Abstract
Pahayokolides A–B are cyanobacteria derived non-ribosomal peptides which exhibit cytotoxicity against a number of cancer cell lines. The biosynthetic origin of the 3-amino-2,5,7,8-tetrahydroxy-10-methylundecanoic acid (Athmu) moiety has been investigated using stable isotope incorporation experiments. While α-ketoisocaproic acid (α-KIC), α-hydroxyisocaproic acid (α-HIC) and leucine all serve as precursors to Athmu, the feeding of [1-13C] α-KIC results in more than threefold greater 13C enrichment than the other precursors. This result suggests that α-KIC is the immediate precursor which is selected and activated by the adenylation domain of the loading NRPS module and subsequently reduced in a fashion similar to that of the recently identified pathways for cryptophycins A–B, cereulide and valinomycin.
Keywords: Pahayokolide, Biosynthesis, αγ-hydroxy-β-amino acid, α-KIC, α-HIC
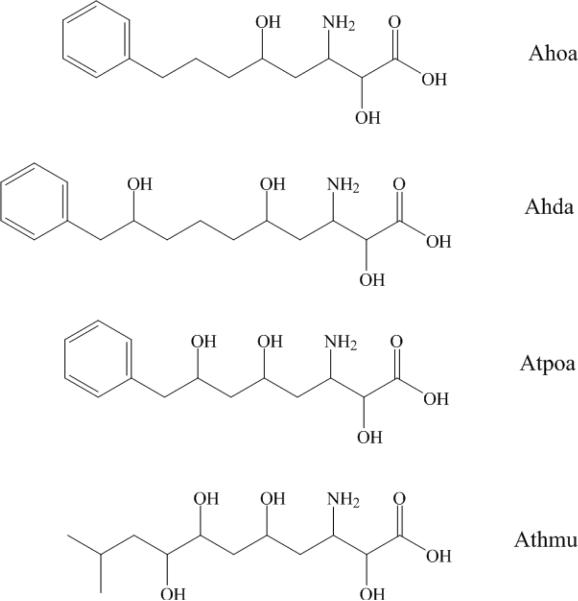
As a group, cyanobacteria have demonstrated the capacity to produce a unique range of defensive secondary metabolites.1,2,3,4 These secondary metabolites exhibit a wide ranging diversity of molecular structure and biological properties including antiproliferative activity, acute cytotoxicity or neurotoxicity.5,6,7 As a result many have earned scientific interest as a source for therapeutic drug leads or pharmacological tools.8,9 Many cyanobacterial peptides feature nonproteinogenic amino acids and/or polyketide chains, which are indicative of NRPS (non-ribosomal peptide synthetase) or hybrid NRPS/PKS (polyketides synthase) biosynthetic pathways. Long-chain α,γ-hydroxy-β-amino acids (Fig. 1) have emerged in cyanobacterial peptides and include Ahoa (3-amino-2,5-dihydroxy-8-phenyloctanoic acid) in nostophycin A, Ahda (3-amino-2,5,9-trihydroxy-10-phenyldecanoic acid) in scytonemin A, Atpoa (3-amino-2,5,7-trihydroxy-8-phenyloctanoic acid) in tychonamides A–B, Athmu (3-amino-2,5,7,8-tetrahydroxy-10-methylundecanoic acid) in schizotrin A, pahayokolides A–B and lyngbyazothrins A–D or portoamides A–D.10~16 The portoamides and the lyngbyazothrins are identical compounds isolated from the freshwater cyanobacteria Lyngbya sp. and Oscillatoria sp, respectively. Tychonamides A–B, schizotrin A, pahayokolides A–B and lyngbyazothrins A–D share several conserved residues. It is interesting that all above cyanobacteria peptides containing long-chain α,γ-hydroxy-β-amino acids were isolated from freshwater cyanobacteria species. Very little is known about the ecological or physiological functions of these cyanobacterial β-peptides. However, a recent report describes the anti-algal properties of the portaamides.16
Figure 1.
Structure of four α,γ-hydroxy-β-amino acids in freshwater cyanobacterial peptides
It was reported that incorporation of β-amino acids into natural peptides can enhance both biological activities and resistance to peptidase hydrolysis.17 These characteristics make peptides of β-amino acids valuable candidates for drug discovery and development. Pahayokolide A–B (1–2) inhibits a number of cancer cell lines over a range of concentrations (IC50 varied from 2.13 to 44.57 μM).18
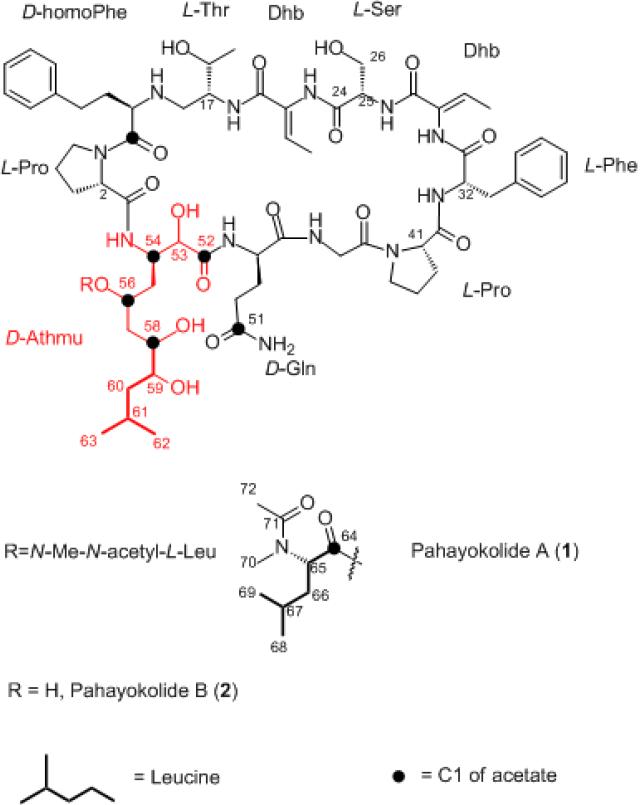
Analysis of the nostophycin biosynthetic pathway indicated that the starter unit for the biosynthesis of the Ahoa moiety in nostophycin is phenylpropanoic acid which must be subjected to a one carbon deletion prior to its extension with three acetate units.19 The one-carbon deletion of the starter unit has been observed in Adda (3-amino-9-methoxy-2,6,8-trimethyl-10-phenyl-4,6-decadienoic acid), a β-amino acid found in the cyanobacterial peptide microcystin.20 The gene cluster coding for the biosynthetic pathway of barbamide includes a one carbon deletion of a leucine unit, however, the mechanism for this process remains uncertain21 In contrast, our earlier studies revealed that the starter unit of the Athmu residue in pahayokolides A–B (1–2) is derived from leucine, which is further extended with three acetate units (Fig. 2).22 Additional required modifications to the leucine moiety include transamination to the ketone followed by reduction. As the biosynthetic pathway for the pahayokolides has not yet been identified, it is unclear if these modification occur prior to or subsequent to loading of the starter unit onto the adenylation domain of the loading module. In this study, we compare the incorporation of (+) and (−) [1-13C] α-HIC (α-hydroxyisocaproic acid) and [1-13C] α-KIC (α-ketoisocaproic acid) with that of leucine into pahayokolide B in order to identify the starter unit.
Figure 2.
Pahayokolides A–B and incorporation of [1-13C] leucine and [1-13C] acetate.
(+) and (−) [1-13C] α-HICs were prepared by selective reduction of [1-13C] α-KICs (99% 13C abundance, purchased from Cambridge Isotope Laboratories, Inc. USA) as previously described.23
Lyngbya sp. strain 15–2 was maintained in 2 L cultures of BG11 medium, buffered with 2-(N-morpholino)-ethanesulfonic acid buffer (2.6 mM) at pH 7.2. Cultures were supplemented with 150 mg of 13C-labeled (+) and (−) [1-13C] α-HIC, and [1-13C] α-KIC (50 mg each on days 30, 34, and 38), respectively. Cultures were harvested after six weeks of growth. Pahayokolides A (1) and B (2) were isolated from the biomass as previously described.14 Pahayokolide A (1) was treated with mild base to provide pahayokolide B (2) in order to avoid the presence of multiple conformers of pahayokolide A (1) and simplify the integration of NMR spectra. 13C NMR spectra were acquired as previously described.22
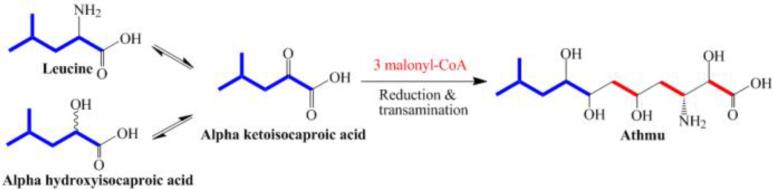
Assignments of 13C NMR signals and results of stable isotope feeding experiments of selected carbons of pahayokolide B (2) are summarized in Table 1. The 13C NMR spectra of pahayokolide B (2) derived from (+) and (−) [1-13C] α-HIC, and [1-13C] α-KIC fed cultures showed significant enrichment at C-58 of pahayokolide B. However, under identical conditions, the incorporation level of [1-13C] α-KIC (1000%) into pahayokolide B was much higher than that of either (+) or (−) [1-13C] α-HIC (291% and 306%, respectively). Furthermore, the incorporation level of [1-13C] leucine (332%) into pahayokolide A under the same conditions was also much lower than that of [1-13C] α-KIC in to pahayokolide B,22 and closer to that of (+) and (−) [1-13C] α-HIC into pahayokolide B. Leucine, α-KIC and α-HIC are biochemically interchangeable through primary metabolic processes. α-KIC, can be derived from both leucine via leucine aminotransferase and from α-HIC via α-keto acid dehydrogenase. Compared with [1-13C] leucine, (+) and (−) [1-13C] α-HIC, [1-13C] α-KIC exhibited greater enrichment by more than threefold, indicating that the immediate precursor for the biosynthesis of Athmu is α-KIC (Fig. 3). We had hoped to resolve the question of absolute configuration of C59 of 2, by comparison of 13C enrichments in (+) and (−) [1-13C] α-HIC feeding experiments. The very similar incorporation level of (+) and (−) [1-13C] α-HIC did not resolve this question. However, it demonstrated that the chirality of C59 likely originates from the stereoselective reduction of the α-keto group of α-KIC after loading onto the biosynthetic assembly line of pahayokolide B. In the biosynthesis of cereulide, α-KIC and α-KIV (α-ketoisovaleric acid) were preferentially activated by adenylation domains of the NRPS CesA and CesB. Embedded ketoreductase domains catalyze the reduction of the α-keto acids to the corresponding α-hydroxy acids (Fig. 4).25 The α-hydroxy acid biosynthesis of valinomycin (Vlm1 and Vlm2), cryptophycin 1 (CrpD), and hectochlorin (HctE) also shared the strategy of α-keto acid incorporation and reduction.25,26, 27 Among known long-chain α,γ-hydroxy-β-amino acids in cyanobacterial peptides (Fig. 1), only the biosynthetic genes of Ahoa have been reported.19 The starter module of the Ahoa biosynthetic genes also contains a ketoreductase domain following an adenylation domain. However, the role of this ketoreductase in Ahoa biosynthesis remains uncertain. The preferential incorporation of α-KIC over α-HIC into pahayokolide B suggests a similar strategy may be at play in the biosynthesis of Athmu. We cannot rule out the possibility that α-KIC is preferentially taken up by Lyngbya cells over α-HIC. There is no report on uptake of α-KIC or α-HIC by Lyngbya. However, α-KIC and α-HIC were transferred into Lactococcus lactis cells at the same rate by the citrate transporter.28 The citrate transporter is also present in cyanobacterial cells.29 It is therefore reasonable to expect that α-KIC and α-HIC reach very similar cellular concentrations. Thus, it is reasonable to assign the direct precursor in the biosynthetic pathway based on incorporation levels of labeled substrates.
Table 1.
Isotopic enrichment of selected carbonsa based on 13C NMR (CD3OD/D2O) for pahayokolide B (2)
| Enrichment ratiob |
||||
|---|---|---|---|---|
| Carbon no.a | δc (ppm) | [1-13C] α-KIC | (+) [1-13C] α-HIC | (−) [1-13C] α-HIC |
| 53 | 73.1 | 104 | 73 | 87 |
| 54 | 49.5 | 78 | 76 | 70 |
| 56 | 64.7 | 72 | 83 | 100 |
| 58 | 71.9 | 1000 | 291 | 306 |
| 59 | 72.6 | 127 | 152 | 167 |
| 2 | 60.7 | 75 | 93 | 100 |
| 17 | 59.3 | 88 | 100 | 73 |
| 24 | 170.8 | 114 | 108 | 153 |
| 25 | 56.8 | 94 | 133 | 74 |
| 26 | 61.4 | 120 | 110 | 147 |
| 32 | 56.1 | 89 | 72 | 98 |
| 41 | 60.3 | 80 | 87 | 82 |
Except C-24, chemical shift of other carbons were in the range of 49.5 – 73.1 ppm
Carbon resonance were integrated relative to the signal for C-24.22
Enriched signals are shown in bold. Enrichment was calculated using the integral values for carbons shown in this table and applying the methods described by Sitachitta.24 Average isotopic enrichment for carbons (excluding signals in bold) = 99 ± 27.
Figure 3.
Proposed biogenic pathway for the α,γ-hydroxy-β-amino acid Athmu.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) Grant S11 ES11181. We acknowledge the support of the Hollings Marine Laboratory NMR Facility. Commercial equipment or materials are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by NIST, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material 13C NMR spectra for natural abundance pahayokolide B, [1-13C] α-ketoisocaproic acid, (+) and (−) [1-13C] α-hydroxyisocaproic acid can be found, in the online version, at
References and notes
- 1.Gerwick WH, Roberts MA, Proteau PJ, Chen JL. J. Appl. Phycol. 1994;6:143–149. [Google Scholar]
- 2.Namikoshi M, Rinehart KL. J. Ind. Microbial. 1996;17:373–384. [Google Scholar]
- 3.Mayer MS, Gustafson KR. Int. J. Cancer. 2003;105:291–299. doi: 10.1002/ijc.11080. [DOI] [PubMed] [Google Scholar]
- 4.Nunnery JK, Mevers E, Gerwick WH. Cur. Opin. Biotechnol. 2010;21(6):787–793. doi: 10.1016/j.copbio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerwick WH, Tan LT, Siachitta N. Nitrogen-containing metabolites from marine cyanobacteria. In The Alkaloids. Academic Press; San Diego, CA, USA: 2001. pp. 75–184. [DOI] [PubMed] [Google Scholar]
- 6.Tan LT. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Rein KS. New peptides isolated from Lyngbya species: a review. Mar. Drugs. 2010;8:1817–1837. doi: 10.3390/md8061817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rastogi RP, Sinha RP. Biotechnol. Adv. 2009;27(4):521–539. doi: 10.1016/j.biotechadv.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Malloy KL, Choi H, Fiorilla C, Valeriote FA, Matainaho T, Gerwick WH. Bioorg. Med. Chem. Lett. 2012;22(1):683–688. doi: 10.1016/j.bmcl.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii K, Sivonen K, Kashiwagi T, Hirayama K, Harada K. J. Org. Chem. 1999;64:5777–5782. [Google Scholar]
- 11.Helms GL, Moore RE, Niemczura WP, Patterson GML, Tomer KB, Gross ML. J. Org. Chem. 1988;53:1296–1307. [Google Scholar]
- 12.Mehner C, Mueller D, Krick A, Kehraus S, Löeser R, Güetschow M, Maier A, Fiebig HF, Brun R, Köenig GM. Eur. J. Org. Chem. 2008;10:1732–1739. [Google Scholar]
- 13.Pergament I, Carmeli S. Tetrahedron Lett. 1994;35:8473–8476. [Google Scholar]
- 14.An T, Kumar TK, Wang M, Liu L, Lay JO, Jr., Liyanage R, Berry J, Gantar M, Marks V, Gawley RE, Rein KS. J. Nat. Pro. 2007;70(5):730–735. doi: 10.1021/np060389p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zainuddin NE, Jansen R, Nimtz M, Wray V, Preisitsch M, Lalk M, Mundt S. J. Nat. Prod. 2009;72:1373–1378. doi: 10.1021/np8007792. [DOI] [PubMed] [Google Scholar]
- 16.Leão PN, Pereira AR, Liuc WT, Ngd J, Pevznere PA, Dorrestein PC, König GM, Vasconcelosa MTSD, Vasconcelosa VM, Gerwick WH. Proc. Natl. Acad. Sci. 2010;107:11183–11188. doi: 10.1073/pnas.0914343107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zubrzak P, Williams H, Coast GM, Isaac RE, Reyes-Rangel G, Juaristi E, Zabrocki J, Nachman RJ. Pept. Sci. 2007;88(1):76–82. doi: 10.1002/bip.20638. [DOI] [PubMed] [Google Scholar]
- 18.Berry JP, Gantar M, Gawley RE, Wang M, Rein KS. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2004;139:231–238. doi: 10.1016/j.cca.2004.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fewer DP, Österholm J, Rouhiainen L, Jokela J, Wahlsten M, Sivonen K. Appl. Environ. Microbiol. 2011;77(22):8034–8040. doi: 10.1128/AEM.05993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks LM, Moffitt MC, Beer LL, Moore BS, Kelleher NL. ACS Chem. Biol. 2006;1(2):93–102. doi: 10.1021/cb500007v. [DOI] [PubMed] [Google Scholar]
- 21.Flatt PM, O'Connell SJ, McPhail KL, Zeller G, Willis CL, Sherman DH, Gerwick WH. J. Nat. Prod. 2006;74(6) doi: 10.1021/np050523q. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Bearden DW, Rein KS. J. Nat. Prod. 2011;69(6):938–944. doi: 10.1021/np200362q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, La B, Fortunak JM. Tet. Lett. 1998;39(31):5501–5504. [Google Scholar]
- 24.Sitachitta N, Marquez BL, Williamson TR, Rossi J, Ann Roberts M, Gerwick WH, Nguyen V-A, Willis CL. Tetrahedron. 2000;56:9103–9113. [Google Scholar]
- 25.Magarvey NA, Ehling-Schulz M, Walsh CT. J. Am. Chem. Soc. 2006a;128(33):10698–10699. doi: 10.1021/ja0640187. [DOI] [PubMed] [Google Scholar]
- 26.Magarvey NA, Beck ZQ, Golakoti T, Ding Y, Huber U, Hemscheidt TK, Abelson D, Moore RE, Sherman DH. ACS Chem. Bio. 2006b;1(12):766–779. doi: 10.1021/cb6004307. [DOI] [PubMed] [Google Scholar]
- 27.Ramaswamy AV, Sorrels CM, Gerwick WH. J. Nat. Prod. 2007;70(6):1977–1986. doi: 10.1021/np0704250. [DOI] [PubMed] [Google Scholar]
- 28.Pudlik AM, Lolkema JS. J.Bacteriol. 2007;194:3627–3635. doi: 10.1128/JB.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirus O, Strauss S, Nicolaisen K, von Haeseler A, Schleiff E. BMC Biology. 2009;7:68. doi: 10.1186/1741-7007-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.