Abstract
Recent genome-wide profiling reveals highly complex regulation networks among ERα and its targets. We integrated estrogen (E2)-stimulated time-series ERα ChIP-seq and gene expression data to identify the ERα-centered transcription factor (TF) hubs and their target genes, and inferred the time-variant hierarchical network structures using a Bayesian multivariate modeling approach. With its recurrent motif patterns, we determined three embedded regulatory modules from the ERα core transcriptional network. The GO analyses revealed the distinct biological function associated with each of three embedded modules. The survival analysis showed the genes in each module were able to render a significant survival correlation in breast cancer patient cohorts. In summary, our Bayesian statistical modeling and modularity analysis not only reveals the dynamic properties of the ERα-centered regulatory network and associated distinct biological functions, but also provides a reliable and effective genomic analytical approach for the analysis of dynamic regulatory network for any given TF.
Reverse engineering of genetic regulatory networks and discovering inherent major interactions within complex biological processes persist as the key tasks of computational systems biology1,2,3,4,5. Barenco et al. proposed a genome-wide transcriptional modeling approach, requiring no prior information in identifying the regulatory network6. This approach ignored different time and space effects existing in cellular transcriptional contents and as a result, it may be ineffective in detecting specific regulatory processes. While many other computational methods for reconstructing transcription networks were also presented during the same period7,8,9,10,11,12,13, most of them focused on a single data source, i.e. ChIP-chip data, cDNA microarray, or combined with other pre-defined transcription factor and motif information to infer network structures7,8,9,11. Others studied network properties such as time-variant, hierarchical and collaborative, correlations between specific network structures and underlying functions10,12,13.
The evolved in vivo genome-wide profiling techniques from ChIP-chip14 to ChIP-seq15,16 enable us to identify thousands of transcription factor binding sites and chromatin modifications at a higher resolution and lower cost. The ChIP-seq technique facilitates accurate identification of transcription factor binding sites and provides the ability to directly ascertain the details of the underlying transcription factors during regulatory processes. As a result, it optimizes the network inference approaches in both accuracy and reliability.
A recent study from Sun et al. proposed a Bayesian error model for identifying regulatory process using single time-point ChIP-chip and cell cycle gene expression data, and they adopted the MCMC sampling technique to infer model parameters, where the model considers the random effects as the error terms in the regulatory process17. However, it is a linear fitting approach, and for most genetic regulatory processes, due to time and space scale differences, linear modeling approach may not reflect those nonlinear and stochastic transcription regulatory processes. Furthermore, due to the single time-point ChIP-chip data, the model can only present a static cell cycle regulatory network.
To our knowledge, the computational methods based on the time-series, or multiple continuous ChIP-seq data have not yet been proposed for the inference of time-varying transcriptional networks. Given the fact that the cellular signaling dynamically responds to an external stimuli, it is essential to integrate both the time-series ChIP-seq and gene expression data in order to capture the dynamic network structures and related biological properties, which are the key to elucidate the inherent regulatory mechanisms.
ERα is an estrogen (E2)-inducible transcription factor (TF) and member of the nuclear receptor superfamily, the dysfunction of which accounts for 70% breast tumors. ERα binds to estrogen response elements (EREs) at target gene's regulatory regions, and works with other signaling components to control downstream transcriptional and translational processes. Many recent genome-wide profiling studies of ERα18,19,20, including ours21,22, have shown a highly complex regulation network involved with both ERα and other TFs23. These studies revealed that many ERα binding sites could be located far away, up to 50-100kb, from a known transcription start site (TSS) and a large number of other TF binding sites (TFBSs) could be co-enriched with ERα binding sites, which constitute a hierarchical regulatory network with target hubs. However, the static network fails to capture dynamic properties of transcriptional regulation responses to estrogens, due to lack of time-series ChIP-seq data.
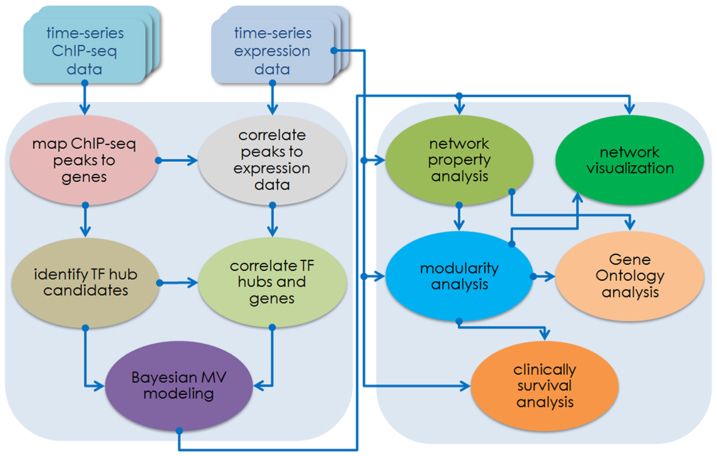
In this study, we integrated both E2-stimulated time-series ERα ChIP-seq data conducted in our laboratory and publically available E2-stimulated time-series gene expression data for reverse engineering the ERα-mediated transcriptional regulatory network. We identified the ERα-centered TF hubs and their target genes from the ERα ChIP-seq data at the four time points after estrogen stimulation, and inferred the time-variant hierarchical network structures using a Bayesian multivariate modeling approach. Furthermore we analyzed the properties of network structures including global connectivity distribution, the correlation between the regulatory coefficients and components' signal-to-noise ratios with respect to absolute rank value distribution of regulatory strength. Finally, we used inherent recurrent motif patterns to determine self-bedded regulatory modules within the hierarchical networks. The Gene Ontology (GO) analyses were also performed to reveal distinct biological functions of ERα genes regulated by each module at different time. Together the survival analysis for the module-regulated targets based on three breast cancer patient data sets revealed statistically significant clinical outcomes. A schematic flowchart (Figure 1) depicts the procedure in analyzing the ERα transcription regulatory network.
Figure 1. The computational analysis flowchart for inferring ERα transcription regulatory network.
It contains two major sections, one is for data processing and the other is for network and modularity analyses. The first section includes ChIP-seq data mapping and peak-calling, correlating ChIP-seq with microarray gene expression data, and Bayesian multivariate modeling for network inference. The second section covers analysis on the inferred network and modularity, the corresponding gene ontology (GO) analysis, and patient survival analysis on the network modules based on three clinical data sets published recently.
Results
Computational identification of ERα-regulated TFs and target genes
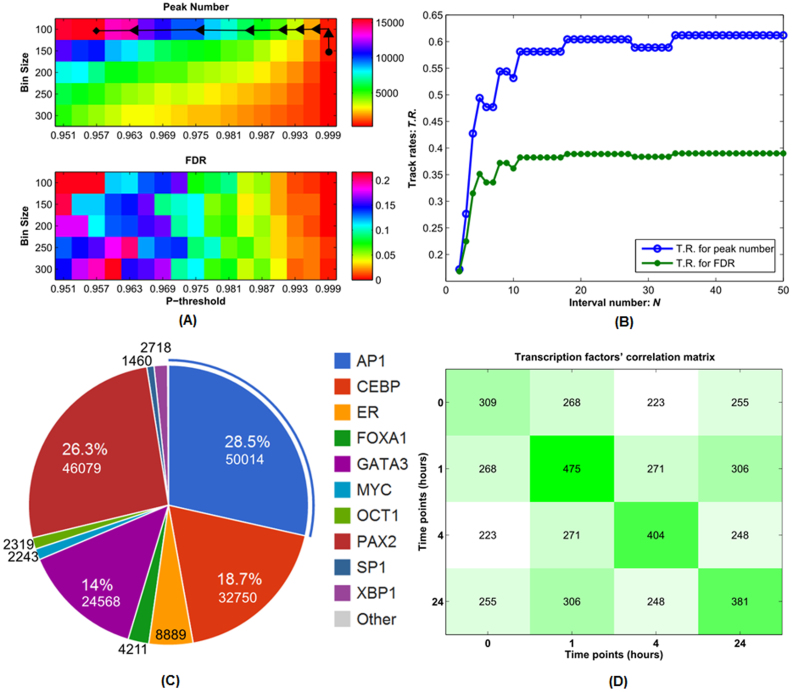
Estrogen-stimulated ERα data (ChIP-seq) at the time 0, 1, 4 and 24 hours were conducted as described in detail in Methods and Supplemental Materials. Since there are several statistical parameters (false discovery rate (FDR) and bin size), in order to derive an optimal set of ERα binding sites at each time point, we propose a flexible data feature detection approach, in which the output parameters can be formulated as a class of optimal track (Methods). Figures 2(A) and (B) illustrate the optimal parameters selection process for ERα ChIP-seq data at the time point 0, where normally it needs to find the highest peak number with a suitable bin-size and a statistically acceptable FDR. Once the optimal parameters for each time point of data are determined, then ERα targeted genes are identified at each time points (see Supplemental Excel file 1).
Figure 2. The selection of optimal parameters for the ERα ChIP-seq data at time point 0 hour.
A distribution of peak numbers (the upper panel) and FDR (the lower panel) vs. the p-threshold (A). A track rate distribution for peak number and FDR with respect to the interval number N (B). The global peak number and FDR distributions, track rate distributions for peak number and FDR plots at other time points are given in the Supplemental Figures S1–3. C and D. The identification of TF hubs according to the occurrence frequency of those candidates (C). The percentages denote the corresponding occurrence distribution among all candidates identified from the time-series ChIP-seq data. TF candidates' pairwise intersection matrix across all the four time points (D). The diagonal entries denote the candidate counts at the corresponding time points, and other non-diagonal entries denote those candidate counts of intersection identified between any two different time points.
We identified ERα-regulated TF hubs by scanning all possible TF candidates through TRANSFAC database which collects more than ~1,000 positional weight matrices (PWM)24,25. We determined the final TF hubs based on the quantitative selective criteria: (1) frequency of occurrence of each TF in all binding sites, see Figure 2(C), where the frequency of occurrence range varies from 0.832% (SP1) to 28.5% (AP1), and the total occurrence counts for those identified TF candidates are 3,997,164 (0 hour), 4,184,256 (1 hour), 2,174,429 (4 hours) and 3,912,712 (24 hours); (2) continuity of occurrence (or TF's overlap information) across four time points, i.e. whether or not those TF hubs have been detected at each time point; and (3) further manually selecting those major TF hubs functionally associating with ERα based on our experiment emphasis and published results from recent literatures.
From recent associated literatures, FOXA1 (also named as HNF3α) has been reported as a pioneer protein for ERα transcriptional process, and its function has potential regulatory roles in ERα activities and relative endocrine responses23,26. The TFs such as AP1, SP1 and NFκB have already been discovered to be regulated by ERα through nontraditional pathways and meanwhile their gene expressions are affected by the ERα regulation27. New binding site event of approximately 67 kbp upstream from MYC was discovered recently, and accordingly OCT1 and CEBP were claimed as nuclear receptor-interacting TF and putative regulator of estrogen target genes in the recent experiments of MCF-7 cell line28. GATA3 and FOXA1 were found with cooperative roles in mediating the ERα transcriptional network29. PAX2, without any previously recognized role, was discovered with a crucial mediator of ER repression30. X-box binding protein, XBP1, is an estrogen-regulated gene and also recognized as being strongly correlated with ERα expression in breast cancers31.
On Figure 2(C), AP1, PAX2, CEBP and GATA3 are the four TFs with the top 4 frequencies of occurrence, approximately 28.5%, 26.3%, 18.7% and 14%, respectively. Others' frequencies of occurrence range from 0.832% to 2.721%. Although other genes have also been detected in those scanning, their frequencies of occurrence or continuity of occurrence in each time point may not satisfy the above criteria.
As above, a total of 11 TFs, i.e. ERα, AP1, CEBP, GATA3, FOXA1, MYC, NFκB, OCT1, PAX2, SP1 and XBP1 were determined as the TF hubs and used for inferring a Bayesian statistical model (See Methods and Supplemental Materials).
We further performed correlation analysis for those TF candidates at each time point by calculating a pairwise TF intersection matrix across the four time points (Figure 2(D)). Our analysis revealed that the correlation between those TF candidates differs in any two time points, i.e. the number of the identified TF candidates at each time point, and the number of identified pairwise common TF candidates among those time points. Those facts also indicated the underlying time-variant property during the ERα regulatory process.
After determining a final set of 11 ERα-regulated TF hubs from the time-series ChIP-seq data, we identified each TF hub regulated target genes by re-scanning each hub TF's PWMs within all target genes (binding regions) at each of the four time points. We found that the TF hubs, PAX2, GATA3, CEBP, OCT1 and ERα itself, regulate relatively larger numbers of target genes, while the TF hubs NFκB, SP1 and MYC only regulate a smaller number of target genes (Table 1). The common target genes for each TF hub for all four time points were also identified (Table 1). A list of the TF hub target genes and their associated gene annotation (Entrez IDs) for each TF hub are provided in the Supplemental Excel file 2.
Table 1. The statistics of the 11 TF hubs' regulated genes and their corresponding commonly-regulated genes across the four time points, 0, 1, 4 and 24 hours, respectively.
| Hubs | 0 hour | 1 hour | 4 hours | 24 hours | Common |
|---|---|---|---|---|---|
| ERα | 169 | 241 | 206 | 178 | 105 |
| MYC | 92 | 107 | 83 | 91 | 59 |
| GATA3 | 276 | 422 | 320 | 278 | 142 |
| XBP1 | 107 | 134 | 108 | 108 | 80 |
| AP1 | 218 | 322 | 252 | 211 | 119 |
| CEBP | 288 | 452 | 368 | 274 | 143 |
| HNF3α | 132 | 172 | 123 | 135 | 84 |
| NFκB | 45 | 57 | 35 | 48 | 22 |
| OCT1 | 198 | 289 | 203 | 192 | 107 |
| PAX2 | 340 | 533 | 449 | 313 | 173 |
| SP1 | 65 | 71 | 60 | 62 | 40 |
Bayesian statistical modeling of the ERα regulatory network
In order to dissect the dynamics of ERα transcriptional regulatory networks, we proposed a Bayesian multivariate statistical approach32,33,34 to reconstruct a hierarchical structure for the ERα-centered time-variant networks. In addition to ERα regulated genes and the identified 11 TF hubs from the time-series ERα ChIP-seq data, we collected a publically available time-series gene expression data18 which monitors gene expression level changes at 20 time-points after estrogen stimulation. In the research work, we selected the expression data at the four time points (0, 1, 4 and 24 hours) to match the corresponding ChIP-seq datasets. The transcription rate is defined by Equation (15) (Supplemental Section 3), the time interval Δt for the transcription rate is determined based on the selected ChIP-seq and microarray data points.
For the statistical modeling, the gene expression values were not only for the input matrix, but also were used for approximating their corresponding transcription rates, see the Methods and Supplemental Materials, and the modeling errors were assumed to follow a normal distribution with zero mean. The transcription rates were formulated as a conditional probability distribution on the a priori distribution of the regulatory coefficient matrix and the corresponding error term distribution. After the regulatory coefficients were normalized within the range −1 to 1, we acquire a regulatory strength distribution.
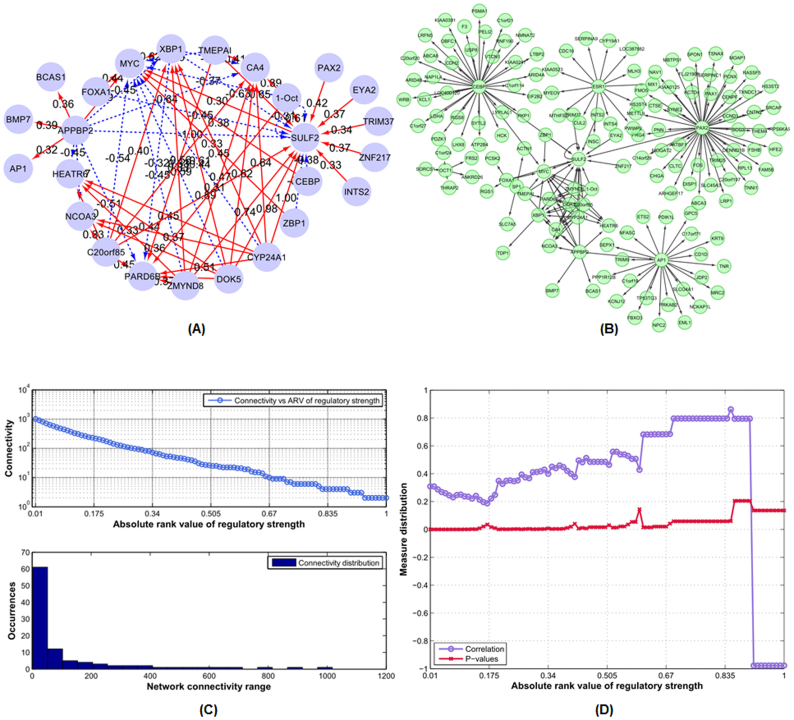
Figure 3(A) illustrates the ERα-centered regulatory network at time 4 hours, and a topology of the hierarchical structure of the network is shown on Figure 3(B), where hierarchical refers to a regulatory network that has an ERα-centered structure as the network core; the ERα directly-regulated hubs and genes form its secondary network structure; and the third layer contains genes directly regulated by other hubs. Other inferred ERα-centered regulatory networks at time points 0, 1, and 24 hours are shown on the Supplemental Figures S4–S6 (A) and (B), respectively.
Figure 3. The ERα transcription regulatory network structure and related analysis at time 4 hours.
(A) The inferred ERα transcription regulatory network structure at time 4 hours. The red edges denote positive activation, and dashed blue edges denote negative inhibition. (B) The hierarchical topological structure of the inferred ERα transcription regulatory network at time 4 hours. (C) and (D) illustrate the connectivity distribution, Pearson correlation and p-value distributions (between the regulatory coefficients and SNRs) as the functions of absolute rank value of regulatory strength for the network structure at time 4 hours. The plots for other time points, i.e. 0, 1, and 24 hours are given in the Supplemental Figures S4–S6.
And to quantitatively characterize the inferred network properties, we analyzed the network structural property, i.e. the global network (edge) connectivity with respect to the absolute rank value distribution of regulatory strength. We found that the global connectivity for each inferred network was approximately following a log-linear trend regarding the absolute rank value distribution of regulatory strength at all the four time points, Figure 3(C) and Supplemental Figures S4–S6 (C).
Furthermore, we analyzed the correlation distribution between the inferred regulatory strength and signal-to-noise ratio (SNR, see Methods) for the network components (TF and target genes), together with the corresponding p-values were also given.
By measuring the SNR values, we captured the major characteristics of the signal and noise levels of the network component expression process during the transcriptional regulation. In this study, we calculated the SNR values by using the time-series E2-treated gene expression profiles. Based on its definition, the higher the SNR value, the higher the signal density and the less noise content will be contained in the corresponding gene expression process. We discovered the inherent statistical trend by increasing the absolute rank value of regulatory strength from 0 to 1, the absolute correlation values between the regulatory coefficient and related SNR also increased, and vice versa (Figure 3(D)).
Figures 3(C) and (D) are the statistical analysis on the inferred network at time 4 hours. Figure 3(C) gives the node connectivity characteristics of the inferred network. Figure 3(D) depicts the correlation distribution between the inferred regulatory coefficient and SNR measure of the network nodes. Both statistical network analyses are based on the inferred regulatory coefficients of the whole network nodes. Horizontal axis (absolute rank value) denotes the threshold of the absolute value of the regulatory coefficients between nodes, from 0 to 1, while the actual regulatory coefficient ranges from −1 (negative regulation) to 1 (positive regulation). Using the absolute rank value as the threshold, we statistically characterize the inferred regulatory node distribution from the whole transcription regulatory network.
Together with the analysis results at the other time points in the supplementary Figures S4, S5 and S6, those analyses are to quantitatively characterize the network difference and dynamic properties across the whole transcription regulatory process. Meanwhile from the analysis results at time points 1, 4 and 24 hours, we also observed the evidently correlative trends, although there exists some abrupt points along those trends across the four time points (Supplemental Figures S4–6D).
Our analysis illustrated that the SNR distribution had certain statistical association with the global network connectivity in the ERα regulation network, i.e. network component with a high SNR value tended to have a high regulatory strength on its potential target components during its regulatory processes. Thus the SNR measure is a meaningful index to determine which component in the ERα transcription network, compared with other ones, has the most prominent roles (regulatory strength) on regulatory processes.
Modularity analysis on the ERα transcriptional regulatory network
Modularity is recognized as a ubiquitous feature in biochemical networks, and it acts as basic building block in genetic regulatory processes through inherent biological functionality, genetic mechanisms and regulatory patterns35,36,37.
Thus, to illustrate the corresponding network characteristics, we further investigated the inferred ERα regulatory network by employing a quantitative modularity analysis approach, and uncovered the inherent design principles and regulatory mechanisms in ERα transcriptional network.
The modularity analysis was based on the following criteria: (1) each identified module should contain the key TF hub, ERα, since we attempted to discover mediatory or regulatory mechanisms of ERα on different TFs within those modules; (2) each identified module should contain one or more regulatory patterns detected from the core regulatory network structure, provided in the Supplemental Figure S7. Such regulatory patterns are important for collecting, dispatching and feedbacking biological signals (regulation or mediation). And it is commonly recognized that they are basic building blocks of regulatory modules or pathways; (3) each identified module should be of topologically reasonable structure. From the topology perspective, we may view the identified module as a directed graphic model with one or more feedback loops. As such, specific regulatory signals can propagate within the models and certain feedback loops will ensure source TFs can receive corresponding feedbacks or reaction signals from other TFs and targets. From the perspective of biological functions, such modularity design principles are essential to maintain their stable structure characteristics.
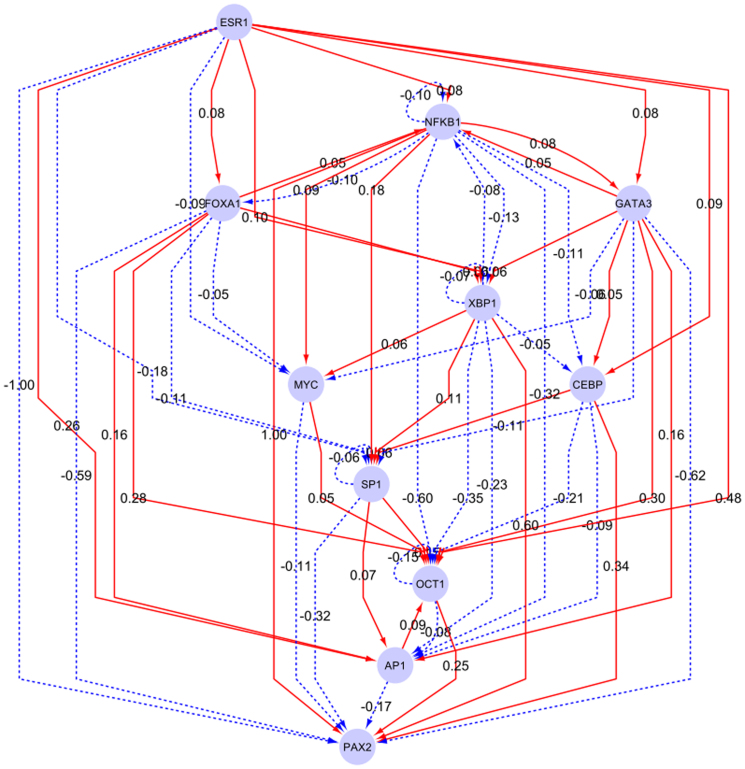
We utilized the above considerations and recurrent motif patterns (see Supplemental Figure S7 and Supplemental Materials) to determine three self-embedded regulatory modules from the core transcription network structure (Figure 4).
Figure 4. The ERα-centered core regulatory module structure.
The hub nodes were selected based on their respective occurrence frequency of transcription binding activities, identified from the time-series ChIP-seq data sets, together with the manually-selected ones, which have already been validated and acknowledged in experiments and literatures published recently. The red edges denote positive activation, and dashed blue ones denote negative inhibition.
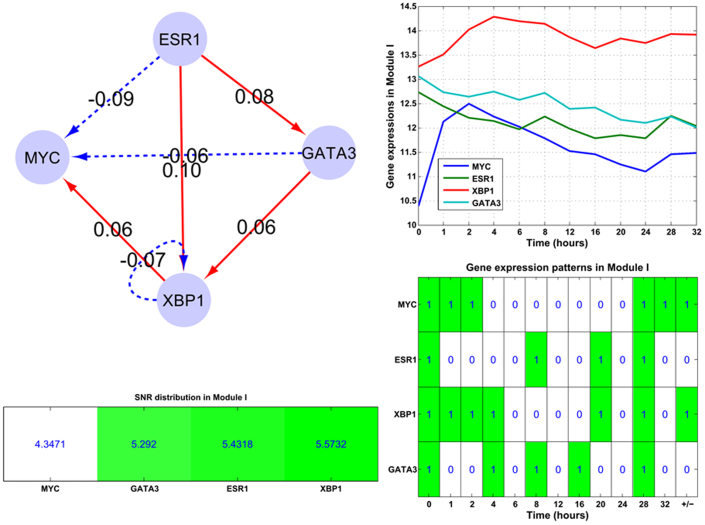
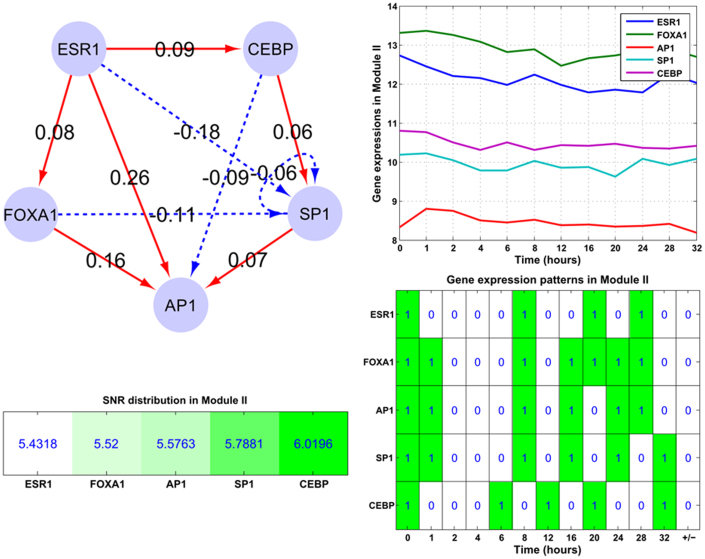
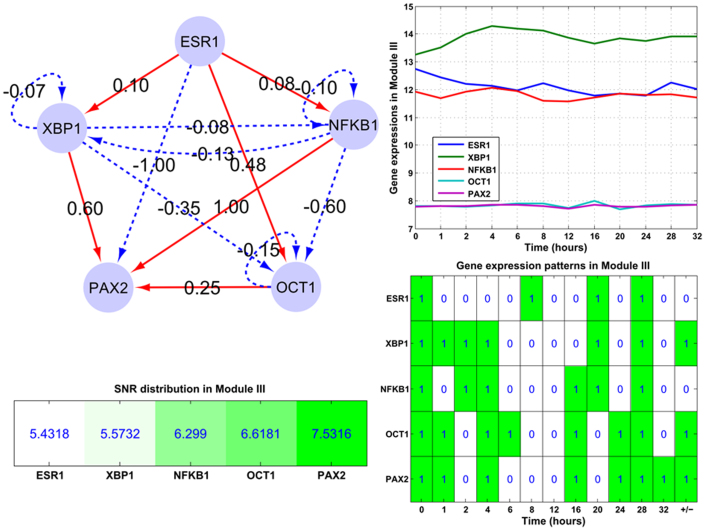
Module I (ERα-GATA3-XBP1-MYC): In this regulatory module, the major events are: ERα activates XBP1 and GATA3, while inhibits MYC; GATA3 activates XBP1 while inhibits MYC. Module II (ERα-SP1-CEBP-AP1-FOXA1): In this module ERα only inhibits SP1 while activates other components. This module is mainly composed of feed-forward loops, e.g. ERα-CEBP-SP1 and ERα-FOXA1-AP1. Module III (ERα-NFκB-PAX2-XBP1-OCT1): This module mainly contains one interactive loop (NFκB and XBP1), two bi-span loop (ERα-NFκB:XBP1-OCT1, i.e. ERα activates while NFκB inhibits both XBP1 and OCT1; ERα-XBP1:NFκB-PAX2, i.e. ERα and XBP1 both have activation and inhibition on NFκB and PAX2), feed-forward loops (e.g. ERα-XBP1-PAX2) and self-inhibitory loops (NFκB, OCT1 and XBP1).
Quantitative models interwoven with major network components significantly contribute to the understanding of cellular networks and underlying biological mechanisms. For example, a small set of recurring regulation patterns, known as motifs, are the basic building elements in diverse organisms from bacteria to humans, and such specific recurrent motif patterns relate directly to certain functions of those models38,39,40. As such, we further analyzed the underlying biological design principles and related properties of the ERα-centered regulatory network and its embedded modules with several major considerations: the identified motif patterns, module's SNR distributions, and the target gene's expression patterns. Thus based on those results, we could determine whether or not there existed any interdependency among those patterns.
In Module I (ERα-GATA3-XBP1-MYC), we found the underlying feed-forward and self-inhibitory motif patterns (top left panel of Figure 5). And the gene expression patterns of those module components showed ERα and GATA3 are on down-regulating trends across all the time points, while MYC and XBP1 have the up-down-up wavy trend, partially because both undergo positive and negative activities. For Module I, its components' SNR statistics are given on the left bottom panel of Figure 5, and the module's average SNR is 5.161 dB.
Figure 5. The structure of the embedded Module I (ERα-GATA3-XBP1-MYC).
The subplots illustrate gene expression patterns and corresponding SNR values for those module components. Module I contains only one self-inhibition loop (XBP1). And the corresponding time-course gene expression plots and inherent expression patterns are given in right panels, together with the up-/down-regulated information (+/−) for each gene on the rightmost column of the bottom plot. MYC and XBP1 are up-regulated in the module.
In Module II (ERα-SP1-CEBP-AP1-FOXA1), the feed-forward loops contributed the most to the gene expression patterns and we also found one bi-span loop. The five components constitute two bi-span loops and other interwoven feed-forward loops, and the components are all down regulated (right panel of Figure 6). For SP1 and AP1, they mainly receive inhibitory activities by self-inhibition and feed-forward loops. For other down-regulated components (FOXA1 and CEBP), they mainly respond to the down-regulation of ERα by the feed-forward loops. Module II's SNR statistics are provided on the left bottom panel of Figure 6, with the average SNR 5.667 dB.
Figure 6. The structure of the embedded Module II (ERα-CEBP-AP1-GATA3-SP1-FOXA1).
The subplots illustrate the gene expression patterns and corresponding SNR values for those module components. Module II mainly contains feed-forward loops, while only one bi-span (ERα-CEBP:AP1-SP1) and self-inhibition loop (SP1). And the corresponding time-course gene expression plots and inherent expression patterns are given in right panels, together with the up-/down-regulated information (+/−) for each gene on the rightmost column of the bottom plot. All genes are down-regulated in the module.
Within Module II, those TFs as AP1, SP1 and NFκB have already been discovered regulated by ERα through nontraditional pathways, and meanwhile their gene expressions are affected by the ERα regulation27. On Figures 6 and 7, those regulatory activities are mainly implemented by the bi-span (ERα-CEBP:AP1-SP1, Figure 6) and feed-forward loops from ERα.
Figure 7. The structure of the embedded Module III (ERα-XBP1-NFκB-OCT1-PAX2).
The subplots illustrate gene expression patterns and corresponding SNR values for those module components. Module III mainly contains four bi-span loops, self-inhibition loops (NFκB, OCT1 and XBP1) and one co-inhibitory loop (XBP1 and NFκB). And the corresponding time-course gene expression plots and inherent expression patterns are given in right panels, together with the up-/down-regulated information (+/−) for each gene node on the rightmost column of the bottom plot. XBP1 is up-regulated, and NFκB is down-regulated in Module III.
In the work by Kong et al.29, ERα, FOXA1 and GATA3 have been discovered to form a functional enhanceosome regulating the genes that shape core ERα function and cooperatively modulate the transcriptional network. In the dissected Modules I and II, such a cooperative mode is implemented through feed-forward loop.
In Module III (ERα-NFκB-PAX2-XBP1-OCT1), the self-inhibition loops contribute less explicitly than bi-span and feed-forward loops to the components' expression patterns, i.e. such components as OCT1 and XBP1 with self-inhibition loops are up-regulated except NFκB (top left panel of Figure 7). For PAX2, the positive feed-forward loop from OCT1, XBP1 and NFκB contributes explicitly in its gene expression pattern. Module III's SNR statistics are provided on the left bottom panel of Figure 7, with the average SNR 6.291 dB, see the supplemental Tables S1, S2 and S3.
Within Module III, PAX2's crucial role as a mediator of ERα repression has been depicted30, mainly by means of feed-forward and bi-span loops with XBP1 and OCT1. Together, X-box binding protein, XBP1, is an estrogen-regulated gene recognized as being strongly correlated with ERα expression in breast cancers31, in this module, the roles have been clearly illustrated through bi-span and feed-forward loops with PAX2 and NFκB.
Furthermore from the association analysis between the SNR values and expression patterns of those module components, on Figures 5 and 6, we find that MYC with a low SNR value (4.3471 dB) is up-regulated; components with high SNR values (from 5.4318 to 6.0196 dB) tend to be down-regulated, see ERα, GATA3, CEBP, SP1, AP1 and FOXA1, except for XBP1. While for those with much higher SNR values (from 6.6181 to 7.5316 dB), e.g. PAX2 and OCT1 on Figure 7, they are up-regulated. This interesting discovery indicates that SNR measure is relevant to the gene expression to a certain extent.
Gene ontology analysis on the targeted genes in the modules
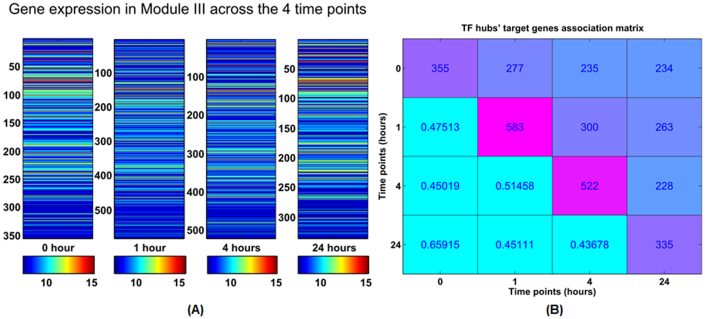
In addition to the identification of three ERα regulatory modules from the network, we further investigated the time-variant biological functions of those regulated targets during the transcription processes. Across the four time points, those regulated targets by those TF hubs also changed in the quantity, e.g. in Module III, the ERα target genes cover 355 genes at time point 0; at 1 hour those genes rise to 583; at 4 hours these regulated genes begins falling to 522; and at time 24 hours, the genes fall to 335 (Figures 8(A) and 8(B), Supplemental Figures S8(A) and (B)).
Figure 8. The statistical analysis of the regulated genes by Module III across the four time points.
(A) The subplots ilustrate the regulated gene expression by Module III at each time, respectively. At time 0 hour, Module III directly regulates the 355 genes; at 1 hour, Module III regulates 583 genes; for time 4 hours, the regulated genes fall down to 522, and 335 at time 24 hours. (B) The right panel depicts the association matrix of those gene regulated by Module III across the four time points, the diagonal entries denote the individual gene number regulated by Module III across all four time points, and off-diagonal entries denote common gene number (percentage) between the corresponding two time points.
We then performed the GO analysis on those target genes regulated by each module. Through the GO functional comparison among those modules across the different time points, we found that Module III has the most significant time-variant properties during the whole transcriptional regulatory process; see the results on Table 2.
Table 2. The GO analysis on Module III across the four time points, respectively. Each table lists the top 10 GO annotation terms at each time, respectively, with their statistically acceptable p-values (<0.001).
| Time 0 hour | Time 4 hours | ||
|---|---|---|---|
| GO annotation | p-value | GO annotation | p-value |
| phosphoprotein | 6.80E-10 | phosphoprotein | 1.34E-09 |
| calcium binding, subdomain | 7.85E-05 | calcium ion binding | 3.40E-05 |
| nucleus | 2.02E-04 | calcium-dep. membrane targeting | 2.75E-04 |
| short sequence motif: DEAH box | 6.25E-04 | cell adhesion | 3.65E-04 |
| cell cycle | 1.24E-03 | Golgi apparatus | 5.78E-04 |
| cellular response to stress | 1.31E-03 | short sequence motif: DEAH box | 6.05E-04 |
| cytoplasm | 2.33E-03 | alternative splicing | 1.39E-03 |
| response to DNA damage stimulus | 4.05E-03 | response to drug | 1.64E-03 |
| meiosis I | 5.16E-03 | sequence-specific DNA binding | 1.74E-03 |
| mutagenesis site | 5.34E-03 | response to DNA damage stimulus | 1.91E-03 |
From the GO analysis results, we found such terms as calcium binding, phosphoprotein and short sequence motif: DEAH box were the common ones throughout the four time points. At time point 0, several events were found relevant to the response to bacterium or other organism; at time point 1 hour, calcium-dependent membrane targeting, response to DNA damage stimulus and cell adhesion events were found. At time point 4 hours, splice variant, mutagenesis site, sequence variant and polymorphism events were also discovered. And for the last time point of 24 hours, helicase ATP-binding, ATPase activity and alternative splicing were also identified. The alternative splicing events were found at all other time points with the exception of time point 0 hour. The GO analysis results for other modules (Modules I and II) are provided in Supplemental Figures S9 and S10.
Target gene signatures of selection and their clinical outcome analysis
One of the most evident advantages of genome-wide analysis via diverse cell lines is to globally identify gene signatures and corresponding clinical information with prognostic functions.
To address the association analysis on the ERα target genes in each of three modules and derive underlying clinical outcomes of breast cancers regarding patients' histological grade and survival rate, we adopt the Kaplan-Meier survival analysis approach and investigated the estrogen receptor status, histological grade (stage) of three breast cancer patient groups composed of 337, 251 and 137 patients41,42,43,44, respectively.
Normally Kaplan-Meier survival probability is adopted for the analysis purpose and differences in survival are further statistically estimated by the log-rank test45,46,47. The analysis results can provide statistically meaningful insights into the relationship between diverse patients' treatment results and certain gene signatures.
For each patient cohort, we adopted an unsupervised clustering approach (k-means) and clustered the patients into 4 different subgroups (PGs) based on their gene signatures, and then associated the targeted genes in each modules with corresponding clinical pathological features, i.e. survival years, histological stages and estrogen receptor status.
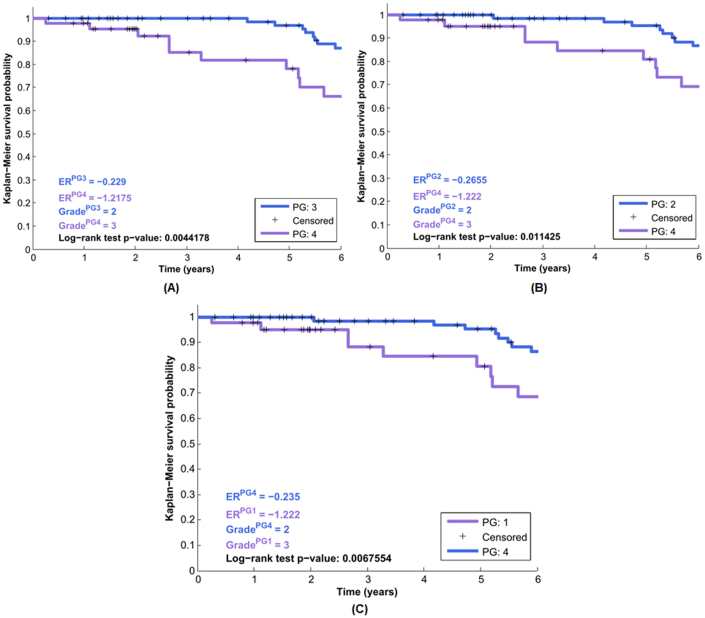
Through analyzing the clinical cohort of 337 patients from van de Vijver, et al.41,42, we found that the targeted genes in each modules had the reasonable predictive abilities to render a significant survival correlation with log-rank p-values much less than 0.05, see Figure 9. Figure 9(A) gives clinical prediction for the patient subgroups PG:3 vs PG:4 (patient subgroups 3 and 4) with the log-rank test p-value < 0.005 (see more details for selecting gene signatures in Supplemental Materials).
Figure 9. The Kaplan-Meier survival analysis based on the target genes regulated by the three modules.
The clinical survival information of 337 breast cancer patients is selected from van de Vijver et al.41,42. The subplot (A) gives the statiscally siginificant result between the patient groups PG:3 vs PG:4 (Module I), and their corresponding group estrogen receptor status and survival stage (grade) information are also provided on the left bottom (log-rank test p-value: 0.0044178). The subplots (B) and (C) depict the analysis results on the patient groups PG:2 vs PG:4 (Module II), and PG:1 vs PG:4 (Module III), respectively.
The patient group under the investigation has the predicative characteristics of the early survival period (< = 6 years) compared with other patient groups, provided in the Supplemental Figures S12 and S13, respectively. And those patient subgroups in Figure 9 mainly came from the tumor grades (stages) II and III, with their estrogen receptor status −0.22 to −1.22 (ER-, negative ER status), respectively.
The clinical analysis results on the other two patient cohorts are provided in the Supplemental Figures S12 and S13, all of which have statically significant log-rank test p-values < 0.05.
Discussion
We proposed a Bayesian multivariate modeling approach for inferring the ERα-centered transcription network, and together the statistical properties of the inferred transcription network were analyzed. Our method utilized the E2-stimulated time-series ChIP-seq data for identifying TF hubs and their targets, and the time-series gene expression data for capturing transcription rates. Our approach is different from traditional linear modeling, fitting or regression approaches for analyzing transcriptional regulation, which is a hierarchical, nonlinear and dynamic-evolving process.
Given that the cellular signaling dynamically responds to external stimuli, it is reasonable to integrate time-series ChIP-seq and gene expression data to capture the time-variant network structures and further elucidate the inherent regulatory mechanisms. To our knowledge, the proposed method by integrating time-series ChIP-seq data and gene expression data has not yet been proposed for analyzing time-varying transcriptional networks.
Through analyzing the statistical properties of the inferred ERα network structure, we found that the SNR measure has statistical association with the inferred regulatory strength, i.e. the components with the higher SNRs tend to have the higher regulatory strength to any possible downstream targets. As a matter of fact, genes with the higher signal densities (SNR value) contributed much more to the inferred ERα-centered regulatory networks than those with the lower ones. Across all the four time points, we noticed this meaningful property from the inferred ERα-centered regulatory networks. Thus, the SNR measure can be an index for the transcriptional regulatory activities among the network components.
Furthermore from the association analysis between the SNR values and expression patterns of those module components, on Figures 5 and 6, we found that MYC with a low SNR value (4.3471) is up-regulated; components with high SNR values (from 5.4318 to 6.0196) tend to be down-regulated, see ERα, GATA3, CEBP, SP1, AP1 and FOXA1, except for XBP1. While for those with much higher SNR values (from 6.6181 to 7.5316), e.g. PAX2 and OCT1 on Figure 7, they are up-regulated. This interesting discovery indicates that to a certain degree the SNR measure is also relevant to the gene expression. The inference error statistical plots for those TF hubs' transcription rates are provided in the supplemental Figure S11.
And with the inherent recurrent motif patterns, we further discovered three self-embedded ERα regulatory modules. From the modularity properties and quantitative analysis, we found that feed-forward and bi-span loops contributed the most to the gene expression patterns in the ERα-centered transcription regulatory network. And other patterns, such as self-loop (activating or inhibitory) and interactive loop also played important roles in the ERα-centered transcription regulatory network.
The GO analyses on those regulatory modules and their targets indicated that distinct functions of ERα target genes were associated with each of the modules at different time points, demonstrating that our modularity analysis was indeed capable of discovering the functional association of the embedded modules together with their target genes.
Furthermore, to address the association analysis on the ERα target genes in each modules and potential clinical outcomes of breast cancer patients in different stages, we adopted the Kaplan-Meier survival analysis approach and further examined the estrogen receptor status, histological grade (stage) of the tumor. Based on the patient survival time, histological grade (stage) and the estrogen receptor status (ER negative/positive) information, the survival analysis results on three cancer patient data sets proved the embedded modules and their targets could render the reasonable clinical outcomes of statistical meanings.
In summary, through the Bayesian statistical modeling and modularity analysis, we not only revealed the dynamic properties of the ERα-centered regulatory network and its associated distinct biological functions, but also discussed the biological design principles from the time-series genomic (binding and expression) data. Furthermore we also performed the GO and clinical survival analysis on the modules and their targets. All the above proved the reliability and effectiveness of our proposed network and modularity analysis approach.
Methods
E2-stimulated time-series ERα ChIP-seq data
The protocol of ERα ChIP-seq was described in detail in our previous study and Supplemental Materials. Briefly, after serum starvation, MCF-7 cells48 were stimulated with 17β-estradiol (E2, 70 nM) for 1, 4 and 24 hours or DMSO (Control) at 0 hour time point.
After crosslinking, cells were treated by lysis buffers and sonicated to fragment the chromatin to a size range of 500 bp - 1 kbp. Chromatin fragments were then immunoprecipitated with 10 ug of antibody/magnetic beads. The antibodies against ERα were purchased from Santa Cruz Biotechnology (Santa Cruz, sc-8005 X). After immunoprecipitation, washing, and elution, ChIP DNA was purified by phenol:chloroform:isoamyl alcohol and solubilized in 70 μl of water. The library was constructed using an Illumina genomic DNA prep kit by following its protocol (Illumina, cat# FC-102-1002). DNA samples (20 nM per sample) quantified by an Agilent Bioanalyzer, were loaded onto an Illumina Genome Analyzer IIx (GAIIx) for sequencing according to the manufacturer's protocol. Reads generated from the Illumina GAIIx pipeline were aligned to the Human Genome Assembly (NCBI build 36.1/hg18) using the ELAND algorithm.
All raw and processed ChIP-seq datasets under this study have been deposited in the Gene Expression Omnibus (GEO) database at National Center for Biotechnology Information (www.ncbi.nlm.gov/geo, accession number: GSE35109).
E2-stimulated time-series gene expression microarray data
E2-stimulated time-series gene expression microarray data in this study were obtained from a publicly available resource and downloaded from the EMBL-EBI (www.ebi.ac.uk, accession number: E-TABM-742).
The raw microarray expression data were preprocessed with the quantile normalization, and then normalized by the log2-transformation.
Parameter-optimization in ChIP-seq data analysis
Due to the direct relationship between peak number and enrichment level of transcription regulation binding sites, parameter-optimization is a key pre-process step for the further ChIP-seq data analysis. There are several statistical parameters constraining the peak number output, e.g. FDR and bin-size, we need to determine an optimal set of binding events (peak number). Thus, we propose a flexible data feature detection algorithm which can be formulated as an optimal track process, illustrated as,
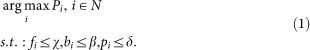 |
where Pi denotes a set of optimal peak numbers under corresponding argument constraints (FDR, bin-size and p-threshold), fi stands for the argument FDR, bi for the bin-size and pi for the p-threshold, χ, β and δ represent presupposed up-bound argument values, respectively.
Based on the optimized results from Equation (1), herein we define a track rate function (TR) to quantitatively characterize the inherent data features from diverse argument pair sets (peak number and FDR), depicted as,
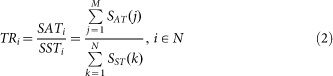 |
where M denotes the interval number for the scoring steps in numerator (actual track), N for the step number for the shortest track from the initial to the destination point in denominator. The shortest track is defined as one path from the initially-selected start (normally the point of minimum peak number with the most stringent FDR and bin-size constraints) to the optimal destination of maximum peak number, similar to the shortest path definition in graph theory, i.e. one path that connects two adjacent nodes but traverses the least intermediate nodes. The shortest track is programmed according to the existing path blocks, e.g. on the upper panel of Figure 2(A) the p-threshold interval is 0.003 and bin-size interval is 50 bp, where the track moves across one block, no matter which direction, then the corresponding score function increases one, denoted by SST. Its score function SST(·) is based on the steps needed for one shortest path, i.e. if n available steps are needed from an initial to a terminal point, then a score n will be assigned to the SST.
SAT represents an actual track score based on the interval M. The procedure for computing SAT is firstly to set an interval number, M and divide the range between minimum to maximum FDR into M intervals, denoted as δ, then from the minimum FDR, πn, one may search the suitable peak number subject to the current FDR threshold,  , the corresponding steps taken are scoring into the SAT function. The pseudo-code for detecting a track set for SAT is depicted as,
, the corresponding steps taken are scoring into the SAT function. The pseudo-code for detecting a track set for SAT is depicted as,
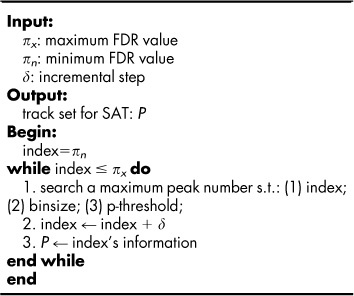 |
Equation (1) illustrates the maximization of peak number subject to argument constraints, FDR, bin-size and p-threshold. It is a parameter optimization since the whole process is to determine the satisfactory parameter pairs by means of adjusting constraints, see Figure 2(A). Equation (2) (track rate function) is introduced to quantitatively characterize the time-series ChIP-seq data features based on the two arguments, FDR and peak number.
Figure 2(B) depicts the relationship between the track rate function TR and interval number N, where we may discover that for this time point 0 hour, the track rate function TR reaches its equilibrium when N is larger than 35. The related plots at other time points are provided at the supplementary Figures S1, S2 and S3.
Furthermore, through comparing both TR measures across the four time points (0.33/0.23 → 0.78/0.72 → 0.61/0.39 → 0.46/0.35) with the common TF candidate counts (309 → 475 → 404 → 381) in Figure 2(D), we found such a parabolic trend in both TR values similar to the common TF candidate counts. Further analysis revealed a strong statistical correlation among those counts, i.e. the Pearson correlation 0.9811 (p-value: 0.0189) between the peak number TR values and the common TF candidates, and 0.9545 (p-value: 0.0455) between the FDR TR values and the common TF candidates across four time points, respectively. The strong statistical correlation between the defined TR function value and the identified TF candidates across the four time points indicates the predictive capability of the defined TR function for the TF candidate counts.
Bayesian multivariate statistical modeling of genetic transcription rates
In the Bayesian statistical modeling schema, prior information about the model parameter is denoted by the probability density function p(θ), the likelihood function is represented as p(X|θ), and the inference purpose is to derive the posterior density function p(θ|X). According to the Bayes theorem, the general inference can be formulated as,
For the multivariate case, p(X) can be specified by the decomposition,
where Pa(·) denote the parental node. It can be regarded as a normalized constant by integrating over all values of θ in the product p(X|θ)p(θ). Thus, the equation above can be formulated as,
The strength of the observation data and corresponding prior knowledge influence those diverse weights on the beliefs inferred from the multiple sources.
Due to a relatively small sample size of E2-stimulated time-series gene expression data which contain no knowledge of transcription factors, binding information and direct target genes, the conventional Bayesian modeling has its limitation. In order to infer ERα-centered regulatory network, we integrate the time-series E2-stimulated ERα ChIP-seq data, where it can detect transcription factors and hubs, and facilitate the further reverse-engineering of the regulatory network by means of inferring parameters in the Bayesian statistical framework.
Herein we propose a Bayesian multivariate statistical approach for modeling the time-variant ERα transcriptional regulatory network. The basic model framework is illustrated as follows,
where 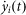 denotes the ith gene's transcription rate, xj(t) for the jth gene's expression level at the investigated time, αij for the corresponding regulatory strength of the jth gene that has any possible transcription regulatory activity on the ith gene, and ε represents the potential stochastic effects during the transcription regulatory process, which normally follows a normal distribution, i.e.
denotes the ith gene's transcription rate, xj(t) for the jth gene's expression level at the investigated time, αij for the corresponding regulatory strength of the jth gene that has any possible transcription regulatory activity on the ith gene, and ε represents the potential stochastic effects during the transcription regulatory process, which normally follows a normal distribution, i.e.
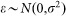 .
.
Thus for a genetic regulatory network containing M transcription factors at T time points, the above equation can be organized as,
where 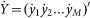 denotes the transcription rate matrix of M transcription factors,
denotes the transcription rate matrix of M transcription factors, 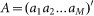 denote the regulatory coefficient matrix,
denote the regulatory coefficient matrix, 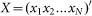 gene matrix and
gene matrix and 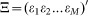 the error term. Thus, inferring the coefficient matrix A of the above equation is to acquire concrete knowledge about the transcription regulatory strength of transcription factors over diverse target genes under investigation.
the error term. Thus, inferring the coefficient matrix A of the above equation is to acquire concrete knowledge about the transcription regulatory strength of transcription factors over diverse target genes under investigation.
The detailed analysis for deriving the posterior mean estimation is given in the Supplemental Materials, Section 3.
Signal power-based criteria for defining regulatory strength
In consideration of the large p-value problem incurred by conventional correlation analysis on short observation samples, we introduce a signal-to-noise (SNR) measure for characterizing the difference of those investigated genes' expression levels. By definition, the SNR measure is used for quantifying the corruption degree of a signal by the inherent systematic noise, defined as49,
where P denotes the average power operator. For most periodic signals, the average power is defined equally to the square of the root-mean-square (RMS), that is,
where T denotes the signal period. For characterizing the expression process, here we consider one gene's mean expression level as the useful signal content and the deviation of the gene's global expression level as the noise factor. The SNR measure can be expressed using the logarithmic decibel scale in standard unit dB.
In our work, we considered ChIP-seq and microarray datasets across the four time points, thus the SNR value for each network node (gene) was calculated on the selected microarray expression values across the four time points.
Author Contributions
Conceived and designed the model and method: BHT and VXJ. Performed the experiment: HKH and PYH. Analyzed the data: BHT. Suggestions on the scripts and the method: RB and SSC. Wrote the paper: BHT and VXJ. Led and coordinated the project: VXJ and THMH.
Supplementary Material
Suppl. Methods and Materials
Suppl. File 1
Suppl. File 2
Acknowledgments
This work has been supported by the research grants from the Ohio Cancer Research Associates (OCRA) and Department of Biomedical Informatics, The Ohio State University Medical Center, Columbus, Ohio, USA. We sincerely thank editors and anonymous referees for their help and suggestions that led to the improvement of our manuscript.
References
- Sprinzak D. & Elowitz M. B. Reconstruction of genetic circuits. Nature 438, 443–448 (2005). [DOI] [PubMed] [Google Scholar]
- Goymer P. Systems biology: Merging data means more powerful networks. Nat Rev Genet 9, 501–501 (2008). [Google Scholar]
- Swami M. Gene regulation: Modelling by building blocks. Nat Rev Genet 10, 3–3 (2009). [Google Scholar]
- Blow N. Systems biology: Untangling the protein web. Nature 460, 415–418 (2009). [DOI] [PubMed] [Google Scholar]
- Davidson E. H. Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenco M. et al. Dissection of a complex transcriptional response using genome-wide transcriptional modelling. Mol Syst Biol 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-C., Huang H.-C., Juan H.-F. & Chen S.-T. GeneNetwork: an interactive tool for reconstruction of genetic networks using microarray data. Bioinformatics 20, 3691–3693 (2004). [DOI] [PubMed] [Google Scholar]
- Xing B. & van der Laan M. J. A causal inference approach for constructing transcriptional regulatory networks. Bioinformatics 21, 4007–4013 (2005). [DOI] [PubMed] [Google Scholar]
- Lemmens K. et al. Inferring transcriptional modules from ChIP-chip, motif and microarray data. Genome Biology 7, R37 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Vainas O., Harbison C. T., Simon I. & Bar-Joseph Z. Reconstructing dynamic regulatory maps. Mol Syst Biol 3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Jensen S. & Stoeckert C. Clustering of genes into regulons using integrated modeling-COGRIM. Genome Biology 8, R4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R. et al. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N., Yan K.-K. & Gerstein M. B. Analysis of diverse regulatory networks in a hierarchical context shows consistent tendencies for collaboration in the middle levels. Proceedings of the National Academy of Sciences 107, 6841–6846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G. M. et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Research 17, 898–909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P. J. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10, 669–680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D. et al. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-seq data. PLoS Comput Biol 7, e1002111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Carroll R. J. & Zhao H. Bayesian error analysis model for reconstructing transcriptional regulatory networks. Proceedings of the National Academy of Sciences 103, 7988–7993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicatiello L. et al. Estrogen receptor α controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and MicroRNAs. The American Journal of Pathology 176, 2113–2130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R. et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M. et al. Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes & Development 24, 2219–2227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V. X. et al. Identifying estrogen receptor α target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Research 32, 6627–6635 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F. et al. Inference of hierarchical regulatory network of estrogen-dependent breast cancer through ChIP-based data. BMC Systems Biology 4, 170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D. & Carroll J. S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V. X., Rabinovich A., Squazzo S. L., Green R. & Farnham P. J. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data - A case study using E2F1. Genome Research 16, 1–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V. X., Apostolos J., Nagisetty N. S. V. R. & Farnham P. J. W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data. Bioinformatics 25, 3191–3193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S. et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FOXA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- DeNardo D. G. et al. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: identification of estrogen-induced/activator protein-1-dependent genes. Molecular Endocrinology 19, 362–378 (2005). [DOI] [PubMed] [Google Scholar]
- Carroll J. S. et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38, 1289–1297 (2006). [DOI] [PubMed] [Google Scholar]
- Kong S. L., Li G., Loh S. L., Sung W.-K. & Liu E. T. Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A. et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 456, 663–666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Sharma C. G. N. & Jordan V. C. Estrogen regulation of X-box binding protein-1 and its role in estrogen induced growth of breast and endometrial cancer cells. Horm Mol Biol Clin Investig 2, 235–243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. J. Bayesian methods in bioinformatics and computational systems biology. Briefings in Bioinformatics 8, 109–116 (2007). [DOI] [PubMed] [Google Scholar]
- van Steensel B. et al. Bayesian network analysis of targeting interactions in chromatin. Genome Research 20, 190–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan A. & Forster J. J. Kendall's advanced theory of statistics: Bayesian inference. 2nd edn, (Wiley, John & Sons, 2004). [Google Scholar]
- Bar-Joseph Z. et al. Computational discovery of gene modules and regulatory networks. Nat Biotech 21, 1337–1342 (2003). [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Pavlicev M. & Cheverud J. M. The road to modularity. Nat Rev Genet 8, 921–931 (2007). [DOI] [PubMed] [Google Scholar]
- Barabasi A.-L., Gulbahce N. & Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002). [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nature Reviews Genetics 8, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- Pe'er D. & Hacohen N. Principles and strategies for developing network models in cancer. Cell 144, 864–873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002). [DOI] [PubMed] [Google Scholar]
- van de Vijver M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- Miller L. D. et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. PNAS 102, 13550–13555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. Journal of the National Cancer Institute 98, 262–272 (2006). [DOI] [PubMed] [Google Scholar]
- Kaplan E. L. & Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association 53, 457–481 (1958). [Google Scholar]
- Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemotherapy Reports 50, 163–170 (1966). [PubMed] [Google Scholar]
- Harrington D. Linear rank tests in survival analysis. Encyclopedia of Biostatistics (2005). [Google Scholar]
- Cheng A. et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell 21, 393–404 (2006). [DOI] [PubMed] [Google Scholar]
- Oppenheim A. V. & Schafer R. W. Discrete-time signal processing. 3rd edn, (Prentice Hall, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Methods and Materials
Suppl. File 1
Suppl. File 2