Abstract
Objective
Diminished serum paraoxonase and arylesterase activities (measures of paraoxonase-1 [PON-1] function) in humans have been linked to heightened systemic oxidative stress and atherosclerosis risk. The clinical prognostic utility of measuring distinct PON1 activities has not been established, and the genetic determinants of PON-1 activities are not known.
Methods and Results
We established analytically robust high throughput assays for serum paraoxonase and arylesterase activities and measured these in 3,668 stable subjects undergoing elective coronary angiography without acute coronary syndrome, and were prospectively followed for major adverse cardiac events (MACE = death, myocardial infarction, stroke) over 3 years. Low serum arylesterase and paraoxonase activities were both associated with increased risk for MACE, with arylesterase activity showing greatest prognostic value (Q4 versus Q1, Hazard Ratio [HR] 2.63, 95%CI 1.97–3.50, p<0.01). Arylesterase remained significant after adjusting for traditional risk factors, C-reactive protein, and creatinine clearance (HR 2.20, 95%CI 1.60–3.02, p<0.01), predicted future development of MACE in both primary and secondary prevention populations, and reclassified risk categories incrementally to traditional clinical variables. A genome-wide association study (GWAS) identified distinct SNPs within the PON-1 gene that were highly significantly associated with serum paraoxonase (1.18×10−303) or arylesterase (4.99×10−116) activity but these variants were not associated with either 3-year MACE risk in an angiographic cohort (n=2,136) or history of either coronary artery disease or myocardial infarction in the CARDIoGRAM consortium (n~80,000 subjects).
Conclusions
Diminished serum arylesterase activity, but not the genetic determinants of PON-1 functional measures, provides incremental prognostic value and clinical reclassification of stable subjects at risk of developing MACE.
Keywords: paraoxonase 1 gene, coronary artery disease, oxidative stress, arylesterase activity
INTRODUCTION
Heightened oxidative stress in the form of oxidation of lipids and proteins by reactive oxidant species adversely contributes to disease progression in cardiovascular disease (CVD)1. Paraoxonase-1 (PON-1) belongs to a family of high-density lipoprotein (HDL)-associated enzymes that show hydrolytic activity towards a variety of substrates, including toxins in the environment2 and oxidized lipids in the body3. Consequently, diminished activities of PON-1 and other paraoxonases have been associated with the development of CVD4, 5. Paraoxonase-1 (EC 3.1.1.2) activity in serum is classically named after the substrate used to monitor enzymatic function, namely, paraoxonase activity (using paraoxon as substrate) and arylesterase activity (using phenyl acetate as substrate) 6. Our group has recently observed the relationship between a specific PON-1 genotype and functional activity with multiple systemic measures of oxidative stress and CVD risk in humans 7, 8. Based upon these encouraging findings, we developed analytically validated semi-automated high throughput methods for arylesterase and paraoxonase activity assays amenable to large scale clinical and genetic studies. We sought to expand and validate our findings in an independent cohort of stable patients undergoing cardiac evaluation in order to examine and contrast the potential role of distinct circulating PON-1 activity measures to predict adverse disease progression. In addition, we sought to identify genetic loci controlling paraoxonase and arylesterase activity by carrying out an unbiased genome-wide association study (GWAS) and determine whether these genetic factors were associated with incident risks of adverse cardiac events or prevalent coronary artery disease (CAD).
METHODS
Study population
The Cleveland Clinic GeneBank study is a large, prospective cohort study from 2001–6 that established a well-characterized clinical repository with clinical and longitudinal outcomes data comprised from consenting subjects undergoing elective diagnostic cardiac catheterization procedure. All GeneBank participants gave written informed consent approved by the Cleveland Clinic Institutional Review Board.
This study involved a total of 3,668 subjects in the GeneBank study who underwent coronary angiography in the absence of acute coronary syndrome and confirmed by including only those with cardiac troponin I (cTnI) <0.03 ng/mL, with no history of revascularization within 30 days before enrollment, and with at least 3 years of long-term follow-up. Adjudicated outcomes were ascertained over the ensuing 3 years for all subjects following enrollment. Framingham risk factors include age, gender, cigarette smoking, low-density lipoprotein and high-density lipoprotein cholesterol, systolic blood pressure, and diabetes mellitus were identified. We defined secondary prevention cohort by a known history of CAD (including stenosis of any coronary artery ≥50% at the time of catheterization), prior myocardial infarction (MI), known history of peripheral artery disease, history of transient ischemic attack, stroke or known cerebrovascular disease, or previous percutaneous or surgical revascularization. Those who did not fulfill secondary prevention cohort criteria were assigned to primary prevention cohort.
An estimate of creatinine clearance (CrCl) was calculated using the Cockcroft-Gault equation, since the majority of patients have preserved renal function. High sensitivity C-reactive protein (hsCRP), cTnI, serum creatinine, fasting blood glucose, and lipid profiles were also measured on the Architect ci8200 platform (Abbott Laboratories, Abbott Park IL). Absolute neutrophil counts were analyzed by the Advia 120 Automated Hematology Analyzer (Siemens Healthcare Diagnostics, Deerfield IL). Major adverse cardiovascular events (MACE) were defined as death, non-fatal MI, or non-fatal cerebrovascular accident following enrollment.
Serum paraoxonase activity and arylesterase activity assays
Serum paraoxonase and arylesterase activities were measured by spectrophotometry in an open channel on the aforementioned Architect ci8200 platform, and in a 96-well plate format (Spectramax 384 Plus, Molecular Devices, Sunnyvale, California), respectively. For serum paraoxonase activity, the rate of generation of para-nitrophenol was determined at 405 nm in 40-fold diluted serum (final) in reaction mixtures compose of 1.5mM paraoxon (Sigma-Aldrich, St Louis, Missouri), 10mM Tris hydrocholoride, pH 8, 1M sodium chloride, and 2mM calcium chloride at 24°C. An extinction coefficient (at 405 nm) of 17 000 M−1•cm−1 was used for calculating units of paraoxonase activity, which is expressed as nanomoles of para-nitrophenol produced per minute per milliliter of serum. The intra-assay and inter-assay coefficients of variance for the high throughput paraoxonase activity assay were 1.9% and 3.3%, respectively, on 30 replicates performed on 15 different days. For serum arylesterase activity measurement, initial hydrolysis rates were determined at 270 nm in 50-fold diluted serum (final) in reactions mixtures composed of 3.4mM phenylacetate (Sigma-Aldrich, St Louis, Missouri), 9mM Tris hydrocholoride, pH 8, and 0.9mM calcium chloride at 24°C. An extinction coefficient (at 270 nm) of 1310M−1•cm−1 was used for calculating units of arylesterase activity, which are expressed as micromoles of phenyl acetate hydrolyzed per minute per milliliter of serum. The intra-assay and inter-assay coefficients of variance for arylesterase activity assay were 3.4% and 3.9%, respectively, on 20 replicates performed on 10 different days.
Genotyping
Genotyping was performed on the Affymetrix Genome-Wide Human SNP Array 6.0 platform. Using these data and those from 120 phased chromosomes from the HapMap CEU samples (HapMap r22 release, NCBI build 36), genotypes were imputed for untyped SNPs across the genome using MACH 1.0 software9. All imputations were done on the forward (+) strand using 562,554 genotyped SNPS that had passed quality control (QC) filters. QC filters for the imputed dataset excluded SNPs with HWE p-values < 0.0001 or minor allele frequencies < 1%, and individuals with less than 95% call rates. This resulted in 2,421,770 autosomal SNPs that were available for analysis.
Statistical analyses
The Student’s t-test or Wilcoxon-Rank sum test for continuous variables and chi-square test for categorical variables were used to examine the difference between the groups. Kaplan–Meier analysis with Cox proportional hazards regression was used for time-to-event analysis to determine Hazard ratio (HR) and 95% confidence intervals (95%CI) for MACE. Adjustments were made for individual traditional cardiac risk factor (including age, gender, diabetes mellitus, hypertension, former or current cigarette smoking, prior coronary artery disease), log-transformed hsCRP, and CrCl. The R package “Mclust” was used for discriminant analysis. The clustering process is based on multivariate normal mixture models. The optimal model parameters and the number of clusters were determined via Bayesian information criterion (BIC). All analyses were performed using R 2.13.1 (Vienna, Austria) and p-values <0.05 were considered statistically significant.
Genome-wide linear regression analyses were used to identify loci associated with serum paraoxonase and arylesterase activity after adjustment for age and gender under an additive model. Genetic analyses were carried out with PLINK (v1.07) using both untransformed (arylesterase activity) and inverse-normal transformed (paraoxonase activity) values. Relative risk for experiencing a MACE as a function of genotype was assessed using Cox proportional hazard models with adjustment for age, sex, Framingham ATP-III risk score (which includes diabetes status), and medication use (aspirin and/or statins). Adjusted hazard ratios (HR) and 95% CI are reported with 2-sided p-values. A haplotype score test was also used to test all haplotypes with greater than 1% frequency, as implemented in the Haplo.Stats package. All genetic analyses were performed using SAS version 9.2 (SAS Institute Inc, Cary, NC) or R 2.10.1 (http://www.R-project.org).
Associations with coronary artery disease (CAD)
The Coronary Artery Disease Genome-wide Replication And Meta-Analysis (CARDIoGRAM) Consortium represents a GWAS meta-analysis of CAD comprising a discovery set of ~22,000 cases and ~65,000 controls10. For each cohort in CARDIoGRAM, logistic regression was first used to test for association with CAD using a log-additive model with adjustment for age and gender and taking into account the uncertainty of possibly imputed genotypes. Subsequently, a meta-analysis was performed separately for every SNP from each study that passed the quality control criteria using a fixed effect model with inverse variance weighting10. The results of this default meta-analysis were used to determine whether SNPs influencing PON-1 functional activity measures were associated with CAD.
RESULTS
Study population
Table 1 describes the baseline characteristics of our primary study population of 3,668 subjects. Serum arylesterase activity levels were normally distributed, with a mean of 104 ±25 µmol/min/mL. However, serum paraoxonase activity levels were not normally distributed, with a median of 562 µmol/min/mL (interquartile range 315–1,045 µmol/min/mL). Both serum paraoxonase and arylesterase levels were lower in men than in women. Compared to those in the highest quartile, subjects in the lowest quartile of serum paraoxonase and arylesterase activity were more likely to have significantly obstructive (≥50% stenosis) coronary artery disease (odds ratio 1.86 [95% CI 1.53, 2.27] p<0.01 for arylesterase, and 1.48 [95% CI 1.22, 1.81] p<0.01 for paraoxonase). However upon adjustment of cardiovascular risk factors, such differences were no longer apparent. In addition, there were very weak (but statistically significant) correlations between serum arylesterase activity and hsCRP (r=−0.09, p<0.001), estimated creatinine clearance (eCrCl, r= 0.15, p<0.001), and absolute lymphocyte count (r=0.09, p<0.001). In contrast, there was no statistically significant relationship between serum arylesterase activity and leukocyte count (r=0.02, p=0.28) or absolute neutrophil count (r=0.002, p=0.91). These findings were similar when correlations were performed within the Q192R genotype subgroups (data not shown).
Table 1.
Baseline Subject Characteristics from GeneBank cohort (n=3,668)
| Variable | Value |
|---|---|
| Age (years) | 63 ± 11 |
| Gender (% male) | 65 |
| Body mass index (kg/m2) | 29.6 ± 6 |
| Race (% Caucasian) | 97 |
| Diabetes mellitus (%) | 29 |
| Hypertension (%) | 70 |
| Smokers (Former/current, %) | 65 / 11 |
| Prior coronary artery disease (%) | 67 |
| LDL cholesterol (mg/dL) | 95 (78, 117) |
| HDL cholesterol (mg/dL) | 34 (28, 41) |
| Triglycerides (mg/dL) | 114 (83, 162) |
| hsCRP (mg/L) | 2.00 (0.91, 4.51) |
| Creatinine clearance (ml/min) | 99.9 (76.8, 126.3) |
| Total leukocyte count (×109 cells/L) | 6.1 (5, 7.4) |
| Absolute neutrophil count (cells/µL) | 3.9 (3.1, 5) |
| Baseline medications (%) | |
| ACE inhibitors/ARBs | 49 |
| Beta-blockers | 61 |
| Statin | 59 |
| Aspirin | 73 |
| Serum paraoxonase activity (nmol/min/mL) | 562 (315–1,045) |
| Serum arylesterase activity (µmol/min/mL) | 104 ± 25 |
Values expressed in mean ± standard deviation or median (interquartile range). Abbreviations: LDL = low-density lipoprotein cholesterol; HDL = high-density lipoprotein cholesterol; hsCRP = high-sensitivity C-reactive protein; ACE = angiotensin converting enzyme; ARB = angiotensin receptor blocker..
PON-1 activities and major adverse cardiac outcomes
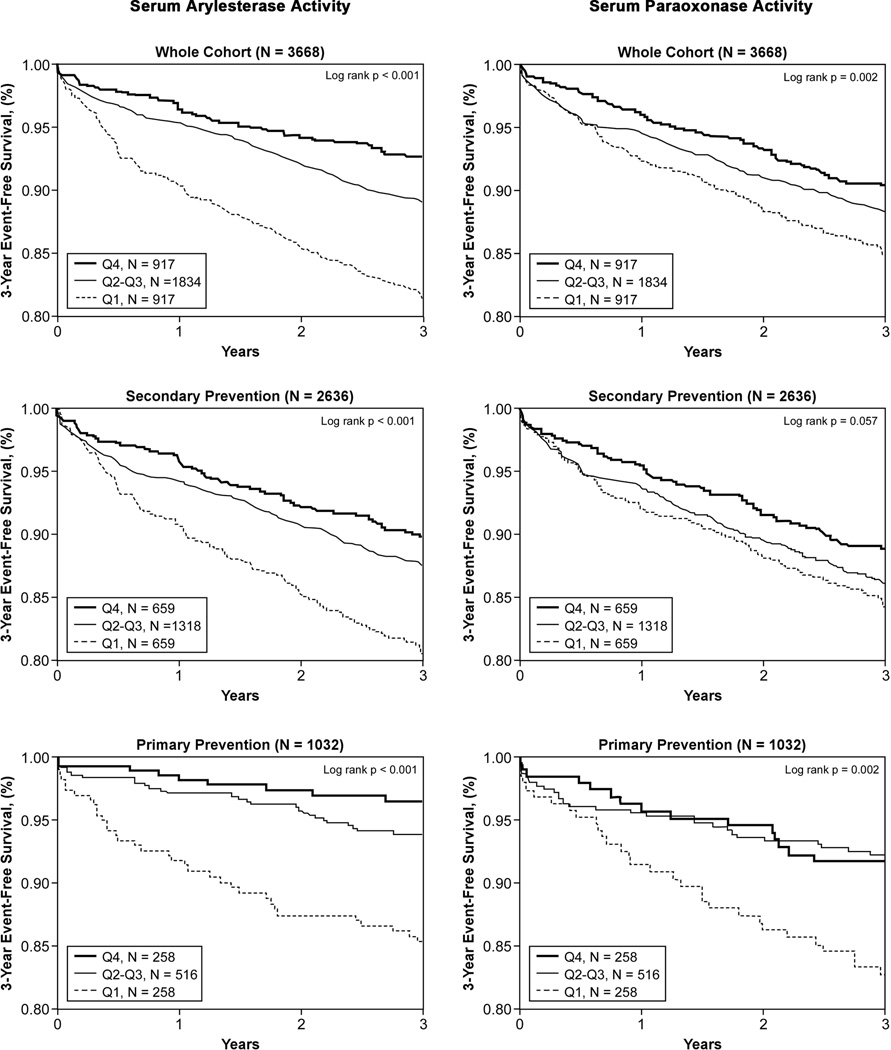
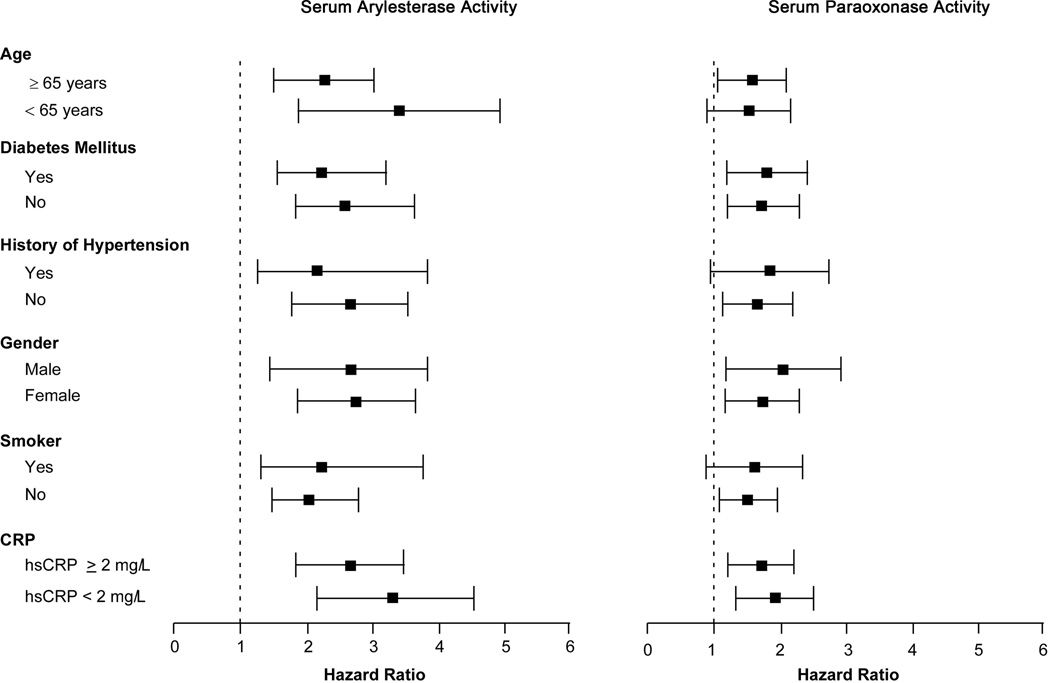
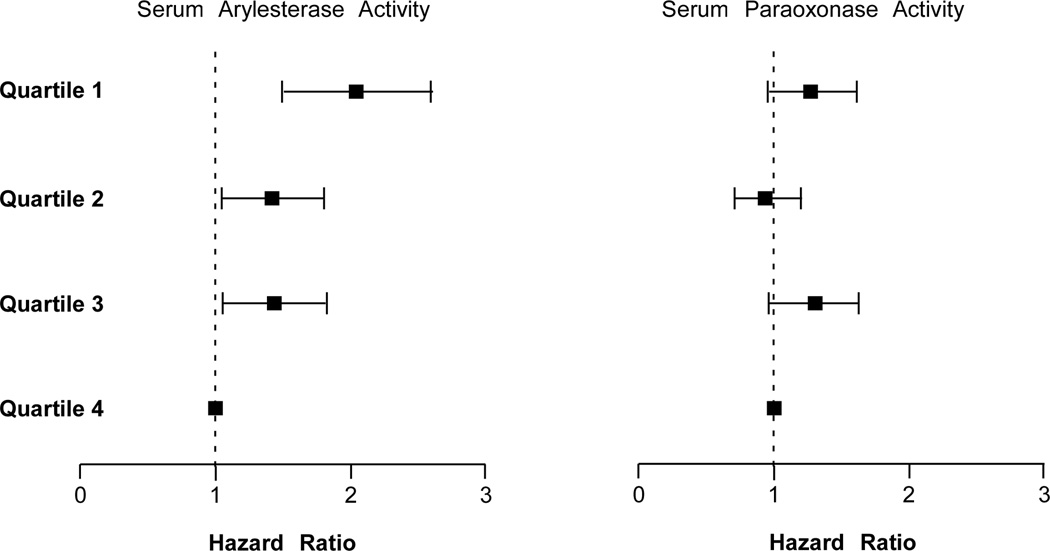
In the 3,668 subjects, a total of 417 cardiac events were recorded within the 3-year period of follow-up. Lower serum paraoxonase and arylesterase activity levels were associated with poorer long-term outcomes when stratified by quartiles (Table 2, Figure 1). After adjusting for Framingham risk factors, eCrCl, diabetes mellitus and log-transformed hsCRP, lower serum arylesterase activity (HR 2.20 [95%CI 1.60–3.02], p<0. 01) and to a lesser extent lower serum paraoxonase activity (HR 1.39 [95%CI 1.04–1.85], p<0.05) demonstrated increased risk in developing future MACE. Even when cTnI levels were added to the model (within the "normal" range of 0.001–0.029 mg/dL), lower serum arylesterase levels still maintain a 2-fold increased risk in MACE at 3 years (HR 2.04 [95%CI 1.49–2.79], p<0.01). The separation is particularly apparent between the lowest quartile and the upper three quartiles at the cut-off of 87 µmol/min/mL (Figure 1). The addition of serum arylesterase activity or serum paraoxonase activity to the model results in significant improvement in risk classification with net reclassification index (NRI) of 7.9% for arylesterase activity (p=0.003) and 7.2% for paraoxonase activity (p=0.002). The prognostic value of serum arylesterase activity was observed within the secondary prevention cohort (Table 2), as well as within subjects who demonstrated a recent “normal catheterization” (i.e., no significant (>50%) angiographic evidence of CAD in any major vessel or preceding history of CAD – the primary prevention cohort, Table 2). Serum arylesterase activity also remained a prognostic indicator within each Framingham risk factor sub-cohort and among those with low hsCRP (Figure 2), or in those without statin therapy (n=1,512). In addition to serum arylesterase remaining a significant predictor of MACE after addition of higher sensitivity troponin testing to traditional risk factors and laboratory risk markers in the models, we further observed an increased risk of developing subclinical myocardial necrosis (troponin levels that are detectable but remain below the 99th percentile diagnostic cutoff among healthy subjects used to define cutoff for MI) with decreasing quartiles of serum arylesterase levels (odds ratio 2.01 [95%CI 1.53–2.64], p<0.001, Figure 3) – this trend was not observed with serum paraoxonase activity levels (Figure 3).
Table 2.
Unadjusted and adjusted hazard ratio for major adverse cardiac events (MACE) at 3-year follow-up according to serum arylesterase activity and paraoxonase activity quartiles, stratified according to primary versus secondary prevention
| Serum Arylesterase Activity (µmol/min/mL) | Serum Paraoxonase Activity (µmol/min/mL) | |||||||
|---|---|---|---|---|---|---|---|---|
| Quartile 4 | Quartile 3 | Quartile 2 | Quartile 1 | Quartile 4 | Quartile 3 | Quartile 2 | Quartile 1 | |
| All Subjects (n=3,668) | ||||||||
| Range | ≥ 121 | 103–121 | 87–103 | <87 | ≥ 1045 | 562–1045 | 315–562 | <315 |
| Unadjusted HR | 1 | 1.50 (1.1–2.05)* | 1.45 (1.06–1.99)* | 2.63 (1.97–3.50)** 2.20 (1.60–3.02)** |
1 | 1.39 (1.04–1.84)* | 1.1 (0.82–1.48) | 1.63 (1.24–2.14)** 1.39 (1.04–1.85)* |
| Adjusted HR (1) | 1 | 1.44 (1.05–1.97)* | 1.34 (0.96–1.86) | 1 | 1.19 (0.89–1.6) | 1.04 (0.77–1.4) | ||
| Adjusted HR (2) | 1 | 1.40 (1.02–1.92)* | 1.27 (0.91–1.77) | 1.85 (1.35–2.55)** | 1 | 1.1 (0.82–1.49) | 1.00 (0.74–1.36) | 1.27 (0.95–1.70) |
| Secondary Prevention Subjects (n=2,636) | ||||||||
| Range | ≥119 | 101–119 | 86–101 | <86 | ≥1022 | 549–1022 | 308–549 | <308 |
| Unadjusted HR | 1 | 1.24 (0.89–1.73) | 1.26 (0.9–1.75) | 2.01 (1.49–2.72)** 1.78 (1.28–2.48)** |
1 | 1.35 (0.98–1.84) | 1.2 (0.87–1.65) | 1.45 (1.07–1.97)* 1.30 (0.94–1.79) |
| Adjusted HR (1) | 1 | 1.23 (0.88–1.72) | 1.21 (0.86–1.72) | 1 | 1.2 (0.87–1.66) | 1.14 (0.83–1.57) | ||
| Adjusted HR (2) | 1 | 1.17 (0.83–1.63) | 1.13 (0.80–1.61) | 1.51 (1.08–2.11)* | 1 | 1.12 (0.81–1.56) | 1.12 (0.81–1.55) | 1.18 (0.86–1.64) |
| Primary Prevention Subjects (n=1,032) | ||||||||
| Range | ≥125 | 107–125 | 91–107 | <91 | ≥1107 | 644–1107 | 335–644 | <335 |
| Unadjusted HR | 1 | 0.91 (0.35–2.35) | 2.63 (1.21–5.72)* | 4.38 (2.11–9.09)** 4.00 (1.82–8.82)** |
1 | 1.20 (0.61–2.38) | 0.71 (0.33–1.55) | 2.15 (1.16–3.97)* 1.85 (0.99–3.45) |
| Adjusted HR (1) | 1 | 0.83 (0.32–2.18) | 2.50 (1.08–5.79)* | 1 | 1.09 (0.55–2.18) | 0.71 (0.33–1.51) | ||
| Adjusted HR (2) | 1 | 0.84 (0.32–2.21) | 2.44 (1.02–5.84)* | 3.02 (1.35–6.80)** | 1 | 0.92 (0.46–1.84) | 0.65 (0.30–1.42) | 1.68 (0.88–3.19) |
Model 1: adjusted for traditional risk factors (include age, gender, systolic blood pressure, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, cigarette smoking, diabetes mellitus)
Model 2: Adjusted for traditional risk factors (Model 1), plus log-transformed hsCRP levels, creatinine clearance, race, body mass index, and statin.
p<0.01 and
p<0.05
Figure 1.
Kaplan-Meier analysis for long-term major adverse cardiac events stratified by serum arylesterase and paraoxonase activities quartiles in overall (A, B), secondary prevention (C, D), and primary prevention (E, F) populations.
Figure 2.
Forest plot regarding Hazard ratios of serum arylesterase and paraoxonase activities according to traditional cardiac risk factors in subgroups of patients
Figure 3.
Adjusted odds ratio across serum arylesterase and paraoxonase activity quartiles with prevalent subclinical myocardial necrosis (defined as cTnI ≥0.009 ng/mL).
GWAS for PON-1 activities
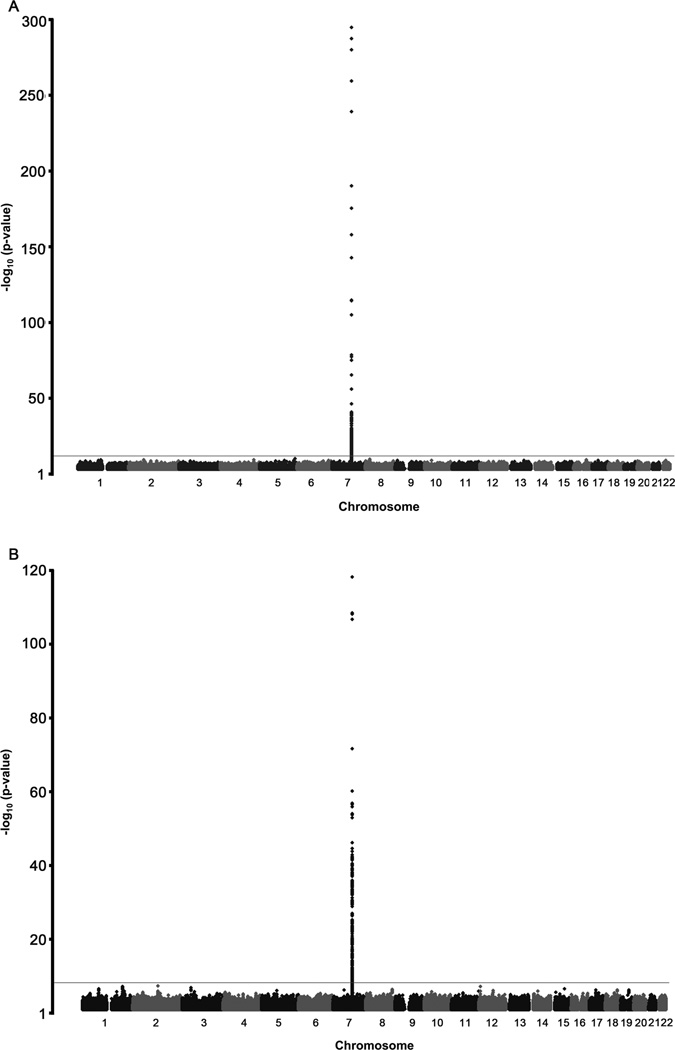
We next performed a GWAS for serum paraoxonase and arylesterase activity in 2,136 GeneBank subjects (all of Caucasian ancestry) for whom both genotype and PON-1 functional data were available. The genomic inflation factors for paraoxonase and arylesterase activity were 1.015 and 1.013, respectively, indicating that the GWAS results were not confounded by underlying population stratification, and the Q-Q plots are shown in Supplemental Figure 1. Serum paraoxonase activity was controlled by a major locus on chromosome 7 containing the PON-1, PON-2, and PON-3 genes (Figure 4A). The lead SNP for serum paraoxonase activity at this locus (rs2057681) is located within the PON-1 gene and yielded a highly significant p-value of 1.18 × 10−303 (Figure 4A, Table 3). Based on our genotype data, rs2057681 is in near complete linkage disequilibrium (LD) (r2=0.99) with a functional amino acid substitution in PON-1 (rs662; Q192R), which is also associated with increased paraoxonase activity (p=3.31 × 10−295; Table 3). Of interest, while rs2057681 and rs662 are associated with significant increase in paraoxonase activity, they are also associated with significant decrease in arylesterase activity (Table 3). Another known coding SNP in PON-1 (rs854560; L55M) exhibited significant association with paraoxonase activity as well (p=1.27 × 10−140; Table 3) but is in relatively weak LD (r2=0.20) with the aforementioned lead SNPs (rs2057681 and rs662). The GWAS analysis for serum arylesterase activity also revealed that this chromosome 7 region was the major locus controlling this measure of PON-1 functional activity (Figure 4B). For example, the lead SNP for serum arylesterase activity, rs854572, is located in the promoter region of PON-1 and yielded a highly significant p value of p=4.99 × 10−116 (Table 3). This SNP (rs854572) also was associated with increase in paraoxonase activity. Of note, the four lead SNPs for arylesterase activity differ from those identified for paraoxonase activity, although they do exhibit some association with paraoxonase activity and vice versa (Table 3). However, the lead SNPs for arylesterase and paraoxonase activity are in weak LD (r2<0.23), suggesting that the reciprocal associations are for the most part independent. Further, the lead SNPs for both paraoxonase (rs2057681) and arylesterase (rs854572) activities are localized in the PON-1 gene and not in LD with variants in either PON-2 or PON-3 genes. We next determined whether the lead SNPs on chromosome 7 influenced other CAD risk factors but did not observe any significant evidence for such associations (Supplemental Table 1). Since BMI and statin use have been shown to potentially affect serum paraoxonase activity11–13, we also adjusted the genetic analyses for these potential confounders. However, the strength of the association of the lead SNPs with paraoxonase and arylesterase activities was not diminished when either BMI or statin use alone was included with age and gender in the genetic models or in a fully adjusted model that included all four covariates (Supplemental Table 2). Thus, the effect of the lead SNPs on paraoxonase and arylesterase activities are robust and independent of traditional cardiovascular risk factors.
Figure 4.
Manhattan plots for Genomewide Association Studies identifying highest SNPs associated with serum arylesterase (A); and paraoxonase (B) activity levels.
Table 3.
Mean Serum Paraoxonase and Arylesterase Activity as a Function of Genotype for Lead SNPs on Chromosome 7.
| Serum Paraoxonase Activity (nmoles/min/ml) | Serum Arylesterase Activity (µmoles/min/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position (bp)a |
SNP | Allelesb | MAFc | 0 | 1 | 2 | p-valuec | 0 | 1 | 2 | p-valuec |
| 94,774,065 | rs2269829 | A/G | 0.28 | 389±269 (n=1078) | 934±361 (n=905) | 1434±492 (n=153) | 3.27×10−288 | 105±25 (n=1106) | 100±24 (n=930) | 93±20 (n=157) | 4.22×10−11 |
| 94,775,382 | rs662 (Q192R) | A/G | 0.29 | 382±261 (n=1062) | 930±358 (n=914) | 1424±497 (n=160) | 3.31×10−295 | 105±25 (n=1088) | 100±24 (n=940) | 94±21 (n=165) | 9.43×10−11 |
| 94,776,193 | rs2057681 | A/G | 0.29 | 377±253 (n=1055) | 929±353 (n=919) | 1433±501 (n=162) | 1.18×10−303 | 105±25 (n=1080) | 100±24 (n=946) | 94±21 (n=167) | 2.11×10−10 |
| 94,784,020 | rs854560 (L55M) | A/T | 0.36 | 906±489 (n=877) | 620±398 (n=1020) | 311±280 (n=269) | 1.27×10−140 | 109±23 (n=868) | 99±24 (n=1053) | 90±22 (n=272) | 2.03×10−38 |
| 94,790,628 | rs854570 | A/C | 0.36 | 670±470 (n=898) | 711±469 (n=987) | 721±455 (n=251) | 2.90×10−09 | 91±21 (n=920) | 107±23 (n=1014) | 122±21 (n=259) | 5.10×10−106 |
| 94,792,632 | rs854572 | G/C | 0.46 | 572±407 (n=645) | 716±474 (n=1075) | 831±494 (n=416) | 1.23×10−35 | 88±21 (n=659) | 104±22 (n=1104) | 119±22 (n=430) | 4.99×10−116 |
| 94,793,157 | rs705382 | G/C | 0.36 | 670±470 (n=897) | 710±469 (n=989) | 723±455 (n=250) | 2.92×10−09 | 91±21 (n=919) | 107±23 (n=1016) | 122±21 (n=258) | 1.98×10−106 |
| 94,793,464 | rs757158 | C/T | 0.42 | 569±403 (n=725) | 731±478 (n=1053) | 843±501 (n=358) | 3.97×10−38 | 89±22 (n=741) | 105±22 (n=1084) | 120±22 (n=368) | 1.04×10−104 |
Data are shown as mean ± SD as a function of carrying 0, 1, or 2 copies of the minor alleles for lead GWAS SNPs.
Base pair positions on chromosome 7 are given according to NCBI build 36.1 of the reference human genome sequence.
Major/minor alleles are given for Caucasians based on the forward (+) DNA strand. MAF, minor allele frequency.
p-values are obtained from multiple linear regression using inverse normal-transformed values for paraoxonase activity and untransformed values for arylesterase activity, adjusted for age and gender.
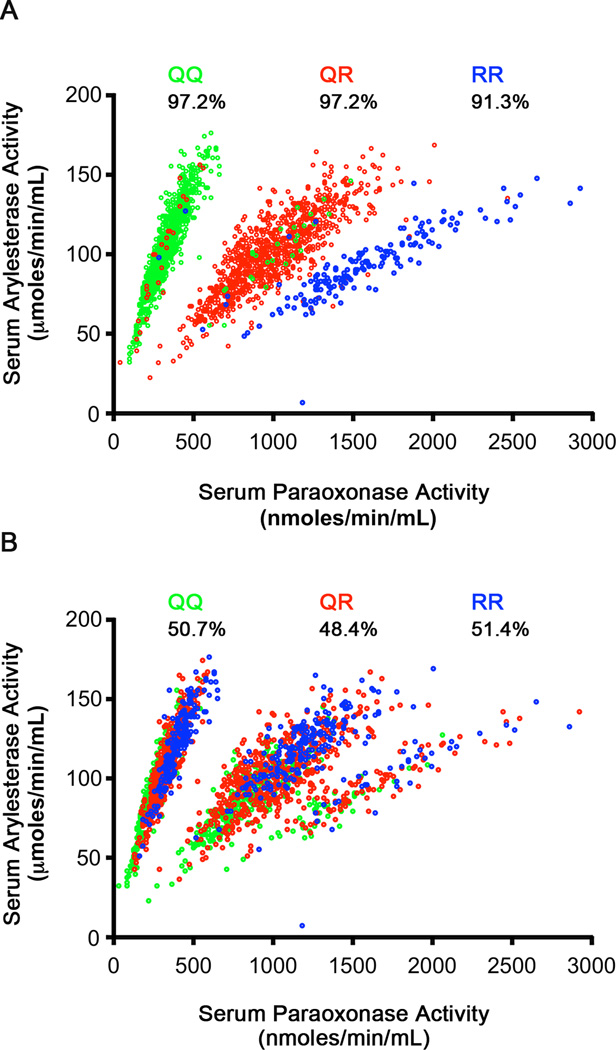
Given the strong effect of the chromosome 7 locus on PON-1 function, we also carried out GWAS analyses for paraoxonase and arylesterase activity conditioned on the lead SNP for each respective trait. These analyses did not reveal other loci in the genome that were significantly associated with either measure of PON-1 activity. To gain further insight into the effect of the chromosome 7 variants on PON-1 function, we also examined the relationship between serum paraoxonase and arylesterase activities in our cohort. As illustrated in Figure 5, both activities were significantly correlated overall (r=0.30, p<0.001) and exhibited a striking grouping of 3 distinct patterns when plotted against each other. These three clusters were found to correspond nearly exactly with the three genotype groups of the Q192R (rs662) polymorphism (Figure 5A), with 97.2% with genotype QQ in the top cluster, 97.2% with genotype QR in the middle cluster, and 91.3% with genotype RR in the bottom cluster. However, such a pattern was not observed with the three genotype groups of the lead SNP for serum arylesterase activity (rs854572; G>C, Figure 5B). Quantitative analysis of the separation of the three genotypes, GG, GC, and CC, across the 3 clusters showed 50.7% with genotype CG in the top cluster, 48.4% with Genotype CG in the middle cluster, and 51.4% with genotype CG in the bottom cluster.
Figure 5.
Relationship between serum arylesterase and paraoxonase activity levels stratified according to (A) the lead SNP for serum paraoxonase activity (rs662; Q192R); (B) the lead SNP for serum arylesterase activity (rs854572; G>C). Percentages reflect separation of the three genotypes within each stratified cluster of each genotype.
Association of PON-1 variants with incident MACE and prevalent CVD
We next sought to determine whether the lead variants influencing paraoxonase and arylesterase activity were associated with the development of MACE. These analyses included the 2136 subjects used in the quantitative association described above plus an additional 567 subjects for whom genotype and MACE data were also available (total n=2703). As shown in Table 4, there were no individual effects of the Q192R (rs662) and L55M (rs854560) substitutions on incident risk of MACE in GeneBank subjects. Given the differential effects of the associated variants on PON-1 function (e.g. rs662 is associated with increase in paraoxonase activity but decrease in arylesterase activity), we also constructed haplotypes with rs662, rs854560, rs854570, and rs854572 and specifically tested the ATAG haplotype, which leads to modest decreases in both paraoxonase and arylesterase activity (data not shown). However, risk of future MACE was not significantly increased in subjects carrying one (HR=0.96, 95%CI 0.68–1.35) or two (HR=0.91, 95% CI 0.50–1.63) copies of this haplotype (p=0.72; Table 4). Similarly, an analysis using a haplotype score test that included all haplotypes with greater than 1% frequency also did not reveal any associations with MACE (data not shown).
Table 4.
Association of Variants Controlling Plasma Paraoxonase or Arylesterase Activity with MACE.
| HR (95% CI) | ||||
|---|---|---|---|---|
| SNP/Haplotype | 0 | 1 | 2 | p–value |
| rs2057681 | 1 (n=1341) | 1.18 (0.94 – 1.49) (n=1155) | 0.94 (0.60 – 1.49) (n=207) | 0.48 |
| rs854572 | 1 (n=794) | 1.18 (0.91 – 1.54) (n=1358) | 1.12 (0.81 – 1.56) (n=551) | 0.44 |
| ATAG | 1 (n=1212) | 0.96 (0.68 – 1.35) (n=1236) | 0.91 (0.50 – 1.63) (n=255) | 0.72 |
HRs are shown as a function of carrying 0, 1, or 2 copies of the minor allele for the lead SNP for paraoxonase (rs2057681) and arylesterase (rs854572) activity or the ATAG haplotype of rs662, rs854560, rs854570, and rs854572. HR are adjusted for age, gender, Framingham ATP–III risk score, and medication use (aspirin and/or statins). 2703 subjects were used in these analyses, of which 311 experienced a MACE (death, MI, or stroke) over 3 years of follow up.
To further evaluate the genetic contribution of these SNPs to cardiovascular risk, we used the large CARDIoGRAM consortium, which is comprised of ~80,000 case-controls subjects. As shown in Table 5, no significant evidence for association of these SNPs was observed with risk of prevalent CAD. A sub-analysis with only cases that had a positive history for MI also did not reveal any associations (Table 5).
Table 5.
Association of Identified SNPs with Risk of Prevalent CAD and Risk of MI in the CARDIoGRAM Consortium.
| Risk of CAD | Risk of MI | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Allele | Frequency | OR (95% CI) | p–value | n | OR (95% CI) | p–value | n |
| rs2269829 | G | 0.28 | 0.99 (0.97–1.03)) | 0.74 | 83,324 | 0.97 (0.94–1.01) | 0.17 | 52,973 |
| rs662 (Q192R) | G | 0.30 | 0.99 (0.96–1.02) | 0.60 | 79,262 | 0.98 (0.94–1.02) | 0.24 | 52,306 |
| rs2057681 | G | 0.28 | 0.99 (0.96–1.02) | 0.44 | 84,106 | 0.97 (0.93–1.00) | 0.09 | 53,649 |
| rs854560 (L55M) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| rs854570 | C | 0.41 | 1.02 (0.99–1.05) | 0.14 | 81,019 | 1.01 (0.98–1.05) | 0.46 | 51,143 |
| rs854572 | C | 0.52 | 1.01 (0.99–1.04) | 0.33 | 83,486 | 1.00 (0.97–1.04) | 0.74 | 53,204 |
| rs705382 | C | 0.41 | 1.02 (1.0–1.05) | 0.09 | 83,367 | 1.02 (0.98–1.05) | 0.40 | 53,035 |
| rs757158 | C | 0.54 | 0.99 (0.96–1.02) | 0.50 | 78,275 | 0.99 (0.96–1.03) | 0.71 | 51,559 |
N/A, not available
DISCUSSION
Paraoxonase-1 is an atherosclerosis protective enzyme associated with HDL and systemic anti-oxidant function. Its catalytic activity within crude serum mixtures has traditionally been measured by quantifying enzymatic hydrolysis rates of two known in vitro substrates, with functional activities named paraoxonase and arylesterase activities4, 5. There are several novel findings in this report that describe the clinical and genetic associations of PON-1 activities with cardiovascular risks that differ from prior observations from smaller cohorts. First, we demonstrated prognostic value of serum arylesterase activity (and to a lesser extent serum paraoxonase activity) in predicting long-term cardiovascular risk in a wide range of subjects already treated with contemporary medical therapy. We observed that diminished serum arylesterase activity, particularly within the lowest quartile range, was predictive of adverse long-term cardiac events independent of standard clinical and biochemical risk factors, and provided incremental value in reclassifying subjects who are at higher risk of long-term MACE. Furthermore, the prognostic value of serum arylesterase activity was evident in both primary as well as secondary prevention subjects. The fact that arylesterase activity provided prognostic value consistently within subjects with or without underlying angiographic evidence of significant CAD implies that a lack of systemic anti-oxidant defense mechanisms (a primary function of PON-1) may both promote greater vulnerability to oxidative stress, as well as increase risk for development and progression of CAD in subjects. Moreover, the ability of low serum arylesterase activity to identify those at significant increased risk of MACE, even among primary prevention subjects with recent coronary angiographic data showing < 50% stenosis in all major coronary vessels, suggests this assay has prognostic value, and can identify a vulnerable cohort of subjects who otherwise are not identified as being at high risk. Taken together, the present results suggest that enzymatic activity measures with arylesterase, more so than paraoxonase, serve as a powerful prognostic indicator of cardiovascular risk in a broad spectrum of subjects.
Studies on PON-1 activities have relied on quantifying its wide ranges of enzymatic activities in breaking down in vitro substrates like paraoxon (paraoxonase activity) and phenylacetate (arylesterase activity). These activities are often reported together and the findings are commonly concordant. Hence, the distinction between serum paraoxonase and arylesterase activities in predicting future adverse cardiac events is somewhat unexpected, even though historically the correlation between these two measures has not been particularly tight. The unique relationship between the two PON-1 activity measurements is largely the result of a very strong association between serum paraoxonase activity and its underlying genetic determinants, which may also explain why serum arylesterase activity is normally distributed while serum paraoxonase activity was not. Meanwhile, the strong association between the PON gene cluster and serum arylesterase activity confirms the long-standing assumption that this locus harbors important genetic determinants of serum arylesterase activity.
The significant genetic associations revealed by the GWAS analyses for paraoxonase and arylesterase activities are of further interest for several reasons. First, there was a stronger relationship between serum paraoxonase activity levels and its genetic determinants than those with serum arylesterase activity. Second, our results confirm the strong association of PON-1 variants with serum paraoxonase and arylesterase activities and demonstrate that the two measured enzymatic functions are associated in a Mendelian-like fashion by a single, major locus on chromosome 7 containing the PON gene cluster. This is supported by the GWAS analyses conditioned on the lead SNPs, which did not identify other genomic regions associated with either paraoxonase or arylesterase activity. Furthermore, the lead SNPs for either enzymatic activity localize to the PON-1 gene, are distinct from each other, and are not in LD with SNPs in the neighboring PON-3 and PON-2 genes. However, while the PON-1 variants exhibited strong (if not opposed) effects on PON function, they were not associated with future risk of MACE, either individually or as a haplotype that modestly decreased both paraoxonase and arylesterase activity. These results are consistent with our analyses from the CARDIoGRAM consortium, which did not reveal an association of these SNPs with either prevalent CAD or history of MI in ~80,000 subjects. It should be noted, however, that since the lead SNPs only contributed to an estimated ~15% of PON-1 activity variation, other processes, such as post-translational modifications, could also play a role in determining the ultimate functionality of PON-1. For example, PON1 is known to be sensitive to post-translational oxidative modification and inactivation14. It is also possible that our study was underpowered to detect genetic effects on prospective risk of MACE since only 311 subjects out of the ~2,700 subjects included in these analyses experienced a MACE over three years of follow up.
The lack of associations between the lead SNPs for PON and arylesterase activities with CAD and MI risks in humans in our study are in direct contrast to what has been observed in mice where PON-1 deficiency leads to increased aortic lesion formation3, and transgenic mice over expressing PON-1 are protected 15, 16. It is of interest that the PON-1 transgenic mouse models demonstrating protective effects have 50–400% increased serum arylesterase activity 15, 16, far in excess of the modest changes in activity associated with the peak SNP identified in the arylesterase GWAS. If arylesterase activity is the more important atheroprotective aspect of PON-1 function, as our clinical associations with MACE suggest, then it may not be surprising that an association was not observed with the SNPs identified. For example, the lead SNP for arylesterase activity (rs854572) only increases activity by ~16% per minor allele copy. It is thus possible that the genetic effects on arylesterase activity from this SNP are too weak to observe, especially if a minimum biological threshold of activity change is needed to influence risk of prevalent CAD, history of MI, or incident risk of MACE.
The ability for serum arylesterase activity to identify a high-risk population in the primary prevention cohort that just underwent cardiac catheterization and showed no significant evidence of stenoses in any major vessel may have important clinical implications. Even though our subset analysis was limited by the relatively smaller sample size and low event rates, a significant 4-fold increase risk in long-term MACE in these otherwise "low risk" subjects was identified. Use of serum arylesterase activity may thus have clinical utility, and help identify an important patient cohort that may warrant more aggressive risk factor reduction treatment strategies who might otherwise not be targeted for aggressive preventive intervention. In addition to low serum arylesterase activity predicting increased risk for MACE in primary prevention subjects, there was a direct association also noted between lower serum arylesterase activity level and increased prevalence of significantly obstructive CAD by angiography. Of note, we did not find a strong relationship between systemic inflammatory biomarkers, such as C-reactive protein or leukocyte parameters, with serum arylesterase levels in this group of stable cardiac patients. This observation is consistent with prior reports that distinguished systemic inflammatory from oxidative stress processes.
Several limitations of our study should be noted. Serum paraoxonase and arylesterase activities utilized substrates that are not the endogenous substrates for PON-1, but are used because they are not readily influenced by other esterases/lactonases in serum, and are presumed to reflect the underlying catalytic activity of PON-1. The GWAS results observed demonstrate these assumptions are reasonable, as only genetic variations in the PON-1 gene were observed to be associated with variations in paraoxonase and arylesterase activities. A further potential limitation is that the measurements were only made under fasting conditions at a single time-point. Hence, we are unable to determine the variability and prognostic value of level changes over time and the impact of dietary or therapeutic interventions on serum arylesterase activity level. Selection bias may also be present for those undergoing cardiac catheterization for symptomatic evaluation and management of cardiac diseases at a tertiary care setting, but the large sample size and event rates of the patient population, together with careful phenotypic evaluation including angiographic data, provides unique insights and adequate power to adjust for clinical and biomarker variables.
CONCLUSIONS
Diminished serum arylesterase activity can provide incremental prognostic value and clinical reclassification of stable patients at risk of developing MACE, even amongst primary prevention subjects who just demonstrated no significant coronary stenoses by angiography and might otherwise be dismissed as low risk. Despite the strong genetic effects of the PON locus on serum paraoxonase and arylesterase activities, the identified variants were not associated with risk of incident MACE or prevalent CAD.
Supplementary Material
Acknowledgments
SOURCES OF FUNDING
This research was supported by National Institutes of Health grants P01HL076491-055328, 5P01HL103453, and the Fondation Leducq. The GeneBank study has been supported by NIH grants P01HL098055, R01HL103866, 1P20HL113452, 1R01HL103931, R01ES021801 (H.A.), and the Cleveland Clinic Clinical Research Unit of the Case Western Reserve University CTSA (UL1TR 000439-06). SLH is also partially supported by a gift from the Leonard Krieger Fund. The John & Jennifer Ruddy Canadian Cardiovascular Genetics Centre investigators are supported by CIHR #MOP--82810, CFI #11966, HSFO #NA6001, CIHR #MOP172605, and CIHR #MOP77682.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
Dr. Tang has received research grant support from Abbott Laboratories, Inc. Drs. Hartiala, Fan, Wu, Stewart, Erdmann, Kathiresan, Roberts, McPherson and Allayee reported no relationships to disclose. Dr. Hazen reports being listed as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. Dr. Hazen reports having been paid as a consultant or speaker for the following companies: Abbott Diagnostics, Cleveland Heart Lab, Esperion, Lilly, Liposcience Inc., Merck & Co., Inc., and Pfizer Inc. Dr. Hazen reports receiving research funds from Abbott, Cleveland Heart Lab, Liposcience Inc., and Pfizer Inc. Dr. Hazen reports having the right to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from the companies shown below: Abbott Laboratories, Inc., Cleveland Heart Lab., Esperion, Frantz Biomarkers, LLC, Liposcience Inc., and Siemens.
AUTHOR CONTRIBUTIONS
Drs Tang and Hazen had full access to all of the data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Tang, Hazen
Acquisition of data: Tang, Hazen, Erdmann, Kathiresan, Allayee
Analysis and interpretation of data: Tang, Hartiala, Fan, Wu, Patel, Allayee, Hazen.
Drafting of the manuscript: Tang
Critical revision of the manuscript for important intellectual content: Tang, Stewart, Roberts, McPherson, Kathiresan, Allayee, Hazen
Statistical analysis: Fan, Wu, Hartiala, Allayee
Obtained funding: Tang, Roberts, McPherson, Hazen
Study supervision: Tang, Hazen
There are no medical writers or editors involved in the preparation of the manuscript.
REFERENCES
- 1.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 2.Costa LG, Li WF, Richter RJ, Shih DM, Lusis A, Furlong CE. The role of paraoxonase (pon1) in the detoxication of organophosphates and its human polymorphism. Chem Biol Interact. 1999;119–120:429–438. doi: 10.1016/s0009-2797(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 3.Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 4.Shih DM, Lusis AJ. The roles of pon1 and pon2 in cardiovascular disease and innate immunity. Curr Opin Lipidol. 2009;20:288–292. doi: 10.1097/MOL.0b013e32832ca1ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durrington PN, Mackness B, Mackness MI. Paraoxonase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2001;21:473–480. doi: 10.1161/01.atv.21.4.473. [DOI] [PubMed] [Google Scholar]
- 6.Furlong CE, Richter RJ, Seidel SL, Costa LG, Motulsky AG. Spectrophotometric assays for the enzymatic hydrolysis of the active metabolites of chlorpyrifos and parathion by plasma paraoxonase/arylesterase. Anal Biochem. 1989;180:242–247. doi: 10.1016/0003-2697(89)90424-7. [DOI] [PubMed] [Google Scholar]
- 7.Tang WH, Wu Y, Mann S, Pepoy M, Shrestha K, Borowski AG, Hazen SL. Diminished antioxidant activity of high-density lipoprotein-associated proteins in systolic heart failure. Circ Heart Fail. 2011;4:59–64. doi: 10.1161/CIRCHEARTFAILURE.110.958348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, Fu X, Shao M, Brennan DM, Ellis SG, Brennan ML, Allayee H, Lusis AJ, Hazen SL. Relationship of paraoxonase 1 (pon1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preuss M, Konig IR, Thompson JR, Erdmann J, Absher D, Assimes TL, Blankenberg S, Boerwinkle E, Chen L, Cupples LA, Hall AS, Halperin E, Hengstenberg C, Holm H, Laaksonen R, Li M, Marz W, McPherson R, Musunuru K, Nelson CP, Burnett MS, Epstein SE, O'Donnell CJ, Quertermous T, Rader DJ, Roberts R, Schillert A, Stefansson K, Stewart AF, Thorleifsson G, Voight BF, Wells GA, Ziegler A, Kathiresan S, Reilly MP, Samani NJ, Schunkert H. Design of the coronary artery disease genome-wide replication and meta-analysis (cardiogram) study: A genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deakin S, Guernier S, James RW. Pharmacogenetic interaction between paraoxonase-1 gene promoter polymorphism c-107t and statin. Pharmacogenet Genomics. 2007;17:451–457. doi: 10.1097/FPC.0b013e3280925716. [DOI] [PubMed] [Google Scholar]
- 12.Ferretti G, Bacchetti T, Masciangelo S, Bicchiega V. Hdl-paraoxonase and membrane lipid peroxidation: A comparison between healthy and obese subjects. Obesity (Silver Spring) 2010;18:1079–1084. doi: 10.1038/oby.2009.338. [DOI] [PubMed] [Google Scholar]
- 13.Himbergen TM, van Tits LJ, Voorbij HA, de Graaf J, Stalenhoef AF, Roest M. The effect of statin therapy on plasma high-density lipoprotein cholesterol levels is modified by paraoxonase-1 in patients with familial hypercholesterolaemia. J Intern Med. 2005;258:442–449. doi: 10.1111/j.1365-2796.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- 14.Besler C, Heinrich K, Rohrer L, Doerries C, Riwanto M, Shih DM, Chroni A, Yonekawa K, Stein S, Schaefer N, Mueller M, Akhmedov A, Daniil G, Manes C, Templin C, Wyss C, Maier W, Tanner FC, Matter CM, Corti R, Furlong C, Lusis AJ, von Eckardstein A, Fogelman AM, Luscher TF, Landmesser U. Mechanisms underlying adverse effects of hdl on enos-activating pathways in patients with coronary artery disease. J Clin Invest. 2011;121:2693–2708. doi: 10.1172/JCI42946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She ZG, Zheng W, Wei YS, Chen HZ, Wang AB, Li HL, Liu G, Zhang R, Liu JJ, Stallcup WB, Zhou Z, Liu DP, Liang CC. Human paraoxonase gene cluster transgenic overexpression represses atherogenesis and promotes atherosclerotic plaque stability in apoe-null mice. Circ Res. 2009;104:1160–1168. doi: 10.1161/CIRCRESAHA.108.192229. [DOI] [PubMed] [Google Scholar]
- 16.Tward A, Xia YR, Wang XP, Shi YS, Park C, Castellani LW, Lusis AJ, Shih DM. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation. 2002;106:484–490. doi: 10.1161/01.cir.0000023623.87083.4f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.