Background: Stimulation of calcineurin phosphatase activity is required for diverse cellular responses to calcium signals.
Results: Disruption of calcineurin regulatory subunit myristoylation causes constitutive activation of calcineurin-dependent gene transcription in yeast.
Conclusion: Myristoylation of calcineurin B limits phosphatase activation by intracellular calcium signals.
Significance: Our findings reveal that N-myristoyltransferase activity regulates the calcium sensitivity of calcineurin activation.
Keywords: Calcineurin, Calcium Signaling, Cell Signaling, Protein Myristoylation, Protein Phosphatase, Crz1, N-Myristoyltransferase
Abstract
The Ca2+/calmodulin-stimulated protein phosphatase calcineurin is a critical component of Ca2+ signaling cascades in eukaryotic cells. Myristoylation of the regulatory subunit of calcineurin (CNB) is conserved from yeast to humans. Here, we show that CNB myristoylation antagonizes phosphatase activation in yeast. Disruption of CNB myristoylation by mutation of the myristoylated glycine triggered constitutive expression of a calcineurin-dependent reporter gene and enhanced calcineurin-dependent phenotypes. Basal phosphatase activity was also increased in nmt1–181 yeast with reduced N-myristoyltransferase activity. Our findings are the first demonstration of a functional role for CNB myristoylation and reveal the importance of Nmt1 in modulating cellular calcineurin activation.
Introduction
Ca2+ is a ubiquitous second messenger utilized by cells to orchestrate responses to diverse environmental stimuli and developmental cues. Cellular responses to Ca2+ signals are determined by the amplitude, duration, frequency, and localization of Ca2+ transients via activation of downstream Ca2+-dependent signaling molecules (1–3). An important determinant of Ca2+ signaling output in eukaryotes is the Ca2+- and calmodulin-stimulated protein phosphatase calcineurin. Calcineurin regulates numerous cellular processes, including cell proliferation, differentiation, survival, and death, via its ability to regulate the activity of transcription factors, ion channels, and cell signaling molecules (4).
A key target of calcineurin in mammals is the nuclear factor of activated T cells (NFAT)2 family of transcription factors. Activation of NFAT by calcineurin-mediated dephosphorylation is necessary for lineage specification in embryonic stem cells (5). Calcineurin-NFAT signaling is also essential for the formation and function of the cardiovascular, nervous, immune, and musculoskeletal systems (6, 7). Calcineurin-NFAT signaling must be appropriately regulated in vivo. Mice expressing an NFAT allele with increased affinity for calcineurin exhibit developmental abnormalities (8). Alterations in calcineurin activity have been linked to multiple human diseases including cardiac hypertrophy (9, 10), cancer (11, 12), and neurodegeneration (13). Inhibition of calcineurin phosphatase activity underlies the therapeutic action of the immunosuppressants tacrolimus and cyclosporine A, which have been extensively administered to patients to prevent organ transplant rejection (14). Binding of immunosuppressant/immunophilin complexes to calcineurin inhibits phosphatase activity, thereby preventing T-cell activation.
Calcineurin is a heterodimer composed of a catalytic subunit CNA and a smaller regulatory subunit CNB that confers Ca2+ responsiveness to phosphatase activation (15). Full phosphatase activation further requires the recruitment of Ca2+/calmodulin to displace the autoinhibitory domain of CNA from the catalytic core. Thus, calcineurin activation by intracellular Ca2+ transients requires the coordinated action of two Ca2+-binding proteins. Given the diversity of Ca2+ signals occurring in specific cell types during development, additional mechanisms must exist to control the spatial and temporal dynamics of calcineurin signaling in vivo. Endogenous factors such as regulator of calcineurin (RCAN), A-kinase-anchoring protein 79 (AKAP79), and superoxide dismutase 1 (SOD1) interact with calcineurin to modulate phosphatase activity, localization, and stability (16, 17). Substrate competition also determines the output of calcineurin signaling in vivo (18). The regulation of calcineurin function via calcineurin-substrate, calcineurin-scaffolding protein, and calcineurin-inhibitory factor interactions has recently been reviewed (19). In contrast, the contribution of calcineurin protein modifications in shaping phosphatase activity in vivo remains largely unknown.
Ca2+ signaling cascades are highly conserved from yeast to humans. As observed for mammalian calcineurin, stimulation of phosphatase activity in yeast requires Ca2+/calmodulin and is sensitive to inhibition by immunosuppressant/immunophilin complexes or regulator of calcineurin overexpression (4, 21). The regulatory subunit of yeast calcineurin is encoded by a single gene, CNB1 (22). The catalytic subunit is encoded by two genes, CNA1 and CNA2 (23). Yeast lacking functional calcineurin due to genetic deletion of either CNB1 (cnb1Δ) or both CNA1 and CNA2 (cna1Δ cna2Δ) are viable, but exhibit reduced survival following exposure to environmental stress (24). Calcineurin-mediated responses to cell stress requires activation of the transcription factor Crz1 (calcineurin-responsive zinc finger) (25, 26), which is regulated in a manner similar to mammalian NFAT. In the absence of Ca2+ signaling, Crz1 is phosphorylated and cytoplasmic. Ca2+ signaling stimulates calcineurin-dependent Crz1 dephosphorylation, leading to Crz1 accumulation in the nucleus where it activates transcription of genes required for calcium homeostasis and stress response (27). We are taking advantage of the yeast model system to identify cellular mechanisms regulating phosphatase activity. As a consequence of our mutational analysis of the evolutionarily conserved serine residue located within the Cnb1 myristoylation consensus site sequence, we have discovered a role for Cnb1 myristoylation in preventing constitutive calcineurin activity in yeast.
EXPERIMENTAL PROCEDURES
Yeast Culture
All yeast culture and transformations were conducted using standard techniques. Yeast rich medium (YPD) or synthetic medium (SD) were purchased from Clontech. Agar, amino acids, and salts were purchased from Difco and Sigma. YPD medium was buffered to pH 5.5 by the addition of 0.5 m succinic acid as indicated. Yeast strains used in this study are noted in Table 1.
TABLE 1.
Yeast strains used in this study
Cloning and Mutagenesis
Yeast CNB1 genomic DNA plasmid pYDZ3 was a kind gift of J. Heitman (28). CNB1 genomic region was cloned into pRS313 BamHI-XhoI to generate pTJK100. QuikChange site-directed mutagenesis (Stratagene) of pTJK100 was conducted according to the manufacturer's instructions to generate mutant CNB1 alleles: CNB1-S6A, GACCATCCACAATTTTGGCAGGAGCAGCACCCAT; CNB1-S6D, GACCATCCACAATTTTGTCAGGAGCAGCACCCAT; CNB1G2A, CATTTTTATTTCTTAAAATGGCTGCTGCTCCTTCCAAAATTG; CNB1-G2E, CATTTTTATTTCTTAAAATGGAAGCTGCTCCTTCCAAAATTG. Constructs were verified by DNA sequencing.
Ion Tolerance Assays
Yeast strains were inoculated into YPD supplemented with a range of LiCl or MnCl2 concentrations as indicated. Yeast were grown as 0.18-ml cultures in 96-well flat bottom dishes in a 30 °C incubator without shaking for 2 days. The A600 was measured using a BioTek μQuant plate reader following resuspension of yeast.
β-Galactosidase Assays
Yeast cultures were grown overnight in SD lacking histidine and uracil, pelleted, and resuspended in YPD (pH 5.5) medium supplemented with CaCl2 as indicated and plated in 24-well dishes. Following 4 h incubation at 30 °C, β-galactosidase activity was measured as described previously (21).
Protein Analysis
Yeast were grown overnight in SD-His medium, pelleted, resuspended in YPD, and cultured an additional 4 h. Yeast were quantitated by A600, and an equal number of yeast were harvested for protein extraction as described previously (21). Proteins were separated by SDS-PAGE on 4–20% Precise protein gels (Thermo Scientific) and subjected to Western blotting using anti-Cnb1 (28). For membrane association studies, cnb1Δ yeast expressing wild type Cnb1, Cnb1G2A, or Cnb1G2E were grown in YPD (pH 5.5) for 4 h before harvesting by bead beating in ice-cold Hepes buffer (20 mm, pH 7.2) with 2 mm EDTA, 1 mm DTT, 1 mm PMSF, and 1× Halt protease and phosphatase inhibitor mixture. Lysates were clarified at 5000 rpm microcentrifugation for 5 min and then subjected to high-speed ultracentrifugation (100,000 × g) for 30 min. Samples were separated by SDS-PAGE and probed with anti-Cnb1p. Total extract sample was collected after initial clarification, prior to 2-fold dilution of extracts for ultracentrifugation, and is thus loaded on gel at 2×.
RESULTS
To identify cellular mechanisms regulating calcineurin activity, we initiated a mutational analysis of the regulatory subunit of calcineurin in yeast. Initially, we focused on mutating the 8 serine residues conserved between yeast Cnb1 and human CNB because serine residues are potential sites for protein phosphorylation. During our analysis, we observed that mutation of the serine residue located within the myristoylation consensus sequence (Ser-6) increased basal calcineurin activity. Site-directed mutagenesis of CNB1 was conducted to mutate Ser-6 of Cnb1 to either alanine (Cnb1-S6A) or aspartic acid (Cnb1-S6D) to generate potential phosphoryl-defective and phosphoryl-mimic alleles. The functional consequence of the Ser-6 mutations was subsequently determined by assaying the ability of the mutant Cnb1 to restore calcineurin activity to yeast in which the chromosomal CNB1 gene has been deleted (cnb1Δ).
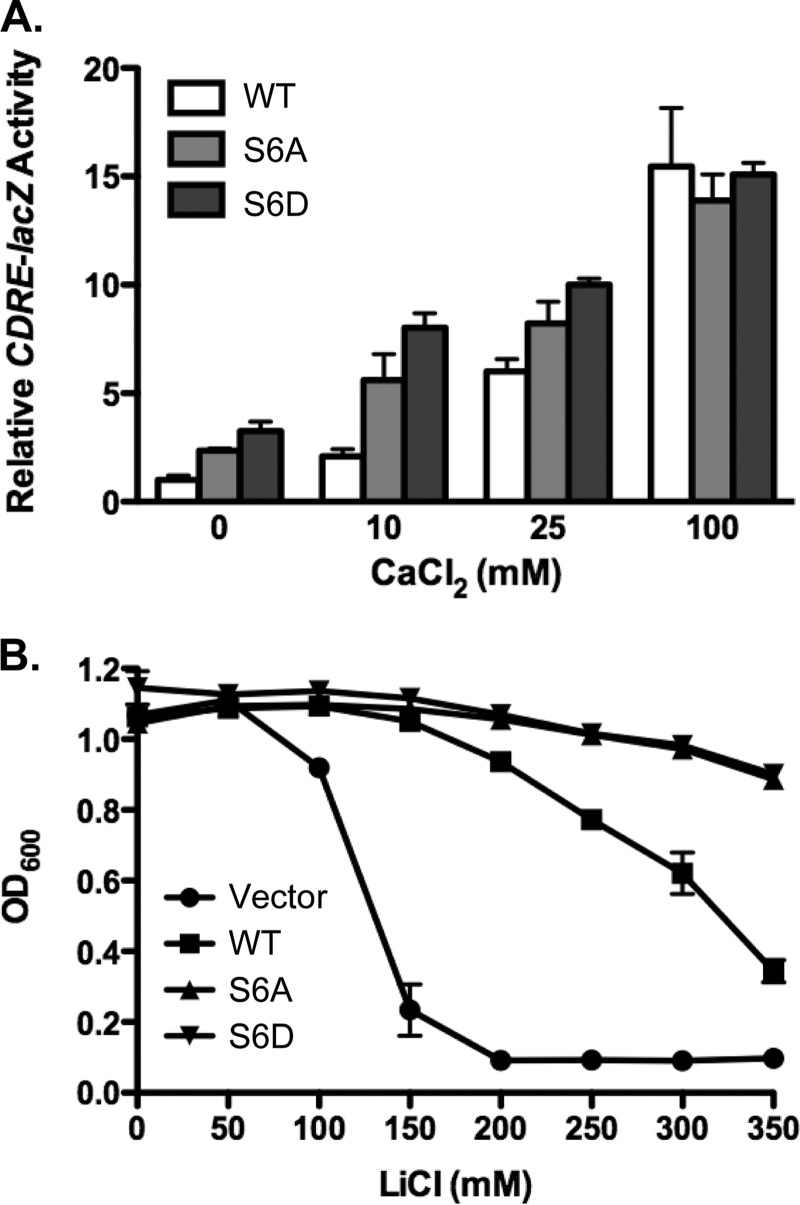
Calcineurin activity in yeast is readily quantitated by measuring expression of the calcineurin-dependent reporter gene CDRE-lacZ (calcineurin-dependent response element), composed of four copies of the minimal binding domain of the calcineurin-dependent transcription factor Crz1 (26). Ca2+ influx triggered by the addition of extracellular CaCl2 leads to robust stimulation of CDRE-lacZ activity. Cnb1Δ yeast harboring the CDRE-lacZ reporter gene were transformed with low-copy centromere-based plasmids expressing wild type Cnb1, Cnb1-S6A, or Cnb1-S6D under control of the endogenous CNB1 promoter. Calcineurin activity was assessed by stimulating yeast with a range of added extracellular CaCl2 (0–100 mm) to initiate Ca2+ signaling. Four hours after stimulation, yeast were harvested for quantitative β-galactosidase assays to measure CDRE-lacZ activity. As expected, Ca2+ influx triggered by the addition of extracellular CaCl2-stimulated CDRE-lacZ activity in a dose-dependent manner in yeast expressing wild type Cnb1 (Fig. 1A). Interestingly, yeast expressing Cnb1-S6A or Cnb1-S6D exhibited 2–3-fold increased CDRE-lacZ activity relative to wild type Cnb1 in response to either 0 mm or 10 mm added extracellular CaCl2. In contrast, no significant difference in CDRE-lacZ activity was observed between yeast expressing wild type Cnb1, Cnb1-S6A, or Cnb1-S6D in response to a strong Ca2+ signaling stimulus of 100 mm extracellular CaCl2. Thus, mutation of Ser-6 specifically enhanced calcineurin activity at low intracellular Ca2+ levels, resulting in constitutive phosphatase activation.
FIGURE 1.
Conserved serine S6 modulates calcineurin activity. The ability of Cnb1-S6A and Cnb1-S6D mutants to complement cnb1Δ yeast was assayed using calcineurin-dependent reporter gene assays and calcineurin-mediated ion-resistant growth assays. A, Cnb1Δ yeast harboring the CDRE-lacZ reporter gene were transformed with centromere-based plasmids expressing wild type Cnb1, Cnb1-S6A (S6A) or Cnb1-S6D (S6D). Yeast were subsequently grown in YPD (pH 5.5) in the presence or absence of added extracellular CaCl2 (0, 10, 25, or 100 mm). Four hours after CaCl2 stimulation, yeast were harvested for quantitative β-galactosidase assays. Data plotted are average of four independent yeast transformants ± S.D. B, Cnb1Δ yeast expressing the indicated CNB1 alleles were grown in YPD supplemented with increasing concentrations of LiCl (in mm) at 30 °C for 2 days. Growth was quantitated by measuring A600. Yeast transformed with empty vector were included as a negative control. Data points are average of four independent yeast transformants assayed in parallel ± standard error.
Because the primary role of calcineurin signaling in yeast is to mediate stress responses, we tested whether the enhanced calcineurin activity observed in yeast expressing Cnb1-S6A or Cnb1-S6D could confer a selective advantage in response to environmental stress. The ability of yeast to grow in the presence of high extracellular Li+ is dependent upon calcineurin signaling (29). We therefore compared the ability of cnb1Δ yeast transformed with Cnb1, Cnb1-S6A, or Cnb1-S6D to grow in the presence of a range of extracellular Li+. Yeast transformed with empty vector, and therefore lacking functional calcineurin, were included as negative control. Li+-resistant growth was assayed by measuring the A600 2 days after inoculation of yeast into medium supplemented with increasing concentrations of LiCl (Fig. 1B). As expected, growth of cnb1Δ yeast transformed with empty vector was inhibited by the presence of extracellular LiCl, exhibiting 75% inhibition of growth at 150 mm LiCl. In contrast, the growth of yeast transformed with wild type Cnb1 was resistant to LiCl, retaining greater than 50% growth in the presence of 300 mm extracellular LiCl. Consistent with the increased calcineurin activity observed in CDRE-lacZ reporter gene assays, yeast expressing Cnb1-S6A or Cnb1-S6D exhibited even more resistance to extracellular Li+ than yeast with wild type calcineurin. Cnb1-G2A- and Cnb1-G2E-expressing yeast maintained greater than 90% cell growth at 350 mm LiCl, the highest concentration of ion tested. These observations suggest that Ser-6 limits calcineurin activity in response to endogenous intracellular Ca2+ signaling triggered by environmental stress.
Although our initial mutations were designed to generate phosphoryl-defective and phosphoryl-mimic versions of Cnb1, our observation that both Cnb1-S6A and Cnb1-S6D enhance calcineurin-dependent phenotypes is not consistent with Ser-6 modulating calcineurin activation via a phosphorylation-dependent switch. Closer examination of the Cnb1 sequence revealed that Ser-6 (underlined) is located within the Cnb1 myristoylation consensus site Met-Gly-X-X-X-Ser-X-X-X. N-Myristoylation refers to the covalent attachment of myristate, a short 14-carbon fatty acid, to the amino-terminal glycine residue following removal of the initiating methionine residue during protein translation (30, 31). Myristoylation of CNB is conserved from yeast to humans (28, 32), but a functional consequence of this modification had not previously been reported.
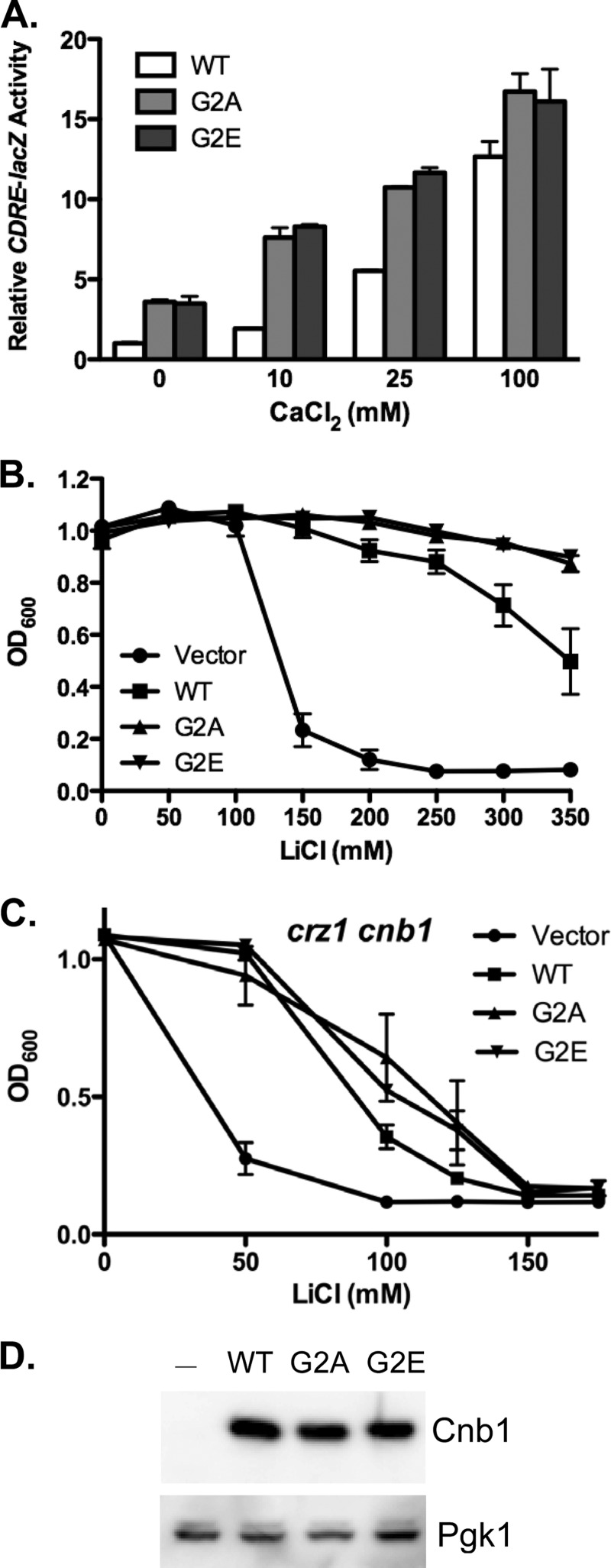
To test the possibility that the increased calcineurin activity observed in yeast expressing Cnb1-S6A and Cnb1-S6D was due to the loss of Cnb1 myristoylation, we conducted site-directed mutagenesis to mutate the myristoylated glycine residue to alanine (Cnb1-G2A). The G2A mutation has previously been shown to disrupt Cnb1 myristoylation in yeast (28). Plasmids expressing either wild type Cnb1 or Cnb1-G2A were transformed into cnb1Δ yeast harboring the CDRE-lacZ reporter gene. The functional consequence of the Cnb1-G2A mutation on calcineurin activity was assayed by quantitating CDRE-lacZ activity following stimulation by the addition of a range of extracellular CaCl2. Similar to our observations for Cnb1-S6A and Cnb1-S6D, expression of Cnb1-G2A increased CDRE-lacZ activity in unstimulated (0 mm CaCl2) and submaximally (10 and 25 mm CaCl2) stimulated yeast (Fig. 2A). In contrast, similar levels of CDRE-lacZ activity were observed for wild type Cnb1 and Cnb1-G2A following the addition of 100 mm extracellular CaCl2. Thus, disruption of Cnb1 myristoylation by the G2A mutation stimulates calcineurin activity in yeast under submaximal Ca2+ signaling conditions.
FIGURE 2.
Cnb1 myristoylation antagonizes calcineurin activation. The myristoylated glycine residue of Cnb1 was mutated to either alanine (Cnb1-G2A) or glutamic acid (Cnb1-G2E). Mutants were then tested for their ability to restore calcineurin-mediated gene expression and ion-resistant growth to cnb1Δ yeast. A, Cnb1Δ yeast harboring the CDRE-lacZ reporter gene were transformed with plasmids expressing Cnb1, Cnb1-G2A, or Cnb1-G2E. Yeast growing in YPD (pH 5.5) were stimulated for 4 h by the addition of extracellular CaCl2 prior to harvesting for quantitative β-galactosidase assays. Data plotted are average of four independent yeast transformants ± S.D. B and C, ion tolerance assays. cnb1Δ (B) or cnb1Δ crz1Δ (C) yeast were transformed with Cnb1, Cnb1-G2A, Cnb1-G2E, or empty vector as indicated. Yeast were inoculated into YPD containing increasing concentrations of LiCl (in mm) and grown at 30 °C for 2 days. Growth was quantitated by measuring the A600. Data points are average of three independent yeast transformants assayed in parallel ± S.E. D, anti-Cnb1 Western blot analysis of protein extracts prepared from cnb1Δ yeast transformed with wild type Cnb1, Cnb1-G2A, Cnb1-G2E, or empty vector. Anti-Pgk1 was used for loading control comparison.
Examination of the dbSNP database revealed an SNP (rs61757747) in which the myristoylated glycine of CNB is mutated to glutamic acid. The CNB SNP was reported from a large-scale analysis of cancer patients,3 but the functional significance or clinical relevance of the SNP is unknown. To determine whether the glycine to glutamic acid mutation would similarly stimulate calcineurin activity, we performed site-directed mutagenesis to generate a CNB1-G2E allele. As observed for Cnb1-G2A, expression of Cnb1-G2E resulted in constitutive expression of CDRE-lacZ and enhanced CDRE-lacZ expression in response to submaximal Ca2+ signaling.
To test the ability of Cnb1-G2A and Cnb1-G2E to enhance calcineurin activity in response to endogenous Ca2+ signaling, we conducted ion tolerance growth assays. As observed for yeast expressing Cnb1 Ser-6 mutants, yeast expressing Cnb1-G2A or Cnb1-G2E exhibited increased resistance to extracellular Li+, retaining 90% growth even at 350 mm LiCl (Fig. 2B). A similar enhancement of calcineurin-mediated Mn2+ resistance was also observed in yeast expressing Cnb1-G2A or Cnb1-G2E as compared with wild type Cnb1 (supplemental Fig. 1). Thus, introduction of the G2A or G2E mutation into Cnb1 increases calcineurin-dependent ion-tolerant growth in yeast. Comparison of Cnb1, Cnb1-G2A, and Cnb1-G2E protein by Western blot revealed similar levels of expression (Fig. 2C), indicating that the increased calcineurin activity observed in yeast expressing Cnb1-G2A or Cnb1-G2E was not simply due to increased Cnb1 expression. These observations suggest that disruption of Cnb1 myristoylation leads to increased calcineurin activity in yeast in response to endogenous Ca2+ signaling.
Dephosphorylation and activation of Crz1 are a critical component of calcineurin-mediated stress responses. To determine whether calcineurin activity toward additional targets is also enhanced in the absence of Cnb1 myristoylation, crz1Δ cnb1Δ yeast were transformed with Cnb1, Cnb1-G2A, or Cnb1-G2E and assayed for Li+-resistant growth. In the absence of Crz1, calcineurin promotes a modest level of Li+ resistance (Fig. 2C). Crz1Δ yeast transformed with empty vector exhibit 50% growth inhibition at 40 mm LiCl. In contrast, crz1Δ yeast expressing wild type Cnb1 did not exhibit 50% growth inhibition until ∼90 mm LiCl. Yeast expressing either Cnb1-G2A or Cnb1-G2E had slightly higher Li+ resistance, retaining 50% growth at 110 mm LiCl. Thus, the enhanced calcineurin activity observed for Cnb1-G2A and Cnb1-G2E is not unique to Crz1.
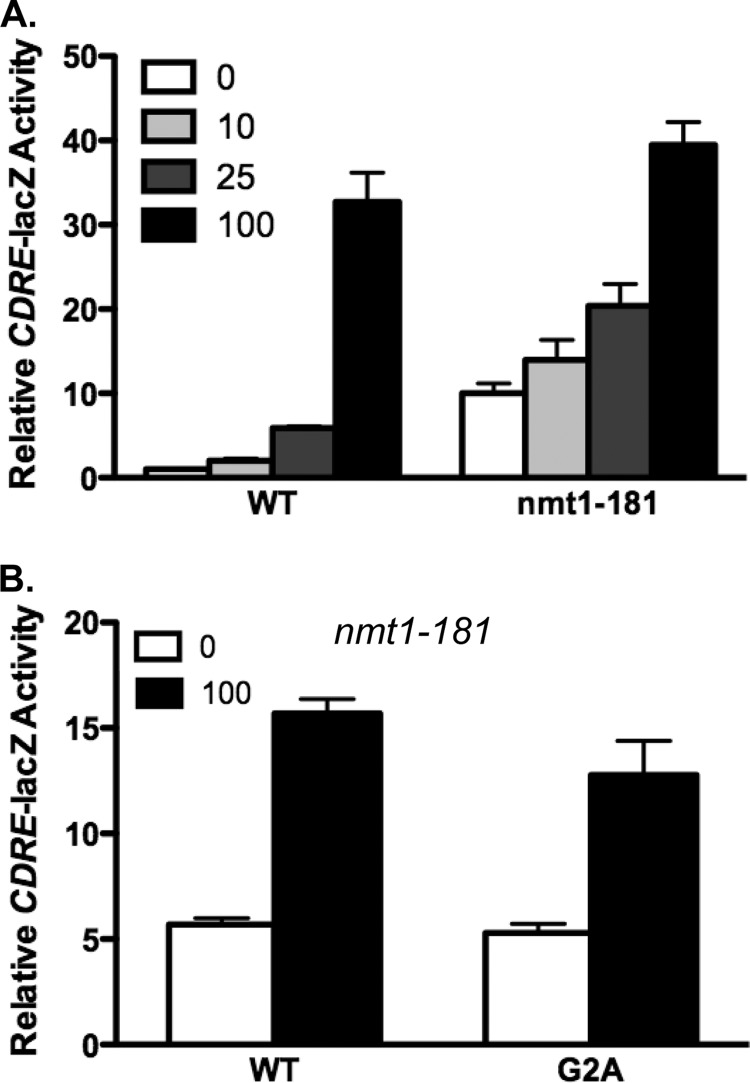
N-Myristoylation of proteins is mediated by N-myristoyltransferase (33, 34). If the enhanced calcineurin activity we observe in Cnb1-G2A- and Cnb1-G2E-expressing yeast is due to the loss of myristoylation, then reduced N-myristoyltransferase activity should similarly lead to enhanced basal calcineurin activity. N-Myristoyltransferase is encoded by the NMT1 gene in Saccharomyces cerevisiae (35). Although deletion of the NMT1 gene leads to the loss of viability in yeast, a temperature-sensitive allele (nmt1–181) with reduced activity has been reported (36). The nmt1–181 mutant and isogenic wild type yeast were transformed with the CDRE-lacZ reporter gene. Calcineurin activity was subsequently assayed by quantitating CDRE-lacZ activity at a semi-permissive temperature of 30 °C in medium supplemented with a range of CaCl2 from 0 to 100 mm. As shown in Fig. 3, nmt1–181 mutant yeast exhibited increased CDRE-lacZ activity in unstimulated or submaximally stimulated yeast as compared with wild type yeast. CDRE-lacZ activity was similar in wild type and nmt1–181 at 100 mm CaCl2. We subsequently generated nmt1–181 cnb1Δ yeast harboring the CDRE-lacZ reporter gene to test whether the increased basal activity observed for Cnb1-G2A calcineurin as compared with wild type Cnb1 could be explained solely by disruption of myristoylation. Following transformation of nmt1–181 cnb1Δ yeast with plasmids expressing either wild type Cnb1 or Cnb1-G2A, we quantitated CDRE-lacZ expression. Basal CDRE-lacZ activity was indistinguishable between nmt1–181 cnb1Δ expressing either wild type Cnb1 or Cnb1-G2A, indicating that the loss of myristoylation was responsible for the increased calcineurin activity associated with Cnb1-G2A.
FIGURE 3.
N-Myristoyltransferase antagonizes calcineurin activity. A, the CDRE-lacZ reporter gene was transformed into nmt1–181 or isogenic wild type yeast. Quantitative β-galactosidase assays were conducted following overnight growth at 30 °C in YPD (pH 5.5) supplemented with CaCl2 (in mm) as indicated. B, nmt1–181 cnb1Δ yeast harboring the CDRE-lacZ reporter gene were transformed with plasmids expressing wild type Cnb1, Cnb1-G2A, or Cnb1-G2E. Yeast were grown in YPD (pH 5.5) in the presence or absence of 100 mm added extracellular CaCl2 for 4 h prior to harvesting for quantitative b-galactosidase assays. Data shown are average of quadruplicates ± S.D.
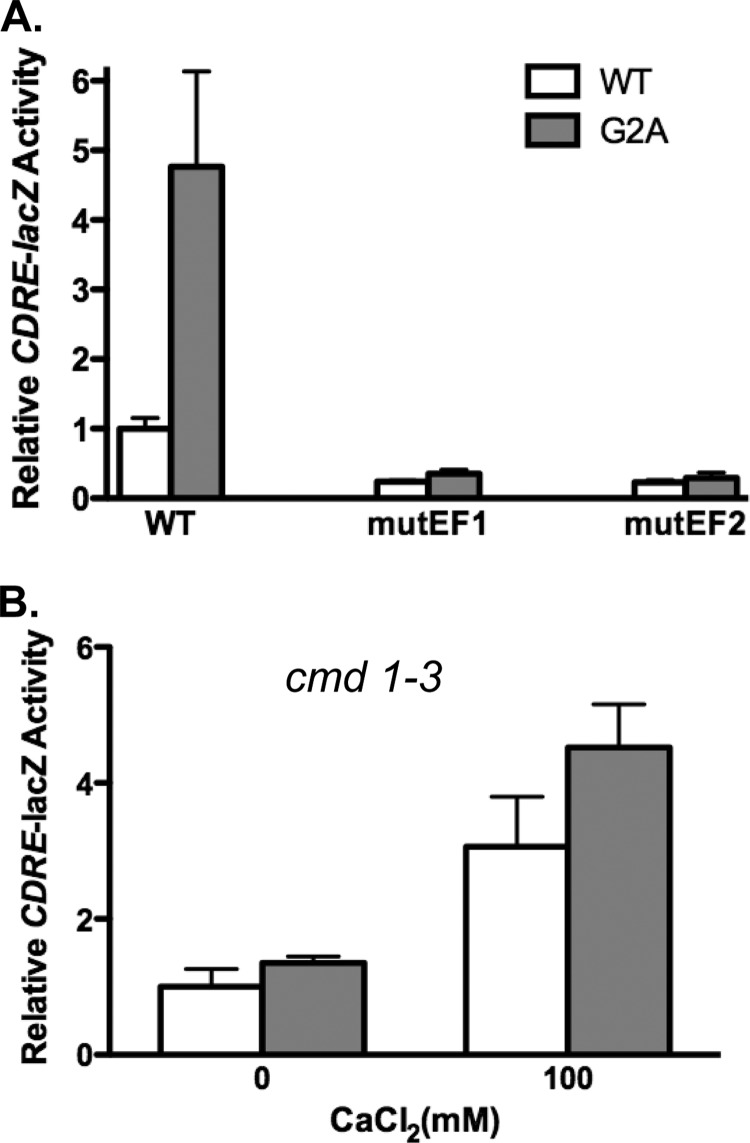
Activation of calcineurin requires Ca2+ binding to Cnb1 and recruitment of Ca2+/calmodulin. To determine whether the increased basal activity of Cnb1-G2A calcineurin reflected either enhanced Ca2+-dependent or enhanced Ca2+-independent phosphatase activation, we tested the consequences of mutating the EF hand Ca2+ binding domains of either Cnb1 or calmodulin. Cnb1, like its mammalian counterpart CNB, contains four EF hand Ca2+ binding domains. Mutation of either the first or the second Ca2+ binding EF hand domain of mammalian CNB inhibits calcineurin activation in vitro (37). We have recently obtained similar results when EF hand mutations are introduced into yeast Cnb1.4 Site-directed mutagenesis was conducted to independently mutate the first or second EF hand calcium binding domain of Cnb1 and Cnb1-S6A to generate Cnb1mutEF1, Cnb1mutEF2, Cnb1-G2AmutEF1, and Cnb1-G2AmutEF2. If disruption of myristoylation stimulates calcineurin activity by increasing the sensitivity of calcineurin to Ca2+, then mutation of the EF hand domains will prevent the enhanced phosphatase activity observed in Cnb1-G2A relative to Cnb1. Plasmids expressing either mutant or control Cnb1 or Cnb1-G2A were transformed into cnb1Δ yeast harboring the CDRE-lacZ reporter gene, and basal CDRE-lacZ activity was measured. As shown in Fig. 4A, introduction of the EF hand mutations reduces basal CDRE-lacZ expression. Comparison of yeast expressing Cnb1mutEF1 versus Cnb1-G2AmutEF1 or Cnb1mutEF2 versus Cnb1-G2AmutEF2 revealed that the presence of the EF hand mutations blocked the ability of the G2A mutation to stimulate basal calcineurin activity. Cnb1 and Cnb1-G2A EF hand mutant-expressing yeast exhibited similar levels of CDRE-lacZ activity. Thus, the increased phosphatase activity observed for Cnb1-G2A calcineurin does not bypass the requirement for Ca2+ binding. Enhanced Ca2+-dependent calcineurin activation upon the loss of Cnb1 myristoylation is consistent with our observation that the elevated basal activity of Cnb1-G2A calcineurin is inhibited by FK506 (supplemental Fig. 2A).
FIGURE 4.
Ca2+ and calmodulin requirements of myristoylation-defective calcineurin. A, site-directed mutagenesis of wild type Cnb1 and Cnb1-G2A was conducted to mutate the first or second EF hand Ca2+ binding domain as indicated. Plasmids expressing either wild type or mutant Cnb1 were transformed into cnb1Δ yeast harboring the CDRE-lacZ reporter gene. Quantitative β-galactosidase assays were conducted on yeast grown in YPD (pH 5.5) in the absence of added extracellular CaCl2 to measure basal CDRE-lacZ activity. B, Cnb1 or Cnb1-G2A expression plasmids were transformed into cmd1–3 cnb1Δ yeast harboring the CDRE-lacZ reporter gene. CDRE-lacZ activity was measured after 4 h of growth in YPD (pH 5.5) in the presence or absence of 100 mm CaCl2. Data plotted are average of quadruplicates ± S.D.
To further test the Ca2+ requirements of Cnb1-G2A basal activity, we tested the consequence of expressing Cnb1-G2A in cmd1–3 mutant yeast, which expresses a mutant form of calmodulin that cannot bind Ca2+ or stimulate calcineurin activity (38). If the enhanced basal calcineurin activity observed in Cnb1-G2A-expressing yeast reflects increased Ca2+-dependent phosphatase activation, then the loss of calmodulin function should disrupt the increase in basal activity. Comparison of basal CDRE-lacZ activity between Cnb1 and Cnb1-G2A calcineurin in cnb1Δ cmd1–3 yeast revealed similar levels of calcineurin activity (Fig. 4B). Thus, Ca2+/calmodulin is required to promote the increased basal calcineurin activity observed for Cnb1-G2A calcineurin. Taken together, our findings suggest that Cnb1 myristoylation limits Ca2+- and Ca2+/calmodulin-mediated activation of cellular calcineurin.
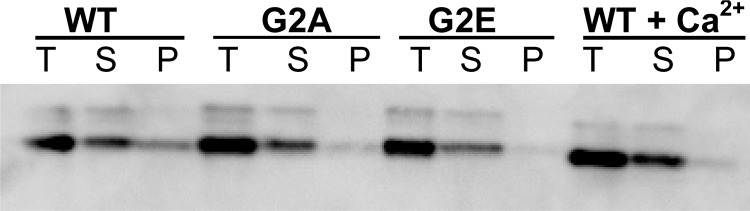
Because myristoylation of proteins can promote interaction with membranes, we next tested the consequences of disrupting Cnb1 myristoylation on calcineurin membrane association. High-speed ultracentrifugation of protein extracts prepared from yeast expressing wild type Cnb1, Cnb1G2A, or Cnb1-G2E was conducted to separate membrane-bound versus cytosolic calcineurin as described previously (28). As shown in Fig. 5, Cnb1-G2A and Cnb1-G2E have reduced membrane association as compared with wild type Cnb1. Quantification of the ratio of calcineurin observed in the pellet versus supernatant (membrane fraction versus soluble) revealed a 2.5- and 2.8-fold decrease for Cnb1-G2A and Cnb1-G2E calcineurin as compared with wild type enzyme. Cnb1 myristoylation therefore appears to promote membrane association of a fraction of calcineurin in yeast. Because disruption of Cnb1 myristoylation also increased phosphatase activation, we tested the possibility that Ca2+ signaling might modulate calcineurin membrane association. Yeast expressing wild type Cnb1 were stimulated with 100 mm CaCl2 for 15 min prior to harvesting extracts for ultracentrifugation. As observed for the myristoylation mutants, Ca2+ signaling reduced membrane association of calcineurin (Fig. 5). Quantification of Cnb1 revealed a 3-fold decrease in the ratio of calcineurin in the pellet versus supernatant following stimulation of Ca2+ signaling as compared with unstimulated yeast.
FIGURE 5.
Myristoylation promotes calcineurin membrane association. Cnb1Δ yeast expressing wild type Cnb1 (WT), Cnb1-G2A (G2A), or Cnb1-G2E (G2E) were grown in YPD (pH 5.5) for 4 h prior to lysis and ultracentrifugation to yield supernatant versus pellet fractions. Wildtype yeast were stimulated by the addition of 100 mm CaCl2 to media 15 min prior to harvesting as indicated (+Ca2+). Protein samples were separated by SDS-PAGE and probed with anti-Cnb1. T, total protein; S, supernatant; P, pellet.
DISCUSSION
Myristoylation of the calcineurin regulatory subunit is conserved from yeast to humans (4). In this study, we used a combination of Cnb1 mutational analysis and mutant yeast strains to demonstrate that Cnb1 myristoylation reduces calcineurin activity in response to submaximal Ca2+ signals. Disruption of Cnb1 myristoylation via mutation of the critical glycine residue or inhibition of N-myristoyltransferase activity resulted in constitutive phosphatase activity. Our findings are the first demonstration of a functional role of Cnb1 myristoylation in modulating calcineurin activity. Previous studies, conducted in yeast and in vitro using recombinant mammalian calcineurin, had demonstrated that myristoylation was not essential for phosphatase activity (28, 39). These studies had focused on testing the requirement for myristoylation in promoting calcineurin activity and were not designed to detect enhanced activity.
The increased basal activity observed in yeast expressing nonmyristoylated calcineurin was blocked when the Ca2+ binding domains of either Cnb1 or calmodulin were mutated. Thus, disruption of Cnb1 myristoylation appears to stimulate calcineurin activity by enhancing the Ca2+ sensitivity of phosphatase activation rather than promoting Ca2+-independent enzyme activation. The enhanced Ca2+ sensitivity of nonmyristoylated calcineurin could reflect increased affinity for Ca2+. Such a mechanism would be consistent with our observation that Cnb1-G2A specifically increased calcineurin activity in response to submaximal Ca2+ signals. N-Myristoylation of the EF hand proteins recoverin, neuronal calcium sensor (NCS-1), and guanylyl cyclase-activating protein (GCAP1) has previously been shown to modulate Ca2+ binding (40–42). Myristoylation of recoverin alters the conformation of the protein, causing an apparent reduction in Ca2+ affinity (40). N-Myristoylation of NCS-1 and GCAP1 increases Ca2+ affinity (41, 42). Interestingly, myristoylation of GCAP1 also increases the affinity of GCAP1 for retinal guanylyl cyclase (RetGC) and modulates the Ca2+-induced conformational changes in GCAP1 critical for regulating RetGC activation (42, 43). Myristoylated GCAP1 consequently stimulates RetGC activation to a higher maximal level than nonmyristoylated GCAP1. Myristate-mediated conformational changes in calcineurin activation may similarly be important for triggering catalytic subunit activation upon Cnb1 Ca2+ binding. Unfortunately, available calcineurin crystal structures were obtained using recombinant proteins lacking the amino terminus of CNB and therefore do not address the localization of the myristate group. Determining whether the myristate interacts with other domains within calcineurin will be critical in developing molecular models of how regulatory subunit myristoylation regulates enzyme activation. Structural details may reveal an alternative role of the Cnb1 myristoylation in reducing enzyme activation, such as binding to a hydrophobic pocket near the catalytic domain to prevent enzyme activation, as has been demonstrated for c-Abl tyrosine kinase 1b isoform (44).
Protein myristoylation is now recognized as a key determinant of signaling pathway function via regulation of protein-protein and protein-membrane interactions (45, 46). Myristoylation of the neuronal calcium sensor proteins influences their interactions with cellular factors and targets (47). N-Myristoylation of neurocalcin δ is required for Ca2+-dependent interaction with clathrin heavy chain, tubulin, and actin (48). Increased proteasome-substrate interaction in the absence of the Rpt2 orthologue myristoylation has been proposed as a potential mechanism to explain the observation that disruption of N-myristoyltransferase leads to enhanced proteasome activity in Aspergillus nidulans (49). Increased substrate affinity may contribute to the elevated basal activity of nonmyristoylated calcineurin. Yeast expressing Crz1 with increased affinity for calcineurin have previously been shown to exhibit enhanced Crz1 dephosphorylation and transcriptional activation in the absence of Ca signaling, similar to our findings in yeast expressing Cnb1-G2A (18). Because changes in calcineurin complex composition and protein interactions may differentially affect the interaction of calcineurin with specific substrates, myristoylation may ultimately help shape the output of calcineurin signaling via influencing the hierarchy of substrate preference. Consistent with altered calcineurin conformation and protein interactions, we have observed that Cnb1-G2A calcineurin exhibited increased sensitivity to the calcineurin inhibitor FK506 (supplemental Fig. 2B).
We also observed that nonmyristoylated calcineurin exhibited reduced membrane association. The finding that reduced membrane association correlated with increased calcineurin activation suggests that sequestration of the phosphatase at the membrane may contribute to the regulation of calcineurin signaling in yeast. Preincubation of purified bovine brain calcineurin with phospholipids has previously been shown to inhibit phosphatase activity in vitro (50). We further demonstrated that membrane association of wild type calcineurin was reduced by Ca2+ transients. Thus, in yeast, Ca2+ transients not only activate calcineurin, but also alter the subcellular localization of a fraction of the phosphatase. The development of new tools to monitor the local activation of calcineurin signaling in real time will be required to determine whether dynamic membrane interactions are important in the regulation of cellular phosphatase activation and to determine whether distinct phosphatase pools can be activated by spatially restricted Ca2+ transients. Despite the correlation between reduced membrane association and increased phosphatase activity, the majority of calcineurin localized to the soluble fraction even in the absence of Ca2+ signaling. Thus, additional mechanisms must also regulate cellular phosphatase activity.
Our findings demonstrate that Cnb1 myristoylation functions to limit calcineurin signaling in yeast and implicate cellular N-myristoyltransferase activity levels as a critical determinant of the Ca2+ responsiveness of phosphatase activation. Myristoylation may regulate calcineurin activity via multiple mechanisms, which are not mutually exclusive. It will be important to determine whether myristoylation of CNB similarly antagonizes calcineurin activity in mammalian cells. If the role of myristoylation in regulating calcineurin is conserved, differences in N-myristoyltransferase activity could contribute to cell-specific or developmental stage-specific regulation of calcineurin activity. Changes in N-myristoyltransferase activity would provide a novel mechanism by which calcineurin activity could be altered in disease development and progression.
Supplementary Material
Acknowledgments
We thank M. Cyert for CDRE-lacZ, J. Heitman for anti-Cnb1 antibody and pYDZ3, J. Gordon for nmt1–181 yeast strain, and K. W. Cunningham for K603 yeast strain. We also thank P. Hogan, K. W. Cunningham, and B. Rothermel for helpful discussions and manuscript comments.
This work was supported, in whole or in part, by National Institutes of Health Grant R021 NS058464 (to T. K.) and Subaward 3P30CA134274 from the NCI.

This article contains supplemental Figs. 1 and 2.
Cancer Genome Project, The Wellcome Trust, Sanger Institute.
S. Connolly and T. Kingsbury, manuscript in preparation.
- NFAT
- nuclear factor of activated T cells
- CDRE
- calcineurin-dependent reporter gene
- CNB
- calcineurin B subunit
- CNA
- calcineurin A subunit
- Nmt
- N-myristoyltransferase
- RCAN
- regulator of calcineurin
- Crz1
- calcineurin-responsive zinc-finger
- YPD
- yeast extract peptone dextrose medium
- SD
- synthetic complete medium
- cmd
- calmodulin
- GCAP1
- guanylyl cyclase-activating protein
- RetGC
- retinal guanylyl cyclase.
REFERENCES
- 1. Parekh A. B. (2011) Decoding cytosolic Ca2+ oscillations. Trends Biochem. Sci. 36, 78–87 [DOI] [PubMed] [Google Scholar]
- 2. Boulware M. J., Marchant J. S. (2008) Timing in cellular Ca2+ signaling. Curr. Biol. 18, R769–R776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berridge M. J. (2006) Calcium microdomains: organization and function. Cell Calcium 40, 405–412 [DOI] [PubMed] [Google Scholar]
- 4. Rusnak F., Mertz P. (2000) Calcineurin: form and function. Physiol. Rev. 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 5. Li X., Zhu L., Yang A., Lin J., Tang F., Jin S., Wei Z., Li J., Jin Y. (2011) Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell 8, 46–58 [DOI] [PubMed] [Google Scholar]
- 6. Wu H., Peisley A., Graef I. A., Crabtree G. R. (2007) NFAT signaling and the invention of vertebrates. Trends Cell Biol. 17, 251–260 [DOI] [PubMed] [Google Scholar]
- 7. Hogan P. G., Chen L., Nardone J., Rao A. (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 8. Müller M. R., Sasaki Y., Stevanovic I., Lamperti E. D., Ghosh S., Sharma S., Gelinas C., Rossi D. J., Pipkin M. E., Rajewsky K., Hogan P. G., Rao A. (2009) Requirement for balanced Ca/NFAT signaling in hematopoietic and embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilkins B. J., Molkentin J. D. (2004) Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem. Biophys. Res. Commun. 322, 1178–1191 [DOI] [PubMed] [Google Scholar]
- 10. Vega R. B., Bassel-Duby R., Olson E. N. (2003) Control of cardiac growth and function by calcineurin signaling. J. Biol. Chem. 278, 36981–36984 [DOI] [PubMed] [Google Scholar]
- 11. Buchholz M., Ellenrieder V. (2007) An emerging role for Ca2+/calcineurin/NFAT signaling in cancerogenesis. Cell Cycle 6, 16–19 [DOI] [PubMed] [Google Scholar]
- 12. Medyouf H., Ghysdael J. (2008) The calcineurin/NFAT signaling pathway: a novel therapeutic target in leukemia and solid tumors. Cell Cycle 7, 297–303 [DOI] [PubMed] [Google Scholar]
- 13. Mukherjee A., Soto C. (2011) Role of calcineurin in neurodegeneration produced by misfolded proteins and endoplasmic reticulum stress. Curr. Opin. Cell Biol. 23, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu J., Farmer J. D., Jr., Lane W. S., Friedman J., Weissman I., Schreiber S. L. (1991) Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815 [DOI] [PubMed] [Google Scholar]
- 15. Klee C. B., Ren H., Wang X. (1998) Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273, 13367–13370 [DOI] [PubMed] [Google Scholar]
- 16. Musson R. E., Smit N. P. (2011) Regulatory mechanisms of calcineurin phosphatase activity. Curr. Med. Chem. 18, 301–315 [DOI] [PubMed] [Google Scholar]
- 17. Liu J. O. (2003) Endogenous protein inhibitors of calcineurin. Biochem. Biophys. Res. Commun. 311, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 18. Roy J., Li H., Hogan P. G., Cyert M. S. (2007) A conserved docking site modulates substrate affinity for calcineurin, signaling output, and in vivo function. Mol Cell 25, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H., Rao A., Hogan P. G. (2011) Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson D. R., Duronio R. J., Langner C. A., Rudnick D. A., Gordon J. I. (1993) Genetic and biochemical studies of a mutant Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase, nmt72pLeu99 → Pro, that produces temperature-sensitive myristic acid auxotrophy. J. Biol. Chem. 268, 483–494 [PubMed] [Google Scholar]
- 21. Kingsbury T. J., Cunningham K. W. (2000) A conserved family of calcineurin regulators. Genes Dev. 14, 1595–1604 [PMC free article] [PubMed] [Google Scholar]
- 22. Cyert M. S., Thorner J. (1992) Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12, 3460–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cyert M. S., Kunisawa R., Kaim D., Thorner J. (1991) Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. U.S.A. 88, 7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cyert M. S. (2003) Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311, 1143–1150 [DOI] [PubMed] [Google Scholar]
- 25. Matheos D. P., Kingsbury T. J., Ahsan U. S., Cunningham K. W. (1997) Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11, 3445–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stathopoulos A. M., Cyert M. S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11, 3432–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stathopoulos-Gerontides A., Guo J. J., Cyert M. S. (1999) Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 13, 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu D., Cardenas M. E., Heitman J. (1995) Myristoylation of calcineurin B is not required for function or interaction with immunophilin-immunosuppressant complexes in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 24831–24838 [DOI] [PubMed] [Google Scholar]
- 29. Nakamura T., Liu Y., Hirata D., Namba H., Harada S., Hirokawa T., Miyakawa T. (1993) Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 12, 4063–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin D. D., Beauchamp E., Berthiaume L. G. (2011) Post-translational myristoylation: fat matters in cellular life and death. Biochimie 93, 18–31 [DOI] [PubMed] [Google Scholar]
- 31. Boutin J. A. (1997) Myristoylation. Cell. Signal. 9, 15–35 [DOI] [PubMed] [Google Scholar]
- 32. Aitken A., Cohen P., Santikarn S., Williams D. H., Calder A. G., Smith A., Klee C. B. (1982) Identification of the NH2-terminal blocking group of calcineurin B as myristic acid. FEBS Lett. 150, 314–318 [DOI] [PubMed] [Google Scholar]
- 33. Farazi T. A., Waksman G., Gordon J. I. (2001) The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276, 39501–39504 [DOI] [PubMed] [Google Scholar]
- 34. Rajala R. V., Datla R. S., Moyana T. N., Kakkar R., Carlsen S. A., Sharma R. K. (2000) N-myristoyltransferase. Mol. Cell. Biochem. 204, 135–155 [DOI] [PubMed] [Google Scholar]
- 35. Duronio R. J., Towler D. A., Heuckeroth R. O., Gordon J. I. (1989) Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science 243, 796–800 [DOI] [PubMed] [Google Scholar]
- 36. Duronio R. J., Rudnick D. A., Johnson R. L., Johnson D. R., Gordon J. I. (1991) Myristic acid auxotrophy caused by mutation of S. cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Cell Biol. 113, 1313–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng B., Stemmer P. M. (2001) Ca2+ binding site 2 in calcineurin-B modulates calmodulin-dependent calcineurin phosphatase activity. Biochemistry 40, 8808–8814 [DOI] [PubMed] [Google Scholar]
- 38. Geiser J. R., van Tuinen D., Brockerhoff S. E., Neff M. M., Davis T. N. (1991) Can calmodulin function without binding calcium? Cell 65, 949–959 [DOI] [PubMed] [Google Scholar]
- 39. Kennedy M. T., Brockman H., Rusnak F. (1996) Contributions of myristoylation to calcineurin structure/function. J. Biol. Chem. 271, 26517–26521 [DOI] [PubMed] [Google Scholar]
- 40. Ames J. B., Porumb T., Tanaka T., Ikura M., Stryer L. (1995) Amino-terminal myristoylation induces cooperative calcium binding to recoverin. J. Biol. Chem. 270, 4526–4533 [DOI] [PubMed] [Google Scholar]
- 41. Jeromin A., Muralidhar D., Parameswaran M. N., Roder J., Fairwell T., Scarlata S., Dowal L., Mustafi S. M., Chary K. V., Sharma Y. (2004) N-terminal myristoylation regulates calcium-induced conformational changes in neuronal calcium sensor-1. J. Biol. Chem. 279, 27158–27167 [DOI] [PubMed] [Google Scholar]
- 42. Peshenko I. V., Olshevskaya E. V., Dizhoor A. M. (2012) Interaction of GCAP1 with retinal guanylyl cyclase and calcium: sensitivity to fatty acylation. Front. Mol. Neurosci. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peshenko I. V., Olshevskaya E. V., Lim S., Ames J. B., Dizhoor A. M. (2012) Calcium-myristoyl Tug is a new mechanism for intramolecular tuning of calcium sensitivity and target enzyme interaction for guanylyl cyclase-activating protein 1: dynamic connection between N-fatty acyl group and EF-hand controls calcium sensitivity. J. Biol. Chem. 287, 13972–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hantschel O., Superti-Furga G. (2004) Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 5, 33–44 [DOI] [PubMed] [Google Scholar]
- 45. Hayashi N., Titani K. (2010) N-myristoylated proteins, key components in intracellular signal transduction systems enabling rapid and flexible cell responses. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 494–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 47. Ames J. B., Lim S. (2012) Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta 1820, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ivings L., Pennington S. R., Jenkins R., Weiss J. L., Burgoyne R. D. (2002) Identification of Ca2+-dependent binding partners for the neuronal calcium sensor protein neurocalcin δ: interaction with actin, clathrin and tubulin. Biochem. J. 363, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee S. C., Shaw B. D. (2007) A novel interaction between N-myristoylation and the 26S proteasome during cell morphogenesis. Mol. Microbiol. 63, 1039–1053 [DOI] [PubMed] [Google Scholar]
- 50. Politino M., King M. M. (1987) Calcium- and calmodulin-sensitive interactions of calcineurin with phospholipids. J. Biol. Chem. 262, 10109–10113 [PubMed] [Google Scholar]
- 51. Cunningham K. W., Fink G. R. (1994) Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124, 351–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.