Abstract
Mitogen activated protein kinases (MAPKs) have a docking groove that interacts with linear motifs in binding partners. To determine the structural basis of binding specificity between MAPKs and docking motifs, we quantitatively analyzed the ability of fifteen linear motifs from diverse MAPK partners to bind to c-Jun N-terminal kinase 1 (JNK1), p38α and extracellular signal-regulated kinase 2 (ERK2). Classical docking motifs mediated highly specific binding only to JNK1, and only motifs with a sequence pattern distinct from the classical MAPK binding docking motif consensus could differentiate between the topographically similar docking grooves of ERK and p38. We also solved the crystal structures for four MAPK-docking peptide complexes that represented JNK-specific, ERK-specific or ERK- and p38-selective binding modes. These structures revealed that the regions located in between consensus positions in the docking motifs showed conformational diversity. Although the consensus positions in the docking motifs served as anchor points that bound to common MAPK surface features and mostly contributed to docking in a non-discriminatory fashion, specificity was determined mainly by the conformation of the intervening region between the anchor points. These insights enabled us to successfully design peptides with tailored MAPK binding profiles by rationally changing the length and amino acid composition of docking motif regions located between anchor points. We present a coherent structural model underlying MAPK docking specificity that reveals how short linear motifs binding to a common kinase docking groove can mediate diverse interaction patterns and contribute to correct MAPK partner selection in signaling networks.
Introduction
It is important to organize physical protein-protein interactions for correct physiological function in intracellular signaling networks. Linear binding motifs that play a crucial role in this process are normally defined based on their consensus motif sequence and are less than 10-20 amino acids in length (1). They enable interactions between unstructured protein regions and globular protein domains and it is becoming accepted that their role in mediating protein-protein interactions parallels the importance of classical interactions formed between globular domains (2-3). Because they bind to “open” protein-protein interaction surfaces and because only a handful of consensus amino acid positions are required for binding, it is enigmatic how they can topographically distinguish often similar binding surfaces. How can intracellular networks in particular rely on them so broadly (4)?
Mitogen activated protein kinases (MAPKs) regulate diverse aspects of cellular life such as cell division, differentiation or apoptosis and interact with proteins through linear binding motifs (5-6). MAPKs are part of multi-tiered kinase cascades in which ERK1/2, JNK and p38 are activated by dedicated mitogen-activated protein kinase kinases (MAP2Ks) MKK1/2, MKK4/7 and MKK3/4/6, respectively (5). These upstream activators bind to the same surface on their cognate MAPK as downstream MAPK substrates, inactivating phosphatases and protein scaffolds by their independently evolved linear motifs (7). Within the diverse repertoire of physical links observed in signaling networks, the use of linear motifs directly binding to signaling enzyme’s catalytic domains, known as docking motifs, is the simplest solution to increase the specificity of promiscuous active sites (8). Similarly to many signaling enzymes, MAPKs contain promiscuous active sites. Therefore, additional protein interaction sites are critical for determining the wiring of MAPK pathways. In this type of interaction, which is referred to as docking for protein kinases and phosphatases, the binding surface is separate from the active site (7). For example, MAPKs phosphorylate serine and threonine residues that are followed by proline (called a S/TP target site or motif) and they also bind to docking (D)-motifs, which are ~8-12 amino acid long fragments satisfying a loose consensus sequence (9), thus enabling these enzymes to form physical connections with other signaling proteins (10).
The docking grooves of paralogous MAPKs show a high degree of similarity, likely because each major group of MAPKs in animals - typical MAPKs such as extracellular signal-regulated kinase (ERK) 1/2; c-Jun N-terminal kinases (JNKs); p38s; ERK5; and atypical MAPKs such as ERK3, 4, and 7 - emerged through whole genome or individual gene duplication events and because three members of this family - ERK2, p38α and JNK1 - share ~40-50% sequence identity (11). Moreover, most docking motifs located in MAPK partners are thought to fit the same consensus: Ψ1-3x3-7ΦxΦ, in which Ψ,Φ and x mark positively charged, hydrophobic or any intervening residues, respectively. What factors then determine the ligand binding space of a given MAPK paralog?
Here, we probed the specificity of linear motifs binding to MAPK docking grooves in vitro to compare the biochemical specificity of these protein-peptide interactions with the biological specificity of MAPKs and their signaling partners. We found that classical D-motif docking peptides efficiently distinguished only JNK from ERK and p38, and only motifs containing a reversed N-to-C-terminal consensus sequence compared to classical D-motifs could discriminate between the latter two MAPKs. Thus, peptides interacting with the MAPK docking groove conformed to two distinct consensus motif sequences (classical D-motifs and reverse D (revD)-motifs): residues in consensus positions interacted with the negatively charged common docking groove and with small hydrophobic pockets. However, the nature of the direct contacts observed in various MAPK-docking peptide complex structures did not explain MAPK specificity. We found that linear motif regions in-between consensus motif positions determine the binding mode in the docking groove and thus the MAPK binding profile of a given docking motif. Thus linear motifs with different intervening region lengths and distinct compositions are specific and simple protein-protein interaction tools. In many instances, they underlie or at least greatly contribute to the in vivo signaling logic between MAPKs and their partners. At the structural level, MAPK docking is a well characterized protein-peptide interaction system (9, 12-16). Due to the lack of systematic and comparative studies, the structural basis of MAPK docking specificity is still unknown. This is in contrast to the systematically studied interactions between SH2 domains and linear motifs for which a coherent structural model of SH2 domain binding specificity has been suggested (17-18). Here, we present a coherent structural model for MAPK-docking motif specificity that explains how docking motifs can be specific or promiscuous and why previously characterized docking peptides bind to MAPKs with their reported specificity.
Results
MAPK binding specificity of classical D-motifs
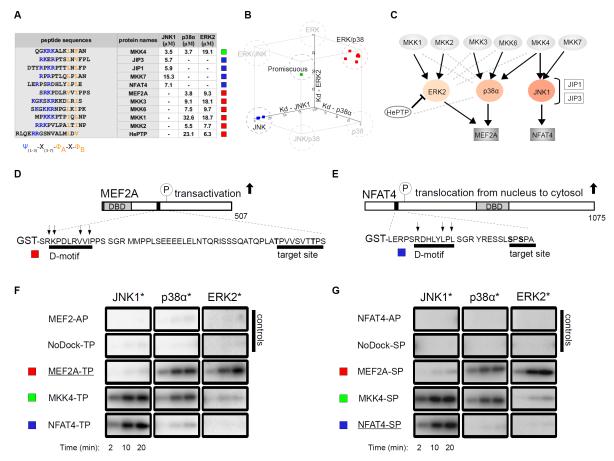
D-motifs play an important role in MAPK signaling networks by facilitating binding between specific MAPKs and their partner proteins (16, 19-21). To quantitatively analyze the MAPK binding properties of these linear motifs, we determined the binding affinities of 11 chemically synthesized D-motif peptides to three MAPKs (ERK2, p38α and JNK1) (Fig. 1A). (Note that chemically synthesized peptides used in this study are indicated with a “pep” prefix throughout the text so that to distinguish them from the full-length MAPK partner proteins in which they occur.) We used reporter peptides from the phosphatase HePTP (hematopoietic protein tyrosine phosphatase) and the transcription factors MEF2A (myocyte enhancer factor 2A) and NFAT4 (nuclear factor of activated T cells 4) to monitor the binding of ERK2, p38α and JNK1 to peptides in fluorescence polarization (FP) based measurements, respectively (fig. S1). Docking motifs from these three proteins bind the MAPK docking groove (9,12,22).
Figure 1. Biochemical specificity of classical D-motifs.
(A) Binding affinities of chemically synthesized D-motif peptides with JNK1, p38α and ERK2. “-” indicates no detectable binding, Kd > 100 μM; N=3 experiments. (B) These binding affinities are plotted on a three-dimensional scatter plot in which axes represent dissociation constants (Kd) for the three different MAPKs and squares correspond to individual peptides listed in the table on the left. (C) The in vivo wiring diagram for MAPKs with their partners. Black connections indicate physiologically relevant links; gray dashed lines indicate binding that does not concur to physiologically relevant connections. (D-G) Docking motifs govern the phosphorylation of critical transcription factor regions in MEF2A (D) and NFAT4 (E) by MAPKs. A representative set of phosphor imaging results of SDS-PAGE gels are shown from at least two in vitro kinase experiments using activated (*) MAPKs to phosphorylate control, “wild-type” or D-motif MEF2A (F) and NFAT4 (G) chimera reporters. AP constructs: Thr or Ser residues in target S/TP site were mutated to alanines; NoDock constructs: the basic and ΦxΦ motif residues, indicated by arrows, were mutated to alanines; DBD, DNA-binding domain.
We grouped peptides into three distinct sets based on their binding affinities: (i) promiscuous peptides bound with biologically relevant affinities (0-100 μM) to all three examined MAPKs; (ii) JNK-specific peptides bound only to JNK1; and (iii) p38/ERK selective peptides bound to p38α and ERK2 with similar affinities (Fig. 1B). Because several peptides showed a more promiscuous binding pattern than expected, the MAPK binding profiles determined in this way showed only a limited agreement with in vivo MAPK signaling network diagrams (Fig. 1C). Peptides derived from JNK partners (MKK7, JIP1, JIP3 and NFAT4) were confirmed to be JNK-specific. Because the MAPK kinase (MAP2K) MKK4 participates in both JNK and p38 pathways (23), a peptide derived from MKK4 (pepMKK4) bound to JNK1 and p38α with similar affinities, as expected. Unexpectedly, D-motif peptides from proteins with documented interactions with either p38α or ERK2 could bind both MAPKs and had modest (less than four-fold) discrimination factors, which is the ratio of binding affinities for cognate compared to non-cognate MAPKs.
Many MAPK docking motifs have been previously characterized in transcription factors whose activity is controlled by MAPK phosphorylation (20). The regulatory phosphorylation sites in transcription factors are often found in unstructured regions outside of the DNA-binding domains. Phosphorylation of two residues in MEF2A (Thr312 and Thr319) by p38α and presumably ERK2 stimulates MEF2A-mediated transactivation in cells, whereas phosphorylation of Ser163 and Ser165 in NFAT4 by JNK promotes its translocation from the nucleus to the cytosol (22,24). To assess the extent to which the biochemical interaction specificity of D-motifs controls the phosphorylation of associated transcription factors, we designed phosphorylation reporters for MEF2A (Fig. 1D) and NFAT4 (Fig. 1E). Indeed, phosphorylation of these transcription factor based reporters in vitro corresponded with the MAPK binding specificity of their D-motifs (Fig. 1F,G).
The inability of pepMKK6 to discriminate between ERK2 and p38α prompted us to explore the role of D-motifs in MKK6-mediated p38α activation. First, we assessed to the extent to which linear motifs contribute to the binding affinity of the full-length protein. Surface plasmon resonance-based measurements of the binding affinity for the MKK6-p38α interaction suggested that the formation of this MAP2K-MAPK complex was quantitatively determined by the linear motif in the unstructured N-terminal region of MKK6 (fig. S2A-E). The importance of the docking motif in full-length protein constructs was also confirmed by in vitro kinase assays monitoring MAP2K-mediated MAPK activation in which a form of MKK6 lacking a docking motif could not phosphorylate its MAPK substrate (fig. S2F-H). These experiments also showed that MKK6 could phosphorylate p38α but not ERK2. This suggested that additional mechanisms apart from the specificity of D-motifs were also involved in governing MAP2K-MAPK signaling. An important factor is the specificity of the MAP2K active site because the sequence of MAPK activation loops adjacent to the phosphorylated threonine and tyrosine residues are strictly conserved within specific MAPK groups but differ among MAPK paralog groups. Indeed, when the amino acid located between the two MAP2K target residues in the p38α activation loop was changed to that found in the corresponding position in the ERK2 activation loop, MAPK phosphorylation by the MAP2K kinase domain was impaired (fig. S2I-K).
Thus, our in vitro MAPK activation results suggest that D-motif binding from non-cognate MAP2Ks may not necessarily result in illicit MAPK activation: Kinase catalytic domains may cooperate with independent linear binding motifs to formulate the specificity of a given signaling event. In summary, docking motifs facilitate signaling in MAP2K-mediated MAPK activation, although specificity here is also influenced by other factors. However, specificity of docking motifs may fully govern the phosphorylation of critical, presumably unstructured regions in transcription factors by promiscuous MAPK enzymes.
MAPK binding specificity of MAPKAPK docking peptides
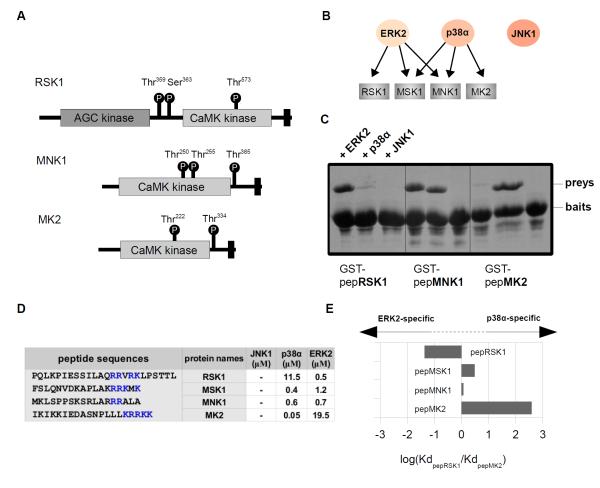
MAPKAPKs are MAPK activated protein kinases that are comprised of a conserved calcium/calmodulin-dependent (CaMK)-type protein kinase domain and a C-terminal extension that enables activation by various MAPKs (ERK, p38 or both) (25). Furthermore, RSKs also possess an additional N-terminal AGC-type kinase domain regulated by its C-terminal CaMK-type protein kinase domain (Fig. 2A) (25). MAPKAPKs form specific complexes with their activating enzyme through short (<20 amino acid) C-terminal docking motifs (25). This region mediates binding to the MAPK docking groove located on ERK or p38 and this association is indispensable for MAPKAPK activation in cells (25). MAPKAPK docking motifs, however, do not bind and are not activated by JNKs. Mammals have the stress-responsive MAPKAPK2, 3, and 5; MNK1 and 2 and MSK1 and 2 which mediate both stress and mitogenic stimuli, and RSK1 to 4 which primarily mediate mitotic and growth responses. The activity of prototypic MAPKAPKs depends upon p38 MAPKs (26). The two MNKs are substrates both for p38 and for ERK (27). MSKs, similarly to MNKs, can also be activated by both MAPKs whereas RSKs are generally thought to be the substrate of ERKs (28) (Fig. 2B).
Figure 2. MAPK binding specificity of MAPKAPK docking regions.
(A) Domain architecture of MAPKAPKs. MAPK docking regions and phosphorylation sites on MAPKAPKs that are important for regulation of MAPKAPK activity regulation in cells are indicated with black rectangles and circles, respectively. (B) The in vivo MAPK interaction profile of the C-terminal MAPK docking regions in select MAPKAPKs. (C) Results of the GST pull down experiments (N=2) are shown on Coomassie stained SDS-PAGE gels. (D) Results of binding affinity measurements with chemically synthesized peptides are shown in table at the bottom. “-” indicates no detectable binding or Kd > 100 μM. N=3 experiments. (E) Discrimination factors of peptides were calculated by taking the logarithm of the ratio of binding affinities for ERK2 and p38α and were plotted in a bar graph.
MAPKAPKs were previously suggested to contain D-motifs that are indispensable for their activation by MAPKs (29). The MAPK interaction region in MAPKAPKs is located in their C-terminal region next to their (CaMK)-type protein kinase domain and this region determines the specificity of MAPK-mediated MAPKAPK activation (30). Therefore we analyzed the MAPK-binding specificity of the C-terminal regions of various MAPKAPKs: pepRSK1, pepMNK1, pepMSK1 and pepMK2. GST pull-down results indicate (Fig. 2C), and quantitative fluorescence polarization measurements confirm (Fig. 2D) that the binding of C-terminal MAPKAPK peptides to MAPKs corresponded with the specificity of MAPK-mediated MAPKAPK activation. Peptides that were expected to bind both ERK2 and p38α bound to these kinases with similar affinities (for example, pepMSK1 or pepMNK1) in the same assay that was used for D-motif peptide measurements (fig. S1B). PepRSK1 and pepMK2, which reside in ERK and p38-specific MAPKAPKs, respectively, show a more than tenfold preference (~20 and 400, respectively) for their cognate MAPK (Fig. 2E). Thus, although they do not have the classical D-motif consensus, MAPKAPK docking peptides efficiently discriminated between ERK2 and p38α docking grooves.
Distinct linear motif binding modes in the MAPK docking groove
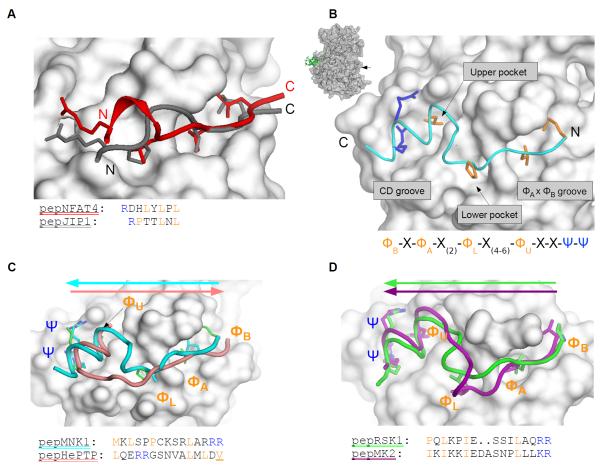
To gain insight into the apparent specificity of three different MAPK docking grooves, we solved the crystal structures of ERK2, p38α and JNK1 in complex with peptides mediating MAPK binding with different degree of specificity. Because the crystal structure of a MAPK-docking peptide complex with the p38α-specific pepMK2 peptide has been determined (31), we focused on complexes with peptides that show JNK-specific, ERK2/p38α-selective and ERK2-specific binding and determined the crystal structure of JNK-pepNFAT4, p38α-pepMKK6, ERK2-pepMNK1 and ERK2-pepRSK1 complexes (table 1 and fig. S3).
Table 1.
Data collection and refinement statistics for MAPK-docking peptide complexes
| JNK1- pepNFAT4 |
p38α- pepMKK6 |
ERK2- pepMNK1 |
ERK2- pepRSK1 |
ERK2- pepSynth-revD |
|
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21 | P3121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) | 46.23, 94.13, 47.84 |
81.92, 81.92, 122.59 |
65.36, 65.88, 95.05 |
41.70, 58.98, 155.73 |
45.03, 65.66, 117.23 |
| α, β, γ(°) | 90.00, 110.74, 90.00 |
90.00, 90.00, 120.00 |
90.00, 90.00, 90.00 |
90.00, 90.00, 90.00 |
90.0, 90.0, 90.0 |
| Resolution (Å) | 1.33-47.07 (1.33-1.40) |
1.95-70.95 (1.95-2.00) |
1.55-53.86 (1.55-1.63) |
2.40-77.865 (2.40-2.47) |
2.10-57.28 (2.10-2.15) |
| Rmerge | 0.03 (0.68) | 0.08 (0.80) | 0.07 (0.65) | 0.095 (0.84) | 0.12 (0.55) |
| I / σI | 17.45 (1.98) | 13.18 (2.04) | 15.82 (1.92) | 18.06 (2.97) | 10.75 (3.77) |
| Completeness (%) | 99.2 (99.3) | 100.0 (100.0) | 98.6 (91.4) | 96.8 (99.0) | 88.1 (90.4) |
| Redundancy | 3.7 (3.6) | 4.9 (4.9) | 5.3 (3.3) | 9.92 (9.83) | 4.5 (4.2) |
| Refinement | |||||
| Resolution (Å) | 1.33-25.69 | 1.95-46.38 | 1.55-53.86 | 2.40-47.02 | 2.10-42.04 |
| No. reflections | 86986 | 35282 | 59398 | 15190 | 18343 |
| Rwork / Rfree | 0.165/0.186 | 0.176/0.209 | 0.155/0.177 | 0.1969/0.2351 | 0.1801/0.2244 |
| No. atoms | |||||
| Protein | 2857 | 2761 | 2919 | 2726 | 2864 |
| Peptide | 100 | 56 | 136 | 133 | 112 |
| Ligands | 39 | - | 31 | 31 | 31 |
| Water | 422 | 293 | 546 | 115 | 200 |
| B-factors | |||||
| Protein | 30.00 | 20.75 | 19.82 | 55.7 | 23.2 |
| Peptide | 32.88 | 38.25 | 28.22 | 81.1 | 48.9 |
| Ligands | 28.93 | - | 10.74 | 84.5 | 20.5 |
| Water | 40.95 | 26.62 | 30.37 | 47.9 | 32.3 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.011 | 0.010 | 0.009 | 0.005 | 0.003 |
| Bond angles (°) | 1.357 | 1.145 | 1.327 | 1.171 | 0.842 |
Values in parentheses are for highest-resolution shell.
To examine how JNK-specific peptides bind to the JNK-docking groove, we compared our JNK1-pepNFAT4 crystal structure to the JNK1-pepJIP1 complex (13) (Fig. 3A). The two docking peptides adopted different main chain conformations between the contact points corresponding to the D-motif consensus sequence. Thus, these structures displayed two unrelated solutions to bind to the JNK docking groove: pepNFAT4 contacted the CD groove with an arginine side-chain from a short α-helical region, which is not present in pepJIP1, and its arginine side chain directly extended from a proline residue located in the lower pocket.
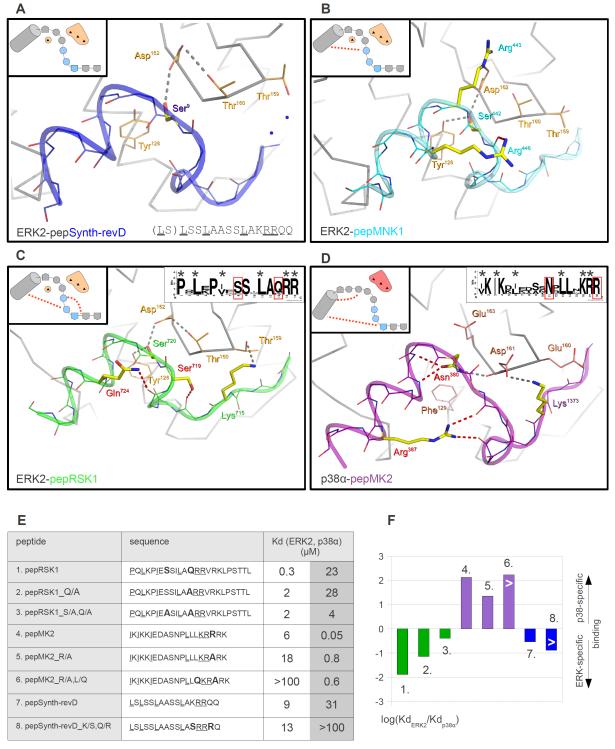
Figure 3. MAPK docking grooves are compatible with different linear motif binding modes.
(A) Overlay of the JNK1-pepNFAT4 and the JNK1-pepJIP1 (PDB ID: 1UKH) crystal structures (13). (B) Crystal structure of the ERK2-pepMNK1 complex. The MAPK docking groove is located on the opposite side of the kinase relative to the active site (arrow in the inset). N and C, N- and C-termini of the peptide, respectively. (C) Comparison of the ERK2-pepMNK1 complex with the ERK2-pepHePTP complex (PDB ID: 2GPH) (12). Colored arrows indicate the N→C-terminal direction of the docking peptides. The last hydrophobic contact point in the pepHePTP D-motif consensus is only inferred because this residue was modified and the slight mis-alignment at this region is likely to be the result of the forced covalent attachment of pepHePTP to ERK2 through an artificial disulfide bond. (D) Superposition of the ERK2-pepRSK1 complex with the p38α-pepMK2 (PDB ID: 2OKR) crystal structure (30). (C) and (D) show the ERK2 or the p38α surface from the ERK2-pepMNK1 and p38α-pepMK2 crystal structures, respectively. Non-cognate protein-peptide pairs do not show any clash, indicating that surface complementarity alone does not explain the selective formation of ERK2-pepRSK1 and p38α-pepMK2 complexes. New structures shown on this figure are JNK1-pepNFAT4, ERK2-pepMNK1 and ERK2-pepRSK1.
These structures also revealed how some classical D-motifs with a highly positively charged N-terminal cluster bind to the CD groove. Similarly to the previously determined structures of the p38α-pepMEF2A and p38α-pepMKK3 complexes, the N-terminal region of the MKK6 D-motif cannot be seen on the electron density map of the pepMKK6-p38α complex (9). This suggests that the positively charged residues may not need to adopt a single conformation when bound in the negatively charged CD groove (fig. S3).
Four peptides derived from MAPKAPK substrates did not contain the classical D-motif consensus because their basic residues are located on their C-termini. Nevertheless, these peptides competed with D-peptide reporters in binding assays, suggesting that they also bound to the same MAPK docking groove (fig. S1B). To explore the binding mode of these motifs, we determined the crystal structure of the ERK2-pepRSK1 and the ERK2-pepMNK1 complex. These peptides bound in a reversed N- to C-terminal orientation compared to D-motif peptides (Fig. 3B-D), similar to the binding of pepMK2 to p38α (31). Based on the structures of these MAPK-docking peptide complexes, the MAPK docking groove can be divided up into the hydrophobic ΦxΦ groove, lower and upper hydrophobic pockets, and the negatively charged CD groove (Fig. 3B). These are all contacted by residues that define the MAPK docking MAPKAPK consensus or “reverse” D-motif (revD-motif): ΦBxΦAx2ΦLx4-6ΦUxxΨΨ; in which U, L, A and B denote residues binding into the upper or lower pockets, or in the A or B position of the ΦxΦ groove; Φ,Ψ and x denote hydrophobic, basic, or any amino acids, respectively. Amino acids that form the consensus revD-motif contact the MAPK docking groove similarly; however, the intervening regions between ΦU and ΦA differ in the two peptides.
Structural basis of binding specificity of JNK compared to ERK/p38 and ERK2 compared to p38α
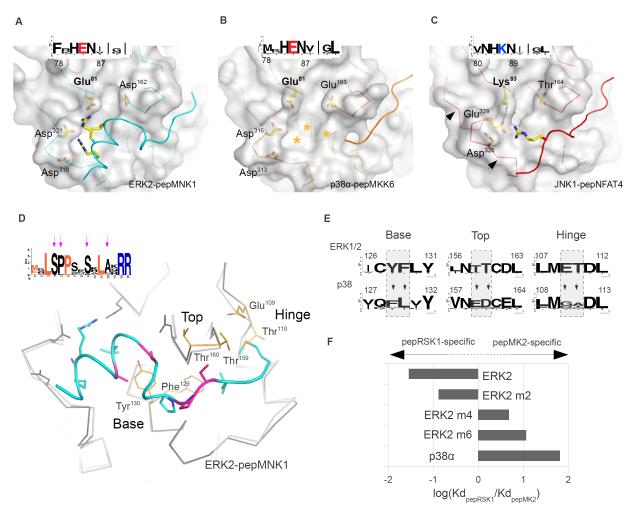
The docking grooves in JNK1, p38α and ERK2 are comprised of the negatively charged CD groove and some small hydrophobic pockets. Overall, the three MAPK docking grooves appear similar; however, the CD groove lies closer to the hydrophobic pockets in JNK1 compared to those in ERK2 (Fig. 4A) or p38α (Fig. 4B) because JNK1 has a positively charged residue (Lys83) where p38α and ERK2 have a negatively charged residue (Glu) in the corresponding position, resulting in a JNK-specific charge-inversion (Fig. 4C). Because of this charge inversion, a salt bridge forms between Lys83 and Glu329 in JNK1. In addition to Lys83, another charged residue in ERK or p38 MAPKs is replaced by an evolutionarily conserved small polar residue (Thr164) in JNK MAPKs. Therefore the JNK docking groove is less charged and narrower and these evolutionarily conserved differences in surface topography and charge distribution sets JNK MAPKs apart from ERK and p38 enzymes. These differences are exploited by JNK-specific peptides: The intervening region between the lower pocket and the CD groove interacting amino acids in JIP1 and NFAT4 D-motifs are shorter compared to those in MKK6- and HePTP-type D-motifs. Furthermore, revD-motifs with their longer, conformationally well-defined intervening regions were not compatible with the narrower CD groove in JNK.
Figure 4. Topography of paralogous MAPK docking grooves.
(A-C) Structural basis for the ability of JNK to discriminate between certain D-motifs and RevD-motifsPeptide-bound docking surfaces of ERK2 (A), p38α (B) and JNK1 (C) are presented in the same orientation. Arrows on (C) indicate the critical region in JNK that determines the width of the CD groove, which in turn also influences the size of the upper pocket. The insets show the sequence logo of the corresponding regions in MAPK homologs from sponges to human. Stars denote the tentative position of the pepMKK6 basic region in the p38α CD groove. (D) Structural determinants of p38-ERK discrimination. Arrows indicate conserved amino acids in the pepMNK1 sequence logo that are not part of the revD-motif consensus, which are colored in magenta on the human MAPK:revD-motif complex structure below. Superimposed p38α is colored in light gray and shown as a Cα-trace. (E) The evolutionarily conserved nature of the base, top and hinge is shown for ERK1/2 and p38 homologs from sponges to human from the KinBase database (73) as sequence logos. (F) The impact of amino acid swaps in the ERK2 docking groove on pepRSK1 or pepMK2 binding is shown as a bar diagram. Amino acid swaps were done in the base (m2), in the base and the top (m4) and in all three regions (m6) of ERK2. New structures shown on this figure are p38α-pepMKK6, JNK1-pepNFAT4 and ERK2-pepMNK1.
MAPKAPK-type revD-motif containing peptides may have a clear binding preference towards either ERK or p38. Unfortunately, a direct comparison of MAPK:revD-motif complexes does not reveal how they can efficiently discriminate between the ERK2 and p38α docking grooves (see Fig. 3D). However, sequence conservation analysis of MAPKAPK revD-motifs showed that some amino acids located in the intervening regions of these motifs are conserved similarly to amino acids that form the revD-motif consensus. The pepMNK1 sequence logo from various species (including C. elegans and human) showed that various amino acid positions besides the revD-motif consensus also show a high degree of conservation (Fig. 4D). Moreover, these conserved positions change in location as well as in amino acid identity between ERK/p38-selective, ERK- and p38-specific groups (Fig 4D). Conversely, sequence analysis and comparison of the ERK2-pepRSK1 and p38α-pepMK2 crystal structures showed that MAPKs harbor evolutionarily conserved differences in key positions of their MAPK docking grooves. MAPK docking grooves have three structural features (base, top and hinge, the latter of which is a flexible connector between the N-terminal and C-terminal kinase lobes), which can be used by docking motifs to discriminate between ERK1/2 and p38. Although the Cα positions of the two proteins superimpose well, some docking groove features in these critical regions differed (Fig. 4E). An ERK2 mutant (ERK2 m6) in which the amino acids at six key positions were changed to those in the corresponding positions in p38α had a ligand discrimination factor for the two most specific ligands (pepRSK1 and pepMK2) resembling that of p38α (Fig. 4F and fig. S4A). Conversely, the p38α m6 chimera with an ERK2-like docking groove was less specific for pepMK2 compared to p38α (fig. S4A). These data suggest that intervening regions of MAPKAPK revD-motifs have adapted to cognate MAPK docking grooves.
H-bond staples in MAPK:revD-motif binding specificity
An artificially designed revD-peptide (pepSynth-revD) bound to both ERK2 and p38α (but not to JNK1) with similar affinities [KdERK2~10 μM, Kdp38α~30 μM]. We also solved the crystal structure of this weakly binding nonspecific peptide and compared the structures of all four known MAPK:revD-motif complexes (Fig. 5A-D). The comparison of less specific peptides with ERK2- or p38α-specific peptides suggested the importance of intra-peptide, sequence specific hydrogen (H)-bonds in MAPK specific binding: Specific peptides tended to contain more of these H-bond “staples”. The artificially designed pepSynth-revD peptide with an arbitrary intervening region did not have H-bond staples; the non-discriminatory pepMNK1 contains one H-bond staple, whereas at least two specific residues in pepRSK1 and pepMK2 are involved in H-bond stapling. Furthermore, these latter residues (Ser719 and Gln724 in RSK1, and Asn380 and Arg387 in MK2) are evolutionarily conserved in most vertebrate MAPKAPK homologs (Fig. 5C-D). Because H-bond stapling amino acids do not necessarily contact the MAPK docking groove directly, they may play an indirect role in MAPK-specific binding by fine-tuning the conformation of the intervening region between anchor points. This could be important for specificity because H-bond stapled intervening docking motif regions are adjacent to evolutionarily highly conserved MAPK docking features such as the top and the base, and the divergent nature of these residues between the ERK and p38 group of MAPKs appear to be particularly important (Fig. 4D,E). Although all three MAPKAPK docking motifs display an alpha-helical conformation in their CD groove binding region or a 310 helical conformation (where the N-H group of an amino acid forms a hydrogen bond with the C=O group of the amino acid three residues earlier) in the intervening region between ΦL and ΦU positions when bound to MAPKs, the C-terminus of MAPKAPKs is unstructured in all of their protein crystal structures (32-33). Because MAPKAPK C-terminal regions contain the revD-motifs and revD-peptides in complex with MAPKs are clearly structured, it is likely that revD-motif containing MAPKAPK regions go through a disorder-to-order transition, which may be assisted both by features located on the MAPK docking groove as well as in the docking motif. Thus, our data suggest that flexible linear motifs may exploit the differences at the MAPK specificity loop (top) and at the base of the docking groove more efficiently by forming inter-molecular H-bonds or van der Waals interactions when they are conformationally held in place (or stapled) by intra-peptide H-bonds. According to this hypothesis, MAPK-specific peptides that lack H-bond stapling should bind to their cognate MAPKs with lower specificity.
Figure 5. Sequence-specific intra-peptide hydrogen bonds in revD-motifs contribute to their MAPK specificity.
(A-D) Sequence-specific inter-molecular and intra-peptide H-bonds in MAPK:revD-motif crystal structures. Peptide sequence specific inter-molecular H-bonds are gray, whereas sequence specific, intra-peptide H-bonds are red. Selected side-chains of the top region and the base are orange for ERK2 and salmon for p38α. The schematic insets on the upper left corners emphasize the intervening region between two anchor points (ΦL and ΦU; hydrophobic anchor points are shown as pentagons). These also depict the top and the base regions because they mediate important differential inter-molecular H-bonding interactions. Light blue circles indicate peptide amino acids adopting a 310 helical conformation and red dotted lines depict H-bond staples. Small insets in the upper right corners of the last two panels show the sequence logo of RSK1/2 or MK2/3 revD-motifs. The logos were based on ten sequences from five different vertebrate organisms (Danio rerio, Xenopus laevis, Gallus gallus, Mus musculus and Homo sapiens). Evolutionarily conserved amino acids that are involved in H-bond stapling are highlighted in red. New structures are ERK2-pepSynth-revD (A), ERK2-pepMNK1 (B) and ERK2-pepRSK1 (C). (E) Role of H-bond staples in mediating MAPK:revD motif binding specificity. The table shows binding affinities of revD-motifs with different H-bond staples to ERK2 and p38α (left). Amino acids involved in H-bond stapling or replaced by alanines are bolded and revD-motif defining amino acids are underlined. (F) Discrimination factors of peptides were calculated by taking the logarithm of the ratio of binding affinities for ERK2 and p38α and were plotted in a bar graph on the right. Heights for bars indicated with a “>” sign are low estimates because the experimental assay cannot measure binding affinities weaker than 100 μM.
To test this hypothesis, we replaced some of the critical residues involved in H-bond stapling with alanines in pepRSK1 and pepMK2. pepRSK1_Q/A contained a replacement for Gln724; pepRSK1_S/A,Q/A contained replacements for Ser719 and Gln724; and pepMK2_R/A contained a replacement for Arg387; however, Asn380 was not replaced because this residue forms inter-molecular H-bonds as well (Fig. 5E and fig. S4B-D). Discrimination factors calculated from the binding affinities of these mutant peptides with ERK2 and p38α showed that alanine replacements in H-bond stapling positions decreased the peptide’s capacity to distinguish between the similar docking grooves in ERK2 and p38α (Fig. 5F). We also investigated the impact of alanine replacements of key amino acids involved in H-bond stapling in a long revD-motif containing RSK1 protein construct (amino acids 411-735 of RSK1c) (fig. S4C). The specificity of wild-type RSK1c and RSK1c_Q/A,S/A constructs with ERK2 and p38α showed a good agreement with measurements carried out with chemically synthesized RSK1 peptides (fig. S4C,D). These results indicate that H-bond stapling residues are important determinants of MAPK specificity not only for chemically synthesized peptides but also for full-length MAPK protein partners.
We also attempted to build new H-bond staples in revD-motif peptides to further assess their role in MAPK binding specificity (Fig. 5E,F and fig. S4E,F). The affinity and specificity of pepMK2_R/A for p38α was lower compared to pepMK2 and we replaced a solvent facing leucine (Leu384) with glutamine in this peptide (pepMK2_R/A,L/Q) to rebuild its original p38α specificity (fig. S4E). Based on the MAPK-pepMK2 crystal structure the introduced glutamine in its preferred side-chain conformation could form a similar H-bond with the main-chain of the peptide compared to the formerly removed one, albeit in a different way (fig. S4E). Although the binding affinity of this latter peptide compared to pepMK2_R/A did not increase substantially, its specificity for p38α compared to ERK2 increased to an extent comparable to that of pepMK2. Similarly, when we tried to build an H-bond staple into pepSynth-revD (pepSynth-revD_K/S,Q/R) the binding specificity for ERK2 was increased largely because of decreased p38α binding (fig. S4F). Overall, these results are consistent with a model in which H-bond staples in the ERK2-pepRSK1 and p38α-pepMK2 complex crystal structures contribute to their MAPK-specific binding.
Designing peptides that bind to MAPK docking grooves
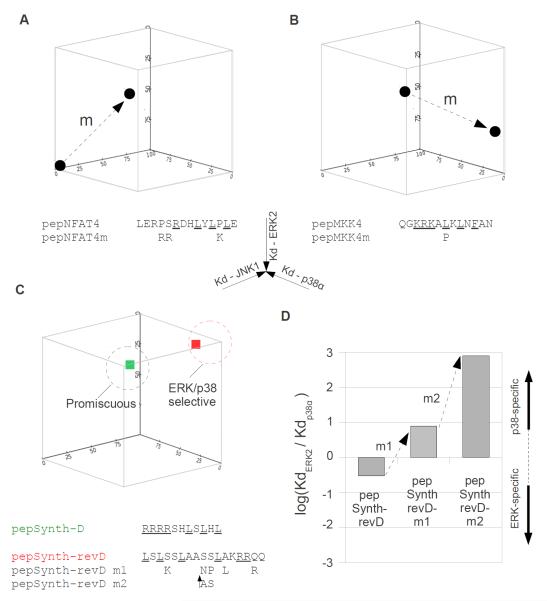
After elucidating the structural basis underlying biochemical specificity for MAPK-docking motif binding, we successfully altered the MAPK binding profiles of naturally occurring motifs and designed novel peptides with tailored MAPK specificity (Fig. 6). Our earlier analysis showed that pepNFAT4 bound JNK1 exclusively, whereas pepMKK4 bound all three tested MAPK paralogs. We used these two peptides as starting points to convert a promiscuous motif into a specific one and vice versa by exploiting the structural and evolutionary sequence conservation information presented above. We changed pepNFAT4, which originally contained a JNK1-specific D-motif motif, to facilitate its binding to ERK/p38 MAPKs as well. The new peptide (pepNFAT4m) had a longer basic motif region to optimize interaction in the wider CD grooves in ERK and p38 and it also contained an additional positively charged residue that could facilitate binding to the top region in ERK2/p38α. This rationally modified peptide indeed behaved in a more promiscuous manner (Fig. 6A and fig. S5A). We could also make pepMKK4 less promiscuous and shift its dual capacity to bind p38α and JNK1 to favor the former MAPK by making the sequence of pepMKK4m more similar to that of pepMKK6. pepMKK6 contains a proline in ΦL and the leucine-to-proline modification in ΦL of pepMKK4 limited its binding to JNK1 to a greater extent than its binding to p38α because this change efficiently interfered with the capacity of pepMKK4 to adopt the JNK1-specific pepNFAT4-type binding mode (Fig. 6B and fig. S5B). Because the ability of pepNFAT4 to bind specifically to JNK1 depended on the formation of a small helical turn at the intervening region between ΦL and Ψ, a proline at this region disfavors an alpha-helical main-chain conformation required to span the distance between ΦL and Ψ in a manner required for the JNK-specific pepNFAT4-type binding mode. In summary, pepNFAT4m became promiscuous presumably because it acquired the pepMKK6-type binding mode in addition to the original JNK1-specific binding mode of pepNFAT4. Conversely, pepMKK4m became p38α-specific because the introduced proline limited the dual capacity of pepMKK4 to bind in a JNK1-specific pepNFAT4-type mode more than its capacity to adopt the p38α compatible pepMKK6-type binding mode.
Figure 6. Modification and design of MAPK-docking motif interaction profiles.
(A-B) Manipulation of MAPK interaction profiles for natural D-motif peptides. The JNK-specific docking peptide of NFAT4 was made more promiscuous (A) and the promiscuous motif found in MKK4 was made more selective (B) with appropriate mutations. Binding affinities of peptides are plotted on a three-dimensional scatter plot where axes represent dissociation constants (Kd) for JNK1, p38α and ERK2. Arrows (m) indicate the impact of amino acid replacements in the MAPK ligand space. Amino acid replacements are indicated below the original peptide sequence. Amino acids in consensus motif defining positions are underlined. (C-D) Design of artificial motifs with tailored MAPK specificity profiles. Peptides containing a minimal D-motif (pepSynth-D) or a revD-motif (pepSynth-revD) are promiscuous or ERK/p38 selective, respectively. Amino acid replacements in the ERK/p38 selective pepSynth-revD are shown under the original peptide sequence (C), and their effect on the discrimination factor for ERK2 compared to p38α binding is shown in the bar graph (D). ↑AS indicates a two amino acid insertion of alanine and serine residues.
Next, we tested the MAPK interaction profile of artificially designed peptides conforming to a classical D-motif consensus (pepSynth-D) or to a classical revD-motif consensus (pepSynth-revD) (Fig. 6C). pepSynth-D contained four arginines as the basic motif to allow unrestricted binding to the CD grooves in MAPKs, three leucines to bind to the lower pocket and the ΦxΦ groove, and serines and histidines in the intervening region positions. pepSynth-revD was comprised of leucines for all hydrophobic positions (ΦB, ΦA, ΦL, and ΦU), two arginines as the basic motif (Ψ), amino acids with large polar groups (lysine or glutamine) at the C-terminus to facilitate α-helix formation at this region, and serine or alanine residues in the intervening regions. Both peptides bound MAPKs with low-to-medium micromolar affinities (1-30 μM) and their MAPK interaction profiles fit well to what we predicted: pepSynth-D was promiscuous, whereas pepSynth-revD was ERK/p38-selective (fig. S5C,D).
pepSynth-revD could not bind to JNK1, presumably because its helical structure around the basic motif was not compatible with the narrower CD groove in JNK1. This artificially designed peptide bound to p38α and ERK2 with modest binding affinities (~30 and 10 μM, respectively) compared to other naturally occurring revD-motifs derived from MAPKAPKs, which normally bind with sub-micromolar affinities to their cognate MAPK. Our earlier observations suggested that the sequence of the intervening region, in addition to the residues located in consensus defining positions, was also important for MAPK-specific binding. Therefore, we changed the intervening region of pepSynth-revD according to an analysis of revD-motifs from MK proteins (Fig. 6C). Important intervening region features from the MK revD-motif sequence logo were introduced in two steps. First, five serine or alanine amino acids in pepSynth-revD were replaced with corresponding amino acids that show a high degree of sequence conservation in the MK logo (pepSynth-revD m1). This new peptide had sub-micromolar affinity for p38α but its affinity for ERK2 was not changed substantially. Next, we introduced a two amino acid insertion (alanine and serine) into the intervening region between the ΦL and ΦU positions of pepSynth-revD m1 (pepSynth-revD m2), because pepMK2 homologous revD-motifs that preferentially bind p38 homologs have two additional amino acids in this region. This second modified peptide still bound to ERK2 with similar affinity as the original Synth-revD motif containing peptide, but its binding affinity for p38α was increased by ~6,000 fold (from 30 μM to 5 nM) (Fig 6D and fig. S5D). Thus, these modifications of an artificially designed minimal revD-motif which originally bound two MAPK paralogs with comparable affinities resulted in a peptide that could discriminate ERK2 from p38α better than pepMK2. We also tested the specificity of pepSynth-revD m2 in the context of a full length protein. JIP1 is a scaffold protein of the JNK MAPK module and recruits JNK through its D-motif (pepJIP1) (34). We replaced the JNK1 specific D-motif (pepJIP1) in JIP1 with the sequence of pepSynth-revD m2 and tested MAPK binding of this chimeric protein in GST pull-down assays using lysates from HEK293T cells that were transiently transfected with ERK2 or p38α constructs (fig. S5E). In contrast to JIP1, the JIP1_pepSynth-revD m2 chimera bound only p38α. This result demonstrated that the binding specificity of small docking peptides may correspond to the binding specificity of full length proteins as well.
Discussion
MAPKs represent a paradigm for understanding how specificity in cellular signaling can be achieved with enzymes that share a common origin (11). Our first analysis of fifteen motifs readily identified peptides that could discriminate three major human MAPK paralogs. However, we unexpectedly found that peptides from MAPK-specific protein partners often behaved less specifically in binding assays. D-motifs from the initial set that bound p38α also bound ERK2, which presumably reflected the highly similar groove topography in these two paralogs. This latter observation highlights the limitations of simple D-motif mediated interactions, although these may be efficiently mitigated in the context of other binary interactions also contributing to MAPK specificity. For example, activation of p38α by MKK6 also depends on the substrate preference of the MKK6 active site even if the MKK6 D-motif autonomously appears to determine the binding affinity between these two proteins. Moreover, the existence of FxFP (Phe-any amino acid-Phe-Pro) motifs in certain MAPK (especially ERK) substrates and a separate docking groove for such motifs on ERK suggest that in some cases two docking grooves on ERK may operate in combination: Elk1 for example contains both an FxFP and a D-motif (35-37). (Note that FxFP motifs are unrelated to D- or revD-motifs.) Efficient phosphorylation of certain ERK/p38 selective MAPK target sites in cells in an ERK- or p38-specific manner might require additional MAPK recruitment events (38).
It appears that MKK4 is unique in its ability to interact both with the highly divergent docking grooves in JNK and ERK/p38, which agrees with its biological function, which is to activate JNK and p38. Unfortunately, our efforts to crystallize MAPK complexes with this peptide have failed so far but the intervening region composition of pepMKK4 is predicted to be compatible with two unrelated binding modes. It bound JNK1 in a similar manner to pepNFAT4 and p38α in a similar manner to pepMKK6. (The weakened but still detectable binding of this peptide to ERK2 was presumably the consequence of the less specific pepMKK6-type binding mode.)
We unexpectedly found that side-chain specific intra-peptide H-bonds contribute to the affinity and to the specificity of MAPK-linear motif interactions. MAPK specificity was decreased when H-bond staples were removed in MAPKAPK revD-motifs by alanine replacements and increased when they were introduced at new positions. Mechanistically, H-bond staples are likely to function by making flexible docking peptides more rigid upon MAPK binding at their intervening region. This could favor binding to cognate MAPKs but H-bond staples may also prevent a peptide from adopting a different conformation compatible with binding to non-cognate MAPKs. H-bond staples, however, are not unique to MAPKAPK docking regions bound to MAPKs and are also used in other linear motif based interaction systems such as those operating in TGFβ signaling (the TGFβ effector SMAD3 interacts with a linear motif in the adaptor SARA) or in DNA repair regulation (the DNA recombinase RAD51 interacts with a short peptide segment from the scaffold protein BRCA2 ) (39-41) (Fig. S6).
The low sequence stringency in linear motif definitions normally hinders an accurate assessment of their frequency in various proteomes. Unfortunately, our current understanding of these interactions is rarely sufficient to segregate motifs into sub-groups representing different binding modes of the same global motif definition. Detailed understanding of the subclasses of linear motifs with more stringent sequence requirements however is likely to promote linear motif discovery at the systems level by decreasing the rate of false positives that may occur by chance. The D-motif consensus sequence was previously loosely defined and it encompassed motif sequences that belonged to different MAPK binding modes and specificity. Our structural and biochemical analysis in combination with other studies clarified the minimal sequence requirements for linear motifs to bind the MAPK docking groove (42-47). We grouped MAPK binding linear motifs into five different classes: pepJIP1, pepNFAT4, pepMKK6, pepHePTP D-motifs and MAPKAPK revD-motif types. A simple motif scan of the human proteome with more stringent consensus sequences representing these five different MAPK-binding classes, after applying a sophisticated filtering and scoring protocol, identified 650 sequence motifs in 595 proteins that match well to known D- or revD-motifs in sequence and in other properties (table S1 and S2). Because of the small number of evolutionarily independent docking motif sequences belonging to a certain motif sub-class, it is likely that additional filtering and scoring protocols will be required to further increase the reliability of this sequence matching approach (for example, by implementing procedures that could also score motif sequence conservation across different organisms or structural compatibility to various MAPK docking grooves). Despite these pitfalls, the lists were statistically significantly enriched in proteins associated with known MAPK signaling functions or with MAPK pathway components. Intervening sequence region information of known MAPK docking motifs representing different docking groove binding modes greatly contributed to this enrichment. This suggests that detailed understanding of the physical nature of these protein-peptide interactions allows the identification of functional linear motifs with greater reliability. This in turn will contribute to the drawing of more extended and more reliable “wiring” diagrams for signaling networks at a system level.
Most of the experiments presented in this work on the MAPK binding specificity of linear motifs were carried out by using chemically synthesized peptides. How relevant are these results to proteins when these fragments function in the contexts of full-length proteins? It was previously shown that the MAPK binding specificity of full length proteins can be altered by swapping motifs that bind different MAPKs in cell based reporter systems (30). In one case the MAPK binding specificity of MAPKAPKs (MK2 and RSK2) was changed from p38 to ERK and vice versa in cells, and another study demonstrated that responsiveness to JNK or p38 in c-JUN and MEF2A could be changed by swapping their docking motifs (48). Similarly, the specificity of ERK2/p38 chimeras had already been tested (19), which showed that the “ED-site” (also known as the MAPK top region) helps to define MAPK-MAPKAPK wiring in cells. In addition, our in vitro docking motif swap experiments using longer fragments from NFAT4 and MEF2A, GST pull down binding experiments with the JIP1_pepSynth-revD m2 chimera and quantitative protein binding assay results with the RSK1c protein construct presented in this study support the notion that binding specificity rules obtained with chemically synthesized peptides can be extended to full-length MAPK partner proteins.
In summary, we showed that the specificity of linear motifs binding to different MAPK paralogs is coded either in the length or in the amino acid composition of intervening regions (or both) of their consensus motifs. Amino acids located in variable intervening regions form intra-motif hydrogen bonds or limit peptide main chain flexibility through prolines (or both). Thus, these residues are mostly responsible for specificity, whereas residues in consensus motif positions bind the general surface features and mostly contribute to these interactions in a non-discriminatory fashion. Because of these properties, docking motifs can stay short (<15-20 aa). They are evolutionarily plastic solutions for forming new connections between signaling proteins because they may freely emerge in unstructured protein regions and their binding specificity for kinases may also be easily changed (49). From an evolutionary standpoint, results of our study help to assess the frequency of MAPK docking groove binding linear motifs in various proteomes (50,51). Moreover, the coherent structural model of MAPK-docking motif binding specificity may enable highly specific interference with various MAPK pathway mediated physiological functions (52). Our results show that small peptides can efficiently discriminate ERK, p38 and JNK docking grooves. The specificity of artificially designed peptides can be fine-tuned and thus they may be used to ablate MAPK-specific signaling in vivo, even if they bind to the docking groove that is common to all MAPKs.
Materials and Methods
Protein expression and purification
Proteins were recombinantly expressed in E. coli Rosetta(DE3) pLysS (Novagen) cells using standard techniques. cDNAs encoding human proteins were produced from mRNA derived from HEK293 cells. Reverse transcription was followed by PCR with protein specific PCR primer pairs. All sequences were verified by DNA sequencing after sub-cloning into pET expression vectors. Recombinant proteins were purified with standard chromatography techniques: the procedure involved a Ni- or GST-affinity chromatography step that was followed by an ion-exchange step. Affinity tags were removed by TEV protease cleavage. Final protein samples were dialyzed into storage buffer containing 20 mM Tris pH=8.0, 100 mM NaCl, 10% glycerol and 2 mM TCEP. All peptides were synthesized on an ABI 431A peptide synthesizer using Fmoc strategy. Activated ERK2, p38α and JNK1 were produced by co-expressing the MAPKs with constitutively active GST-tagged MAP2Ks in E. coli using bi-cistronic modified pET vectors. Dephosphorylated MAPKs were produced by co-expressing the MAPKs with GST-tagged λ phage phosphatase. Activated MAPKs were purified and treated similarly as described above for dephosphorylated MAPKs. The activity of MAPK samples were then characterized in in vitro kinase assays using MBP (myelin basic protein) as the substrate. Double phosphorylation of the activation loop of MAPKs was confirmed in Western blots using anti-phospho MAPK specific antibodies: ERK2, 9101S; p38α, 9215S; and JNK1, 9251S (Cell Signaling). RSK1c, full length JIP1 and constitutively active forms of MKK1 and MKK6 were expressed as N-terminal GST-fusion proteins with C-terminal His-tag, double-affinity purified, then used in binding affinity measurements, GST pull downs or in vitro kinase assays. ERK2 and p38α cDNAs were cloned into a modified pcDNA 3.1 vector which adds a C-terminal FLAG tag to the inserted protein. HEK293T cells were transiently transfected with expression vectors by lipofectamine (Invitrogen), harvested and lysed 48 hours after transfection in Triton lysis buffer (1% Triton, 25mM Tris pH 7.4, 150mM NaCl, 2.5mM DTT, 2mM EDTA + protease inhibitor mix).
Protein-protein binding assays
For fluorescence polarization (FP) based binding affinity measurements reporter peptides were labeled at the N-terminus with carboxy-fluorescein (CF) or tetramethylrhodamine (TAMRA) fluorescent dyes (CF-pepHePTP: RLQERRGSNVALMLDV; TAMRA-pepMEF2A: SRKPDLRVVIPPS and TAMRA-pepNFAT4: LERPSRDHLYLPLE) by Fmoc chemistry. An increase in the FP signal, which indicated complex formation between the MAPK and the labeled peptide, was monitored as a function of increasing concentration of purified MAPKs with an Analyst GT (Molecular Devices) plate reader in 384-well plates. The labeled peptide was used at 10 nM in 20 mM Tris pH=8.0, 100 mM NaCl, 0.05% Brij35P, 2 mM DTT. The resulting binding isotherms were fit to a quadratic binding equation with the OriginPro7 software (OriginLab Corporation). The affinities of the unlabeled peptides to MAPKs were measured in steady-state competition experiments: 10 nM of labeled reporter peptide was mixed with MAPK samples at a concentration to achieve ~60-80% complex formation. Subsequently, increasing amount of unlabeled peptide was added and the FP signal was measured as described earlier for direct titration experiments. The Kd for each MAPK-unlabeled peptide interaction was determined by fitting the data to a competition binding equation. Titration experiments were carried out in triplicates and the average FP signal was used for fitting the data with OriginPro7. (Note that the MAPK interaction profiles determined in this way are based on Ki values because the competition assay measures the extent to which the tested peptide inhibits the binding of reporter peptides.) For GST pull-down experiments GST beads were equilibrated with binding buffer (20mM Tris, 100 mM NaCl, 0.1% IGEPAL, 2mM β-mercaptoethanol). 10 μg immobilized GST-fusion peptides or GST-fusion proteins (and GST protein as negative control) were incubated with 10 μM MAPKs in 200 μl binding buffer for 30 min at room temperature. GST beads were pelleted by centrifugation and washed three times. Retained proteins were eluted from the resin with SDS loading buffer. GST pull-downs with MAPKs from HEK293T cell lysates were performed in a similar manner to that described above but precipitated proteins were detected by Western-blot using anti-FLAG antibody (F1804, Sigma).
In vitro kinase assays
50 nM activated MAPKs were used in each in vitro kinase assay experiment to phosphorylate 0.5 μM control, “wild-type” or D-motif chimera reporters. Reactions were carried out in 50 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2, 0.05% IGEPAL, 5% glycerol, 2 mM DTT using recombinantly expressed and purified proteins, 200-400 μM ATP, and ~5 μCi of ATP(γ32P). Reactions were stopped by protein loading sample buffer containing 20 mM EDTA, boiled and then subjected to SDS-PAGE. Gels were dried before analysis by phosphorimaging on a Typhoon Trio+ scanner (GE Healthcare).
X-ray structure determination
10-15 mg/ml dephosphorylated MAPK samples were mixed with peptide solutions to yield an approximate four-fold peptide excess and the solution was supplemented with 2 mM AMPPNP and 2 mM MgCl2. Crystallization conditions were found by using a custom in-house PEG crystallization screen in standard vapor diffusion set-ups with hanging drops at room temperature. The best crystals for the ERK2-pepMNK1 complex grew in 15% PEG20000 buffered with 100mM MIB composite buffer pH=6.5. p38α-pepMKK6 crystals were obtained in 22% PEG3350, 100 mM HEPES pH=7.5. JNK1-pepNFAT4 crystals with a C-terminal truncated version of JNK1 (JNK1ΔC20) grew in 20% PEG8000 buffered with 100mM sodium citrate pH=5.5. (The diffraction of JNK1-pepNFAT4 crystals with full-length JNK1 was considerably weaker, only to ~ 2.6 Å resolution. This structure is also deposited in the PDB with accession number: 2XS0). Crystals were shock-frozen in liquid nitrogen after supplementing the mother liquor with 15-20% glycerol or 15% ethylene glycol.
The ERK2-pepSynth-revD complex crystallizes in the P212121 space group with cell parameters: a=45.03Å, b=65.66Å, c=117.23Å, in 20-24% PEG6000 buffered with 100mM TRIS pH=8.5. The first two amino acids of pepSynth-revD (LSLSSLAASSLAKRRQQ, underlined; and consensus positions are shown in bold) could not be located in the density because a hydrophobic side-chain from a symmetry-related ERK2 molecule occupied the ΦB pocket. Because earlier attempts to obtain an ERK2-pepRSK1 complex with wild-type ERK2 failed, we designed an ERK2 construct in which amino acids responsible for crystal packing interactions observed in the ERK2-pepSynth-revD crystal were mutated to alanines (ERK2AA: R77A. E314A). This construct readily crystallized in complex with pepRSK1 and crystals grew within 4-5 days with typical dimensions of 0.30mm x 0.10mm x 0.02mm in 27-29% PEG6000 buffered with 100mM MES composite buffer pH=6.5. The ERK2AA-pepRSK1 complex crystallized in the P212121 space group, with cell parameters: a=41.80Å, b=59.08Å, c=156.12Å.
High resolution data on frozen crystals were collected on PXI and PXIII beam lines of the Swiss Light Source (Villigen, Switzerland), and data for the pepMNK1-ERK2 complex was collected on a Rigaku R200 rotating anode X-ray generator at the Institute of Chemistry, Eötvös Loránd University. Structures were solved by molecular replacement. Data was processed with XDS (53). The main chain of ERK2, p38α, and JNK1 from the PDB entry 2GPH, 3GC7, 1UKH, respectively, were used as molecular replacement (MR) models in PHASER (54,55). The MR search identified single MAPK molecules per asymmetric unit in all cases. Structure refinement was carried out in PHENIX (56) and structure remodeling and building was done in Coot (57).
MAPK binding linear motif identification in silico
Putative MAPK binding docking motifs were identified in silico with the following procedure. Human protein sequences and sequence annotations were retrieved from UniProtKB/SwissProt (58). All sequences were scanned for instances of D-motif (pepJIP-, pepNFAT4-, pepMKK6-, or pepHePTP-type) and revD-motif (RSK/MAPKAP-type) subclasses using the regular expressions [RK][P]..[LIV].[LIVMPF] (pepJIP-type), [RK]..[LIVMP].[LIV].[LIVMPF] (pepNFAT4-type), [RK].{3,4}[LIVMP].[LIV].[LIVMPFA] (pepMKK6-type), [LI]..[RK][RK].{5}[LIVMP].[LIV].[LIVMPFA] (pepHePTP-type), and [LIVMPFA].[LIV].{1,2}[LIVMP].{4,6}[LI]..[RK][RK] (RSK/MAPKAP-type). We discarded sequence matches that overlapped with secondary structure elements (alpha-helices, turns, beta-strands, coiled coils), modular protein domains, repeats, signal peptides, propeptides, or mitochondrial transit peptides. Because we were interested in intracellular MAPK signaling, we removed motifs that were located in extracellular or intralumenal proteins or protein regions. To make our search criteria more stringent for D-motifs, we excluded putative MAPK substrates that had no potential S/TP target site located within 10-50 amino acids C-terminal of their D-motif end position, because roughly two-thirds of MAPK S/TP target sites that have been shown to be physiologically relevant are located within this region (table S3). Note that this target site filter essentially limited the D-motif lists for MAPK substrates only. This latter filter, however, also removed some bona fide motifs that may also govern phosphorylation of S/TP target sites falling outside the region specified by this filter (59,60). Because the revD-motif consensus search pattern was inherently longer, the putative revD-motif containing protein list was not further filtered. Moreover, a target site filter would not have been as efficient in removing false positives because revD-motifs mediate phosphorylation of distant SP/TP sites located N-terminal from motifs (29). Moreover, linear motifs binding into MAPK docking grooves may not only contribute to signaling specificity through MAPK substrate partner selection. They may recruit MAPKs into higher order complexes, which may not necessarily involve direct phosphorylation of their binding partners.
Because linear motifs often occur in unstructured regions (49), we used DisEMBL 1.5 (61) to computationally predict amino acids belonging to an unstructured or disordered protein region. To increase the reliability of disorder prediction, putative D-motifs were only considered if more than 60% of the amino acids spanning from the motif start site to the first S/TP target site were annotated as disordered (according to at least one of the DisEMBL disorder criteria); revD-motifs were only analyzed further if more than 90% of their amino acids were disordered. These values were obtained by inspecting a set of benchmark motifs that were either proven to mediate binding or to have a physiological relevance in cells. The final list was further manually curated to remove residual false positives consisting fof intralumenal, vesicular, mitochondrial or extracellular proteins (amounting to less than 5% of the final entries).
We incorporated this information about the intervening regions in-between conserved motif residues by developing position-specific scoring matrices (PSSMs) to rank the remaining lists of putative MAPK D-motifs. We did not perform scoring for revD-motifs due to the lack of our knowledge on revD-motifs apart from those in the RSK/MAPKAPK family. The PSSMs were based on human examples of each particular docking peptide conformation class. To obtain a sample of representative size, we used the high conservation of MAPK docking surfaces in metazoans and collected the motif sequences of non-human proteins orthologous to human examples from UniProtKB. Non-human sequences were only included in the dataset if they were of the same motif subclass as their human correspondent, as determined by pairwise and multiple alignments. The proteins included in the PSSM-generator sets were as follows: JIP-type: JIP(1/2), JIP(3/4), IRS (1/2/4) and their orthologs containing the same motif (n=51); NFAT4-type: NFATC3, JUN(B/C/D), MKK4, Gli(1/2/3) and their orthologs (n=133); MKK6-type: MEF2(A/C), MKK3, MKK6, MKK1, MKK2, MKK4, Gli(1/2/3) and their orthologs (n=87). The HePTP-type set included PTPN5, PTPN7 and PTPRR and their orthologs (including the PTP-ER proteins known from insects, thus yielding n=65). Note that the same motif may be included in more than one subclass, because promiscuous motifs can satisfy more than one consensus at the same time (for example, pepMKK4 is both NFAT4-type and MKK6-type). Consensus motif window size was set to 12 in the case of classic short (≤12 amino acids) D-motifs (JIP, NFAT4 and MKK6-types) to make their scores directly comparable and appropriately longer for those in the HePTP-type D-motif subclass spanning ≥15 amino acids. For each putative D-motif we calculated a score as the log ratio score of observed and expected probabilities (a priori amino acid probabilities were calculated based on human UniProtKB/SwissProt sequences), as previously described (62). This logPSSM-score was used to rank the top candidates and identify further false positives (for example, JIP peptides in the MKK6-type list). After manual list inspections, we set the logPSSM score cutoffs to -3.0 for JIP-type, -3.0 for NFAT4-type and +2.5 for MKK6-type motifs, and removed all motifs with lower scores from the final list. Because the pepHePTP-type PSSM was based only on one related set of proteins, logPSSM scores were used only for ranking, but no cutoff was applied.
The benchmark set in the analysis included 30 previously characterized MAPK binding partners, belonging to 10 independent groups of paralogous proteins: JIP, NFAT, MAP2K, IRS, GLI, JUN, MEF2, MAPK phosphatases and MAPKAPKs (63, 64, 50, 48, 65, 12, 25). Our in silico analysis correctly identified 29 of these benchmark proteins and mostly grouped them correctly into five motif classes, presumably binding MAPKs with different profiles: pepJIP1- and pepNFAT4-type D-motifs bind to JNK, whereas pepMKK6-, pepHePTP-type D-motifs or revD-motifs bind to ERK or p38 with varying degree of specificity, depending on the composition of the intervening regions.
To identify broader biological processes and pathways among the MAPK binding proteins identified in our study, we performed enrichment analyses for each motif subclass and the combined full list. Protein annotations were retrieved from the Gene Ontology (GO) Annotation Database (UniProtKB-GOA) and the Pathway Interaction Database (PID) (66,67). Enrichments were calculated with a hypergeometric test in R (68,69); the background distribution of annotations was calculated based on the human UniProtKB/Swissprot sequences. We adjusted the resulting p-values for multiple testing errors using the false discovery rate (FDR) (70). Only annotations with an adjusted p-value (q-value) of less than 0.05 were considered to be significantly enriched in the respective list. Because GO annotations are organized in a hierarchy, the set of enriched GO annotations can be further summarized by identifying and filtering semantically similar annotations (71). We used ReEViGO to identify semantically similar annotations and only kept the annotation that was most representative for the group (72).
Supplementary Material
Acknowledgements
We thank András Patthy for excellent quality peptide synthesis.
Funding: A. Reményi is a János Bolyai Fellow of the Hungarian Academy of Sciences and a Wellcome Trust International Senior Research Fellow (081665/Z/06/Z). This work was also supported by a Marie Curie International Reintegration Grant (205436) within the 7th European Community Framework Programme, by the European Union and the European Social Fund (TÁMOP) 4.2.1./B-09/KMR-2010-0003, by the German National Genome Research Network (NGFN), by the Cluster of Excellence for Multimodal Computing and Interaction (MMCI) funded by the German Research Foundation (DFG), and by the OTKA NK81950 and NKTH-OTKA H07-A 74216 grants (Hungary).
Footnotes
Author contributions: Á.G and A.Z. participated in the experimental design and carried out the biochemical experiments. and I.T. and G.G. solved the crystal structures. F.F. and T.B. carried out in vitro kinase assays. J.V. and A.A. developed protein expression systems. H.B., D.E. and M.A conducted all bioinformatics analysis. A.R. designed the experiments and wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Atomic coordinates and structure factors for the ERK2-pepMNK1, ERK2-pepSynth-revD, ERK2-pepRSK1, p38α-pepMKK6, and JNK1-pepNFAT4 complexes have been deposited in the Protein Data Bank with the accession numbers 2Y9Q, 4FMQ, 3TEI, 2Y8O and 2XRW, respectively.
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Neduva V, Linding R, Su-.Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 3.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008;19:344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 5.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 6.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 7.Reményi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Ubersax JA, Ferrell JE. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 9.Chang CI, Xu B.-e, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 10.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 11.Caffrey DR, O’Neill LA, Shields DC. The evolution of the MAP kinase pathways: coduplication of interacting proteins leads to new signaling cascades. J. Mol. Evol. 1999;49:567–582. doi: 10.1007/pl00006578. [DOI] [PubMed] [Google Scholar]
- 12.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Heo Y-S, Kim S-K, Seo CI, Kim YK, Sung B-J, Lee HS, Lee JI, Park S-Y, Kim JH, Hwang KY, Hyun Y-L, Jeon YH, Ro S, Cho JM, Lee TG, Yang CH. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Sun J-P, Zhou B, Zhang Z-Y. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma W, Shang Y, Wei Z, Wen W, Wang W, Zhang M. Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase. Structure. 2010;18:1502–1511. doi: 10.1016/j.str.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Reményi A, Good MC, Bhattacharyya RP, Lim WA. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko T, Huang H, Zhao B, Li L, Liu H, Voss CK, Wu C, Shiller MR, Li SSC. Loops Govern SH2 Domain Specificity by Controlling Access to Binding Pockets. Science Signaling. 2010;3:ra34–ra34. doi: 10.1126/scisignal.2000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneko T, Sidhu SS, Li SSC. Evolving specificity from variability for protein interaction domains. Trends Biochem. Sci. 2011;36:183–190. doi: 10.1016/j.tibs.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharrocks AD, Yang SH, Galanis A. A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 21.Bardwell AJ, Frankson E, Bardwell L. Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 2009;284:13165–13173. doi: 10.1074/jbc.M900080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow CW, Rincón M, Cavanagh J, Dickens M, Davis RJ. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 23.Dérijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 24.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 25.Gavin AC, Nebreda AR. A MAP kinase docking site is required for phosphorylation and activation of p90(rsk)/MAPKAP kinase-1. Curr. Biol. 1999;9:281–284. doi: 10.1016/s0960-9822(99)80120-1. [DOI] [PubMed] [Google Scholar]
- 26.Gaestel M. MAPKAP kinases - MKs - two’s company, three’s a crowd. Nat. Rev. Mol. Cell Biol. 2006;7:120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 27.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 29.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JA, Poteet-Smith CE, Lanningan DA, Freed TA, Zoltoski AJ, Sturgill TW. Creation of a stress-activated p90 ribosomal S6 kinase. The carboxyl-terminal tail of the MAPK-activated protein kinases dictates the signal transduction pathway in which they function. J. Biol. Chem. 2000;275:31588–31593. doi: 10.1074/jbc.M005892200. [DOI] [PubMed] [Google Scholar]
- 31.ter Haar E, Prabhakar P, Prabakhar P, Liu X, Lepre C. Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. J. Biol. Chem. 2007;282:9733–9739. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 32.Underwood KW, Parris KD, Frederico E, Mosyak L, Czerwinski RM, Shane T, Taylor M, Svenson K, Liu Y, Hsiao C-L, Wolfrom S, Maguire M, Malakian K, Telliez J-B, Lin L-L, Kriz RW, Seehra J, Somers WS, Stahl ML. Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme. Structure. 2003;11:627–636. doi: 10.1016/s0969-2126(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 33.Malakhova M, Tersko V, Lee S-Y, Yao K, Cho Y-Y, Bode A, Dong Z. Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nat. Struct. Mol. Biol. 2008;15:112–113. doi: 10.1038/nsmb1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney LM, Whitmarsh AJ. Docking interactions in the c-Jun N-terminal kinase pathway. J. Biol. Chem. 2004;279:11843–11852. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]
- 35.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol. Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 36.Dimitri CA, Dowdle W, MacKeigan JP, Blenis J, Murphy LO. Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr. Biol. 2005;15:1319–1324. doi: 10.1016/j.cub.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhard KA, Chen F, Shapiro P. Quantitative analysis of ERK2 interactions with substrate proteins: roles for kinase docking domains and activity in determining binding affinity. J. Biol. Chem. 2011;286:2477–2485. doi: 10.1074/jbc.M110.177899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin BY, Lam SS, Correia JJ, Lin K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 2002;16:1950–1963. doi: 10.1101/gad.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrini L, Yu DS, Lo T, Anand S, Lee MY, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 42.Bardwell L, Shah K. Analysis of mitogen-activated protein kinase activation and interactions with regulators and substrates. Methods. 2006;40:213–223. doi: 10.1016/j.ymeth.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho DT, Bardwell AJ, Abdollahi M, Bardwell L. A docking site in MKK4 mediates high affinity binding to JNK MAPKs and competes with similar docking sites in JNK substrates. J. Biol. Chem. 2003;278:32662–32672. doi: 10.1074/jbc.M304229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho DT, Bardwell AJ, Grewal S, Iverson C, Bardwell L. Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J. Biol. Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardwell AJ, Abdollahi M, Bardwell L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem. J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Z, Zhou B, Zhang Z. Molecular determinants of substrate recognition in hematopoietic protein-tyrosine phosphatase. J. Biol. Chem. 2004;279:52150–52159. doi: 10.1074/jbc.M407820200. [DOI] [PubMed] [Google Scholar]
- 47.Kusari AB, Molina DM, Sabbagh W, Lau CS, Bardwell L. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang SH, Galanis A, Sharrocks AD. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Whisenant TC, Ho DT, Benz RW, Rogers JS, Kaake RM, Gordon EA, Huang L, Baldi P, Bardwell L. Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans P, Sacan A, Ungar L, Tozeren A. Sequence alignment reveals possible MAPK docking motifs on HIV proteins. PLoS ONE. 2010;5:e8942. doi: 10.1371/journal.pone.0008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaestel M, Kracht M. Peptides as signaling inhibitors for mammalian MAP kinase cascades. Curr. Pharm. Des. 2009;15:2471–2480. doi: 10.2174/138161209788682299. [DOI] [PubMed] [Google Scholar]
- 53.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The Uniprot Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nihalani D, Wong HN, Holzman LB. Recruitment of JNK to JIP1 and JNK-dependent JIP1 phosphorylation regulates JNK module dynamics and activation. J. Biol. Chem. 2003;278:28694–28702. doi: 10.1074/jbc.M304212200. [DOI] [PubMed] [Google Scholar]
- 61.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 62.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 63.Chow CW, Rincón M, Cavanagh J, Dickens M, Davis RJ. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 64.Lee YH, Giraud J, Davis RJ, White MF. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 2003;278:2896–2902. doi: 10.1074/jbc.M208359200. [DOI] [PubMed] [Google Scholar]
- 65.Vinciguerra M, Vivacqua A, Fasanella G, Gallo A, Cuozzo C, Morano A, Maggiolini M, Musti AM. Differential phosphorylation of c-Jun and JunD in response to the epidermal growth factor is determined by the structure of MAPK targeting sequences. J. Biol. Chem. 2004;279:9634–9641. doi: 10.1074/jbc.M308721200. [DOI] [PubMed] [Google Scholar]
- 66.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R Development Core Team . R: A language and environment for statistical computing. Vienna, Austria: 2008. [Google Scholar]
- 69.Rivals I, Personnaz L, Taing L, Potier M. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23:401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 71.Schlicker A, Albrecht M. FunSimMat update: new features for exploring functional similarity. Nucleic Acids Res. 2010;38:D244–248. doi: 10.1093/nar/gkp979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Supek F, Bošnjak M, Škunca N, S̆muc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milanesi L, Petrillo M, Sepe L, Boccia A, D’Agostino N, Passamano M, Di Nardo S, Tasco G, Casadio R, Paolella G. Systematic analysis of human kinase genes: a large number of genes and alternative splicing events result in functional and structural diversity. BMC bioinformatics. 2005;6:S20. doi: 10.1186/1471-2105-6-S4-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dérijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 75.Yazgan O, Pfarr CM. Regulation of two JunD isoforms by Jun N-terminal kinases. J. Biol. Chem. 2002;277:29710–29718. doi: 10.1074/jbc.M204552200. [DOI] [PubMed] [Google Scholar]
- 76.Gupta S, Campbell D, Dérijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 77.Diring J, Camuzeaux B, Donzeau M, Vigneron M, Rosa-Calatrava M, Kedinger C, Chatton B. A cytoplasmic negative regulator isoform of ATF7 impairs ATF7 and ATF2 phosphorylation and transcriptional activity. PLoS ONE. 2011;6:e23351. doi: 10.1371/journal.pone.0023351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knauf U, Newton EM, Kyriakis J, Kingston RE. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 79.Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000;20:3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saxena M, Williams S, Brockdorff J, Gilman J, Mustelin T. Inhibition of T Cell Signaling by Mitogen-activated Protein Kinase-targeted Hematopoietic Tyrosine Phosphatase (HePTP) Journal of Biological Chemistry. 1999;274:11693–11700. doi: 10.1074/jbc.274.17.11693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.