconstruction (Fig. 5B), although the gains were relatively minor. For comparison, we also evaluated several other published reconstruction schemes with the identical synthetic data set (Fig. 5C), including back-projection with phase flipping CTF correction (Frank et al., 1996) and an iterative algebraic method also combined with phase-flipping (Sorzano et al., 2004). These reconstructions yielded Cref curves similar or lower than our unfiltered, gridded reconstruction, but falling below the SPW values (Fig. 5C).
Estimating f particle
The basis for our method of estimating fparticle is to find the filter function that maximizes the agreement in a representative “core” region of two half-data set reconstructions (see Theory). To generate a “core” mask containing only particle density, we applied a 30 Å low-pass filter to the initial, unfiltered, gridded reconstruction, then selected a threshold value to define a mask limited to a subset of the protein interior (Fig. 2H; mask volume was ~20% of the protein envelope volume). We generated a series of half-data set reconstructions using our synthetic data set, applying the integrated SPW filter to one half-data set reconstruction (Eq. (2)) but scaling the fparticle term systematically from 0 to 1. The second half-data set reconstruction was generated using the gridded Fourier inversion algorithm without the SPW filter (Eq. (A2.6)). The SSNR of the data was estimated via. Eq. (6). As shown in Fig. 5D, maximizing the CCC between the “core” density of the two half-data-set maps (defined by the central ~20% of the kinesin protein envelope, see Methods) led to the assignment of fparticle = ~0.022. A similar result was seen for the post-processing version of the SPW filter (Fig. 5D). For comparison, the volume contained by the molecular surface defined by the atomic model, which captures the solvent envelope of a high-resolution structure (see Methods), was 0.023. Thus, the simple scheme described here produces an estimate for fparticle that closely agrees with the “true” value expected from basic principles.
Similarly accurate estimates of fparticle were obtained with both the integrated and the post-processing forms of the SPW filter, although the CCC values were slightly lower in the case of the post-processing filter (Fig. 5D). We also experimented with different “core” mask choices by using the molecular surface itself, or subfragments thereof (Fig. 2A-D), for the core mask in the fparticle estimation procedure; these latter experiments (Fig. 5D, upper dashed curves) indicated that the results of the estimation procedure were relatively insensitive to the choice of core region.
Application to an experimental high-resolution data set
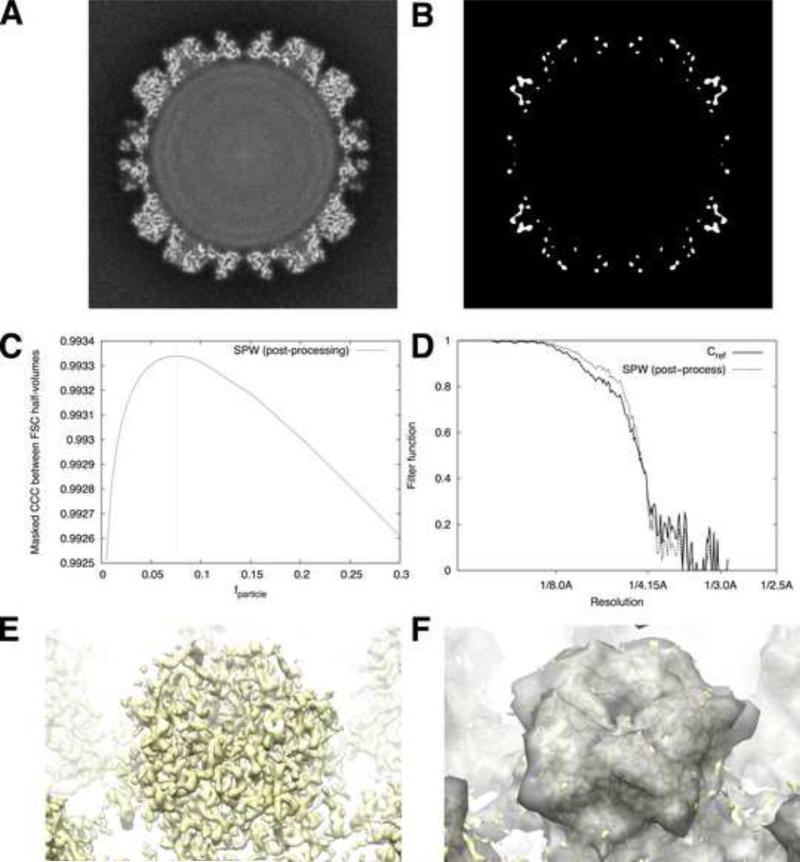
We tested our filter expressions on a set of papillomavirus images that were used to obtain a near-atomic resolution 3D map (Wolf et al., 2010). We used the FREALIGN software (Grigorieff, 2007) to duplicate the methods of Wolf et al., generating a full-data-set gridded (unfiltered) reconstruction (Fig. 6A) and two half-data set reconstructions output by the program for the purpose of computing the FSC function (icosahedral averaging was performed, but no other averaging was done). We then applied our estimation scheme for fparticle, varying fparticle until we observed the maximum real-space correlation (Fig. 6C) between a non-filtered gridded reconstruction (half-data set #1), and the post-filtered SPW map (half-data set #2), restricting the comparison to small core regions within the protein interior (Fig. 6B). For FSC computations, we duplicated the mask parameters of Wolf et al. resulting in a mask in the form of hollow sphere (fmask ≈ 0.26). This strategy yielded an estimated value for fparticle of 0.075 (Fig. 6C). To visualize this value of fparticle, we rendered an isosurface of the low-pass-filtered virus reconstruction, adjusting the threshold until the enclosed volume was equivalent to fparticle. As can be seen in Fig. 6E and 6F, this isosurface tightly encloses the volume occupied by the virus capsid proteins, indicating that our methods find a reasonable approximation to fparticle in this case. We also compared the actual filter function values of the FOM scheme vs. our SPW post-processing filter (Fig. 6D); remarkably, the filter function originally obtained by Wolf et al. using the FOM scheme (solid curve) nearly coincides with the post-processing SPW filter function values (lower dashed curve). Thus, for this particular instance the FOM filter closely matches the SPW post-processing filter, at least for the chosen masking parameters.
Figure 6. Application to an experimental high-resolution data set.
A. Cross-section of the unfiltered and unsharpened papillomavirus map, generated by FREALIGN using the methods described by Wolf et al. (2010).
B. Core mask function for this procedure was defined by first applying a 15 Å low-pass-filter to the reconstruction in panel A. The core mask was then defined by choosing a binary cutoff threshold such that 1% of the total reconstruction volume was included (volume occupied by the virus shell was ~15% of the reconstruction volume).
C. Results of our estimation procedure for fparticle , using the post-processing SPW filter.
D. Comparison of post-processing filter functions. The solid curve depicts the FOM weights used by Wolf et al. (2010) following the scheme of Rosenthal and Henderson (2003), using Eq. (11). The dashed curve depicts the SPW post-processing weights obtained through Eq. (9).
E. Isosurface of the density map from A, depicting an L1 pentamer on the surface of the capsid.
F. Isosurface from E, superposed with semitransparent isosurface of the same map (gray), low-pass-filtered to 15 Å and thresholded such that the volume enclosed by the surface is equivalent to fparticle = 0.075, the value identified by our estimation procedure in panel C.
DISCUSSION
We have used a new theoretical framework to derive a least-squares solution to the single-particle 3D reconstruction problem, specifically accounting for the presence of a noisy solvent region of uniform density. Key to our analysis was the observation that the SSNR of an image or volume of a single particle is linearly related to the fractional area/volume occupied by the particle (Sindelar and Grigorieff, 2011) – a result that enabled us to quantify the effects of masking on FSC calculations, hence permitting much more accurate SSNR estimation. We find that the resulting SPW reconstruction algorithm is closely related to the Wiener filter, from which it was derived. We also find that the SPW method is closely related to an FOM weighting scheme proposed by Rosenthal and Henderson (2003). However, our analysis demonstrates that the SPW method improves on these two earlier methods. Moreover, our theoretical treatment connects the earlier methods to each other, and explains why they fail to produce optimal results under certain circumstances.
SPW method is distinct from a Wiener filter
The least-squares method we have implemented here, as embodied by Eq. (2) (and which we previously described for the treatment of aligned 2D images (Sindelar and Grigorieff, 2011)), differs from the classically defined Wiener filter (Saxton, 1978) in a subtle but important way. In the SPW method, we have introduced the assumption that the density of interest occupies only a fraction of the reconstructed map, which is otherwise occupied by a uniform background value. When this assumption is applied to the problem of 2D or 3D averaging, an approximate least-squares solution results whose form (Eq. (2)) is nearly identical to the Wiener filter, but where the SSNR term is scaled by the inverse of the fractional particle volume, fparticle (Sindelar and Grigorieff, 2011). Given the approximately linear relationship found between image area/volume and the SSNR (Sindelar and Grigorieff, 2011), it is tempting to identify the scaled SSNR function, 1/fparticleSSNR(s), as the signal-to-noise ratio “inside the particle region”. While this identification is appealing intuitively, it is not strictly correct because the scaled SSNR function contains low-frequency terms that describe the overall shape of the particle, not only its interior. Thus, the PSSNR term in the denominator of Eq. (2) does not correspond to the signal-to-noise ratio of an actual image (or volume), indicating that the SPW filter is distinct from a true Wiener filter. As we have shown, fparticle tends to diverge quite far from unity in typical single-particle applications (for example, 0.075 in the papillomavirus data set considered here), leading to substantially different behavior of the SPW filter compared with the Wiener filter.
The SSNR estimate includes contributions from image misalignment and other indirect error sources
Many sources of error can degrade the quality of a 3D reconstruction. Not only does error arise due to noise in the images themselves, but also due to errors in the orientation and translation parameters that have been assigned to the images during the course of structure refinement. Artifacts and errors in the 3D reconstruction algorithm itself will reduce the quality of the final map.
Importantly, the method we have described for estimating the SSNR of the data images, Eq. (6), does not distinguish between these various error sources. Because Eq. (6) is a measure of the consistency between two separate data sets after image processing is completed, this formula therefore yields a composite description of most or all sources of signal attenuation and noise. This feature of Eq. (6) is particularly advantageous in the process of single-particle orientation and translation refinement, because misalignment of images is a major source of signal attenuation (and hence resolution degradation) during single particle structure refinement. Eq. (6) will automatically measure a lower SSNR when images are misaligned. Thus, the SPW filter will behave more aggressively with poorly aligned images, and will do so in a way to “optimize” whatever signal does emerge after summing the current image alignment. Our approach, which parallels the Bayesian approach of Scheres (2012), contrasts with other Wiener filter methods (for example, Ludtke et al. (2001)) where the SSNR is estimated via separate measurements of the signal strength and noise strength, derived from the sample itself (Ludtke et al., 2001). This latter approach may lead to suboptimal behavior of the Wiener filter due to the presence of other, undetected error sources during refinement/reconstruction. On the other hand, our SSNR estimation approach, similar to that of Scheres (2012), is expected to filter away noise in the map due to alignment errors; this could in principle lead to faster and more accurate convergence of alignment parameters during 3D structure refinement.
The optimal SPW filter can be estimated without precise knowledge of the particle volume
Key to the successful application of the SPW reconstruction scheme is knowledge of both the image SSNR characteristics as well as the fractional particle volume, fparticle. We have shown how a combination of masking and FSC computation (Eq. (6)) allows the composite SSNR of the input images to be estimated with high accuracy. Perhaps more surprising was our finding that fparticle can be estimated via a real-space comparison of two half-data-set reconstructions (Fig. 5D), essentially in the absence of any knowledge of the particle/solvent boundary. We note that the accuracy of the estimate for fparticle depends on a number of factors, including the availability of an accurate estimate of the image SSNR (for example, by Eq. (6)). Indeed, some underestimation of the ground-truth image SSNR by Eq. (6) is apparent in Fig. 3D at higher spatial frequencies. A favorable aspect of our estimation scheme for fparticle, however, is that it inherently seeks the value which best optimizes the filter performance (as judged by the measured error between FSC half-data-set reconstructions). Thus, one expects fparticle to be underestimated for the data set in Fig. 3, in order to compensate for the underestimation of the SSNR. Consistent with this prediction, our methods report a value for fparticle that falls slightly below the molecular volume of the particle (Fig. 5D). Thus, within our formalism the fparticle term will function to at least partially compensate for errors in the determination of the image SSNR (insofar as correction is possible by a scalar factor), in order to better approximate the ‘perfect’ SPW filter.
We note that a potential problem occurs when the FSC computation is affected by over-refinement which can artificially increase the FSC (Stewart and Grigorieff, 2004). The increased FSC will increase the estimated SSNR (Eq. (6)) while also artificially increasing the real-space CCC. However, we would argue that once over-refinement has occurred, it is no longer possible to distinguish ‘real’ signal from artifactual signal due to noise correlations. The SPW defines ‘signal’ as the information that is consistently present between two half-data-set reconstructions and optimally represents this information in a least-squares sense, whether it is ‘real’ or artifact. As such, however, the SPW filter can itself be used to reduce the possibility of over-refinement, by suppressing noise in maps produced at intermediate stages during iterative single-particle parameter refinement. A related approach has recently been explored by Scheres (2012) with promising results (see below). In addition, the SPW filter can serve as a tool for the user to diagnose the presence of over-refinement, if the map has reached a resolution where recognizable features such as secondary structure or chain traces would be evident. If the resolution of the refinement has reached 8 Å, for example, alpha helices and beta sheets should be evident in the SPW-filtered map.
Comparison to previous implementations of the Wiener filter for 3D reconstruction
Zhang et al. (Zhang et al., 2008a) incorporated a Wiener filter into their nearest-neighbor Fourier inversion reconstruction algorithm, thus yielding an algorithm very similar to ours but lacking the fparticle term. Thus, although Zhang et al. do not give a detailed analysis of the effects of noise in their reconstruction algorithm, the expectation based on our analysis is that their implementation would produce strongly over-filtered maps. More recently, Scheres presented a 3D reconstruction scheme (Scheres, 2012) within a Bayesian formalism, yielding an algorithm very similar to the filters of Zhang et al. and in the current work. In Scheres’ method, the term corresponding to SSNR is multiplied by an adjustable coefficient T, which was arbitrarily set to 4. Thus, T corresponds to 1/fparticle in our formalism, and so yields a scheme that is expected to yield an approximate least-squares solution for the case of a particle that occupies ¼ of the reconstruction volume. While Scheres does not supply a detailed analysis of the reconstruction error as we have done, the multiplication by T would lead to substantially less over-filtering than the method of Zhang et al., although the implied value of 0.25 for fparticle nevertheless seems too high for many (if not most) cryo-EM images that are analyzed. Importantly, Scheres selected T not on the basis of minimizing the error found in the reconstructed map, but rather on a more indirect measure – T was selected so as to minimize the degree of noise bias that occurred during the course of a refinement loop. In the absence of more sophisticated schemes for minimizing noise bias, over-filtering the reference volume is expected to reduce noise bias during map refinement (Stewart and Grigorieff, 2004), so that T = 4 is probably a reasonable choice for this purpose unless the particle occupies an exceptionally large fraction of the map volume.
Post-processing variant of SPW corrects a previously proposed figure-of-merit scheme
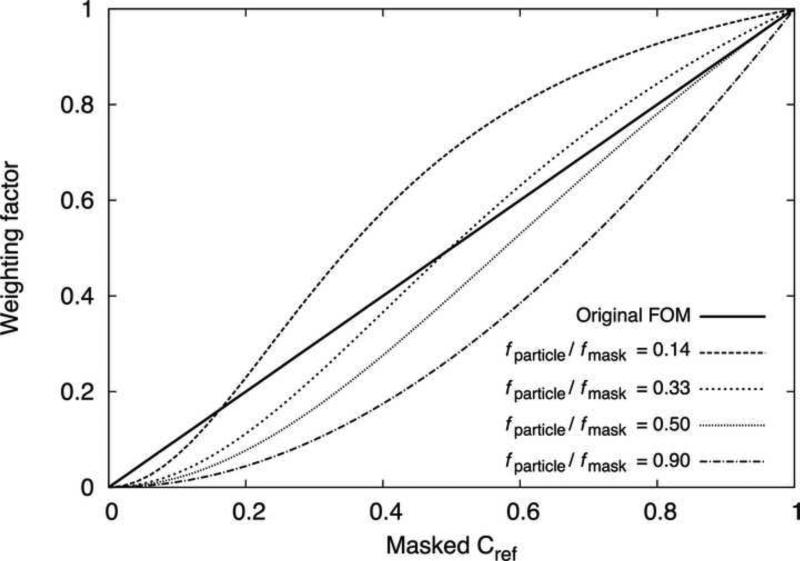
Rosenthal and Henderson (2003) observed that the error in a reconstructed 3D map is reduced when the structure factors are scaled by the FSC curve (Cref, or ‘figure-of-merit’) that would correspond to a comparison between the initially reconstructed map and the true but unknown, noise-free reference volume. In connecting our SPW filter to the weighting scheme of Rosenthal and Henderson, we identified a potentially significant correction to their formula. As seen by comparing Eqs. (10) and (11), in the limit of a particle that entirely fills the reconstruction volume (fparticle=1) our post-processing filter expression converges to the square of theirs. In non-limiting cases where fparticle < 1, the correction factor we derive is more complicated, but easily quantified (Fig. 7). Remarkably, we find that the FOM scheme yields filter values fairly close to our corrected expression when fparticle ≈ 0.33. While this value of fparticle is unrealistic for typical cryo-EM particles (as noted above), Rosenthal and Henderson compute the weighting factor using masked FSC calculations, which implicitly adds a correction factor of 1/fmask to the SSNR estimate for the reconstruction (see Eq. (6)). Thus, when masked volumes are used to compute the FSC, the FOM weighting scheme reasonably approximates a least squares solution when the ratio of the particle volume to the mask volume (fparticle / fmask) is ~0.33. This value is not unreasonable for typical soft-edged masks used in cryo-EM applications; for example, for the papillomavirus data set considered here (Wolf et al., 2010) we determined the particle/mask volume ratio to be ~0.3 (Fig. 6). Accordingly, the density map produced by SPW post-processing filter for this case was virtually indistinguishable from the FOM weighted map (results not shown).
Figure 7. Comparison between ‘Figure-of-Merit’ and SPW post-filtering schemes.
The weighting factor is plotted vs. the corresponding value of the masked Cref . The original FOM scheme of Rosenthal and Henderson (2003) is identical to the masked Cref, resulting in a straight line. The SPW post-processing filter, given by Eq. (9), varies according to the ratio of masked fparticle to fmask; examples for four different ratios are plotted.
Accounting for variability in particle quality and/or noise
One aspect of 3D reconstruction we have not explicitly considered here is the high variability in image quality that is usually inherent in a cryo-EM data set. Notably, our expression for estimating the SSNR of the image data (Eq. (6)) yields a single function that expresses the composite SSNR of the entire data set. In contrast, the experimental papillomavirus data set analyzed here contains particle images with significant variations in quality (Wolf et al., 2010). We addressed this variability using the identical methods as Wolf et al. (2010): within the FREALIGN refinement program, an exponential weighting function was applied to each particle Fourier transform (Grigorieff, 2007). While heuristic in nature, the FREALIGN weighting function adopts a similar mathematical form as the individual noise terms found in Wiener filter implementations where particle-to-particle variations in SSNR were explicitly accounted for (Ludtke et al., 2001; Scheres, 2012). We therefore anticipate that the SPW formalism could be expanded to include a formal treatment of variability in particle SSNR.
CONCLUSIONS
As a variant of the Fourier inversion method, the single-particle reconstruction scheme presented here is among the most computationally efficient. Furthermore, we have demonstrated that its accuracy (by FSC or real-space correlation criteria) exceeds that of other methods under carefully controlled testing conditions. Moreover, the theoretical relationships presented here clarify the relationship between particle size and error minimization, and are sufficiently general to be applied to other forms of image analysis.
METHODS
Generation of 2D Projection Images
A randomized set of viewing orientations was generated by first creating a set of 10000 quasi-uniformly spaced Euler angle triplets using the “VO EA” command from the SPIDER package (Frank et al., 1996). This set of 10000 Euler angles was then randomly sampled 1000 times to simulate 1000 random orientations of the particles. Projections were then generated using the resulting set of Euler angles. In order to avoid artifacts and/or signal loss at high resolutions due to interpolation, the following projection protocol was used. The atomic coordinates of 1MKJ were rotated in 3D space according to specified Euler angles, and subsequently used to generate a 3D Coulomb potential map (using CP FROM PDB from the SPIDER image processing package). We then formed a 2D projection image down the z-axis of the map coordinate system (using the PJ 3Q command from SPIDER). This projection protocol entirely avoids interpolation, and is thus predicted to maintain full signal strength all the way to the Nyquist frequency. This prediction is confirmed by a comparison of the average signal power in the projected images as a function of resolution (Fig. 1) to the average structure factors in the reference volume.
Contrast transfer function modulation for synthetic images
To ensure proper treatment of the simulated contrast transfer function (CTF) of the microscope, images were padded to a final size of 256 × 256 before convolving the noise-free projection images with the simulated CTF, thus allowing for information delocalization (Glaeser, 2007) to a distance of ~1 particle diameter = 96 Å from the boundary of the imaged particle. Each projection image was assigned a random defocus in an approximately uniform distribution between 0.5μm and 1.5μm. Other parameters for CTF simulation were: an accelerating voltage of 400kV, a spherical aberration constant of 4.1 (no CTF envelope function was modeled). Gaussian-distributed white noise images were generated using the MO function of SPIDER, and the noise images were scaled and added to the CTF-modulated molecular projections in order to produce a final signal-to-noise ratio (computed for the image size of 256 × 256) of 0.002.
Mask generation
Envelope mask volumes: For the synthetic data set, the known protein envelope mask volume was computed as the molecular surface of the 1MKJ coordinate set (Connolly, 1983), using a solvent radius parameter of 1.6. Experimental solvent mask volumes for FSC calculations were generated from the reconstructed maps similar to the method described in Grigorieff (2007), by applying a 14 Å low-pass filter to the maps and subsequently defining a binary envelope by selecting a density threshold such that the envelope contained a specified volume. The binary envelope was then smoothed by a cosine edge mask (edge distance was 14 Å).
Fourier inversion reconstruction algorithm
The FREALIGN software (Grigorieff, 2007) was used for all 3D reconstructions, but was modified to separately save to disk the accumulated sum of CTF-multiplied image data (numerator term in Eq. (A2.6)), as well as the accumulated sum of CTF squared terms (denominator term in Eq. (A2.6)). No parameter refinement was done in FREALIGN; all input parameters were either set to default values (for the synthetic data set), or taken from the published refinement (for the virus data set (Wolf et al., 2010)). The intermediate data files from FREALIGN were then read into the Octave open-source numerical analysis package (http://www.gnu.org/software/octave/doc/interpreter/), where subsequent analysis was completed.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the reviewers for providing extremely helpful and thorough comments and suggestions, and we thank Hemant Tagare for sharing his insights on the limitations of the Wiener filter. N.G. was supported by NIH Grant P01 GM- 623580.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Glaeser R. Electron Crystallography of Biological Macromolecules. Oxford University Press; USA: 2007. [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr. Opin. Struct. Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik Optik. 1986;73:146–156. [Google Scholar]
- Ludtke SJ, Jakana J, Song JL, Chuang DT, Chiu W. A 11.5 A single particle reconstruction of GroEL using EMAN. J. Mol. Biol. 2001;314:253–262. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- Penczek PA. Fundamentals of three-dimensional reconstruction from projections. Meth. Enzymol. 2010;482:1–33. doi: 10.1016/S0076-6879(10)82001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Saxton WO. Computer techniques for image processing in electron microscopy. Academic Press; New York: 1978. [Google Scholar]
- Scheres SHW. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV, Grigorieff N. An adaptation of the Wiener filter suitable for analyzing images of isolated single particles. J. Struct. Biol. 2011;176:60–74. doi: 10.1016/j.jsb.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano COS, Marabini R, Velázquez-Muriel J, Bilbao-Castro JR, Scheres SHW, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Van Trees H. Detection, estimation, and modulation theory. John Wiley & Sons; 2001. [Google Scholar]
- Wang BC. Methods in Enzymology. Academic Press; New York: 1985. Resolution of Phase Ambiguity in Macromolecular Crystallography. pp. 90–112. [DOI] [PubMed] [Google Scholar]
- Wiener N. Extrapolation, Interpolation, and Smoothing of Stationary Time Series. Wiley; New York: 1949. [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kimmel M, Spahn CMT, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008a;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. U.S.A. 2008b;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.