Abstract
Pancreas specific transcription factor 1a (Ptf1a), a bHLH transcription factor, has two temporally distinct functions during pancreas development; initially it is required for early specification of the entire pancreas, while later it is required for proper differentiation and maintenance of only acinar cells. The importance of Ptf1a function was revealed by the fact that loss of Ptf1a leads to pancreas agenesis in humans. While Ptf1a is one of the most important pancreatic transcription factors, little is known about the differences between the regulatory networks it controls during initial specification of the pancreas as opposed to acinar cell development, and to date no comprehensive analysis of its downstream targets has been published. In this paper, we use Xenopus embryos to identify putative downstream targets of Ptf1a. We isolated anterior endoderm tissue overexpressing Ptf1a at two early stages, NF32 and NF36, and compared their gene expression profiles using microarrays. Our results revealed that Ptf1a regulates genes with a wide variety of functions, providing insight into the complexity of the regulatory network required for pancreas specification.
Keywords: Xenopus, endoderm, Ptf1a, microarray, pancreas development
Introduction
The pancreas is an endodermal organ comprised of exocrine and endocrine cells. Exocrine tissue includes acinar cells that secrete digestive enzymes and ductal cells that transport the enzymes to the duodenum. Endocrine tissue is composed of five cells types, α, β, δ, ε and PP, which secrete glucagon, insulin, somatostatin, ghrelin and pancreatic polypeptide hormones, respectively, into the bloodstream.
The embryological origin of the pancreas is conserved from mammals to amphibians, where the pancreas develops from separate dorsal and ventral buds (Gittes, 2009; Kelly and Melton, 2000; Pearl et al., 2009). In Xenopus laevis, the dorsal bud arises first at NF35/36 just below the notochord at the level of the pronepheros. The ventral pancreas derives from two anlagen at the junction of the liver bud and developing duodenum at NF37/38. Morphological movements of the gastrointestinal tract during NF39 and 40 lead to the repositioning of the pancreatic buds driving their fusion and the formation of a single discrete organ (Jarikji et al., 2009; Kelly and Melton, 2000; Pearl et al., 2009). Prior to the emergence of the dorsal pancreatic bud, punctate expression of the beta cell differentiation marker insulin is detectable by in situ hybridization as early as NF32; subsequent expression within the ventral pancreas does not occur until NF47 (Horb and Slack, 2002; Kelly and Melton, 2000). Unlike insulin, expression of the other two endocrine markers, glucagon and somatostatin, are not restricted to the pancreas. These two genes are initially expressed in a punctate pattern within the stomach and duodenum at NF41, and are only detected in the pancreas beginning at NF44/45. The rapidity of pancreas development in Xenopus, coupled with its embryological and molecular benefits, makes it a useful system for functional studies on pancreas development (Blitz et al., 2006; Harland and Grainger, 2011; Pearl et al., 2012).
In the last few years our lab has utilized these benefits to characterize the function of several new genes in Xenopus pancreas development. First, in studying liver to pancreas transdifferentiation we found that Ptf1a and Pdx1 were sufficient to convert liver to pancreas in Xenopus transgenics (Horb et al., 2003; Jarikji et al., 2007). Second, we isolated individual dorsal and ventral pancreatic buds prior to fusion and used microarrays to compare their gene expression profiles. From this we characterized the function of three different genes, Insm1 (zinc finger transcription factor), Tm4sf3 (tetraspanin) and Brunol1 (RNA binding protein) and showed that they were involved in endocrine cell differentiation, fusion of the pancreatic buds and proliferation of endodermal progenitor cells, respectively (Horb and Horb, 2010; Horb et al., 2009; Jarikji et al., 2009). Third, we showed that Staufen2, an RNA binding protein, played an essential role in early patterning of the anterior endoderm (Bilogan and Horb, 2012). Fourth, we performed an exhaustive functional analysis of Rfx6, a neonatal diabetes candidate gene, and through microarray analysis identified new target genes involved in development of several anterior endodermal organs (Pearl et al., 2011). Fifth, we recently described a novel protocol to maximize production of pancreatic beta cells over alpha cells by controlled overexpression of the pancreatic endocrine transcription factor Ngn3 in naïve endoderm; using this gain-of-function phenotype we isolated endoderm shortly after Ngn3 expression and identified novel targets that function downstream of Ngn3 (Oropez and Horb, 2012). These studies demonstrate the benefits of using Xenopus to study pancreas development, namely the ability to perform both gain- and loss-of-function studies in naïve endoderm and the ability to make transgenics rapidly for tissue-specific overexpression studies. In this paper we describe a combined gain-of-function approach with microarray analysis to identify new target genes of a pancreatic transcription factor (Ptf1a) in Xenopus endoderm.
The basic helix-loop-helix (bHLH) protein Ptf1a (pancreas specific transcription factor 1a) is one of the first endodermal transcription factors restricted to early pancreatic progenitors before overt morphogenesis of the dorsal and ventral buds (Kawaguchi et al., 2002). Ptf1a has different stage-specific functions that are crucial for proper pancreas organogenesis. Initially, it is required to specify undifferentiated foregut endoderm into a pancreatic fate (Kawaguchi et al., 2002). Later, its expression becomes restricted to the cells at the tip of the pancreas where it functions in the initiation and maintenance of acinar cells (Krapp et al., 1996; Masui et al., 2007). The different functions are largely dependent on protein binding partners of Ptf1a, specifically which RBPJ paralog is bound by Ptf1a. RBPJ, the vertebrate Suppressor of Hairless [Su(H)], is found in the early PTF1 complex (PTF-J) and is required for the epithelial growth and development of the dorsal and ventral pancreatic buds. The PTF1-J complex binds to the Rbpjl promoter, activates its expression, and is eventually replaced by the RBPJL in the PTF1 complex (PTF1-L) during acinar cell differentiation (Masui et al., 2007). The PTF1-L complex then activates the promoters of genes encoding the secretory digestive enzymes (Beres et al., 2006). In humans, loss-of-function gene mutations in PTF1A lead to pancreatic and cerebellar agenesis (Sellick et al., 2004). Similarly, loss-of-function studies in mice and zebrafish revealed that functional Ptf1a is essential for exocrine cell development and the development of a subset of endocrine cells (Kawaguchi et al., 2002; Krapp et al., 1998; Lin et al., 2004; Zecchin et al., 2004).
In Xenopus, we found Ptf1a to be both necessary and sufficient for development of the pancreas. Morpholino knockdown of Ptf1a resulted in a complete loss of exocrine cells and loss of a subset of endocrine cells, while overexpression of Ptf1a was found sufficient to promote ectopic development of pancreatic cells (Afelik et al., 2006; Jarikji et al., 2007). Overexpression of ptf1a mRNA in naïve endoderm was sufficient to respecify early stomach and duodenal cells into both acinar and endocrine pancreas cells, while a superactive version was only capable of promoting an acinar cell fate. Similarly, we showed that transgenic overexpression of Ptf1a at later stages (after the foregut organs had formed) was able to promote ectopic endocrine and exocrine cell fates within the stomach and duodenum. Though we know Ptf1a can specify both endocrine and exocrine cell fates, the transcriptional hierarchy downstream of Ptf1a is still unknown.
In an attempt to elucidate the gene regulatory network activated by Ptf1a in early pancreas development we set out to identify gene expression changes that occurred upon overexpression of Ptf1a at the onset of pancreas development. Taking advantage of the Ptf1a overexpression phenotype, we compared control endodermal tissue to Ptf1a overexpression tissue at two early time-points, NF32 and NF36. This led to the identification of many genes with altered expression levels; the genes identified had varied functions revealing the diverse roles of Ptf1a in early pancreas development. We describe the expression of seven candidate genes with functions ranging from transcription, vesicle fusion and cell adhesion. In addition, we identified novel genes expressed within the developing pancreatic endoderm.
Results
Microarray analysis endodermal tissue overexpressing Ptf1a
We previously showed that overexpression of Pf1a was sufficient to convert stomach and duodenum to pancreas (Jarikji et al., 2007). Based on this phenotype we sought to identify downstream targets of Ptf1a by isolating endodermal tissue overexpressing Ptf1a at two early time-points and comparing the gene expression changes using microarrays. The first microarray was performed at NF32, which is shortly after endogenous Ptf1a expression begins (named MA32); the second microarray was performed at NF36 (MA36), which is 8 hours after the first time-point.
We injected ptf1a and gfp mRNAs together or gfp mRNA alone into the two dorso-vegetal blastomeres of eight-cell embryos, targeting the anterior endoderm. Forty (NF32) to forty-eight (NF36) hours later the anterior endoderm was dissected out and total RNA isolated. 15 endoderm explants were pooled for each RNA preparation, and both control and experimental samples were collected from the same clutch of embryos. This was done in triplicate at NF32 and in quadruplicate at NF36, each replicate coming from sibling embryos (Fig. 1a). The NF36 microarray was performed first using the Affymetrix 3′ Xenopus laevis Genome 1.0 GeneChip, and the NF32 microarray second using the Affymetrix 3′ Xenopus laevis Genome 2.0 GeneChip (the NF36 microarray was performed prior to the release of the 2.0 GeneChip). Primary analysis was done using the Affymetrix algorithm PLIER and globally normalized to 100% of the mean value. Multiple t-tests were used, including a Cyber-T test, a probability interval of 0.9 and 0.95 was set and only genes with a minimal fold change of 2.0 were identified as candidate genes. The analysis yielded 296 probe sets up-regulated at NF32 and 846 probe sets up-regulated at NF36.
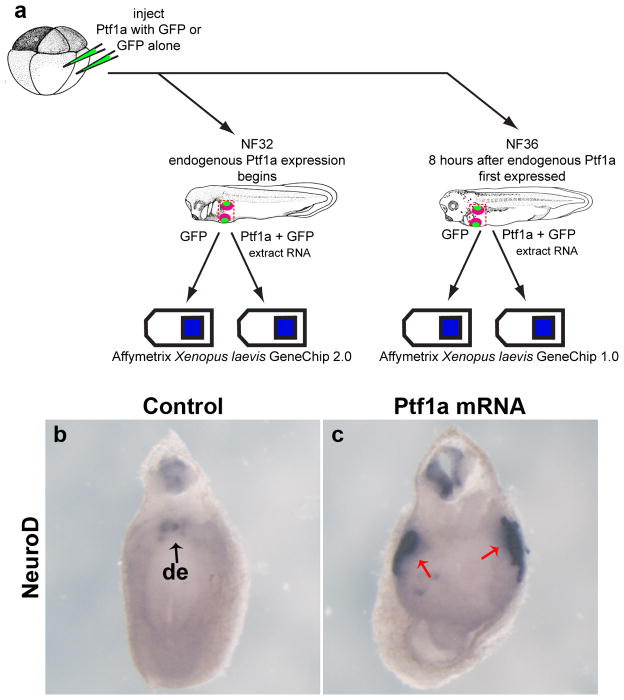
Figure 1.
Microarray experiment schematic and ISH verification. a: Schematic diagram of microarray experiment. Eight-cell stage embryos were injected with either 800pg of Ptf1a mRNA or 400pg of GFP mRNA. Samples were collected at NF32 and NF36; RNA was extracted and hybridized to Affymetrix Xenopus laevis GeneChip 2.0 and Affymetrix Xenopus laevis GeneChip 1.0, respectively. b: NeuroD expression in gfp-injected control embryos at NF32, punctate expression in the dorsal endoderm (black arrow). c: NeuroD expression in Ptf1a-injected embryos at NF32, increased expression in the dorsal endoderm (red arrows). de, dorsal endoderm.
In agreement with our previous data that showed Ptf1a was sufficient to promote ectopic acinar cell development (Jarikji et al., 2007), we found up-regulated expression of numerous exocrine genes in endodermal tissue overexpressing Ptf1a at both time-points. Carboxypeptidase A (CPA) was up-regulated 4-fold in both MA32 and MA36 (Table 1) and Carboxypeptidase B1 (CPB1) was up-regulated 6-fold in MA36 (Table 3). Endocrine genes were also found up-regulated, including Pax6, NeuroD, Carboxypeptidase E (CPE) and Somatostatin. Pax6 and CPE were found up-regulated 2.7-fold and 2.46-fold in MA32 (Table 2). NeuroD and somatostatin were found up-regulated 5-fold and 4.33-fold in MA36 (Fig. 1b,c, Table 3). In conjunction with our previous results that showed loss of stomach and duodenal tissue, we found down-regulated expression of several stomach and duodenal genes, such as villin-1 and intestinal fatty acid binding protein (IFABP) in endoderm overexpressing Ptf1a (data not shown, see GEO data GSE34193). In summary, we found the changes in gene expression identified in the microarrays were consistent with our earlier data, suggesting genes identified in the microarray are involved in promoting the ectopic pancreas phenotype.
Table 1. Genes up-regulated Ptf1a microarray at NF32 and NF36.
Genes found upregulated in both MA32 and MA36 relative to control samples. Genes are listed in descending order based on relative values in MA32. Unigene ID numbers that did not represent a known gene and duplicated genes were removed from this table. p < 0.05.
| Fold Change | |||
|---|---|---|---|
| Gene Title | Unigene ID | MA32 | MA36 |
| Matrix metalloproteinase 1 | Xl.26520 | 5.98 | 10.39 |
| Pancreas-specific transcription factor 1a | Xl.29862 | 5.81 | 7.74 |
| Immunoresponsive gene 1 | Xl.53505 | 5.70 | 10.06 |
| Interleukin-1-beta | Xl.415 | 5.58 | 9.37 |
| Peripherin | Xl.16232 | 5.51 | 5.26 |
| Transmembrane protein TA-2-like | Xl.736 | 5.50 | 9.44 |
| Apolipoprotein O-like | Xl.24724 | 5.30 | 4.97 |
| ELAV-like protein 4 | Xl.1036 | 5.27 | 4.22 |
| Soluble toll-like receptor 5 | Xl.53947 | 5.23 | 8.52 |
| Cholesterol 25-hydroxylase | Xl.18444 | 5.13 | 8.40 |
| Matrix metalloproteinase 18 | Xl.17465 | 4.98 | 9.10 |
| Fast skeletal troponin C beta | Xl.24572 | 4.94 | 4.74 |
| Stathmin-like 2 | Xl.11 | 4.90 | 5.08 |
| Troponin T type 3 (skeletal, fast) | Xl.3362 | 4.89 | 4.96 |
| Dihydropyrimidinase-like 3 | Xl.5161 | 4.76 | 3.03 |
| Similar to Parvalbumin (calcium binding protein) | Xl.9653 | 4.41 | 4.13 |
| Olfactomedin 4 | Xl.17389 | 4.40 | 4.86 |
| Tumor necrosis factor, alpha-induced protein 3 | Xl.1952 | 4.40 | 6.15 |
| Neuronal pentraxin I | XlAffx.54 | 4.37 | 5.99 |
| Oncomodulin | Xl.48817 | 4.31 | 5.77 |
| Synaptotagmin 4 | Xl.34868 | 4.06 | 2.95 |
| Tumor necrosis factor receptor superfamily, member 12A | Xl.34690 | 3.96 | 4.82 |
| Carboxypeptidase A1 | Xl.73839 | 3.96 | 3.85 |
| Jun B proto-oncogene | Xl.52744 | 3.96 | 5.46 |
| Actinin, alpha 3 | Xl.76514 | 3.95 | 3.11 |
| TNFAIP3 interacting protein 2 | Xl.47119 | 3.84 | 6.47 |
| Amphiphysin | Xl.14350 | 3.82 | 2.87 |
| Platelet-activating factor receptor | Xl.4290 | 3.81 | 6.27 |
| Early growth response protein 1-A | Xl.637 | 3.79 | 5.44 |
| Internexin neuronal intermediate filament protein, alpha | Xl.13474 | 3.74 | 3.41 |
| Syntaxin 1B | Xl.49106 | 3.69 | 3.29 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | Xl.78163 | 3.58 | 6.09 |
| Phosphoglycerate mutase 2 | Xl.12227 | 3.55 | 4.81 |
| Proto-oncogene c-Fos | Xl.10283 | 3.48 | 4.39 |
| Interferon regulatory factor 1 | Xl.1419 | 3.45 | 7.13 |
| Suppressor of cytokine signaling 3 | Xl.26512 | 3.45 | 5.73 |
| Tumor necrosis factor receptor superfamily, member 11b | Xl.47506 | 3.44 | 4.37 |
| Pleiotrophin | Xl.900 | 3.34 | 4.41 |
| Early B-cell factor 2 | Xl.547 | 3.19 | 4.15 |
| Olfactomedin 1 | Xl.8515 | 3.16 | 2.40 |
| Matrix metalloproteinase 9 | Xl.25126 | 3.13 | 4.96 |
| Mab-21-like 1 | Xl.29719 | 3.13 | 2.58 |
| NUAK family, SNF1-like kinase, 2 | Xl.8315 | 3.03 | 3.12 |
| Growth arrest and DNA-damage-inducible, gamma | Xl.12125 | 2.98 | 4.01 |
| TRAF interacting protein TANK | Xl.53193 | 2.98 | 3.47 |
| Pleckstrin homology-like domain family A member 2 | Xl.57018 | 2.82 | 2.89 |
| Hes5.2-a | Xl.586 | 2.76 | 3.06 |
| Thrombospondin 3 | Xl.198 | 2.61 | 4.37 |
| Keratin | Xl.76286 | 2.61 | 5.84 |
| Hyaluronan and proteoglycan link protein 3 (Hapln3) | Xl.22996 | 2.52 | 5.35 |
| TRAF-interacting protein with FHA domain-containing protein A | Xl.49742 | 2.51 | 3.57 |
| TNFAIP3 interacting protein 1 | Xl.15972 | 2.48 | 3.73 |
| Sox3 | Xl.22 | 2.48 | 2.59 |
| Claudin 5 | Xl.14214 | 2.47 | 4.47 |
| Rho-related GTP-binding protein Rho6 precursor | Xl.7206 | 2.43 | 4.11 |
| Hairy/enhancer-of-split related with YRPW motif 1 (Hey1) | Xl.469 | 2.43 | 4.68 |
| Adenosine deaminase | Xl.80690 | 2.42 | 4.65 |
| Tribbles homolog 1 | Xl.75411 | 2.41 | 3.26 |
| B-cell translocation gene 5 | Xl.51810 | 2.20 | 3.96 |
| Serpine1 | Xl.25094 | 2.18 | 3.80 |
| B-cell translocation protein 2 | Xl.48530 | 2.13 | 3.57 |
| FXYD domain containing ion transport regulator 6 | Xl.18084 | 2.12 | 3.43 |
| Bone morphogenetic protein 2 A | Xl.1138 | 2.04 | 3.18 |
| ATPase, Na+/K+ transporting, alpha 2 polypeptide | Xl.10662 | 2.03 | 3.98 |
Table 3. Genes up-regulated in Ptf1a microarray at NF36.
Genes have been sorted based on their fold change in MA36 relative to control samples. Only those with greater than 4-fold upregulation are listed. Unigene ID numbers that did not represent a known gene and duplicated genes were removed from this table. p < 0.05.
| Gene Title | Unigene ID | Fold Change |
|---|---|---|
| Phosphoenolpyruvate carboxykinase 1, cytosolic | Xl.12459 | 8.92 |
| Glucose-6-phosphatase, catalytic | Xl.12866 | 7.44 |
| Ornithine decarboxylase-2 | Xl.8949 | 7.23 |
| Phosphoenolpyruvate carboxykinase 1 (soluble) | Xl.18730 | 7.03 |
| Similar to glucose-6-phosphatase | Xl.5165 | 6.95 |
| Uroplakin III | Xl.25311 | 6.65 |
| Insulin-like growth factor binding protein 1 | Xl.34669 | 6.61 |
| Na+/K+-ATPase beta 2 subunit | Xl.53667 | 6.57 |
| Hydrogen/potassium-exchanging ATPase 12A b | Xl.2659 | 6.46 |
| Uncoupling protein 3 (mitochondrial, proton carrier) | Xl.49433 | 6.46 |
| Arginase, non-hepatic 1 | Xl.892 | 6.43 |
| Carboxypeptidase B1 | Xl.26001 | 6.38 |
| Similar to deoxyribonuclease I-like 3 | Xl.6017 | 6.26 |
| CCAAT-enhancer binding protein delta | Xl.29876 | 6.24 |
| Serine/threonine-protein kinase Sgk1-A | Xl.7842 | 6.22 |
| Anterior gradient 2 | Xl.25847 | 6.21 |
| Paired box protein Pax-9 | Xl.21602 | 6.06 |
| Uroplakin 1B | Xl.8143 | 5.93 |
| Tripartite motif-containing 29 | Xl.9051 | 5.85 |
| Keratin, type I cytoskeletal 47 kDa | Xl.66586 | 5.82 |
| Cornifelin homolog B | Xl.23712 | 5.82 |
| Matrix metalloproteinase 13 | Xl.9058 | 5.73 |
| Caveolin-3 | Xl.53974 | 5.69 |
| Peroxisomal membrane protein 2, 22kDa | Xl.83319 | 5.66 |
| Pyruvate dehydrogenase kinase 2 | Xl.1717 | 5.55 |
| ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 | Xl.15867 | 5.54 |
| Claudin 4 | Xl.53796 | 5.54 |
| Purine nucleoside phosphorylase | Xl.57569 | 5.53 |
| Arrestin domain containing 2 | Xl.19508 | 5.52 |
| Otogelin | Xl.847 | 5.50 |
| V-type proton ATPase subunit G 3 | Xl.34612 | 5.43 |
| Serpin peptidase inhibitor, clade E, member 2 | Xl.12045 | 5.31 |
| Glutathione S-transferase P 1 | Xl.54920 | 5.27 |
| L-lactate dehydrogenase C chain | Xl.4591 | 5.25 |
| Glucose-6-phosphatase, catalytic, 2 | Xl.47697 | 5.21 |
| Guanylyl cyclase-1 | Xl.906 | 5.17 |
| Prostaglandin-endoperoxide synthase | Xl.78035 | 5.09 |
| Serine dehydratase | Xl.72557 | 5.05 |
| Phospholipid scramblase 1 | Xl.60828 | 5.04 |
| Neurogenic differentiation 1(NeuroD) | Xl.26304 | 5.02 |
| Carbonic anhydrase XII | Xl.10362 | 4.97 |
| Amiloride-sensitive sodium channel subunit beta-2 | Xl.1175 | 4.97 |
| Keratin 15 | Xl.57085 | 4.97 |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 | Xl.8104 | 4.95 |
| Claudin 1 | Xl.16310 | 4.94 |
| Sox9 | Xl.1690 | 4.92 |
| Tetraspanin 1 | Xl.39094 | 4.92 |
| X-epilectin | Xl.6048 | 4.91 |
| ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 | Xl.8939 | 4.88 |
| Sciellen | Xl.15658 | 4.88 |
| Calpain 9 | Xl.47406 | 4.87 |
| Pendrin-like anion exchanger | Xl.10887 | 4.78 |
| Lectin, galactoside-binding, soluble, 3 | Xl.1779 | 4.76 |
| Collagen, type II, alpha 1 (col2a1) | Xl.606 | 4.74 |
| ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A, isoform 1 | Xl.54891 | 4.72 |
| Collagen, type I, alpha 1 | Xl.47042 | 4.65 |
| Collagen, type I, alpha 2 | Xl.49126 | 4.65 |
| Sestrin-1 | Xl.12255 | 4.58 |
| Collagen, type II, alpha 1 (col2a1) | Xl.606 | 4.57 |
| Alkaline phosphatase | Xl.1299 | 4.57 |
| Collagen, type VI, alpha 1 (col6a1) | Xl.9863 | 4.55 |
| Cleavage stimulation factor, 3′ pre-RNA, subunit 3, 77kDa, b | Xl.24776 | 4.53 |
| T-box transcription factor TBX1 | Xl.51448 | 4.50 |
| Forkhead box protein E1 | Xl.50069 | 4.48 |
| Versican | Xl.70991 | 4.46 |
| Villin-like | Xl.15156 | 4.45 |
| Osteoglycin | Xl.21905 | 4.45 |
| Synaptotagmin-like 2 | Xl.52466 | 4.38 |
| Fibroblast growth factor binding protein 1 | Xl.48872 | 4.38 |
| Augurin precursor | Xl.21897 | 4.37 |
| ATPase, H+ transporting, lysosomal V0 subunit a4 | Xl.2715 | 4.34 |
| Somatostatin | Xl.13593 | 4.33 |
| NIPA-like domain containing 4 | Xl.5679 | 4.31 |
| Actin, alpha 2, smooth muscle, aorta (acta2) | Xl.83891 | 4.30 |
| Phosphoinositide-3-kinase interacting protein 1 | Xl.77369 | 4.30 |
| Distal-less homeobox 3 (dlx3-a) | Xl.838 | 4.30 |
| Transcription factor AP-2 alpha (tfap2a-a) | Xl.2143 | 4.28 |
| Glutamate-ammonia ligase (glutamine synthase) (glul) | Xl.47079 | 4.25 |
| GDP-mannose 4, 6-dehydratase | Xl.10260 | 4.23 |
| Calpain 1, (mu/I) large subunit | Xl.3776 | 4.23 |
| D-dopachrome decarboxylase-A | Xl.23562 | 4.23 |
| Adenylate kinase 1 | Xl.25877 | 4.22 |
| 5-hydroxytryptamine (serotonin) receptor 3A | Xl.9068 | 4.22 |
| Myosin, heavy chain 6, cardiac muscle, alpha | Xl.3161 | 4.22 |
| Arrestin domain containing 3 | Xl.25985 | 4.19 |
| Annexin A1 | Xl.2308 | 4.18 |
| Macrophage stimulating 1 receptor (c-met-related tyrosine kinase) | Xl.115 | 4.18 |
| Keratin 16 | Xl.15435 | 4.15 |
| Sal-like 4 (sall4) | Xl.77515 | 4.15 |
| Zinc finger protein 36, C3H type, homolog | Xl.26472 | 4.10 |
| Distal-less homeobox 5 (dlx-5) | Xl.18401 | 4.09 |
| Histidine ammonia-lyase | Xl.2256 | 4.08 |
| Fucosyltransferase 6 (alpha (1,3) fucosyltransferase) | Xl.16504 | 4.07 |
| Aquaporin-3 | Xl.34454 | 4.07 |
| Phosphatidylinositol glycan anchor biosynthesis (PIG-S) | Xl.48652 | 4.05 |
| Aquaporin 1 | Xl.5043 | 4.04 |
| Myosin, light chain 4, alkali; atrial, embryonic | Xl.11969 | 4.03 |
| Plasminogen activator, tissue | Xl.14787 | 4.02 |
| Coagulation factor II (thrombin) receptor-like 2 | Xl.23535 | 4.01 |
Table 2. Genes up-regulated in Ptf1a microarray at NF32.
Genes have been sorted based on their fold change in MA32 relative to control samples. Only those with greater than 2-fold upregulation are listed. Unigene ID numbers that did not represent a known gene and duplicated genes were removed from this table. p < 0.05.
| Gene Title | Unigene ID | Fold Change |
|---|---|---|
| Matrix metallopeptidase 8 | Xl.49512 | 5.21 |
| Fatty acid binding protein 7, brain | Xl.48742 | 5.00 |
| Thrombospondin 4 | Xl.9871 | 4.92 |
| Tubulin, beta 3 | Xl.57483 | 4.75 |
| Similar to oncomodulin | Xl.3153 | 4.70 |
| Fast troponin I | Xl.2142 | 4.62 |
| Myosin light chain, phosphorylatable, fast skeletal muscle | Xl.6263 | 4.60 |
| Oncomodulin | Xl.14726 | 4.48 |
| Calcium ATPase at 60A | Xl.11405 | 4.46 |
| Fast skeletal troponin C beta | Xl.1032 | 4.34 |
| SNAP25a | Xl.8686 | 4.32 |
| Slow troponin I | Xl.11074 | 4.30 |
| Neuritin 1-A | Xl.11951 | 4.04 |
| Low molecular weight neuronal intermediate filament | Xl.992 | 3.99 |
| Alpha-actinin-2 | Xl.34825 | 3.95 |
| Growth associated protein 43 | Xl.9994 | 3.90 |
| Similar to Parvalbumin (calcium binding protein) | Xl.25492 | 3.87 |
| Neuritin-B precursor | Xl.34198 | 3.85 |
| Neuro-oncological ventral antigen 1 | Xl.47571 | 3.81 |
| Solute carrier family 1 | Xl.20736 | 3.80 |
| Troponin I type 2 (skeletal, fast) | Xl.18396 | 3.78 |
| Myosin binding protein H | Xl.6142 | 3.77 |
| Guanine nucleotide binding protein (G protein), gamma 3 | Xl.24553 | 3.67 |
| Tropomyosin 2 a isoform 1 | Xl.21893 | 3.58 |
| Solute carrier family 1 | Xl.20736 | 3.58 |
| Muscle-related coiled-coil protein | Xl.57797 | 3.50 |
| Phosphoglycerate mutase 2 | Xl.84002 | 3.36 |
| Enolase 1 alpha | Xl.66912 | 3.34 |
| Calcium-dependent secretion activator 1 | Xl.26179 | 3.34 |
| Growth associated protein 43 | Xl.1133 | 3.33 |
| Enolase 3, beta muscle | Xl.19056 | 3.32 |
| Beta-synuclein | Xl.14705 | 3.26 |
| Sox10 | Xl.1588 | 3.24 |
| Actin, alpha skeletal muscle | Xl.70068 | 3.22 |
| Synuclein, gamma (breast cancer-specific protein 1) | Xl.11348 | 3.22 |
| Middle molecular weight neurofilament protein NF-M(1) | Xl.173 | 3.19 |
| Aspartyl beta-hydroxylase | Xl.23399 | 3.10 |
| RUN and FYVE domain containing 3 | Xl.8516 | 3.01 |
| Acetylcholine receptor subunit alpha-1-A | Xl.1118 | 3.00 |
| Desmin | Xl.2665 | 2.99 |
| Syntaxin binding protein 1 | Xl.26111 | 2.96 |
| Jun oncogene | Xl.541 | 2.93 |
| Dihydropyrimidinase-like 4 | Xl.77348 | 2.90 |
| Jun proto-oncogene | Xl.542 | 2.90 |
| Similar to calsequestrin 2 (cardiac muscle) | Xl.15700 | 2.87 |
| Protein tweety homolog 1-A | Xl.11254 | 2.85 |
| Collapsin response mediator protein 1 | Xl.50837 | 2.83 |
| Chondromodulin-I precursor | Xl.21971 | 2.79 |
| Synaptophysin | Xl.230 | 2.79 |
| Transcription factor Sp8 | Xl.49163 | 2.77 |
| Neural cell adhesion molecule L1 | Xl.21565 | 2.76 |
| Tubulin | Xl.8919 | 2.76 |
| Protocadherin 8 | Xl.72042 | 2.75 |
| CUGBP Elav-like family member 3-B | Xl.12160 | 2.73 |
| CUGBP Elav-like family member 3-A | Xl.986 | 2.73 |
| Pax6 | Xl.647 | 2.69 |
| Synaptophysin | Xl.232 | 2.66 |
| Dihydropyrimidinase-like 2 | Xl.3001 | 2.65 |
| Kinesin family member 5A | Xl.18417 | 2.64 |
| Middle molecular weight neurofilament protein NF-M(2) | Xl.174 | 2.61 |
| VENT homeobox 1 | Xl.1420 | 2.57 |
| MICAL-like 2 | Xl.34034 | 2.56 |
| Synaptotagmin 4 | Xl.48319 | 2.55 |
| Neuritin 1-A | Xl.11951 | 2.53 |
| Tubulin beta-2 chain | Xl.121 | 2.50 |
| Diacylglycerol kinase iota | Xl.16337 | 2.49 |
| Carboxypeptidase E | Xl.8483 | 2.46 |
| Cell adhesion molecule 3 precursor | Xl.23549 | 2.45 |
| Neuronal pentraxin-2 precursor | Xl.11864 | 2.45 |
| Reticulon-1-B | Xl.8555 | 2.45 |
| Actin, alpha skeletal muscle 3 | Xl.24656 | 2.43 |
| Lysyl oxidase homolog 3 precursor | Xl.13606 | 2.42 |
| Similar to myozenin 1 | Xl.21921 | 2.41 |
| Trimeric intracellular cation channel type A | Xl.45053 | 2.39 |
| Seizure protein 6 homolog precursor | Xl.51942 | 2.36 |
| Guanine nucleotide-binding protein G(O) alpha subunit 1 | Xl.20976 | 2.36 |
| Fox-1 homolog C | Xl.57891 | 2.35 |
| Coiled-coil domain containing 28B | Xl.9739 | 2.35 |
| Zinc finger protein ZIC 1 | Xl.1796 | 2.33 |
| RAB33A, member RAS oncogene family | Xl.47462 | 2.33 |
| Enah/Vasp-like | Xl.12544 | 2.32 |
| Wnt-8 | Xl.49 | 2.29 |
| Stathmin-like 3 | Xl.21810 | 2.29 |
| Hes-4-A | Xl.25977 | 2.20 |
| Sodium/potassium-transporting ATPase subunit beta-1- interacting protein 4 | Xl.55497 | 2.20 |
| Zinc finger protein 36, C3H type, homolog | Xl.24023 | 2.17 |
| Calcium channel, voltage-dependent, beta 3 subunit | Xl.53876 | 2.09 |
| Hes5.1 | Xl.48575 | 2.03 |
Identification of temporally regulated genes
To compare differential gene up-regulation in the two different time-points, we separated the genes based on whether they were up-regulated in both MA32 and MA36 or only in one time-point. Using MA32 as the base we identified 297 genes to be up-regulated greater than 2-fold in both MA32 and MA36 (Table 1). At NF32, we identified 143 genes to be up-regulated only at this stage, while at NF36 there were 702 up-regulated genes (Tables 2 & 3). Due to the extended amount of time between onset of endogenous Ptf1a expression and sample collection, we believe those genes up-regulated at the later stage represent secondary or tertiary targets of Ptf1a and not direct targets.
To provide insight into the different genes regulating pancreas specification downstream of Ptf1a at NF32 versus NF36, we used DAVID (Database for Annotation, Visualization and Integrated Discovery) to map the up-regulated genes from both microarrays to enriched GO Biological Process categories (Huang da et al., 2009a; Huang da et al., 2009b). Gene annotation enrichment analysis revealed the top three functions at MA32 were regulation of transcription, regulation of RNA metabolic process and biological adhesion. At this stage there appears to be enrichment of genes that may function in cell fate specification and in cell biological processes, such as adhesion for tissue patterning. In comparison, at MA36 the top three functions were ion transport, nitrogen compound biosynthetic process and purine nucleotide metabolic process. As expected, genes at this later stage appear to be enriched in functions indicative of differentiated cells in comparison to naïve cells undergoing specification.
Temporally regulated genes were also mapped using SwissProt to determine enriched functions. The top 3 enhanced keywords at MA32 were developmental proteins, calcium and intermediate filaments. At MA36, the top 3 enhanced functional similarities were transmembrane related proteins, calcium and intermediate filaments. The enrichment of developmental proteins at MA32 suggested Ptf1a was regulating the expression of genes required to define the pancreatic fate at this early stage. At MA36, the most enriched protein family was transmembrane related proteins, which included many proteins with functions in signal transduction pathways (e.g., guanylyl cyclase 1, EPH receptor B1, Syndecan-4). The difference between the enriched protein families across the time-points correlates with our analysis that up-regulated genes at NF32 are functioning in cell fate specification, while at NF36 the cells are more differentiated and these putative secondary or tertiary Ptf1a target genes are involved in maintaining proper cell function.
Analysis of the 1142 probe sets up-regulated in both microarrays showed the most enriched GO Biological Process category was transcriptional regulation. Of the 45 transcription factors that were up-regulated, we identified known pancreatic transcription factors that function downstream of Ptf1a (NeuroD1, Pax6, Sox9), which have been shown to be expressed in the pancreas at these two stages. We also identified many genes with known functions in pancreas development that have yet to be identified as Ptf1a targets (JunB, IRF1, Hey1). However, the majority of up-regulated genes have unknown functions in pancreas development (Sox3, Hairy2, FoxC1), though related family members have been shown to play a role in pancreas development. The identification of both known and unknown pancreatic genes suggested that these new transcription factors might be part of the initial Ptf1a gene regulatory network. However, further functional analysis is required to validate and understand the roles of these potential pancreatic transcription factors.
Genes up-regulated in Ptf1a microarray are expressed during early pancreas specification
To validate genes involved in early pancreatic specification from the up-regulated probe sets, we chose to examine the spatial and temporal expression of 94 genes by whole-mount in situ hybridization (ISH). We confirmed 50 of these genes to be expressed in anterior endoderm, but we focused our attention on the 32 that showed specific expression within the mature pancreas (between NF40 and NF46). Of the 32 genes expressed in the mature pancreas, 9 genes showed expression within the anterior endoderm at the level of the developing pancreas between NF32-36. The 9 endodermally expressed genes were: syntaxin binding protein 1 (Stxbp1), putative transmembrane protein TA-2, cholesterol 25-hydroxylase (C25H), Insulin-like growth factor binding protein 1 (Igfbp1), interferon regulatory factor 1 (Irf1), hyaluronan and proteoglycan link protein 3 (Hapln3), hairy/enhancer-of-split related with YRPW motif 1 (Hey1), sestrin1, syndecan-4.
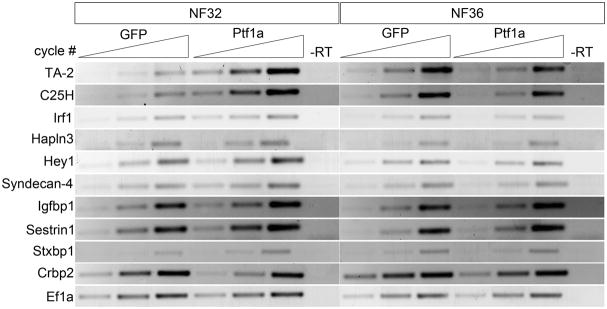
According to the microarray analysis six of the genes, TA-2, C25H, Irf1, Hapln3, Hey1 and Syndecan-4, were found significantly up-regulated at both NF32 and NF36; Igfbp1 and Sestrin1 were only up-regulated at NF36, whereas Stxbp1 was only up-regulated at NF32. Consistent with the microarray data, we confirmed relative expression for seven of these genes by RT-PCR (Fig. 2). However, we found expression of C25H and TA-2 to be reduced in the Ptf1a samples at MA36 (Fig. 2). As a control, we examined expression changes for one of the genes found decreased in both MA32 and MA36, cellular retinol binding protein 2 (Crbp2). In agreement with the microarray data, we found it to be decreased in Ptf1a overexpressing endoderm (Fig. 2). These results suggested that genes identified in the analysis were targets of Ptf1a, and based on the confirmation of the RT-PCR and their known functions and putative roles in pancreas development, we examined seven genes in more detail, Hey1, Irf1, Stxbp1, Syntaxin 1B, Igfbp1, Hapln3 and Syndecan-4.
Figure 2.
RT-PCR verification of selected differentially expressed genes from the microarray analyses. GFP and Ptf1a sample data provided from three different cycle time-points. GFP, gfp mRNA-injected control; Ptf1a, ptf1a mRNA-injected; -RT, control reaction without reverse transcriptase.
Hey1
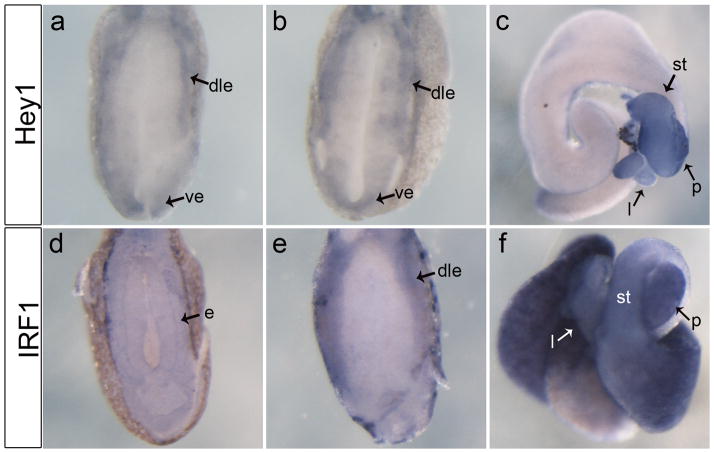
Hey1 is a bHLH transcriptional repressor protein that was shown to interact with Ptf1a and is expressed in both endocrine and exocrine cells of the human pancreas (Ghosh and Leach, 2006; Johansson et al., 2008). In Xenopus embryos, we first detected hey1 expression at NF32 along the periphery of the endoderm (Fig. 3a,b). At later stages expression became restricted to the stomach and pancreas (Fig. 3c). To confirm the RT-PCR data we compared the spatial expression of hey1 in control and ptf1a-injected embryos. At NF32, expression of hey1 in ptf1a-injected embryos was stronger particularly in the ventral and dorsal region of the endoderm where the pancreatic buds will arise (Fig. 6a,b). At NF36, expression of hey1 remained much stronger in the endoderm in comparison to the controls (Fig. 6c,d). Expression of hey1 in the developing endoderm and up-regulation in the region of the prospective pancreatic buds suggests it is a downstream target of Ptf1a in vivo and likely plays a role in pancreatic cell specification.
Figure 3.
Expression pattern of transcription factors Hey1 and Irf1 in the developing endoderm. a: Transverse section through anterior endoderm at NF32 showing expression of hey1 restricted to the outer periphery of the endoderm. b: Expression of hey1 is more diffuse within the endoderm of the NF36 embryo, transverse section. c: NF44 whole gut showing expression of hey1 throughout the stomach, liver and pancreas. d: Transverse section through anterior endoderm at NF32 showing expression of irf1 throughout the entire endoderm. e: Transverse section at NF36 shows restricted expression of irf1 along the dorsolateral endoderm. f: NF44 whole gut showing expression of irf1 throughout the stomach, liver, pancreas and in the intestine. dle, dorsolateral endoderm; ve, ventral endoderm; e, endoderm; l, liver; st, stomach; p, pancreas.
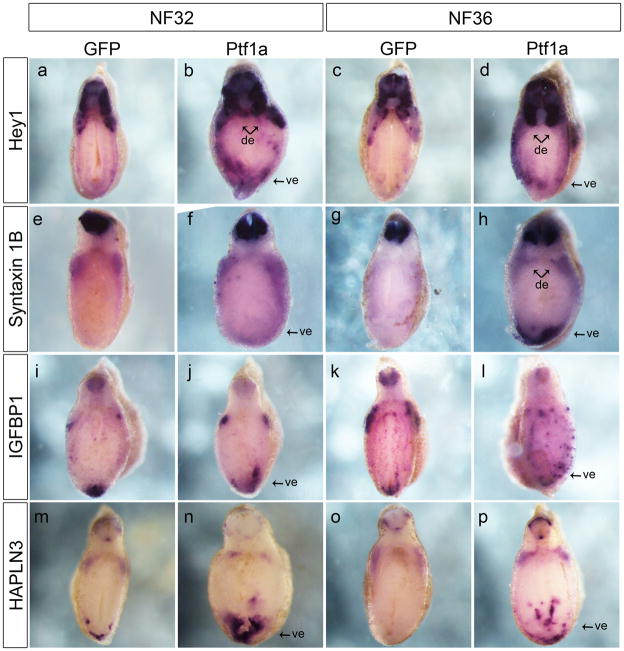
Figure 6.
Ptf1a overexpression promotes ectopic expression of candidate genes in the developing endoderm. All images are transverse section through the anterior endoderm. a,b: Stronger expression and expanded domain of hey1 in the anterior endoderm of a ptf1a mRNA injected embryo at NF32 (7/10). c,d: At NF36, expression of hey1 is stronger along the periphery of the endoderm (8/11). e,f: At NF32, syntaxin 1B expression is stronger in the dorsal endoderm and expands ectopically towards the ventral endoderm (7/10). g,h: Expression of syntaxin 1B is much stronger in the dorsal and ventral endoderm of ptf1a-injected embryos at NF36 (6/10). i,j: igfbp1 expression is expanded in the ventral endoderm but unchanged in the dorsal endoderm of ptf1a-injected embryos at NF32 (9/9). k,l: At NF36 ptf1a-injected embryos the punctate expression of igfbp1 is stronger and more abundant (8/11). m,n: Expression domain of hapln3 in the ventral endoderm is stronger and broader in ptf1a-injected embryos at NF32 (6/10). o,p: At NF36, expression of hapln3 is ectopically expressed in the ventral endoderm but unaffected in the dorsal endoderm (5/10). de, dorsal endoderm; ve, ventral endoderm.
Irf1
Irf1 was originally identified as a regulator of interferon (IFN)-β and is a key factor in the transcriptional regulation of the IFN response, but it has also been studied for its role in cell growth regulation (Fujita et al., 1988; Kröger et al., 2002). In the domain of the developing pancreas in Xenopus at NF32, irf1 was expressed along the ventrolateral endoderm; this expression expanded laterally by NF36 (Fig. 3d,e). Within the mature gut, the expression was dispersed throughout the duodenum and liver (Fig. 3f). The developing anterior endoderm not only gives rise to the pancreas, but also the liver, stomach and intestine. Since overexpression of Ptf1a is sufficient to convert stomach and duodenum to pancreas it is possible that Ptf1a may activate genes, such as Irf1, that inhibit proper stomach and duodenum development.
Stxbp1
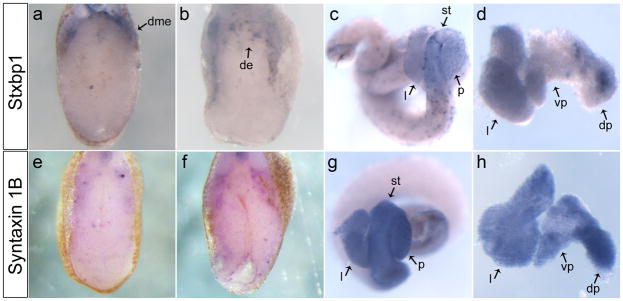
In our microarray, we also identified several proteins involved in vesicle fusion. As examples, we focused our analysis on Stxbp1 and Syntaxin 1B, as they were both up-regulated over two-fold. Stxbp1 was originally identified for it role in exocytosis through yeast genetics as Sec1 (Ferro-Novick and Jahn, 1994). Homologs have been identified in many species from Caenorhabditis elegans (UNC-18) to mammals (Munc-18) (Hata et al., 1993; Hosono et al., 1992). Stxbp1 is known to function in neuronal cells as well as in pancreatic islets and insulin-producing cells (Jacobsson et al., 1994). In Xenopus embryos, stxbp1 was expressed within the dorsal mesendoderm border at NF32 (Fig. 4a), by NF36 it was expressed in a punctate fashion within the dorsal endoderm at the level of the developing pancreatic bud (Fig. 4b). In the mature gut, stxbp1 expression was punctate throughout the stomach, duodenum and pancreas (Fig. 4c,d). These results indicate that stxbp1 was expressed in a punctate pattern in the developing dorsal pancreatic bud, similar to the expression of insulin in the dorsal anlagen prior to pancreas morphogenesis.
Figure 4.
Expression of SNARE complex proteins in the developing endoderm. a: Stxbp1 expressed along the dorsal mesendoderm periphery in the transverse section at NF32. b: Transverse section at NF36 shows punctate expression of stxbp1 restricted to the dorsal endoderm. c: Whole gut at NF44, stxbp1 has a punctate expression throughout the gut. d: NF42 liver and pancreas showing stxbp1 punctate expression in the dorsal pancreas and more diffuse expression in the liver. e: syntaxin 1B has minimal punctate expression within the endoderm at NF32. f: At NF36 there is an increase in the punctate expression of syntaxin 1B in the endoderm, transverse section. g: NF44 whole gut showing expression of syntaxin 1B in the stomach, liver and duodenum. h: NF42 liver and pancreas with expression throughout the liver and pancreas. dme, dorsal mesendoderm; de, dorsal endoderm; l, liver; st, stomach; p, pancreas; dp, dorsal pancreas; vp, ventral pancreas.
Syntaxin 1B
Syntaxin 1B is a member of the t-SNARE family, these proteins are located in the plasma membrane and play a fundamental role in vesicle docking and fusion (Ferro-Novick and Jahn, 1994). Stxbp1 can directly bind Syntaxin, this interaction has been shown to result in negative regulation of the insulin secretory machinery in insulin-secreting HIT-T15 cells (Hata et al., 1993; Zhang et al., 2000). In Xenopus embryos syntaxin 1B was not expressed within the developing pancreatic buds (Fig. 4e,f), but it was uniformly expressed through the mature stomach, liver and pancreas (Fig. 4g,h). At both NF32 and 36 in ptf1a-injected embryos, syntaxin 1B expression was ectopically expressed in the dorsal and ventral endoderm (Fig. 6e-h). At this early stage insulin mRNA is only beginning to be expressed, and it is unclear what role these genes, which are implicated in insulin-release, might play at such an early stage in development.
Igfbp1
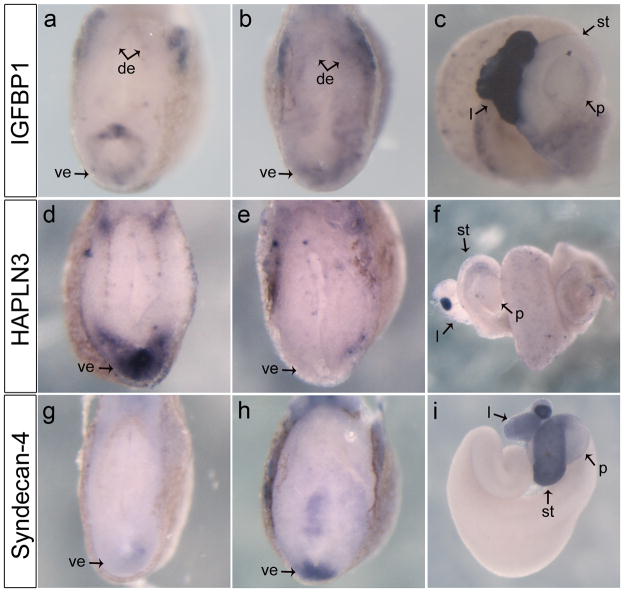
Insulin-like growth factor-binding proteins (IGFBPs) are secreted proteins that bind to Insulin-like growth factors (IGFs) and sequester them from binding to the IGF receptor, which plays a crucial role developmental growth and metabolism (Duan, 2002; Hwa et al., 1999). In confirmation of the microarray, we found igfbp1 to be expressed within the developing endoderm at all stages examined. At NF32 it was expressed in the ventral endoderm and dorsolateral mesoderm (Fig. 5a). Eight hours later at NF36 expression within the endoderm increased, and was now found to extend into the dorsal endoderm (Fig. 5b). In isolated guts at NF45, expression was strongest in the liver, but was also found diffuse throughout the posterior part of the gut (Fig. 5c). In ptf1a overexpressed embryos expression was expanded in the ventral endoderm at NF32 (Fig. 6i,j). Similarly at NF36 igfbp1 expression was stronger in the ventral endoderm in comparison to the control and the punctate pattern was more abundant (Fig. 6k,l). Concentrated igfbp1 expression in the ventral endoderm was amplified in ptf1a-injected embryos, and based on these observations we consider that it may play a role specific for only ventral pancreas development.
Figure 5.
Expression of cell shape/adhesion genes in the developing endoderm. a: Transverse section through anterior endoderm at NF32 showing expression of igfbp1 restricted to the dorsal and ventral endoderm. b: At NF36 igfbp1 is expressed most highly in the dorsal and ventral endoderm, but diffusely throughout the rest of the endoderm, transverse section. c: NF45 whole gut showing igfbp1 strongly expressed in the liver, and lighter in the duodenum. d: Hapln3 is expressed in the ventral endoderm in the transverse section at NF32. e: Transverse section at NF36, hapln3 expression is lost in the ventral endoderm, but is punctate through the endoderm. f: NF45 whole gut showing strong expression of hapln3 in the gall bladder, and punctate expression through the pancreas and duodenum g: Syndecan-4 is only expressed in the most ventral region of the endoderm shown in the transverse section of NF32. h: Transverse section at NF36 is showing expression of syndecan-4 only in the most ventral part of the endoderm, but stronger than at NF32. i: NF44 whole gut shows expression of syndecan-4 in the stomach and liver, and slightly in the pancreas. de, dorsal endoderm; ve, ventral endoderm; l, liver; st, stomach; p, pancreas.
Hapln3
Hyaluronan and proteoglycan-binding link protein 3 (Hapln3) is a member of the link-protein family HAPLN, in Xenopus Hapln3 was identified as an essential extracellular matrix protein that stabilizes hyaluronan matrix formation during cardiogenesis (Ito et al., 2008). Our analysis of hapln3 expression showed it to be highly expressed within the most ventral endoderm at NF32 (Fig. 5d). At later stages, hapln3 expression was barely detectable in the endoderm (Fig. 5e,f). In embryos overexpressing ptf1a mRNA, hapln3 was expressed in broader and stronger pattern in the ventral endoderm at NF32 (Fig. 6m,n). At NF36, ectopic expression of hapln3 was observed in the ventral endoderm in ptf1a-injected embryos (Fig. 6o,p). These results indicate that hapln3 is expressed in the ventral endoderm prior to morphogenesis of the pancreatic buds, and this ventral expression was up-regulated in embryos overexpressing ptf1a mRNA in the developing anterior endoderm.
Syndecan-4
Lastly, we focused on Syndecan-4, which as a member of the Syndecan family is a heparan sulfate bearing transmembrane protein involved in cell adhesion and linkage to the cytoskeleton (Whiteford et al., 2008). At NF32 faint syndecan-4 expression was detected in the ventral endoderm (Fig. 5g). By NF36, expression within the most ventral portion of the endoderm was increased substantially (Fig. 5h). In NF44 guts syndecan-4 was most strongly expressed within the stomach, but expression was also detected in the liver (Fig. 5i). These observations indicate that syndecan-4 is expressed in the ventral endoderm during stages when the ventral pancreatic cells are being specified.
Discussion
Numerous studies have addressed the transcription factor hierarchy involved in pancreas development; none has examined in detail the Ptf1a transcriptional network. Ptf1a is a particularly interesting gene in this cascade, as it is one of the earliest transcription factors to have restricted expression within the developing pancreatic buds. Ptf1a plays two roles, early in initial pancreas specification and later in maintenance of mature pancreatic exocrine cells. While a greater understanding of its function in exocrine cell maintenance has been achieved, much less is known about the genes activated downstream of Ptf1a during initial stages of pancreas development. In this paper we have used mRNA overexpression and microarray analysis to identify early downstream targets of Ptf1a that may play a role in initial specification of the pancreas. From this study, we identified numerous genes that were not previous known to function in pancreas development
Through microanalysis of Ptf1a overexpression over two time-points we have identified over 800 genes that are temporally regulated within the anterior endoderm during the initial stages of pancreas development, and have validated a subset of these which show expression within the anterior endoderm. Many of these have no prior known function in pancreas development. A total of 45 transcription factors were identified in our analysis, and while the majority of these have no known role in pancreas development, several established regulators of pancreas development were observed (NeuroD1, Atf3, Sox9). This suggests the other up-regulated transcription factors in our data sets are likely to also play a role in pancreas development.
Using gene annotation enrichment analysis we have identified differences between the two time-points. At NF32 the most enhanced gene sets are involved in transcriptional regulation, biological adhesion and have known functions during development. This suggests that Ptf1a directly activates genes involved specification of the pancreatic cells and developing tissue. Genes up-regulated at NF36 are likely secondary or tertiary targets of Ptf1a, and at this stage the pancreas has formed into buds that will soon fuse to form a discrete organ. From this data set, we identified many genes enriched in transmembrane function and ion transport.
Although Ptf1a has been shown to be essential for development of a subset of endocrine cells, little is know about how it specifies these cells. To begin to define the Ptf1a endocrine regulatory circuit we sought to identify overlap between the Ptf1a and endocrine Ngn3 regulatory network, which we recently published (Oropez and Horb, 2012). This analysis identified a list of 14 genes potentially involved in endocrine cell development (Data not shown). Five of the identified genes have previous known pancreatic function (CPE, Hey1, Hes5, Ptgs2, NeuroD4). The identification of all these genes in both the Ngn3 and Ptf1a microarrays strongly suggest they function downstream of these key transcription factors in early pancreas specification, but exactly what role they might play in early pancreas specification downstream of Ptf1a is not known. However, this overlap provides a starting point for further evaluation of these genes to define the overlap between the Ptf1a and Ngn3 pathways.
Endodermally expressed candidate genes
The specification of pancreatic cell lineages involves sequential activation of several classes of transcription factors, in this study we identified two transcription factors expressed within the developing endoderm and up-regulated by Ptf1a: Irf1 and Hey-1. Irf1 has been studied for its involvement in immune response, including islet inflammation, but has other known roles in tumor suppression, apoptosis and cell growth regulation (Fujita et al., 1988; Kröger et al., 2002). Irf1 was up-regulated 3-fold at NF32 and 7-fold at NF36, and given its previous known functions it is possible the role of Irf1 downstream of Ptf1a may be to inhibit growth of naïve duodenal cells into a mature organ.
Early in pancreas development the Notch signaling pathway regulates cell fate differentiation. Ultimately it activates target genes, including the Hairy Enhancer of Split (HES) family of bHLH transcriptional repressors, which maintain cells in an undifferentiated state. Later in development Notch has been shown to delay the onset of the exocrine lineage; effector proteins of Notch, including Hes1, Hey1 and Hey2 are capable of interacting with Ptf1a and preventing the PTF1 complex from binding to target DNA (Esni et al., 2004; Ghosh and Leach, 2006). It is likely that Hey1 is interacting with Ptf1a to maintain pancreas progenitor cells in an undifferentiated state leading to an increase in cell growth and pancreatic cells.
We also identified genes with known functions in mature pancreatic cells, such as Stxbp1 and Igfbp1, but their role in pancreas development has not yet been studied. In mice there are three Munc18 (the mammalian stxbp1 homolog) isoforms: Munc18a, Munc18b and Munc18c, all of which are expressed in islet β-cells and function in insulin release (Oh and Thurmond, 2009; Spurlin et al., 2004; Tomas et al., 2008). These cytosolic proteins are also capable of binding to Syntaxin with high affinity, and this interaction is known to have both inhibitory and positive roles in vesicle transport (Lehtonen et al., 1999). Not only is binding partner Syntaxin 1B up-regulated in the microarrays, but so are many other components of the SNARE complex, such as SNAP-25, Synaptotagmin-like 1, Synaptotagmin 4, Synaptophysin A and B and VAMP-2. At this early stage in pancreas development at which our microarrays are performed insulin is not yet being released, suggesting that Stxbp1 and components of the SNARE complex may have other functions early during pancreas development, such as generation of polarization of specific cells leading to a differentiated cell fate (Lehtonen et al., 1999).
Igfbp1 has been shown to function independently of IGF, it can inhibit the mitogenic activity of epidermal growth factor (EGF) when bound to certain integrins and it also enhances migration of smooth muscle cells, extravillous trophoblast cells and Chinese hamster ovary cells (Cavaillé et al., 2006; Chakraborty et al., 2002; Gockerman et al., 1995; Jones et al., 1993). Recently Igfbp1 has been shown to activate kinases and small monomeric GTPases to affect cell migratory behavior of oligodendrocytes (Chesik et al., 2010). It may be possible that Igfbp1 has IGF-independent effects during pancreas development, involving the migration of precursor cells into defined anlagen.
Finally, we identified two genes with novel expression patterns within the early pancreatic endoderm, Hapln3 and Syndecan-4. Hapln3 is an extracellular matrix protein that stabilizes hyaluronan matrix formation. Hyaluronan is shown to play roles cell-cell adhesion, migration, proliferation and differentiation (Comper and Laurent, 1978; Shimabukuro et al., 2005). Recently, Hapln3 has been shown to be necessary for cardiogenesis through its role in hyaluronan matrix formation around the developing heart in X. laevis (Ito et al., 2008). Has2, the catalytic enzyme which synthesizes hyaluronan, was shown to be expressed in the same cardiac region as Hapln3 (Ito et al., 2008), but was also increased in procine neonatal pancreas cells stimulated with EGF, where it was suggested it may play a role in the proliferation and migration of pancreatic duct cells (Jeon et al., 2004). It is possible that Hapln3 up-regulation in Ptf1a injected embryos may aid in hyaluronan matrix formation of the ectopic hyaluronan synthesized by Has2 leading to proliferation of Ptf1a cells resulting in ectopic pancreas.
Our results are the first to show expression of hapln3 and syndecan-4 at such an early stage in the developing anterior ventral endoderm. To our knowledge, this is also the first time that hey1, syntaxin 1B, hapln3 and igfbp1 have been shown to be ectopically expressed in the developing endoderm in response to ptf1a mRNA overexpression in vivo. The fact that we found all of these genes to be up-regulated only in the ventral endoderm at the level of the developing ventral pancreatic buds suggests that Ptf1a function is more pronounced in this region of the endoderm than in more central, dorsal or posterior areas.
While all these genes were identified as up-regulated in the early endoderm in response to Ptf1a overexpression, many do not have restricted expression within the pancreas and have been previously identified in other cell types. This is not uncommon; several key pancreatic genes are expressed in multiple tissues, such as Pdx1 which is also expressed in the duodenum, or Ngn3, NeuroD and Ptf1a, which are also expressed in neural tissue (Lee et al., 1995; Miller et al., 1994; Sellick et al., 2004; Sommer et al., 1996). The ability of these genes to be involved in cell fate specification in multiple tissues rely on networks of other tissue-specific genes, it is possible that the genes we have identified in our microarray may function in pancreatic specification when expressed with their pancreatic-specific co-factors.
In conclusion, we are the first to report results from a microarray analysis of Ptf1a. We have found that Ptf1a modulates the expression of genes with a range of functions, from transcription to cell adhesion. Ptf1a is a key transcription factor necessary for pancreas development, and genes identified through this microarray are likely to play critical roles downstream of Ptf1a in defining cells along a particular pancreatic lineage.
Methods
Microarray analysis
All mRNA for microinjection was created using the Ambion mMessage mMachine kit. To confirm targeting, experimental mRNAs were injected along with 400pg gfp mRNA and targeting verified by observing appropriate fluorescence. For functional analysis we selected only samples for which the entire anterior endoderm was targeted. 800pg ptf1a and gfp mRNAs were injected into the dorsal vegetal blastomeres at the eight-cell stage. Anterior endoderm explants were collected at NF32 and NF36. RNA extraction of the sample sets (15 explants/set) was performed using TRIzol (Invitrogen) and purified using the RNeasy Micro Kit (Qiagen). RNA analysis, cDNA preparation and hybridization were performed by Genome Québec (McGill University, Montréal).
Microarray results were analyzed using Affymetrix Expression Console and normalized using the Probe Logarithmic Intensity ERror estimation (PLIER) algorithm. Differential gene expression was analyzed using consecutive sampling with bin size of 25 (Guilbault et al., 2006; Novak et al., 2006a; Novak et al., 2006b; Novak et al., 2002). Representative standard deviations in each bin were calculated using non-linear regression to determine the boundaries of probability intervals. Candidate genes were selected as genes that lay beyond the probability interval of 0.9. The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE34193.
RT-PCR
RT-PCR was performed on isolated anterior endoderm explants from ptf1a expressing and non-expressing embryos and normalized to EF1-α. PCR conditions using Taq DNA polymerase (Invitrogen) included 1 min at 94°C preincubation, followed by 26, 28 or 30 amplification cycles (except Ef1-α which only had 20, 22 or 24 cycles) comprising 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and one cycle at 72°C for 10 min. The following primers were used:
-
Stxbp1 Fwd 5′-TCTGCATCGGGTATCCGTTTCCTT-3′ and
Stxbp1 Rev 5′-ATAAAGGTCGGGAGTGGTCGTTGT-3′
-
TA-2 Fwd 5′-ACAGTTATACCCAGCACAGCCCTT-3′ and
TA-2 Rev 5′-ACTGAACCACAGAGCTGACACAGT-3′
-
Crbp2 Fwd 5′-CCGTGGGCGTTGAATTCGATGAAA-3′ and
Crbp2 Fwd 5′-TTCCAGCCACGGTTGTTCTTCTCT-3′
-
C25H Fwd 5′-ACCTTTGCACTGGCCACTCAGTAT-3′ and
C25H Fwd 5′-AAATGGTACCAGCTTGTGGGAGGA-3′
-
Igfbp1 Fwd 5′-TTACGGCCTAGTGAAGCAGCAGAT-3′ and
Igfbp1 Rev 5′-TTCTGCGCTCAAGCATCTTTCGTG-3′
-
Irf1 Fwd 5′-GCACAGGCCCTTAATGTGATCCAA-3′ and
Irf1 Rev 5′-GCAAACGAGTGGCAAACTGGTC-3′
-
Hapln3 Fwd 5′-GATGGCTGGTTGAGACTCATGTCT-3′ and
Hapln3 Rev 5′-AGAAAGCTCACTCACAAGGGACG-3′
-
Hey1 Fwd 5′-CAGTAAAGCTGTGCATGGAAGGG-3′ and
Hey1 Rev 5′-GTTGTATAGTCCGGGTTCATGTGC-3′
-
Sestrin1 Fwd 5′-CCTCCCCCGGATATAGATGT-3′ and
Sestrin1 Rev 5′-CCTGCACTCGAAAAGTAGGC-3′
-
Syndecan-4 Fwd 5′-GCTAAGCCCAAAACATTGGA-3′ and
Syndecan-4 Rev 5′-AAGGAAACTGCAACGACCAG-3′
Probes and whole mount in situ hybridizations
All genes were cloned using cDNA from wild-type NF35 embryos with the following primers:
Hapln3 Fwd 5′-GACCCAATTGTTTTGGGGCTACCT-3′ and Hapln3 Rev 5′-GGGAGCGCAGTGAGTGCAGG-3′ designed based on BC046259. Hey1 Fwd 5′-TCCTCCGCTGCTCTCCTCCA-3′ and Hey1 Rev 5′-GGGTGATGTCGCACCCAAGCA-3′ designed based on BC084410. Igfbp1 Fwd 5′-CTGGATCCCCTGAAAACAGA-3′ and Igfbp1 Rev 5′-GGACCTGGGATTTTCTGGAT-3′ designed based on BC060008. Irf1 Fwd 5′-TGCCATTGCCTGACAGCACA-3′ and Irf1 Rev 5′-GGGCCTGTGCAAACCGATGC-3′ designed based on BC059984. Syndecan-4 Fwd 5′-TCCTGCTGCTTTTAGCGCTGGTT-3′ and Syndecan-4 Rev 5′-ACCAGGCCAACCAACGTGCC-3′ designed based on DQ116029. Syntaxin 1B Fwd 5′-CAGAGCATCTCTGAGCATCT-3′ and Syntaxin 1B Rev 5′-GATTCTTTCAGAGTCCCAGG-3′ designed based on BC084156. Syntaxin binding protein 1 ordered from XDB (http://xenopus.nibb.ac.jp/) contig number xl103a06. PCR products were cloned into pCRII (Invitrogen) and confirmed by sequencing. Whole mount in situ hybridizations were performed as described using BM purple (Horb et al., 2003).
Acknowledgments
We are grateful to Zeina Jarikji for her valuable assistance in the microinjections and in situ hybridizations and to Frédéric Bourque for his care of the frogs. Special thanks go to Lori Horb for the Tnt assay, Dr. Jaroslav P. Novak of GenexAnalysis (http://genexanalysis.net) for his mathematical analysis of microarray data.
Grant: National Institutes of Health (DK077197)
References
- Afelik S, Chen Y, Pieler T. Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes Dev. 2006;20:1441–1446. doi: 10.1101/gad.378706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilogan CK, Horb ME. Xenopus staufen2 is required for anterior endodermal organ formation. Genesis. 2012;50:251–259. doi: 10.1002/dvg.22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Andelfinger G, Horb ME. Germ layers to organs: using Xenopus to study “later” development. Semin Cell Dev Biol. 2006;17:133–145. doi: 10.1016/j.semcdb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Cavaillé F, Neau E, Vouters M, Bry-Gauillard H, Colombel A, Milliez J, Le Bouc Y. IGFBP-1 inhibits EGF mitogenic activity in cultured endometrial stromal cells. Biochem Biophys Res Commun. 2006;345:754–760. doi: 10.1016/j.bbrc.2006.04.169. [DOI] [PubMed] [Google Scholar]
- Chakraborty C, Gleeson LM, McKinnon T, Lala PK. Regulation of human trophoblast migration and invasiveness. Can J Physiol Pharmacol. 2002;80:116–124. doi: 10.1139/y02-016. [DOI] [PubMed] [Google Scholar]
- Comper WD, Laurent TC. Physiological function of connective tissue polysaccharides. Physiol Rev. 1978;58:255–315. doi: 10.1152/physrev.1978.58.1.255. [DOI] [PubMed] [Google Scholar]
- Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C. Specifying the cellular responses to IGF signals: roles of IGF-binding proteins. J Endocrinol. 2002;175:41–54. doi: 10.1677/joe.0.1750041. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, Yu X, MacDonald RJ, Civin CI, Real FX, Pack MA, Ball DW, Leach SD. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-beta gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Leach SD. Interactions between Hairy/Enhancer of Split-related proteins and the pancreatic transcription factor Ptf1-p48 modulate function of the PTF1 transcriptional complex. Biochem J. 2006 doi: 10.1042/BJ20051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Gockerman A, Prevette T, Jones JI, Clemmons DR. Insulin-like growth factor (IGF)-binding proteins inhibit the smooth muscle cell migration responses to IGF-I and IGF-II. Endocrinology. 1995;136:4168–4173. doi: 10.1210/endo.136.10.7545099. [DOI] [PubMed] [Google Scholar]
- Guilbault C, Novak JP, Martin P, Boghdady ML, Saeed Z, Guiot MC, Hudson TJ, Radzioch D. Distinct pattern of lung gene expression in the Cftr-KO mice developing spontaneous lung disease compared with their littermate controls. Physiol Genomics. 2006;25:179–193. doi: 10.1152/physiolgenomics.00206.2005. [DOI] [PubMed] [Google Scholar]
- Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Horb LD, Horb ME. BrunoL1 regulates endoderm proliferation through translational enhancement of cyclin A2 mRNA. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horb LD, Jarkji ZH, Horb ME. Xenopus Insm1 is essential for gastrointestinal and pancreatic endocrine cell development. Dev Dyn. 2009;238:2505–2510. doi: 10.1002/dvdy.22071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horb ME, Shen CN, Tosh D, Slack JM. Experimental conversion of liver to pancreas. Curr Biol. 2003;13:105–115. doi: 10.1016/s0960-9822(02)01434-3. [DOI] [PubMed] [Google Scholar]
- Horb ME, Slack JM. Expression of amylase and other pancreatic genes in Xenopus. Mech Dev. 2002;113:153–157. doi: 10.1016/s0925-4773(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Hosono R, Hekimi S, Kamiya Y, Sassa T, Murakami S, Nishiwaki K, Miwa J, Taketo A, Kodaira KI. The unc-18 gene encodes a novel protein affecting the kinetics of acetylcholine metabolism in the nematode Caenorhabditis elegans. J Neurochem. 1992;58:1517–1525. doi: 10.1111/j.1471-4159.1992.tb11373.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherma BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- Ito Y, Seno S, Nakamura H, Fukui A, Asashima M. XHAPLN3 plays a key role in cardiogenesis by maintaining the hyaluronan matrix around heart anlage. Dev Biol. 2008;319:34–45. doi: 10.1016/j.ydbio.2008.03.042. [DOI] [PubMed] [Google Scholar]
- Jacobsson G, Bean AJ, Scheller RH, Juntti-Berggren L, Deeney JT, Berggren PO, Meister B. Identification of synaptic proteins and their isoform mRNAs in compartments of pancreatic endocrine cells. Proc Natl Acad Sci USA. 1994;91:12487–12491. doi: 10.1073/pnas.91.26.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarikji Z, Horb LD, Shariff F, Mandato CA, Cho KW, Horb ME. The tetraspanin Tm4sf3 is localized to the ventral pancreas and regulates fusion of the dorsal and ventral pancreatic buds. Development. 2009;136:1791–1800. doi: 10.1242/dev.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarikji ZH, Vanamala S, Beck CW, Wright CV, Leach SD, Horb ME. Differential ability of Ptf1a and Ptf1a-VP16 to convert stomach, duodenum and liver to pancreas. Dev Biol. 2007;304:786–799. doi: 10.1016/j.ydbio.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SY, Baek KH, Kim YS, Park CG, Kwon HS, Ko SH, Song KH, Yoo SJ, Son HS, Cha BY, Lee KW, Son HY, Kang SK, Yoon KH. Differentially up-regulated genes in proliferating porcine neonatal pancreas cells caused by epidermal growth factor. J Cell Biochem. 2004;91:354–364. doi: 10.1002/jcb.10752. [DOI] [PubMed] [Google Scholar]
- Johansson T, Lejonklou MH, Ekeblad S, Stålberg P, Skogseid B. Lack of nuclear expression of hairy and enhancer of split-1 (HES1) in pancreatic endocrine tumors. Horm Metab Res. 2008;40:354–359. doi: 10.1055/s-2008-1076695. [DOI] [PubMed] [Google Scholar]
- Jones JI, Gockerman A, Busby WHJ, Wright G, Clemmons DR. Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the alpha 5 beta 1 integrin by means of its Arg-Gly-Asp sequence. Proc Natl Acad Sci U S A. 1993;90:10553–10557. doi: 10.1073/pnas.90.22.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kelly OG, Melton DA. Development of the pancreas in Xenopus laevis. Dev Dyn. 2000;218:615–627. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1027>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger A, Köster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lehtonen S, Lehtonen E, Olkkonen VM. Vesicular transport and kidney development. Int J Dev Biol. 1999;43:425–433. [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CP, McGehee RE, Jr, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JP, Kim SY, Xu J, Modlich O, Volsky DJ, Honys D, Slonczewski JL, Bell DA, Blattner FR, Blumwald E, Boerma M, Cosio M, Gatalica Z, Hajduch M, Hidalgo J, McInnes RR, Miller MC, 3rd, Penkowa M, Rolph MS, Sottosanto J, St-Arnaud R, Szego MJ, Twell D, Wang C. Generalization of DNA microarray dispersion properties: microarray equivalent of t-distribution. Biol Direct. 2006a;1:27. doi: 10.1186/1745-6150-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JP, Miller MC, 3rd, Bell DA. Variation in fiberoptic bead-based oligonucleotide microarrays: dispersion characteristics among hybridization and biological replicate samples. Biol Direct. 2006b;1:18. doi: 10.1186/1745-6150-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JP, Sladek R, Hudson TJ. Characterization of variability in large-scale gene expression data: implications for study design. Genomics. 2002;79:104–113. doi: 10.1006/geno.2001.6675. [DOI] [PubMed] [Google Scholar]
- Oh E, Thurmond DC. Munc18c depletion selectively impairs the sustained phase of insulin release. Diabetes. 2009;58:1165–1174. doi: 10.2337/db08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oropez D, Horb ME. Transient expression of Ngn3 in Xenopus endoderm promotes early and ectopic development of pancreatic beta and delta cells. Genesis. 2012;50:271–285. doi: 10.1002/dvg.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Bilogan CK, Mukhi S, Brown DD, Horb ME. Xenopus pancreas development. Dev Dyn. 2009;238:1271–1286. doi: 10.1002/dvdy.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. Genesis. 2012;50:155–163. doi: 10.1002/dvg.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Jarikji Z, Horb ME. Functional analysis of Rfx6 and mutant variants associated with neonatal diabetes. Dev Biol. 2011;351:135–145. doi: 10.1016/j.ydbio.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, Garrett C, Gloyn AL, Edghill EL, Hattersley AT, Wellauer PK, Goodwin G, Houlston RS. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Shimabukuro Y, Ueda M, Ichikawa T, Terashi Y, Yamada S, Kusumoto Y, Takedachi M, Terakura M, Kohya A, Hashikawa T, Murakami S. Fibroblast growth factor-2 stimulates hyaluronan production by human dental pulp cells. J Endod. 2005;31 doi: 10.1097/01.don.0000158242.44155.49. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Spurlin BA, Park SY, Nevins AK, Kim JK, Thurmond DC. Syntaxin 4 transgenic mice exhibit enhanced insulin-mediated glucose uptake in skeletal muscle. Diabetes. 2004;53:2223–2231. doi: 10.2337/diabetes.53.9.2223. [DOI] [PubMed] [Google Scholar]
- Tomas A, Meda P, Regazzi R, Pessin JE, Halban PA. Munc 18-1 and granuphilin collaborate during insulin granule exocytosis. Traffic. 2008;9:813–832. doi: 10.1111/j.1600-0854.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- Whiteford JR, Ko S, Lee W, Couchman JR. Structural and cell adhesion properties of zebrafish syndecan-4 are shared with higher vertebrates. J Biol Chem. 2008;283:29322–29330. doi: 10.1074/jbc.M803505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Zhang W, Efanov A, Yang SN, Fried G, Kolare S, Brown H, Zaitsev S, Berggren PO, Meister B. Munc-18 associates with syntaxin and serves as a negative regulator of exocytosis in the pancreatic beta-cell. J Biol Chem. 2000;275:41521–41527. doi: 10.1074/jbc.M005479200. [DOI] [PubMed] [Google Scholar]