Abstract
miRNAs are short noncoding RNA molecules that are important in regulating gene expression. Due to the correlation of their expression levels and various diseases, miRNAs are being investigated as potential biomarkers for molecular diagnostics. The fast-growing miRNA exploration demands rapid, accurate, low-cost miRNA detection technologies. This article will focus on two platforms of nanopore single-molecule approach that can quantitatively measure miRNA levels in samples from tissue and cancer patient plasma. Both nanopore methods are sensitive and specific, and do not need labeling, enzymatic reaction or amplification. In the next 5 years, the nanopore-based miRNA techniques will be improved and validated for noninvasive and early diagnosis of diseases.
Keywords: biomarker, biosensor, diagnostics, lung cancer, miRNA, nanopore, noninvasive detection, PCR
Every aspect of cellular activities, including cell proliferation, differentiation, metabolism and apoptosis, can be regulated by a class of tiny but very important nucleic acid fragments termed miRNAs. The binding of miRNAs to specific mRNAs causes mRNA degradation or inhibits its translation, thereby regulating gene expression. While understanding miRNA regulatory mechanisms remains the main focus of miRNA research, rapidly increasing efforts are being made in exploring the correlation of miRNAs with disease development. The downstream aim of disease correlation is to discover miRNAs that can be used as biomarkers for disease diagnostics and prognostics. For example, in cancer cells, dysregulated miRNAs disrupt the homeostasis of the normal biological processes. Aberrant expression of miRNAs has been found in all types of tumors, including lung cancer, and different cancer types have distinct miRNA expression patterns and profiles. Therefore, miRNAs (either the whole spectrum or a specific set of miRNAs) in tissues or biofluids such as blood can be utilized as a diagnostic biomarker for cancer early detection, diagnosis, staging and monitoring.
The fast-growing area of miRNA research demands rapid, accurate and low-cost miRNA detection technologies. The current main technologies for miRNA measurements include quantitative reverse-transcription PCR (qRT-PCR; gold standard), microarrays and next-generation sequencing. In this review, we start with a brief discussion of the biogenesis of miRNAs in humans, and then summarize two nanoporebased single-molecule techniques for quantitative measurement of miRNAs. The nanopore method is highly sensitive and specific; the measurement procedure does not require labeling, an enzymatic reaction or amplification. The nanopore sensors can not only detect miRNAs, but also detect various nucleic acid fragments, genetic alterations and pathogenic DNAs/RNAs. They could prove a useful tool for the discovery of disease biomarkers, which are important for noninvasive screening and early diagnosis of diseases such as cancer. In the section titled ‘Expert commentary’, we will discuss the mechanism for the nanopore sensor’s sensitivity and selectivity, and in the section titled ‘Five-year view’, highlight the pathway to the translation of nanopores into a robust, clinically usable miRNA detector.
miRNAs, gene regulation & diseases
Small RNAs are important regulators of gene expression in eukaryotic cells. miRNAs are the best-studied group of endogenous, small noncoding RNA. They are approximately 18–24 nucleotides (nts) in length, acting as post-transcriptional regulators of mRNA stability and protein synthesis in many eukaryotic organisms [1,2]. The altered levels of these small RNAs can be important for both the diagnosis and prognosis of human disease. Since its initial discovery in Caenorhabditis elegans, a nematode, in 1993 [3,4], over 17,000 mature miRNAs have been identified across different species [5]. At present, the human miRNA database contains 1424 miRNAs that represent approximately 2–3% of the total number of genes in the human genome (see miRBase [101]). Approximately half of miRNA genes are located in the introns of protein-coding genes or long noncoding RNA transcripts, whereas the remainder are independent transcription units [2].
Biogenesis of miRNAs in eukaryotic cells is a multistep process involving several distinct enzymes in protein complexes [1,2,6]. Like mRNA, miRNA is transcribed mainly by RNA polymerase II as long primary transcripts of variable size characterized by hairpin structures and capped and polyadenylated at the ends (pri-miRNA) [7]. The pri-miRNAs are recognized by the RNAse III enzyme Drosha and a protein DGCR8 (microprocessor) in the nucleus, and are cleaved into 70–100-nt-long precursor miRNAs (pre-miRNAs) [8]. The precursor miRNA molecule has a short stem with a 2–3-nt overhang that is recognized and exported by the exportin 5 nuclear export factor to the cytoplasm [9], where another RNAse III – Dicer with its partner TRBP and Argonaut proteins 1–4 – excises the terminal loop and generates a 22–24-nt duplex, named miR/miR* [10]. Only one strand of miRNA duplex (guide strand or mature miRNA) is incorporated in the complex known as the RNA-induced silencing complex, whereas the other strand (passenger strand or miR*) is subjected to degradation. Nucleotides 2–7 of the mature miRNA sequence create the ‘seed region’ that determines the specificity of binding to its target mRNA. As part of this complex, the mature miRNA is able to regulate gene expression by binding to the 3′ untranslated region of target mRNAs by Watson–Crick base pairing, leading to mRNA degradation in the case of perfect matching or translation inhibition if it is not a perfect match [11,12].
Since one miRNA can target multiple mRNAs and there are hundreds of miRNA species in a given cell, it is not surprising that miRNAs regulate many genes involved in almost all aspects of cell biological functions, including normal development, cell proliferation and differentiation, cell cycle, apoptosis and signaling pathways [13,14]. Accordingly, aberrant expression of miRNA by either downregulation or upregulation has been demonstrated in many disease conditions including cancer [15,16]. Genome-wide expression profiling of miRNAs in different types of cancer revealed specific signatures of abnormal miRNAs [17]. miRNAs can function as potential oncogenes or tumor suppressor genes (TSGs), depending on the property of individual miRNAs, the cellular context and the target genes they regulate in many types of cancer [18]. The first evidence of alterations of miRNA genes in human cancer came from studies of chronic lymphocytic leukemia (CLL) [19]. CLL is the most common leukemia of adults in Western countries and is a chronic lymphoproliferative disorder of mature B cells that is characterized by the progressive accumulation of functionally incompetent malignant B cells in the bone marrow and blood. Although the leukemic cells are homogenous in morphology and immunophenotype, CLL is a remarkably diverse disorder in its genetics and epigenetics, with different clinical courses and prognoses. Deletions of chromosome 13 at band q14 are detected by cytogenetic studies in half of CLL patients. Calin and colleagues aimed to identify a possible TSG in this critical region of 13q14. However, they did not find any TSG coding sequences, but rather two miRNA genes, miR-15a and miR-16-1, the repressors of the BCL-2 oncogene [19]. A large study conducted by the same group indicated that a signature of 13 miRNAs is capable of distinguishing between indolent and aggressive CLL forms [20]. This difference contributes to the clinical decision-making in managing CLL patients. Notably, it was found that miR-155 was overexpressed in aggressive CLLs [20]. miR-155 is an oncogenic miRNA that is overexpressed in different lymphomas such as Burkitt’s lymphoma, an aggressive form of diffuse large B-cell lymphoma and Hodgkin’s lymphoma, as well as in solid tumors such as lung cancer [18]. Many other miRNAs have been identified to be dysregulated in virtually all types of cancer and other diseases [15,16,18].
Interestingly, miRNAs are constantly released from primary tumors into body fluids and blood [17]. Circulating miRNAs are present as an incredibly stable form and are detectible with various molecular methods [21]. These features render miRNAs suitable noninvasive biomarkers for cancer detection [22]. For example, Zheng et al. has utilized circulating miRNAs as biomarkers for early detection of lung cancer and molecular monitoring [23]. By screening plasma samples of lung cancer patients for 15 miRNAs that are frequently overexpressed in primary lung cancer tissues, three miRNAs, including miR-155, miR-197 and miR-182, have been identified that are significantly elevated in lung cancer patients compared with cancer-free controls. The combination of the three miRNAs yielded 81% sensitivity and 87% specificity in discriminating lung cancer from controls. The levels of the triplet markers showed no significant difference in tissue types of lung cancer and between clinical stages, but it clearly differentiated early-stage lung cancer (stage 1) from normal controls. Furthermore, the levels of miR-155 and miR-197 were significantly higher from lung cancer patients with metastasis than in those without metastasis. These results indicated that miR-155, miR-197 and miR-182 may become potential noninvasive molecular biomarkers for lung cancer screening, early diagnosis and clinical follow-up. miRNAs as biomarkers for cancer detection have been extensively studied in all types of cancer and are becoming a rapidly evolving field [22,24].
Current miRNA detection methods
To explore miRNA functions, it is important to have sensitive technologies that can accurately determine their levels in tissues and biofluids. Currently, main technologies for miRNA measurement are qRT-PCR, microarrays and next-generation sequencing. The detection of miRNAs is complicated by their small size (18–24 nts). When a longer RNA such as mRNA is detected by reverse transcription PCR (RT-PCR), it requires two opposing DNA primers. The first primer is used to synthesize cDNA with reverse transcriptase, and the second primer to copy cDNA to dsDNA. However, the short length of miRNA precludes the two-primer approach. This is because the shortness of the miRNA sequences makes it difficult to design two opposing primers. Detection by PCR amplification needs to be controlled for bias in annealing of primers and for differential amplification of miRNAs. To obtain quantitative data, the internal miRNA controls are required [25–27]. One approach for PCR amplification uses unligated hairpin primers that have a short 3′ extension [28]. Alternative methods involve ligation of a DNA linker to the miRNA [29] or tailing of miRNA with poly A polymerase to allow cDNA synthesis [30]. The sequencing power of the next-generation sequencing platforms has greatly expanded the discovery of novel miRNAs. The sequencing approaches usually require ligation of a DNA linker to the 3′ end of the RNA, followed by PCR amplification. Ligation methods often exclude some sequences because of the enzyme bias against certain structures [31].
Methods for miRNA detection that do not involve amplification include labeling of endogenous miRNAs with a dye or radioactivity followed by hybridization to an array with miRNA-specific capture probes [32], or capture of the dye-labeled RNA using a bead (Luminex Corporation) that is barcoded for laser detection [17]. Another approach, developed by NanoString, uses a linear string of dyes attached to a reporter probe for detection of the miRNA. Laser detection of the dyes gives quantitative and qualitative information on the number of miRNAs present [33]. This approach requires very-high-resolution microscopy. A novel detection method uses the dsRNA binding protein p19, from a plant virus, to selectively bind a miRNA:RNA probe duplex. The p19 protein does not bind ssRNA probes or long RNA, but has a preference for 19–21-bp dsRNA. The p19 protein is attached to chitin magnetic beads for easy usage. Labeled RNA probes can be detected using radioactivity [34], and unlabeled probes can be detected using a nanopore [35] (see following section). Using p19, the dsRNA of a miRNA:probe duplex can be enriched over 100,000-fold. This approach can be used both for detection of known small RNAs and for the discovery of novel dsRNAs. Other techniques based on colorimetry, bioluminescence, enzyme turnover and electrochemistry have been proposed; nanoparticles, molecular beacons, deep sequencing [25,27] and single-molecule fluorescence [36] have also been applied to miRNA detection. For a detailed review of these technologies, please see [25,27]. A classical technique, in situ hybridization, has been quickly adapted to detect miRNA in a histologic section [37,38]. Although it is not a quantitative assay, it can localize miRNA in the context of tissue or cells. In combination with other methods such as in situ PCR, different chromogenic labeling or with immunohistochemistry, in situ hybridization can detect both precursor and mature miRNA and its target protein at the same tissue section [39,40]. A locked nucleic acid probe is often used to increase the efficiency of hybridization [41,42]. Since the method can detect miRNA in the widely used formalin-fixed paraffin-embedded tissues, it should significantly expand miRNA studies, especially in cancer research.
Recently Wanunu et al. [35] and Wang et al. [43] have independently reported on a nanopore single-molecule method to electrically detect miRNA in tissue and biofluids. Both technologies are label-free and do not require enzyme reactions and amplification. Wanunu et al. has utilized a 3-nm synthetic nanopore to detect the complex of liver miR-122a hybridized with the probe RNA after enrichment using the p19 protein, providing an accurate miRNA quantification method for quantifying miRNA in tissue [35]. It is assumed that the same approach could be used for miRNA detection using serum to make dsRNA. Wang et al. used oncogenic miR-155 and TSG LET-7 miRNAs as testing cases to verify the sensitivity and specificity of a protein–nanopore-based sensor in plasma samples of lung cancer patients [43]. Both methods will be reviewed in the following sections.
Nanopore single-molecule detector
The nanopore is a molecular-scale pore structure that is able to detect with high sensitivity the position and conformation of a single molecule that is present within the pore lumen [44]. From the characteristic change in the nanopore conductance, one can electrically elucidate single-molecule kinetic pathways and quantify the target. Nanopores can be constructed in a variety of ways, including biological pores formed from transmembrane protein channels [45], and synthetic pores in a variety of materials [46]. Various nanopore sensors are being developed with broad biotechnological applications [44,46–53], including the next generation of DNA sequencing technology [54,55]. The development of nanopore-based miRNA detectors is a novel effort in this rapidly evolving field.
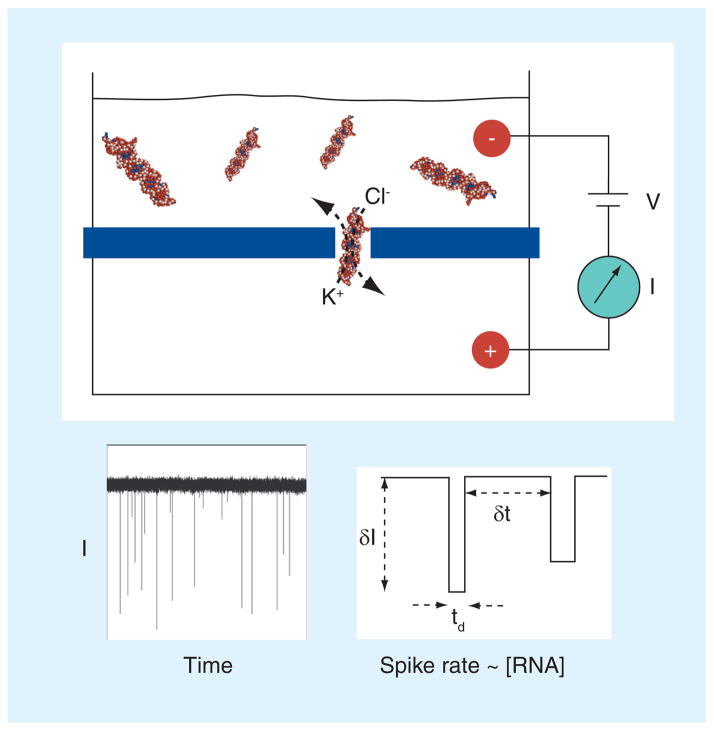
The principle of the nanopore sensor is illustrated in Figure 1. The sensor detects changes in ion current across a nanopore as biomolecules such as RNA occlude the pore’s entrance. A nanopore sensor comprises an insulating membrane that partitions the recording chamber. A single nanometer-scale aperture is formed in the center of the membrane. When both sides of the membrane are exposed to electrolyte solutions, the nanopore serves as the only ion-conducting pathway across the membrane. Two electrodes are placed in solutions on both sides of the membrane, to supply a voltage across the nanopore and to record the picoampere level ionic flux through the pore. Passage of individual analyte molecules causes transient pulses that correspond to a reduction in the ionic conductance of the pore, observable by high-bandwidth measurements of current versus time. The statistical properties of a large set of these pulses are useful for analyzing a biomolecular species in solution. Of specific importance to the miRNA application, the rate of the current pulses corresponds linearly to the concentration of the molecules in the sample. Therefore, one only needs to count the occurrence frequency of the current pulse to calculate the concentration of the target miRNA.
Figure 1. Scheme of nanopore-based single-molecule detection.
Voltage is applied using a pair of electrodes that provide an electrochemical circuit with the solution. The voltage (V) drives ions through the nanopore, which results in a measurable steady-state ‘open pore’ current (I). When a biomolecule in the top chamber diffuses into the pore, it blocks the ion pathway of the pore and the flux of ions is reduced, resulting in a negative spike that signals each molecule. Under the same applied voltage conditions, the rate of spike events has a linear correspondence with the macromolecular concentration. Therefore, miRNA duplexes can be counted and quantified by measuring their rate of passage through the pore. A real current trace is shown for illustration, as well as three parameters that are typically quantified in nanopore experiments. δI and td are used to identify the target blocks, while δt is used to calculate the rate of block occurrence, which is the inverted value of δt.
δI: Current block amplitude; δt: Interval between adjacent blocks; I: Current; td: Block duration; V: Voltage.
miRNA detection using a synthetic nanopore
Recently, sequence-specific detection of mature miRNA from total mammalian RNA was demonstrated using ultrathin solid-state nanopores as single-molecule counters [35]. Success in this work was enabled by overcoming two critical barriers: probe-specific isolation of miRNA from total cellular RNA; and quantification using a stochastic, nanopore-based molecular sensor.
Sequence-specific miRNA isolation
To isolate a specific miRNA sequence, we utilized the p19 protein from the carnation Italian ringspot virus. p19 has been found to bind dsRNAs that are 19–21 nts in length. However, while this protein is nucleic acid length-selective, it exhibits no sequence selectivity, which makes it ideal for isolating probe-specific miRNAs [56]. Furthermore, p19 exclusively binds short dsRNAs, and does not bind ssRNAs or other structured RNAs such as transfer-RNAs or ribosomal RNAs [34]. Therefore, to simplify the isolation process, the C-terminal of p19 was fused with a chitin-binding domain so that the protein can bind to chitin-coated magnetic beads. This approach has already been demonstrated for probe-hybridized miRNA enrichment by five orders of magnitude [34]. The structure of the p19:dsRNA complex is shown in Figure 2A. The use of p19 binding in the duplex has significant advantages over traditional methods to detect nucleic acid hybrids that involve digestion with a nuclease to remove single-stranded probes. The digestion methods result in significant amounts of natural RNA hairpins, mainly from ribosomal RNA. The use of p19 binding is much more selective, as it only binds dsRNA in a narrow size range (19–21 nts). This selectivity evolved so that the plant virus could block RNA interference by sequestering the short siRNAs generated by the plant’s Dicer. This property has been utilized to make a selective binding reagent that can isolate for detection an unlabeled miRNA:RNA probe duplex free of other contaminating RNAs.
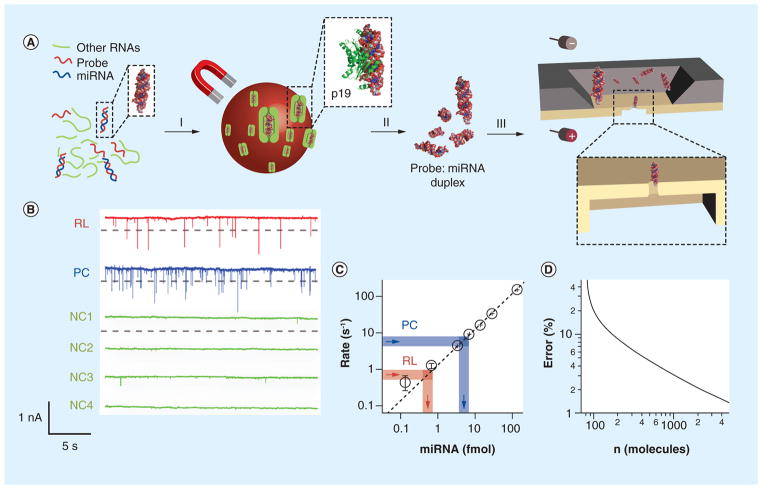
Figure 2. miRNA detection using solid-state molecular counters.
(A) Scheme of the miRNA-specific detection method. First, RNA is extracted from tissue (not shown), and the extract is hybridized to a miRNA-specific oligonucleotide probe (red). In step (I), the probe:miRNA duplex is enriched by binding to p19-functionalized magnetic beads, followed by thorough washing in order to remove other RNAs from the mixture. In step (II), the hybridized probe:miRNA duplex is eluted from the magnetic beads. In step (III), the eluted probe:miRNA duplex is electronically detected using a nanopore. (B) Detection of miR-122a from RL RNA using a 3-nm diameter nanopore in a 7-nm thick membrane. Method shown in panel (A) was applied to the detection of miR-122a from 1 μg of RL total RNA. Representative 30-s current versus time traces are shown for a pore after the addition of the enriched miR-122a (RL), a PC containing a synthetic miR-122a RNA duplex bound to magnetic beads, followed by washing, elution and detection (PC), and four different NCs (NC1–NC4). The NCs did not produce any signal below the threshold, which was set to I0–0.4 nA (see dashed gray lines), where I0 was the baseline current. (C) Quantification of miR-122a from the mean capture rates. A calibration curve of capture rate versus concentration was constructed (dashed black line) using different concentrations of synthetic 22-bp RNA duplex, showing that capture rate scales linearly with concentration over three orders of magnitude. Determination of miR-122a amounts (per μl of solution) is based on the spike rate for sample RL (thick red lines) and the positive control PC (thick blue lines). (D) Relative error in the determined RNA concentration as a function of the number of molecules counted by the nanopore (see text). To achieve 95% accuracy under our conditions, the time required for determination of a 1-fmol RNA sample is 4 min, corresponding to approximately 250 translocation events.
NC: Negative control; PC: Positive control; RL: Rat liver RNA.
Adapted from [35].
miRNA quantification using a stochastic sensor
Following the isolation of a specific miRNA, detection is afforded by loading of the RNA sample onto a stochastic nanopore sensor. The nanopore sensor detects changes in ion current across a nanopore as RNA:probe complexes individually occlude the pore’s entrance. Passage of individual RNA:probe complexes causes transient pulses that correspond to a reduction in the ionic conductance of the pore, observable by high-bandwidth measurements of current versus time. Statistical analysis of a large set of these pulses gives the frequency of the pulse occurrence, which then is transformed into the concentration of the target miRNA owing to their linear correlation.
miRNAs are extremely difficult to detect without RT-PCR amplification. From a physical dimensions perspective, miRNAs are extremely small biomolecules, have a cross-section of approximately 2.5 nm and a contour length of 6–7 nm. Since the nanopore signal from a molecule is only measured during the molecule’s transit through the pore, one has to ensure that transit is slow enough to enable detection, and that it produces a distinct signal. To achieve this, nanopores in ultrathin solid-state membranes were fabricated using a scheme that is detailed in reference [35]. Briefly, approximately 40-nm-thick free-standing silicon nitride membranes supported by a silicon chip were thinned using electron beam lithography and reactive ion etch chemistry. The thinning resolution is 1 nm, dictated by the reactive ion etch time. Using this procedure, ultrathin membranes as thin as 6 nm have been fabricated, the thickness of a biological lipid bilayer. Following the membrane fabrication, approximately 3-nm-diameter nanopores were drilled in the thinned membrane region using the electron beam of a field-emission transmission electron microscope. Current traces of a nanopore under applied bias before and after the addition of RNA molecules to the analyte chamber are shown in Figure 2B. As seen from the figure, the addition of RNA molecules results in a frequent rate of current signal spikes, each corresponding to the passage of a single RNA molecule through the pore.
Since nanopores are molecular counters, it is important to eliminate the possibility that a particular signal for target quantification comes from an impurity in the sample. After isolation of miRNAs, the solution contains various agents that may interfere with nanopore detection. These agents include bovine serum albumin that coats the beads, sodium dodecyl sulphate used for dsRNA elution from the p19, and trace amounts of other RNAs from the total RNA sample. Therefore, control detections need to be conducted. Figure 2B illustrates a series of 30-s current traces for different samples that have been treated following the same p19-bead enrichment procedures. The first trace (red) was for the miR-122a:probe duplex enriched from 1 μg of rat liver RNA (RL). The second trace (blue) was a positive control (PC) detection for 30 ng of synthetic miR-122a:probe duplex that has been bound to and eluted from p19 beads. Both traces for samples PC and RL show spikes with current amplitudes as expected from dsRNA. In addition to the PC sample, four negative controls (NCs) NC1–NC4 were performed. In contrast to RL and PC, no spikes with amplitudes greater than 0.3 nA were presented in any of the four NCs.
The target miRNA can be quantified by measuring the capture rate for the miR-122a:probe duplex in the pore. The calibration curve in Figure 2C shows the capture rate at various concentrations of the synthetic miR-122a:probe complex. When an unknown miRNA sample was introduced to the nanopore, a current threshold of I0–0.4 nA was set (dashed gray lines under the traces), where I0 was the baseline current and only spikes crossing the threshold were counted. By counting approximately 250 current spikes, the mean capture rate was calculated and the miR-122a concentration was determined from the calibration curve. Using this method, the concentration of the 20-fold diluted miR-122a in sample RL was determined to be 0.7 fmol/μl. This concentration was translated to an original abundance of 78 ± 2 pg miR-122a/μg liver RNA in rat liver cells, in close agreement with the previously reported 58–67 pg miR-122a/μg RNA [34]. In addition, sample PC for the control test showed an expected concentration of 5.2 fmol/μl, indicating an efficient loading/unloading from the p19 beads during enrichment. The negative controls do not show any crossed-the-threshold current spikes in 2 min each, indicating very low background noise of <7 amol/μl, which is at least two orders of magnitude lower than sample RL. Figure 2D compares the relative error in concentration at various numbers of detected molecules. The relative error has been generated by computing the standard error in mean capture rates for populations ranging in size from 100 to 4000 spikes.
In summary, this work represents a proof of concept that single-molecule counters can quantify sequence-specific miRNA from cellular RNA. Coupled to p19-based extraction of probe-specific RNA, the nanopore counters utilize very small devices that are approximately 3 nm in all dimensions to count 20-bp miRNA duplex molecules with good signal-to-noise ratios.
Protein nanopore for circulating miRNA detection
Recently, Wang et al. proposed a protein nanopore-based miRNA sensor that followed a different principle and demonstrated potential for a clinical diagnostic test [43]. This assay utilized the α-hemolysin protein pore of Staphylococcus aureus as the sensor element. The focus of the sensor is a DNA probe that hybridizes target miRNA in the solution. The probe has been specially designed such that the miRNA:probe complex, when trapped in the pore, produces a electrical signature that shows drastically different translocation kinetics from free miRNA or probe. Therefore, the target miRNA can be quantified by counting the signature events per unit time. This nanopore sensor can determine diagnostically relevant circulating miRNAs in patient-derived samples, and can distinguish single nucleotide differences between sequence-similar miRNAs.
Electrical signatures of single miRNA molecules
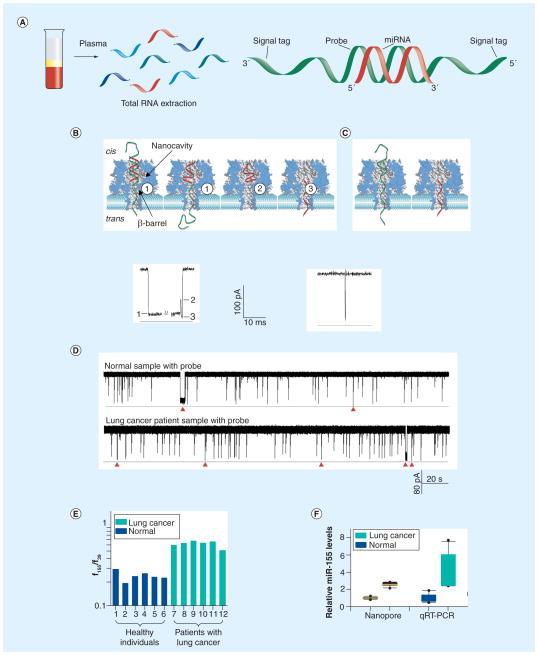
The ion pathway of the α-hemolysin pore possesses a special geometric profile. According to the molecular structure [57], the section from the pore’s cis entrance (2.6 nm) to the nanocavity (4.6 nm) is wide enough to hold a double-stranded nucleic acid fragment (2 nm), whereas the constrictive β-barrel from the middle of the pore (1.5 nm) to the trans mouth (2 nm) only allows single-stranded nucleic acid translocation [45,58–62]. Based on this structural feature, the probe has been designed to include the following domains: a capture domain used to hybridize with target miRNAs in the solution; and a signal tag attached to the 3′ and 5′ terminals to guide the entrapment of the miRNA:probe hybrid from the cis (wide) opening (Figure 3A). Once trapped in the nanopore, the miRNA:probe hybrid can generate a multilevel long block as shown in Figure 3B. Initially, the duplex section of the hybrid is encapsulated in the nanocavity, and the signal tag in the β-barrel; then the transmembrane voltage pulls the signal tag to induce the dissociation of the hybrid. This procedure with the signal tag in the β-barrel is characterized by the long level 1 block (250 ms; +100 mV) that reduces 90% of the pore conductance. Upon dissociation of the hybrid, the probe passes through the pore, but the miRNA can reside in the nanocavity for a short period (410 μs). This configuration generates level 2, which only reduces approximately 40% of the pore conductance. Finally the miRNA in the nanocavity rapidly traverses the pore, driven by the voltage, producing a short level 3 (270 μs) at similar conductance to level 1. Clearly, the dissociation–translocation kinetics comprises a characteristic triple-level current signal that is well distinguished from spike-like single-level events produced by free miRNA or probe translocation (Figure 3C), and therefore serves as a signature for single miRNA identification.
Figure 3. miRNA detection using a protein nanopore sensor (see facing page).
(A) Total RNA was extracted from plasma, then mixed with the probe (green) that hybridizes with the target miRNA (red). The probe bears signal tags on each end. (B) Upper panel shows the molecular mechanism of the miRNA:probe hybrid dissociation and translocation in the nanopore. The mixture in (A) was added to the cis solution, and the miRNA:probe hybrid entered the pore from the cis opening. The lower panel is a typical multilevel long block in the nanopore as a signature generated by the hybrid of miR-155 miRNA and its probe P155 (miR-155:P155). It was from a trace recorded at +100 mV in solutions containing 1 M KCl buffered with 10 mM Tris (pH 8.0). Level 1: trapping of the miRNA:probe hybrid in the pore, unzipping of the miRNA from the probe and translocation of the probe through the pore. Level 2: unzipped miRNA residing in the pore cavity. Level 3: translocation of the unzipped miRNA through the pore. (C) A spike-like short block generated by the translocation of unhybridized miR-155 or P155 from the cis solution. (D) Current traces for total plasma RNAs from healthy volunteers (normal sample) and lung cancer patients in the presence of the probe P155. The traces were recorded in 1 M KCl at +100 mV. Red arrows are signature events, which were seen only in the presence of probe for both healthy volunteers and lung cancer patients. (E) Frequency ratio of miR-155 and spiked-in synthetic miR-39 signature events (f155/f39) from six healthy individuals (1–6) and six patients with lung cancer (7–12). Conditions of patients: 7: metastatic squamous lung carcinoma; 8: recurrent small-cell cancer; 9: early-stage small-cell carcinoma, status postchemotherapy and radiation; 10: early-stage small-cell cancer, status postchemotherapy; 11: late-stage non-small-cell carcinoma, status post-resection and -chemotherapy; 12: late-stage adenocarcinoma, status postchemotherapy. (F) Box and whisker plots of the relative miR-155 levels in healthy and lung cancer groups measured with the nanopore sensor and qRT-PCR. Boxes mark the intervals between the 25th and 75th percentiles. Black lines inside the boxes denote the medians. Whiskers denote the intervals between the 5th and 95th percentiles. Filled circles indicate data points outside of the 5th and 95th percentiles.
qRT-PCR: Quantitative reverse-transcription PCR.
Adapted from [43].
Sensitivity & ability to discriminate miRNAs with similar sequences
The probe in the nanopore sensor determines both the sensitivity and specificity. The probe structure, including its length, composition and directionality, can be optimized to maximize the capture rate of the miRNA:probe complex for high detecting sensitivity. It has been known that that a poly(dC)30 tag at the 3′ end of the probe increases the capture rate over 20-fold compared with the same tag at the 5′ end [61,62]; a poly(dC)30 tag is much more efficient in generating signature events than a poly(dA)30 and poly(dT)30 tag, or a shorter tag such as poly(dC)8. The probe optimization, combined with other approaches such as pore engineering [63], salt gradient [64] and high voltage, can effectively enhance the miRNA sensitivity to 100 fM.
This nanopore sensor has demonstrated high specificity due to the Watson–Crick base pairing in miRNA:probe hybridization. Most notably, the nanopore can discriminate miRNAs with similar sequences. Among over 1400 human miRNAs that have been identified, including many members of the same miRNA family such as the Let-7 tumor-suppressing miRNA family [6,65,66], are only different in one or several nucleotides. Owing to their short lengths, sequence-similar miRNAs are difficult to distinguish using current PCR or hybridization-based methods [25,28,67]. The nanopore, however, can discriminate them because single mismatch in the miRNA:probe complex weakens their hybridization strength, which significantly shortens its dissociation time (i.e., the duration of signature events compared with a fully-matched miRNA:probe complex). For example, Let-7a and -7c only have a one nucleotide difference. When using the probes for Let-7a (Pa) and Let-7c (Pc) to target both miRNAs at +100 mV, the duration of signature events decreased 2.4-fold from 303 ms for Let-7a:Pa to 124 ms for Let-7c:Pa, and 2.0-fold from 342 ms for Let-7c:Pc to 179 ms for Let-7a:Pc. This result is consistent with previous reports on detection of dsDNAs containing single nucleotide mismatch in the nanopore [68–71]. Overall, the nanopore method can be applied to studying both miRNAs with similar sequences and SNPs in genomic detection that are associated with significant biological properties of cancers, such as susceptibility, prognosis and the response to therapeutic agents [72].
Nanopore detection of circulating miRNAs
The protein nanopore sensor has demonstrated ability to detect circulating miRNAs. In the nanopore detection, miR-155 has been selected as the target, among over 100 miRNAs identified to be dysregulated in lung cancer [6,21,27,65,66,72–76]. It has been found that high levels of miR-155 is correlated with significantly poor prognoses and shorter survival times in lung cancer patients [77,78]. Using the nanopore, the miR-155 level in plasma has been compared for lung cancer patients versus healthy individuals. The measurement was conducted in total RNA extracted from each plasma sample using mirVana™ PARIS Kit (Ambion, Austin, TX). When the probe (P155) was mixed with the recording solution containing RNA extract, both distinct short and long current blocks can be identified in the normal group (see upper trace of Figure 3D) and lung cancer group (see lower trace of Figure 3D). As the characteristic long blocks shared the same current profile and properties with the synthetic miR-155 RNA (Figure 3B), they are considered as the single molecular signature for the miR-155:P155 complex. The frequencies of signature events for miR-155 (f155) in the lung cancer group were higher than those in the control group (Figure 3D). To reduce the variability, the spiked-in miR-39 was used as an internal control. The frequencies of spiked-in synthetic C. elegans miRNA miR-39 events (f39) have been found to be independent of the samples. Therefore the ratio f155/f39 was used to normalize the assays. Figure 3E shows that f155/f39 in the lung cancer group (0.62 ± 0.06) was significantly higher than f155/f39 in the control group (0.24 ± 0.05). Collectively, the mean level of miR-155 was increased 2.6-fold in the lung cancer group compared with the control group as measured by the nanopore sensor (Figure 3F). This trend was verified using qRT-PCR, the result of which shows a 4.3-fold increase with a greater variability. In conclusion, both the nanopore and qRT-PCR assays indicated a significant elevation of miR-155 in lung cancer patient samples, but the nanopore method demonstrated higher accuracy with no requirement for labeling or amplification (Figure 3F).
In summary, the protein nanopore-based miRNA sensor utilizes a programmable oligonucleotide probe to generate an electrical signature signal for the direct and label-free detection of target miRNAs in total RNA extracted from clinical plasma samples.
Expert commentary
Both nanopore platforms described above utilize single-molecule strategies for quantitative miRNA detection. In single-molecule measurement, each target molecule interacting with the nanopore can generate a unique electrical response, which can be an electrical pulse – like that in the synthetic nanopore (Figures 1 & 2B) – or a signature current block in the protein nanopore (Figure 3B). The target miRNA at certain concentrations can result in a specific frequency of signal event occurrence (number of events per unit time), which in principle is proportional to the target concentration. Therefore, the target can be quantified from the event frequency. Throughout the procedure, no enzyme or amplification are involved. The detection accuracy relies on the target capture rate (number of events per unit time per unit target amount). With a higher capture rate, one would be able to count more events at the same target concentration and in the same recording time. As a result, both accuracy and efficiency are promoted. The high capture rate in the synthetic nanopore is related to the appropriate pore size. The 3-nm pore diameter is suitable for miRNA detection. Smaller pore size may limit the target access, reducing the capture rate, while larger pore size may be less sensitive to the short oligonucleotide translocation. Meanwhile, the 100,000-fold enrichment by the p19 protein greatly promotes the detection efficiency. The synthetic pore can accurately measure 1 fmol/μl of enriched miRNA in a couple of minutes [35]. Unlike the synthetic nanopore, the protein pore’s diameter is fixed. Therefore the capture rate cannot be adjusted by the pore size. However, it can be greatly enhanced by various approaches, including recording at high voltage, engineering the pore lumen with positively charged amino acids [63], optimizing the probe sequence [43] and establishing a electrolyte gradient across the membrane [64] to attract negatively charged oligonucleotides into the pore. With these approaches, the protein nanopore can detect low miRNA concentrations at sub-picomolar levels [43].
The nanopore is more than a single-molecule counter. One of the intrinsic properties of the nanopore is that the pore conductance is very sensitive to the target molecule residing in the pore. The conductance amplitude of the nanopore block indicates a specific configuration of the target molecule, and the block duration represents the lifetime for the molecule staying in a kinetic state. This means an observed conductance profile (or pattern) could represent a series of sequential molecular configurations. With this capability, the nanopore can be used to investigate kinetic pathways of single-molecule reactions. Such specific conductance changes can serve as a signature. For example, the signature identified in the protein pore sensor (Figure 3B) is produced by the trapped miRNA:probe complex that sequentially unwinds and translocates through the pore. These signatures completely distinguish the miRNA:probe complex from other components that interact with the pore, such as free miRNA or probe translocation, providing high detection specificity in total RNA extract (not enriched miRNAs). The high specificity allows discrimination of miRNAs with similar sequences.
Five-year view
We anticipate that the nanopore technique can be translated into a real-time, clinically usable miRNA quantification tool. But several issues need to be resolved in the next couple of years during the development. For the synthetic nanopore, future devices need to improve on the isolation method and detection limits by miniaturizing the fluidic cell in which the nanopore experiments are carried out, as well as by parallelization of the experiments using multiple, electrically addressable pores [79]. For the protein pore system, it is important to validate the engineered probe for high sensitivity and specificity. For example, the probe can be incorporated into unnatural compounds such as locked nucleic acids and peptide nucleotide acids. These compounds strengthen hybridization of the probe with miRNAs, thus enhancing the target miRNA discriminatory ability. In addition, the barcode probe can be designed to fulfil multiple miRNA detection [59,80]. Another improvement will be the construction of a robust protein pore device. To realize such a device, the membrane in which the protein pore is embedded needs stabilization. Many promising membrane strategies have already shown feasibility, such as a macropolymer-encapsulated lipid bilayer [81–83], supported or suspended lipid bilayer [84–86] and droplet interface bilayer [87]. The droplet interface bilayer is also the first membrane platform showing the potential to form a protein pore array [87], as well as minimized sample consumption down to a microliter level. These advantages make it possible for high throughput miRNA detection and real-time applications.
Key issues.
miRNA is an important indicator for the development of diseases including cancer.
The increasing exploration of miRNAs demands rapid, accurate, low-cost miRNA detection technologies.
One of the main challenges for miRNA-based biomarkers is the need to use purified miRNAs from biological samples. Stability of the RNA and quantitative detection of miRNAs compared with an internal RNA standard is a challenge for the clinical use of miRNAs.
The nanopore single-molecule technology can accurately quantify miRNA in tissue and blood from cancer patients without labeling, enzymatic reaction or amplification.
The nanopore method paves a road to the development of a noninvasive diagnostic tool for early cancer detection.
Issues such as robustness and multiplex detection should be solved in the future, in order to translate the nanopore technique into a clinically favorable miRNA detection method.
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
This investigation was supported by the National Science Foundation (NSF 0546165) and the NIH (NIH 1R01GM079613). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program Grant C06-RR-016489–01 from the National Center for Research Resources, National Institutes of Health. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. A comprehensive review of small RNAs including miRNAs, siRNAs and piRNAs in terms of biogenesis pathways and their regulations, along with their functions at both the genome and the transcriptome level. [DOI] [PubMed] [Google Scholar]
- 3••.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene LIN-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. The first publication that identified a small RNA encoded by the LIN-4 locus that was associated to the developmental timing of the nematode Caenorhabditis elegans by binding in the 3′ untranslated region (UTR) of LIN-14 mRNA and negatively modulating the protein Lin-14. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene LIN-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. MiRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 10.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development (Cambridge) 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 14.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 15.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Gen. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18•.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. A comprehensive review of the involvement of miRNAs in various cancer types, including hematopoietic and solid tumors, and their potential clinical applications such as diagnostic, prognostic and therapeutic tools. Briefly discusses the mechanisms related to miRNA regulations in tumorigenesis, including DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. The first publication to establish the relationship between miRNA and malignancy. Found frequent deletions and downregulation of miRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, a form of mature B-cell malignancy. Since then, many miRNAs have been found to be involved in almost all types of cancer functioning as oncogenic or tumor suppressive genes depending on the cellular context and their target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. Represents an example showing how miRNA profile and signature have been used to predict cancer prognosis. A unique 13-miRNA expression signature was identified to differentiate the cases of chronic lymphocytic leukemia with low levels of ZAP-70 expression associated with mutated IgVH (better prognosis) from those with high levels associated with unmutated IgVH (worse prognosis) [DOI] [PubMed] [Google Scholar]
- 21.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 23.Zheng D, Haddadin S, Wang Y, et al. Plasma micrornas as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 24.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt EA, Goulding AM, Deo SK. Direct detection and quantification of microRNAs. Anal Biochem. 2009;387:1–12. doi: 10.1016/j.ab.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy J, Bustin SA. Reliability of real-time reverse-transcription PCR in clinical diagnostics: gold standard or substandard? Expert Rev Mol Diagn. 2009;9(2):187–197. doi: 10.1586/14737159.9.2.187. [DOI] [PubMed] [Google Scholar]
- 27.Yendamuri S, Kratzke R. MicroRNA biomarkers in lung cancer: MiRacle or quagMiRe? Transl Res. 2011;157:209–215. doi: 10.1016/j.trsl.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 28•.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. An original paper that described a novel PCR method, termed stem-loop reverse-transcription PCR, to quantify small RNA molecules such as miRNAs. This TaqMan®-based (Roche Molecular Systems, Inc) miRNA assay showed a dynamic range of seven orders of magnitude and could quantify from less than ten to more than 30,000 miRNA copies per cell with high specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 30•.Ro S, Park C, Jin J, Sanders KM, Yan W. A PCR-based method for detection and quantification of small RNAs. Biochem Biophys Res Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. An original paper that described another simple and sensitive PCR-based method for small RNA quantification. Small RNAs were polyadenylated using a poly(A) polymerase and were then reverse-transcribed using a primer containing oligo dTs (thymidines) flanked by an adaptor. Conventional SYBR green quantitative PCR was carried out using a small RNA-specific primer and a universal reverse primer that matched the adaptor. The assay showed a dynamic range of five logs and was capable of detecting as little as 0.001 ng of the target cDNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner M, Renwick N, Brown M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reis PP, Waldron L, Goswami RS, et al. MRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin G, Cid M, Poole CB, McReynolds LA. Protein mediated miRNA detection and siRNA enrichment using p19. BioTechniques. 2010;48:xvii–xxiii. doi: 10.2144/000113364. [DOI] [PubMed] [Google Scholar]
- 35.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 36.Neely LA, Patel S, Garver J, et al. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen BS. MicroRNA in situ hybridization. Methods Mol Biol. 2012;822:67–84. doi: 10.1007/978-1-61779-427-8_5. [DOI] [PubMed] [Google Scholar]
- 38.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 39.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2010;52:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen S, Baker A, Moller S, Nielsen BS. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Pena JT, Sohn-Lee C, Rouhanifard SH, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Bayley H, Jayasinghe L. Functional engineered channels and pores (review) Mol Membr Biol. 2004;21:209–220. doi: 10.1080/09687680410001716853. A comprehensive review on the nanopore principle and applications. [DOI] [PubMed] [Google Scholar]
- 45.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. A comprehensive review covering both synthetic nanopores and protein nanopores. [DOI] [PubMed] [Google Scholar]
- 47.Gu LQ, Shim JW. Single molecule sensing by nanopores and nanopore devices. Analyst. 2010;135:441–451. doi: 10.1039/b907735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma L, Cockroft SL. Biological nanopores for single-molecule biophysics. Chembiochem. 2010;11:25–34. doi: 10.1002/cbic.200900526. [DOI] [PubMed] [Google Scholar]
- 49••.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr Opin Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. A comprehensive review focusing on application of protein nanopores in biosensor and DNA sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Olasagasti F, Lieberman KR, Benner S, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu A, Zhao Q, Guan X. Stochastic nanopore sensors for the detection of terrorist agents: current status and challenges. Anal Chim Acta. 2010;675:106–115. doi: 10.1016/j.aca.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 53•.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. A comprehensive review of nanopore nucleic acid detection. [DOI] [PubMed] [Google Scholar]
- 54•.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–637. doi: 10.1016/j.cbpa.2006.10.040. A review on single-molecule DNA sequencing methods. [DOI] [PubMed] [Google Scholar]
- 55••.Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. A comprehensive review on nanopore-based DNA sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vargason JM, Szittya G, Burgyan J, Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 57.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 58.Meller A, Nivon L, Branton D. Voltage-driven DNA translocations through a nanopore. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell N, Howorka S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew Chem Int Ed. 2008;47:5565–5568. doi: 10.1002/anie.200800183. [DOI] [PubMed] [Google Scholar]
- 60.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathé J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 63.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci USA. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 66.Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem. 2009;16:1047–1061. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394:1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 68.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 69.Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys Rev Lett. 2003;90(23):238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 71.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 72.Cho WC. Role of miRNAs in lung cancer. Expert Rev Mol Diagn. 2009;9(8):773–776. doi: 10.1586/erm.09.57. [DOI] [PubMed] [Google Scholar]
- 73.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 75.Rosell R, Wei J, Taron M. Circulating MicroRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer. 2009;10:8–9. doi: 10.3816/CLC.2009.n.001. [DOI] [PubMed] [Google Scholar]
- 76.Donnem T, Eklo K, Berg T, et al. Prognostic impact of miR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 78.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 79.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10:2237–2244. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeon TJ, Malmstadt N, Schmidt JJ. Hydrogel-encapsulated lipid membranes. J Am Chem Soc. 2006;128:42–43. doi: 10.1021/ja056901v. [DOI] [PubMed] [Google Scholar]
- 82.Shim JW, Gu LQ. Stochastic sensing on a modular chip containing a single-ion channel. Anal Chem. 2007;79:2207–2213. doi: 10.1021/ac0614285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang XF, Cheley S, Rice-Ficht AC, Bayley H. A storable encapsulated bilayer chip containing a single protein nanopore. J Am Chem Soc. 2007;129:4701–4705. doi: 10.1021/ja068654g. [DOI] [PubMed] [Google Scholar]
- 84.White RJ, Ervin EN, Yang T, et al. Single ion-channel recordings using glass nanopore membranes. J Am Chem Soc. 2007;129:11766–11775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 85.Cornell BA, Braach-Maksvytis VLB, King LG, et al. A biosensor that uses ion-channel switches. Nature. 1997;387:580–583. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 86.Baaken G, Sondermann M, Schlemmer C, Rühe J, Behrends JC. Planar microelectrode-cavity array for high-resolution and parallel electrical recording of membrane ionic currents. Lab Chip. 2008;8:938–944. doi: 10.1039/b800431e. [DOI] [PubMed] [Google Scholar]
- 87.Bayley H, Cronin B, Heron A, et al. Droplet interface bilayers. Mol Biosyst. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.miRBase. http://microrna.sanger.ac.uk.