Abstract
The NAD(P)H:quinone acceptor oxidoreductase (NQO) gene family belongs to the flavoprotein clan and, in the human genome, consists of two genes (NQO1 and NQO2). These two genes encode cytosolic flavoenzymes that catalyse the beneficial two-electron reduction of quinones to hydroquinones. This reaction prevents the unwanted one-electron reduction of quinones by other quinone reductases; one-electron reduction results in the formation of reactive oxygen species, generated by redox cycling of semiquinones in the presence of molecular oxygen. Both the mammalian NQO1 and NQO2 genes are upregulated as a part of the oxidative stress response and are inexplicably overexpressed in particular types of tumours. A non-synonymous mutation in the NQO1 gene, leading to absence of enzyme activity, has been associated with an increased risk of myeloid leukaemia and other types of blood dyscrasia in workers exposed to benzene. NQO2 has a melatonin-binding site, which may explain the anti-oxidant role of melatonin. An ancient NQO3 subfamily exists in eubacteria and the authors suggest that there should be additional divisions of the NQO family to include the NQO4 subfamily in fungi and NQO5 subfamily in archaebacteria. Interestingly, no NQO genes could be identified in the worm, fly, sea squirt or plants; because these taxa carry quinone reductases capable of one- and two-electron reductions, there has been either convergent evolution or redundancy to account for the appearance of these enzyme functions whenever they have been needed during evolution.
Keywords: NAD(P)H:quinone oxidoreductase gene family, diaphorase, cytochrome b5 reductase, quinone reductase, aminoazo dye reductase, flavoprotein clan, flavodoxins, nitric oxide synthases, ribonucleotide reductases, flavin reductases, melatonin
Introduction
Quinones comprise a large class of aromatic compounds, found widely in plants, in the general (benzoyl) form of C6H4O2, but also found in all organisms as flavonoids, electron-carrying coenzymes and metabolic end-products of oxidation. Commercially, quinones are used in making dyes, tanning hides and in photography. Quinones are potentially dangerous, in that they can be involved in a one-electron reduction to form semiquinones that will elicit oxidative stress in any cell; benzoquinones and naphthoquinones are among the most reactive in this category. Quinones were undoubtedly present in the environment and in unicellular organisms between 2 and more than 3 billion years ago (at the time that partial pressure of ambient oxygen on the planet was rising). Hence, during this time it is very likely there was a strong selection to develop an enzyme that would reduce quinones to hydroquinones by a two-electron step, thereby avoiding free radical formation, which is highly destructive to the living cell.
Background and history
The NAD(P)H:quinone acceptor oxidoreductase (NQO) gene family is therefore believed to be ancient -- probably between 2 and more than 3 billion years old. Cytosolic NQO flavoenzymes catalyse the beneficial two-electron reduction of quinones to hydroquinones. The two-electron reaction prevents the reduction of quinones by one-electron reductases, which would result in the formation of reactive oxygen species (ROS) generated by redox cycling of semiquinones in the presence of molecular oxygen [1,2].
In animals, in addition to the likely role in detoxication of many dietary quinones derived from plants, NQO enzymes have been shown to catalyse the reductive activation of chemotherapeutic quinones such as mitomycins and aziridinyl-benzoquinones [3,4]. The hydroquinones derived from these agents appear to be chemically more reactive, either because of rapid autoxidation or rearrangement -- which, in either case, will produce reactive species that can kill malignant cells [5,6].
Discovery of DT diaphorase
Ernster and Navazioare credited with discovering (in the rat) the first member of the NQO family in the late 1950s [7-9]. This enzyme is now officially named NQO1 (EC 1.6.99.2; previous names included DT diaphorase and aminoazo dye reductase, NQOR1, NMO1, NMOR1 and QR1). It was later concluded [9,10] that this enzyme is probably identicaltoa vitamin K reductase that had been isolated by Martius and colleagues. The mammalian NQO1 enzyme is expressed constitutively in most tissues and is strikingly upregulated during oxidative or electrophilic stress; NQO1 has been shown to be amember of the [AHR] gene battery [11]. The crystal structure [12], enzymic functions [13] and polymorphisms [14] in the NQO1 gene have all recently been summarised.
The human NQO1 gene on chromosome 16q22.1 comprises six exons, spanning approximately18 kilobases (kb), encodes 274 amino acids and contains, to date, 93 single nucleotide polymorphisms (SNPs) within 10 kb 5'-ward of the first exon and 2 kb 3'-ward of the last exon (http://SNPper.chip.org/). Two additional variants, besides the consensus gene, were found in the Entrez Gene database (Figure 1). Compared with the human consensus (reference, 'wild-type') NQO1*1 allelecodingfor normal NQO1 enzyme and activity, the NQO1*2 allele encodes anon-synonymous mutation (Pro187Ser) that has negligible NQO1 enzyme activity. The NQO1*2 allelic frequency ranges between 0.22 (Caucasian) and 0.45 (Asian) in various ethnic populations. Alarge epidemiological investigation of a benzene-exposed population [15] has shown that NQO1*2 homozygotes exhibit as much as a seven-fold greater risk of bone marrow toxicity, leading to diseases such as aplastic anaemia and leukaemia. The NQO1*3 allele (Arg139Try) represents an additional NQO1 polymorphism [16] that has been characterised; allelic frequencies have been reported as high as 0.05 in some ethnic groups [17], and the NQO1*3 allele has been linked to a higher rate of exon 4 deletion -- an alternatively spliced form of NQO1 mRNA [14].
Figure 1.
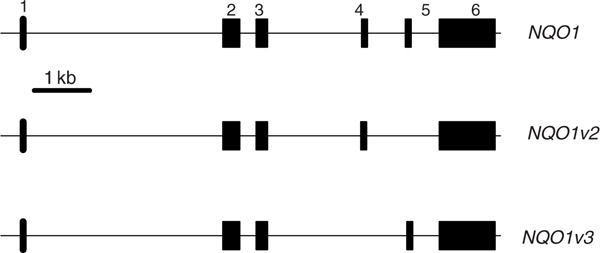
Gene structure of the human NQO1 gene and two splice variants listed in the Entrez Gene database. Variant 2 has exon 5 missing and variant 3 has exon 4 missing. The complete gene and two variants are also included in the phylogenetic analysis of Figure 2.
The other member [18,19] of this mammalian gene family is NQO2, which has recently been demonstrated to be identical to a protein isolated from human kidney in 1962. NQO2 is 43 residues shorter than NQO1 at its C-terminus and shows 49 per cent sequence identity with NQO1. The human NQO2 gene on chromosome 6p25.2 comprises seven exons (first exon non-coding) spanning 19.8 kb, encodes 231 amino acids and contains, to date, 254 SNPs within 10 kb 5'-ward of the first exon and 2 kb 3'-ward of the last exon (http://SNPper.chip.org/). Whereas earlier reports [20,21] had suggested an association between both NQO1 and NQO2 polymorphisms and Parkinson disease, a more recent study [22] showed no association between alleles of either NQO1 or NQO2 and Parkinson's disease.
NQO1 efficiently utilises NAD(P)H as the source of reducing equivalents, but -- because the 43-amino acid deletion in NQO2 contains a portion of the pyridine nucleotide binding site -- NQO2 has been shown to prefer dihydronicotinamide ribosyl, and other similar derivatives, as a cofactor [23]. Similar to NQO1, NQO2 can also bioactivate certain anti-tumour agents, such as the CB1954 nitro compound [13]. A crystal structure of human NQO2 has been published [23], but not many biological functions of this enzyme have so far been well characterised. Curiously, NQO2 contains a melatonin-binding site [24], which might be a reasonable explanation for the anti-oxidant role ascribed to melatonin.
Both NQO1 and NQO2 activities are observed at high levels in many human solid tumours, including lung, colon, liver and breast; this finding has encouraged oncologists to explore the possibility of NQO1 and NQO2 as drug targets for the development of novel anti-tumour agents that can be activated by these enzymes [13].
Definition of 'Pfam' and 'clan'
Pfam is a means of identifying similar domain structures of all proteins that are summarised in UniProt (http://www.ebi.uniprot.org/). Both NQO1 and NQO2 were found to belong to the flavodoxin-2 family (Pfam 02525) because they carry a domain having a flavodoxin-like fold.
The flavodoxin-2 family is a member of the flavoprotein clan. Based on the Pfam database definition:
'A clan contains two or more Pfam families that have arisen from a single evolutionary origin. Evidence of their evolutionary relationship is usually determined by similar tertiary structures or, when structures are not available, by common sequence motifs.'[25]
The category of 'clans' is thus usually introduced in order to encompass two or more gene families that can be globally quite distant evolutionarily but have some segment of the gene products that show a high degree of similarity. Hence in a BLAST search, a sequence may significantly hit more than one member of another family of the clan -- as well as all members within one family.
The flavoprotein clan
The flavoprotein clan includes four Pfam members: flavodoxin-1, flavodoxin-2, flavodoxin NdrI and FMN red (Table 1).
Table 1.
Relationship of the NQO gene family to the flavoprotein clan
| Flavoprotein clan(four Pfam members) |
|---|
| Flavodoxin-1 family |
| Flavodoxins |
| Nitric oxide synthases |
| Flavodoxin-2 family |
| Bacterial NQO3 genes |
| Vertebrate NQO1, NQO2 genes |
| Fungal NQO4 genes |
| Archaebacterial NQO5 genes |
| Bacterial acyl carrier protein phosphodiesterase |
| Flavodoxin Ndrl family |
| Ribonucleotide reductases |
| flavin mononucleotide red family |
| Flavin reductases |
The flavodoxin-1 family includes flavodoxins and nitric oxide synthases. Members of this clan are composed of flavin mononucleotide (FMN)- or flavin adenine dinucleotide-binding redox proteins. Flavoproteins act in various electron transport systems as functional analogues of ferredoxins; they are characterised by an open twisted alpha/beta structure consisting of five parallel beta-sheets connected by alpha-helices which surround the sheet. Flavodoxins are electron-transfer proteins that function in various electron transport systems. They bind one FMN molecule, which serves as a redox-active prosthetic group, and are functionally interchangeable with ferredoxins. Flavodoxin genes are found in prokaryotes, cyanobacteria and some eukaryotic algae. Nitric oxide synthases (EC:1.14.13.39) produce nitric oxide from L-arginine and NADPH. Nitric oxide acts as a second messenger signalling molecule in the body. Five NOS genes have been found in the human genome: NOS1 (neuronal); NOS2A (hepatocyte inducible); NOS2B; NOS2C; and NOS3 (endothelial cell) (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/).
The flavodoxin-2 family includes bacterial and eukaryotic NAD(P)H:quinone reductases (as described above), as well as the bacterial acyl carrier protein (ACP), phosphodiesterase (EC:3.1.4.14). The acyl carrier enzyme converts the holo-ACP into apo-ACP by catalysing the hydrolysis of the phosphodiester linkage between Ser-36 of ACP and the 4'-phosphopantethein prosthetic group. ACP is a small (8.8 kDa) protein that plays a central role in the biosynthesis of fatty acids in bacteria, plant chloroplasts and other organisms. ACP phosphodiesterase is known to be encoded by the acpD gene in Escherichia coli and has been widely distributed in other bacteria, including Azospirillum brasilense, Bacillus halodurans, B. stearothermophilus, B. subtilis, Haemophilus influenzae, Lactococcus lactis, Mesorhizobium loti, Mycoplasma pneumoniae, Pseudomonas aeruginosa and Streptomyces coelicolor.
The flavodoxin_NdrI family contains ribonucleotide reductases, which are a family of complex enzymes that play an essential role in all organisms. These enzymes catalyse de novo synthesis of deoxyribonucleotides required for DNA replication and repair [26].
Finally, members of the FMN red family (flavin reductases) use flavins as substrates and are distinct from flavoenzymes, which have tightly bound flavins. The reduced flavin can serve to provide electrons for ferric ion complexes and iron proteins. In E. coli, reactivation of ribonucleotide reductase is achieved by the formation of reduced flavins by flavin reductase.
Previous confusion about NQO genes and diaphorases
Much of the confusion that has arisen with the nomenclature of NQO1, NQO2 and related proteins can be traced back to the widespread use of the term 'diaphorase' in enzymatic studies more than 50 years ago. In fact, this terminology is still being carried forward [27]. The term 'diaphorase' has generally been used to describe a 'coenzyme' (often a flavoprotein) that can transfer electrons from reduced pyridine nucleotides to electron acceptors [9,28]. In any biological system, therefore, there are many enzymes that can be characterised as 'diaphorases'.
The term 'DT'-diaphorase was originally used by Ernster, to describe what is now known as NQO1, because the enzyme worked with equal facility, employing either DPNH (NADH) or TPNH (NADPH). Edwards et al. [29] discuss a series of 'diaphorases' defined in human tissues. Diaphorase-1 was NADH cytochrome b5 reductase-3 (Table 2) [30,31]. Diaphorase-2 was the major enzyme in erythrocytes catalysing the oxidation of NADPH. Diaphorase-3 was the principal source of oxidised coenzyme in sperm. Diaphorase-4 was characterised by Edwards et al. [29], and this enzyme was found to be identical to the previously characterized DT-diaphorase [32]. Interestingly, Edwards and colleagues also showed that diaphorase-4 activity was absent in approximately 4 per cent of the UK population, which is in agreement with more recent genetic studies of the prevalence of the null NQO1*2 allele [14].
Table 2.
Human CYB5R genes listed in the HUGO gene nomenclature committee database
| Approved gene symbol | Approved gene name | Location | Sequence accession IDs | Previous symbols | Aliases |
|---|---|---|---|---|---|
| CYB5R1 | Cytochrome b5 reductase-1 |
1q32.1 |
AF169481 NM_016243 |
NQO3A2 | humb5R2, 1500005G05Rik |
| CYB5R2 | Cytochrome b5 reductase-2 |
11p15.4 |
AF169802 NM_016229 |
||
| CYB5R3 | Cytochrome b5 reductase-3 |
22q13.31- qter |
M16461 | DIA1 | |
| CYB5R4 | Cytochrome b5 reductase-4 |
6pter- q22.33 |
AF169803 NM_016230 |
NCB5OR | b5+b5R, dJ676J13.1 |
All diaphorases discussed by Edwards [29] are distinct gene products but, because of their similarity based on enzymic function rather than evolutionary divergence, searches for 'NQO1' in some databases will still return descriptions of 'cytochrome b5 reductase'. These cytochrome b5 reductase 'diaphorases' are not evolutionarily related, are very different functionally and are encoded by separate gene loci (Table 2). A second source of confusion involves the use of the generic term 'quinone reductase', because a wide variety of enzymes in prokaryotes, plants and animals can reduce quinones via both one- and two-electron steps.
A search of the HUGO Gene Nomenclature Committee database using 'NQO' provided three hits--two NQO genes plus 'NQO3A2' (which actually encodes cytochrome b5 reductase-1)--the latter appears in the database because of its previous name as a diaphorase (see above). In fact, one of its aliases is 'NQO3A2', which has been listed in the http://SNPper.chip.org/ database; the authors have pointed out this error to this organisation. A search of the SwissProt and TrEMBL databases using 'NQO' provided 168 hits (39 in SwissProt and 129 in TrEMBL), but not all of them belong to the flavodoxin-2 family. The term 'NQO' has therefore been used for other enzymes and care should be taken to avoid such confusion.
Evolutionary divergence of the NQO genes
Figure 2 shows the evolutionary relatedness of a number of selected NQO gene products from mammals, birds, amphibians, fish, fungi and bacteria. Vertebrates carry the NQO1 and NQO2 genes while eubacteria carry the most ancient NQO3 genes. The authors suggest naming the fungal subfamily as NQO4 and the archaebacterial subfamily as NQO5. Four plants (bottom of Figure 2) are complete outliers, being members of the flavodoxin-1 quinone reductase family. Curiously, it was found that frog, zebrafish and puffer fish have NQO1 but not NQO2; conversely, chicken and ray-finned fish have NQO2 but not NQO1. Whether the other homologous gene will be found in any of these genomes in the future remains to be determined. Undoubtedly, the NQO1 and NQO2 genes in vertebrates arose from the prokaryotic NQO3 ancestor. Fungal NQO4 genes also arose from bacterial NQO3, however it is impossible to say whether the eubacterial NQO3 genes or the archaebacterial NQO5 genes represent the original ancestor. One prokaryotic taxon might have captured the gene from the other prokaryotic taxon by horizontal gene transfer.
Figure 2.
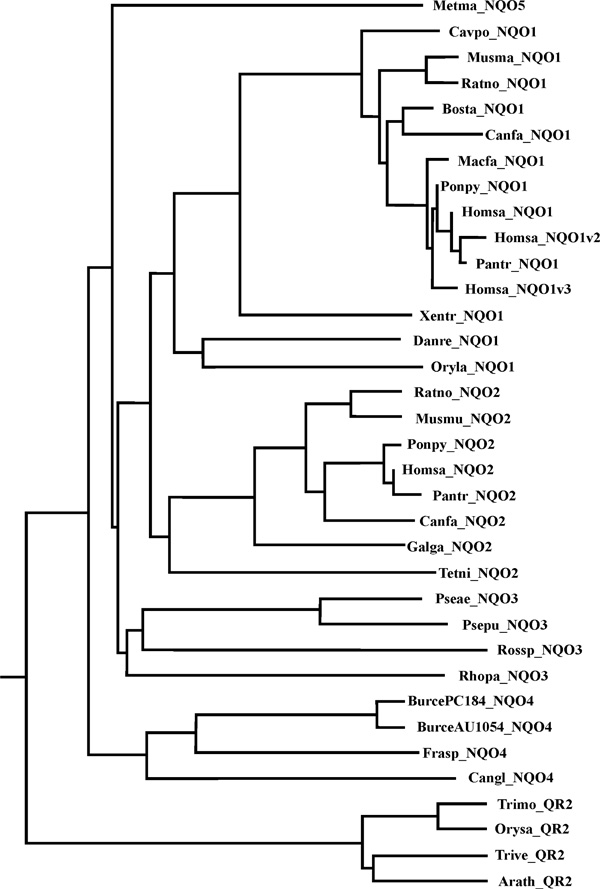
Phylogenetic organisation of selected NQO genes in bacteria and eukaryotes. Metma, Methanosarcina mazei (an archaebacterium); Cavpo, Cavia porcellus (guinea pig); Musmu, Mus musculus; Ratno, Rattus norvegicus; Bosta, Bos taurus; Canfa, Canis familiaris; Macfa, Macaca fascicularis (macaque); Ponpy, Pongo pygmaeus (orangutan); Homsa, Homo sapiens; Pantr, Pan troglodytes (chimpanzee); Xentr, Xenopus tropicalis; Danre, Danio rerio (zebrafish); Oryla, Oryzias latipes (ray-finned fish); Galga, Gallus gallus (chicken); Tetni, Tetraodon nigroviridis (puffer fish); Pseae, Pseudomonas aeruginosa; Psepu, Pseudomonas putida; Rossp, Roseovarius_sp_217; Rhopa, Rhodopseudomonas palustri; BurcePC184, Burkholderia cenocepacia PC184; BurceAU1054, Burkholderia Burkholderia cenocepacia AU 1054; Frasp, Frankia sp; Cangl, Candida glabrata; Trimo, Triticum monococcum; Orysa, Oryza sativa (rice); Trive, Triphysaria versicolor; Arath, Arabidopsis thaliana; QR2, quinone reductase-2, which belongs to the flavodoxin-1 fold, and not to the flavodoxin-2 (NQO) fold. No NQO genes were found in Caenorhabditis elegans, Drosophila melanogaster or Ciona savignyi (sea squirt), nor in several plants whose genomic databases were searched.
The authors performed a series of BLAST searches -- using NQO1, NQO2 or NQO3 sequences -- and no homologous protein of significance in Ciona intestinalis, Caenorhabditis elegans, Drosophila melanogaster, Anopheles gambiae, Oryza sativa (rice) or Arabidopsis thaliana was detected. Wheat plants have been reported to show quinone reductase activity (both the one- and two-electron reductase forms), however, induced in response to powdery mildew infection;[33] infection and inflammation are known to be processes that induce oxidative stress [11]. Plants appear to utilise quinone reductase by way of evolutionarily unrelated classes of quinone oxidoreductases, carrying out both one- and two-electron reduction reactions [34], and also alternative NAD(P)H dehydrogenases [35]. This finding might therefore be an example of convergent evolution: Mother Nature needed a particular enzyme in a particular location within the plant cell, did not have NQO enzymes in this phylum and therefore selected another type of quinone oxidoreductase or an alternative NAD(P)H dehydrogenase to carry out the enzymic function. Alternatively, this finding might be an example of redundancy, in which multiple systems have evolved independently to face a particular selective pressure.
Do plants suffer from oxidative stress? The absence of NQO genes in plants might reflect the fact that plants use carbon dioxide rather than molecular oxygen; therefore, plants may not experience oxidative stress in the way animals do. This is a fascinating bit of speculation that may be worthy of further study. The putative plant NQO equivalents (QR1, QR2) contain the flavodoxin-1 domain and belong to the flavodoxin-1 family. This domain is found in a number of proteins, including flavodoxin and nitric oxide synthase (as described above).
Putative role of NQO in oxidative stress
BCL2 is well known to be anti-apoptotic and therefore prevents apoptosis from proceeding [36]. Proposed functions of BCL2 include mediation of ROS levels and the redox status of the cell, prevention of nucleocytoplasmic trafficking of TRP53 and other cell cycle regulatory factors, neutralisation of BAX, BAD and other proteins that heterodimerise and promote apoptosis and deterrence of mitochondrial cytochrome c release [37-39]. The C. elegans cell death-9 (ced-9) gene is the homologue of the mammalian BCL2 gene, ced-9 is part of a polycistronic locus in C. elegans that includes the cyt-1 gene encoding a protein similar to cytochrome b560 [38]. Cytochrome b560 is an inner mitochondrial membrane protein involved in system II (succinate dehydrogenase), which transfers an electron from succinate to ubiquinone. Interestingly, this dehydrogenase has been described as a member of the NQO gene superfamily;[40] however, the authors found that cytochrome b560 is instead a member of the more expansive flavodoxin-1 family (described above). They suggest that various members of the flavoprotein clan, including mammalian NQO1, probably participate in aiding the cell in preventing unwanted apoptosis [40,41].
Concluding remarks
Isolation and characterisation of a cytosolic enzyme in several rat tissues in the 1950s led to the description of a quinone reductase in at least two independent laboratories. A substantial number of publications, calling this enzyme 'DT-diaphorase', has led to confusion about the evolutionary relatedness of NAD(P)H:quinone oxidoreductases to one another. There are two NQO genes in vertebrates, a homologous NQO3 subfamily of genes in eubacteria, a NQO4 subfamily in fungi and a NQO5 gene subfamily in archaebacteria. The fascinating functions of the human NQO1 enzyme include protection against myeloid leukaemia and other types of leukaemia in workers exposed to benzene and a possible anti-apoptotic role fundamental to the cell cycle. NQO2 appears to participate in the anti-oxidant properties of melatonin. Although many SNPs exist in the human NQO2 gene, little is known about any association with a physiological disorder or protection against environmental toxicants.
Acknowledgements
The authors thank their colleagues and many e-mails for valuable discussions and a critical reading of this manuscript. They appreciate the help of Jadwiga K. Kepa and Marian Miller with the graphics. The bioinformatics searches and writing of this paper were funded, in part, by NIH grants R01 EY11490 (V.V.), R01 ES09554 (D.R.) and P30 ES06096 (D.W.N.).
References
- Ernster L. DT-diaphorase. Methods Enzymol. 1967;10:309–317. [Google Scholar]
- Lind C, Cadenas E, Hochstein P. et al. DT-diaphorase. Purification, properties and function. Methods Enzymol. 1990;186:287–301. doi: 10.1016/0076-6879(90)86122-c. [DOI] [PubMed] [Google Scholar]
- Siegel D, Gibson NW, Preusch PC. et al. Metabolism of mitomycin C by DT-diaphorase: Role in mitomycin C-induced DNA damage and cytotoxicity in human colon carcinoma cells. Cancer Res. 1990;50:7483–7489. [PubMed] [Google Scholar]
- Gibson NW, Hartley JA, Butler J. et al. Relationship between DT-diaphorase-mediated metabolism of a series of aziridinylbenzoquinones and DNA damage and cytotoxicity. Mol Pharmacol. 1992;42:531–536. [PubMed] [Google Scholar]
- Ross D, Siegel D, Beall H. et al. DT-diaphorase in activation and detoxification of quinones. Bioreductive activation of mitomycin C. Cancer Metastasis Rev. 1993;12:83–101. doi: 10.1007/BF00689803. [DOI] [PubMed] [Google Scholar]
- Ross D, Beall H, Traver RD. et al. Bioactivation of quinones by DT-diaphorase. Molecular, biochemical, and chemical studies. Oncol Res. 1994;6:493–500. [PubMed] [Google Scholar]
- Ernster L, Navazio F. Soluble diaphorase in animal tissues. Acta Chem Scand. 1958;12:595–602. [Google Scholar]
- Ernster L. Diaphorase activities in liver cytoplasmic fractions. Fed Proc. 1958;17:216. [Google Scholar]
- Ernster L. DT-diaphorase: A historical review. Chem Scripta. 1987;27A:1–13. [Google Scholar]
- Martius C. Die Stellung des Phyllochinons (Vitamin K1) in der Atmungskette. Biochem Z. 1954;326:26–27. [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ. et al. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/S0006-2952(99)00310-X. [DOI] [PubMed] [Google Scholar]
- Li R, Bianchet MA, Talalay P. et al. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy. Mechanism of the two-electron reduction. Proc Natl Acad Sci USA. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. Quinone reductases. Multitasking in the metabolic world. Drug Metab Rev. 2004;36:639–654. doi: 10.1081/DMR-200033465. [DOI] [PubMed] [Google Scholar]
- Ross D, Siegel D. NAD(P)H:quinone oxidoreductase-1 (NQO1, DT-diaphorase), functions, and pharmacogenetics. Methods Enzymol. 2004;382:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- Rothman N, Smith MT, Hayes RB. et al. An epidemiologic study of early biologic effects of benzene in Chinese workers. Environ Health Perspect. 1996;104(Suppl 6):1365–1370. doi: 10.1289/ehp.961041365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SS, Forrest GL, Akman SA. et al. NAD(P)H:quinone oxidoreductase expression and mitomycin C resistance developed by human colon cancer HCT 116 cells. Cancer Res. 1995;55:330–335. [PubMed] [Google Scholar]
- Gaedigk A, Tyndale RF, Jurima-Romet M. et al. NAD(P)H:quinone oxidoreductase: Polymorphisms and allelic frequencies in caucasian. Chinese and Canadian native Indian and inuit populations. Pharmacogenetics. 1998;8:305–313. doi: 10.1097/00008571-199808000-00004. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK, Burnett P, Adesnik M. et al. Nucleotide and deduced amino acid sequence of a human cDNA (NQO2) corresponding to a second member of the NAD(P)H:quinone oxidoreductase gene family. Extensive polymorphism at the NQO2 gene locus on chromosome 6. Biochemistry. 1990;29:1899–1906. doi: 10.1021/bi00459a034. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Yang XL, Holtzclaw WD. et al. Unexpected genetic and structural relationships of a long-forgotten flavoenzyme to NAD(P)H:quinone reductase (DT-diaphorase) Proc Natl Acad Sci USA. 1997;94:1669–1674. doi: 10.1073/pnas.94.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Fujii C, Hayashi A. et al. An association between idiopathic Parkinson disease and polymorphisms of phase II detoxification enzymes: Glutathione S-transferase M1 and quinone oxidoreductase 1 and 2. Biochem Biophys Res Commun. 2001;288:887–892. doi: 10.1006/bbrc.2001.5868. [DOI] [PubMed] [Google Scholar]
- Shao M, Liu Z, Tao E. et al. Polymorphism of MAOB gene and NAD(P)H:quinone oxidoreductase gene in Parkinson disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2001;18:122–124. [PubMed] [Google Scholar]
- Okada S, Farin FM, Stapleton P. et al. No associations between Parkinson's disease and polymorphisms of the quinone oxidoreductase (NQO1, NQO2) genes. Neurosci Lett. 2005;375:178–180. doi: 10.1016/j.neulet.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Foster CE, Bianchet MA, Talalay P. et al. Crystal structure of human quinone reductase type-2, a metalloflavoprotein. Biochemistry. 1999;38:9881–9886. doi: 10.1021/bi990799v. [DOI] [PubMed] [Google Scholar]
- Vella F, Ferry G, Delagrange P. et al. NRH:quinone reductase-2: An enzyme of surprises and mysteries. Biochem Pharmacol. 2005;71:1–12. doi: 10.1016/j.bcp.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK. et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents E, Roca I, Gibert I. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: Transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology. 2003;149:1011–1020. doi: 10.1099/mic.0.26133-0. [DOI] [PubMed] [Google Scholar]
- Danson S, Ward TH, Butler J. et al. DT-diaphorase: A target for new anticancer drugs. Cancer Treat Rev. 2004;30:437–449. doi: 10.1016/j.ctrv.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Corran HS, Green DE, Straub FB. On the catalytic function of heart flavoprotein. Biochem J. 1939;33:793–801. doi: 10.1042/bj0330793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YH, Potter J, Hopkinson DA. Human FAD-dependent NAD(P)H diaphorase. Biochem J. 1980;187:429–436. doi: 10.1042/bj1870429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JC, Leroux A. Les systemes NADH et NADPH diaphorasiques des cellules sanguines humaines. Bull Soc Chim Biol. 1970;52:1243–1249. [PubMed] [Google Scholar]
- Leroux A, Junien C, Kaplan JC. Generalised deficiency of cytochrome b5 reductase in congenital methaemoglobinaemia with mental retardation. Nature. 1975;258:619–620. doi: 10.1038/258619a0. [DOI] [PubMed] [Google Scholar]
- Shaw PM, Reiss A, Adesnik M. et al. The human dioxin-inducible NAD(P)H:quinone oxidoreductase cDNA encoded protein expressed in COS-1 cells is identical to diaphorase-4. Eur J Biochem. 1991;195:171–176. doi: 10.1111/j.1432-1033.1991.tb15691.x. [DOI] [PubMed] [Google Scholar]
- Greenshields D, Liu G, Selvaraj G. et al. Differential regulation of wheat quinone reductases in response to powdery mildew infection. Planta. 2005;222:867–875. doi: 10.1007/s00425-005-0029-7. [DOI] [PubMed] [Google Scholar]
- Matvienko M, Wojtowicz A, Wrobel R. et al. Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 2001;25:375–387. doi: 10.1046/j.1365-313x.2001.00971.x. [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE. Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol. 2004;55:23–39. doi: 10.1146/annurev.arplant.55.031903.141720. [DOI] [PubMed] [Google Scholar]
- Brustugun OT, Fladmark KE, Doskeland SO. et al. Apoptosis induced by microinjection of cytochrome c is caspase-dependent and is inhibited by BCL2. Cell Death Differ. 1998;5:660–668. doi: 10.1038/sj.cdd.4400399. [DOI] [PubMed] [Google Scholar]
- Kirsch DG, Doseff A, Chau BN. et al. Caspase-3-dependent cleavage of BCL2 promotes release of cytochrome c. J Biol Chem. 1999;274:21155–21161. doi: 10.1074/jbc.274.30.21155. [DOI] [PubMed] [Google Scholar]
- Martinou JC. Apoptosis. Key to the mitochondrial gate. Nature. 1999;399:411–412. doi: 10.1038/20804. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Narita M, Tsujimoto Y. BCL2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunol Today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]


