Background: The ULK complex regulates autophagy, but how it interacts with the basal autophagy apparatus is unknown.
Results: ULK1, -2, ATG13, and FIP200 bind to ATG8 family proteins via LIR (LC3 interacting region) motifs.
Conclusion: ATG8 family proteins act as scaffolds anchoring the ULK complex on autophagosomes.
Significance: We define sequence requirements for the LIR motifs and suggest how the ULK complex interacts with autophagosomes.
Keywords: Autophagy, Molecular Cell Biology, Peptide Arrays, Protein Motifs, Protein-Protein Interactions, ATG13, ATG8, GABARAP, LIR, ULK1
Abstract
Autophagy is a lysosome-dependent degradation system conserved among eukaryotes. The mammalian Atg1 homologues, Unc-51 like kinase (ULK) 1 and 2, are multifunctional proteins with roles in autophagy, neurite outgrowth, and vesicle transport. The mammalian ULK complex involved in autophagy consists of ULK1, ULK2, ATG13, FIP200, and ATG101. We have used pulldown and peptide array overlay assays to study interactions between the ULK complex and six different ATG8 family proteins. Strikingly, in addition to ULK1 and ULK2, ATG13 and FIP200 interacted with human ATG8 proteins, all with strong preference for the GABARAP subfamily. Similarly, yeast and Drosophila Atg1 interacted with their respective Atg8 proteins, demonstrating the evolutionary conservation of the interaction. Use of peptide arrays allowed precise mapping of the functional LIR motifs, and two-dimensional scans of the ULK1 and ATG13 LIR motifs revealed which substitutions that were tolerated. This information, combined with an analysis of known LIR motifs, provides us with a clearer picture of sequence requirements for LIR motifs. In addition to the known requirements of the aromatic and hydrophobic residues of the core motif, we found the interactions to depend strongly on acidic residues surrounding the central core LIR motifs. A preference for either a hydrophobic residue or an acidic residue following the aromatic residue in the LIR motif is also evident. Importantly, the LIR motif is required for starvation-induced association of ULK1 with autophagosomes. Our data suggest that ATG8 proteins act as scaffolds for assembly of the ULK complex at the phagophore.
Introduction
Unc-51 like kinase 1 (ULK1)3 is a multifunctional protein involved in macroautophagy (hereafter referred to as autophagy) and axonal elongation (1–3). Autophagosome formation is initiated by the formation of a crescent-shaped double membrane structure, the phagophore, which subsequently elongates to engulf cytoplasmic components, and finally closes upon itself to form an autophagosome (4–6). The outer membrane of the autophagosome fuses with a lysosome allowing lysosomal hydrolytic enzymes to degrade the intra-autophagosomal components together with its inner membrane. The mammalian ULK complex is present on phagophores (7), and together with the class III phosphatidylinositol 3-kinase complex, consisting of Beclin 1, ATG14L, Vps34, and Vps15, regulates the initial events of autophagosome formation (6). A RalB-induced assembly of active ULK1 and the class III phosphatidylinositol 3-kinase complex on the Exo84 exocyst subcomplex is required for autophagosome formation upon starvation (8). The expansion of the phagophore (elongation) involves additional autophagy proteins, such as the WIPI family proteins (yeast Atg18 homologues) and the transmembrane protein ATG9, as well as ATG2 (9). Elongation of the phagophore also depends on two ubiquitin-like (Ubl) conjugation reactions. In the first reaction, ATG12 is conjugated to ATG5 resulting in the formation of a multimeric complex consisting of the ATG12-ATG5 conjugate and ATG16L. This complex is in turn required for conjugation of ATG8 family proteins to phosphatidylethanolamine on the phagophore membrane.
ATG8 homologues are produced as C terminally extended pro-forms. These are cleaved into form I by ATG4 cysteine proteases, resulting in the exposure of a C-terminal glycine that can be conjugated to phosphatidylethanolamine. Conjugation of ATG8 to phosphatidylethanolamine results in formation of form II that is covalently attached to the membrane and has been suggested to drive the elongation of the phagophore (10). There is only a single Atg8 protein in yeast, but mammals have at least seven ATG8 family members, i.e. LC3A–C, with two isoforms of LC3A, GABARAP and GABARAPL1–2 (11). Both LC3 and GABARAP subfamily members are conjugated to the phagophore (12, 13), and knockdown studies indicate that these two subfamilies have unique roles and are both needed for the formation of autophagosomes (14). ATG8 homologues become conjugated both to the inner and outer surface of the phagophore. Those on the outer surface are removed by ATG4 upon autophagosome closure, but those on the inner surface are not removed and therefore degraded together with the cargo. Conjugated LC3/GABARAP proteins act as scaffolds to recruit various proteins to the phagophore. For many of these proteins, the interaction with ATG8 homologues is mediated by a LIR (LC3-interacting region) motif. This is a short linear motif that was first identified in p62 (15, 16), but has later been found in a growing list of proteins from yeast and mammals (17). The consensus for the core LIR motif is (W/F/Y)XX(L/I/V), and in all characterized LIR motifs, this linear sequence is surrounded by at least one, but often more than one, acidic residue (18). Structural studies have revealed that the conserved hydrophobic residues of the core LIR motif dock into hydrophobic pockets in the Ubl (ubiquitin-like) domain, whereas adjacent acidic residues form electrostatic interactions with the N-terminal arm of the ATG8 homologues. The core LIR motif adopts an extended β-conformation and forms an intermolecular parallel β-sheet with the β2 strand of the ATG8 homologues (17).
Cargo receptors involved in selective autophagy, such as p62, NBR1, and optineurin, are specifically recruited to the inner surface of the phagophore via LIR interactions (16, 19, 20). Recently, the Rab7 effector protein FYCO1 and two Rab guanosine triphosphatase-activating proteins, OATL1 (TBC1D25) and TBC1D5, were shown to associate with the surface of autophagosomes by LIR-dependent interactions with ATG8 family proteins (21–23). Despite their specific binding to ATG8 family proteins, these proteins are not substrates for autophagy. This illustrates the fact that LIR-mediated ATG8 protein interactions are also used to scaffold proteins on the surface of autophagosomes.
The proteins present in the mammalian ULK complex involved in autophagy are ULK1 and ULK2 (2, 24), ATG13 (25, 26), focal adhesion kinase family interacting protein of 200 kDa (FIP200) (7), and ATG101 (27, 28). Together, they form a complex with an estimated size of about 3 MDa (26). Gel filtration analysis of purified proteins indicates that whereas ULK1 and ATG13 are probably monomeric in the absence of other proteins, FIP200 is likely a homo-polymer (29). In cells lacking ULK1 (2, 24), ATG13 (25, 26, 28–30), FIP200 (7), or ATG101 (27, 28), autophagy is impaired, indicating that an intact complex is required to initiate autophagosome formation. ULK2 is strongly related to ULK1 and recruited to the phagophore (7). ULK2 is not needed for autophagy in cells expressing ULK1 (24), but ULK2 can compensate for ULK1 depletion (31). The ULK complex is negatively regulated by mTORC1-mediated phosphorylation of ULK1, ULK2, and ATG13 (26, 29, 30). On the other hand, once ULK1 is autophosphorylated, it phosphorylates FIP200 and ATG13 (26, 29, 30), and the kinase activity of ULK1 is essential for autophagosome formation. However, overexpression of either wild type or kinase-dead ULK1 inhibits autophagy (24). Very recently it was shown that the interaction between ULK1 and the Hsp90-Cdc37 chaperone complex stabilizes ULK1, and disrupting this complex triggers ULK1 degradation by the proteasome (32). In addition, ULK1, ATG13, and ATG101 have all been shown to be destabilized if a functional ULK complex cannot be formed because of the total absence or reduced levels of either one of the components. For instance, ULK1 has been shown to be rapidly degraded in FIP200−/− mouse embryonic fibroblasts (7), and both the level and activity of ULK1 is similarly reduced after RNAi-mediated knockdown of FIP200 (29), ATG13 (27, 29), or ATG101 (27). There is also an interdependent relationship between the levels of ATG13 and ATG101. If one of these proteins is depleted, the level of the other is also reduced (27). However, FIP200 stability does not seem to be affected by ATG13 or ATG101 depletion (27).
ULK1 has previously been found to interact with GABARAP and GABARAPL2 (33). In this study, we characterize the interaction between ULK1 and human ATG8 homologues, and we demonstrate that this interaction is more selective for the GABARAP subfamily and mediated by a single LIR motif in ULK1. We also show that three additional proteins in the autophagy-specific ULK complex, i.e. ULK2, ATG13, and FIP200, contain a LIR motif that preferentially interacts with the GABARAP subfamily. We suggest that a major role of these interactions is to facilitate and/or stabilize association of the ULK complex with the phagophore via ATG8 family proteins.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following primary antibodies were used: rabbit anti-GST antibody (Z-5 SC-459; Santa Cruz Biotechnology), rabbit anti-GFP antibody (ab290, Abcam), mouse monoclonal anti-MBP antibody (Sigma), mouse monoclonal anti-FLAG antibody (200471, Stratagene), rabbit anti-actin antibody (A2066, Sigma), anti-ULK1 antibody (A748, Sigma), rabbit anti-LC3B antibody (L7543, Sigma), rabbit anti-GABARAP L1 antibody (11010-1-AP, Proteintech), and mouse monoclonal anti-WIPI2 antibody (gift from Sharon Tooze). Secondary antibodies used were HRP (horseradish peroxidase)-conjugated goat anti-rabbit Ig (554021) and goat anti-mouse Ig (554002) antibodies were from BD Bioscience Pharmingen. HRP-conjugated anti-GST antibody (clone RPN1236) was purchased from GE Healthcare.
Plasmids
Plasmids used in this work are listed in supplemental Table S1. Point mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and Gateway destination plasmids were made using Gateway LR recombination reactions (Invitrogen) following the manufacturer's instructions. All plasmid constructs made in this study were verified by DNA sequencing (BigDye sequencing kit, Applied Biosystems). The oligonucleotides used for mutagenesis, PCR, and DNA sequencing were purchased from Operon.
Cell Culture and Transfections
HEK293 Flp-In T-Rex cell lines expressing GFP-ULK1 or GFP-ULK1 F357A/V360A from a tetracycline inducible CMV promoter were made according to the Flp-In system manual (Invitrogen). The HEK93-based Flp-In T-Rex host cell line (Invitrogen) contains a single integrated FRT site for insertion of selected constructs. For the formation of stable transfectants, Flp-In plasmids carrying the selected GFP-tagged constructs were co-transfected with pOG44 encoding the Flp-In recombinase (Invitrogen). Expression from the CMV-TetO2 promoter was induced by adding 1 μg/ml of tetracycline (Sigma, T7660). HEK293 and Flp-In T-Rex cell lines were maintained in Dulbecco's modified Eagle's medium (Sigma, D6046) supplemented with 10% fetal calf serum and 1% streptomycin-penicillin (Sigma, P4333). HeLa cells were maintained in Eagle's minimum essential medium supplemented with 10% fetal calf serum, nonessential amino acids, 2 mm l-glutamate, and 1% streptomycin-penicillin. Subconfluent cells were transfected with the different plasmid constructs using either Lipofectamine PLUS (Invitrogen) or Metafectene Pro (Biontex) following the manufacturer's recommendations. Cells were treated as indicated with 0.2 μm bafilomycin A1 (Sigma, B1793) or 25 μm MG132.
Immunoprecipitations and Immunoblots
Transfected cells were rinsed with ice-cold PBS prior to lysis in RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 (v/v), 0.25% Triton X-100) supplemented with Complete Mini EDTA-free protease inhibitor mixture tablets (1 tablet per 10 ml) (11836170001, Roche Applied Science). Lysates were cleared by centrifugation followed by a 30-min incubation with protein A-agarose beads (SC-2001, Santa Cruz Biotechnology). The precleared lysates were then incubated with the indicated primary antibodies overnight at 4 °C and then with protein A-agarose beads for an additional 1 h. Precipitated immunocomplexes were washed five times with lysis buffer and eluted by boiling for 5 min in SDS-PAGE loading buffer. Samples were subsequently resolved by SDS-PAGE and transferred to Hybond-ECL nitrocellulose membranes (Amersham Biosciences). After blocking the membranes for 1 h in 5% (w/v) nonfat dry milk in TBST (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% (v/v) Tween 20), blots were probed with the indicated primary antibodies overnight at 4 °C and then by HRP-conjugated secondary antibodies for 1 h at room temperature. The membranes were washed six times (5 min each) with TBST prior to detection with a Western blotting Luminal Reagent kit (SC-2048, Santa Cruz Biotechnology) and a LumiAnalyst imager (Roche Applied Sciences).
GST- and MBP Pulldown Assays
GST and MBP proteins were expressed in Escherichia coli strains LE392 and DB3.1, respectively. GST and MBP fusion proteins were expressed in E. coli BL21 STAR (DE3)pLysS (Invitrogen). GST and GST fusion proteins were purified and immobilized on glutathione-coupled Sepharose beads (Glutathione-Sepharose 4 Fast Flow, Amersham Biosciences). MBP and MBP-tagged proteins were purified and immobilized using amylose resin (New England Biolabs).
MBP pulldown assays were performed by incubating immobilized MBP or MBP-tagged proteins with recombinant GST or GST fusion proteins. Recombinant GST or GST-tagged proteins were eluted from the beads with 10 mm reduced glutathione in 50 mm Tris-HCl (pH 8.0), pre-cleared by incubating with amylose resin for 30 min, and then incubated with immobilized MBP or MBP fusion proteins for at least 2 h at 4 °C with gentle agitation. Unbound proteins were removed by washing the resins five times with NET-N buffer. The proteins were then eluted by boiling for 5 min in SDS gel loading buffer and separated by SDS-PAGE. Recombinant proteins were detected by Western blotting using anti-MBP and anti-GST antibodies.
GST pulldown assays were performed by incubating immobilized GST or GST-tagged proteins either with 35S-labeled in vitro translated proteins or with recombinant MBP or MBP fusion proteins eluted from the amylose resin with maltose. The 35S-labeled proteins were produced using the TnT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]methionine (Amersham Biosciences). The synthesized proteins were diluted ×20 with NET-N buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) containing Complete Mini EDTA-free protease inhibitor mixture. Diluted in vitro translated proteins or recombinant MBP or MBP-tagged proteins were pre-cleared by incubating with glutathione-coupled Sepharose beads for 30 min prior to incubation with immobilized GST or GST fusion proteins for at least 2 h at 4 °C with gentle agitation. Unbound proteins were removed by washing the resins five times with NET-N buffer. The proteins were then eluted by boiling for 5 min in SDS gel loading buffer and separated by SDS-PAGE. After vacuum drying the gel, 35S-labeled proteins were detected on a Fujifilm bioimaging analyzer BAS-5000 (Fuji). Recombinant proteins were detected by Western blotting.
Confocal Microscopy
Cells were seeded at a density of 104 cells/well in 8-well coverglass slides (Nunc) and transfected with 100 ng of plasmid expression vectors 24 h later using Lipofectamine PLUS (Invitrogen). A day after transfection, cells were analyzed by confocal microscopy using a Zeiss Axiovert 200 microscope with a ×40, 1.2W C-Apochroma objective, equipped with an LSM510-META confocal module using the LSM 5 software version 3.2 (Carl Zeiss Inc.). GFP, Alexa 555, and Alexa 647 were excited using 488, 543, and 633 nm laser lines, respectively. Fluorescence channels were detected sequentially using the following band passes: 500–550 nm for GFP, 565–615 nm for Alexa 555, and 644–700 nm for Alexa 647.
For quantitative analysis of intracellular punctuated structures, two- or three-channel confocal stacks (Ztotal = 3, step size = 0.5 μm) were acquired. For all stacks, the middle Z slice was positioned at the specimens best focus. All images were acquired during the same experiment using identical instrument settings. Image analysis was performed in Volocity version 6.1.1 (PerkinElmer Life Sciences). Using a combination of intensity thresholding and object size discrimination, two-channel confocal stacks were analyzed for punctate localization of GFP-ULK1 (wild type and LIR mutant) and Alexa 555-labeled LC3B, GABARAP L1, or WIPI2. GFP dots were categorized as GFP+/A555+ if the dot perimeter touched an A555+ dot. The number of GFP+ dots was determined for at least 39 stacks for each condition, each comprising between 7 and 25 cells. Bar graphs represent the average number of dots per cell and S.D. for each condition. For each Alexa 555-labeled antibody tested, co-localization was analyzed for at least 9 stacks for each condition and the bar graphs represent the average number of GFP+/A555+ dots per cell.
Flow Cytometry
Flp-In T-Rex cells expressing proteins fused to GFP were trypsinized and analyzed as described (34), using a FACSAria cell sorter and FACSDiva software version 5.0 (BD Biosciences). GFP fluorescence was excited by the blue laser and collected through a 530/30-nm band pass filter. A FSC versus SSC gate was used to discriminate single cells and the median GFP-A value for at least 10,000 events was used for quantification of GFP fluorescence. To account for decreased cell volume after nutrient starvation, GFP fluorescence was multiplied by median FSC-A divided by median FSC-A of control cells grown in normal medium. Presented bar graphs show mean ± S.D. from three independent experiments. FCS files were analyzed using FlowJo version 9.4.11 (Tree Star Inc., Ashland, OR).
SPOT Synthesis of Peptide Arrays and GST Overlay Assay
Synthesis of peptide arrays on cellulose membranes were performed using a MultiPep automated peptide synthesizer (INTAVIS Bioanalytical Instruments AG, Germany) as previously described (55). After blocking the cellulose membranes in TBST with 5% nonfat dry milk, peptide interactions with GST or GST fusion proteins were tested by overlaying the membranes with 1 μg/ml of recombinant protein for 2 h at room temperature. Filters were washed in TBST, and bound proteins were detected with HRP-conjugated anti-GST antibody (1:5000; clone RPN1236; GE Healthcare).
RESULTS
ULK1, ATG13, and FIP200 Interact with Human ATG8 Family Proteins
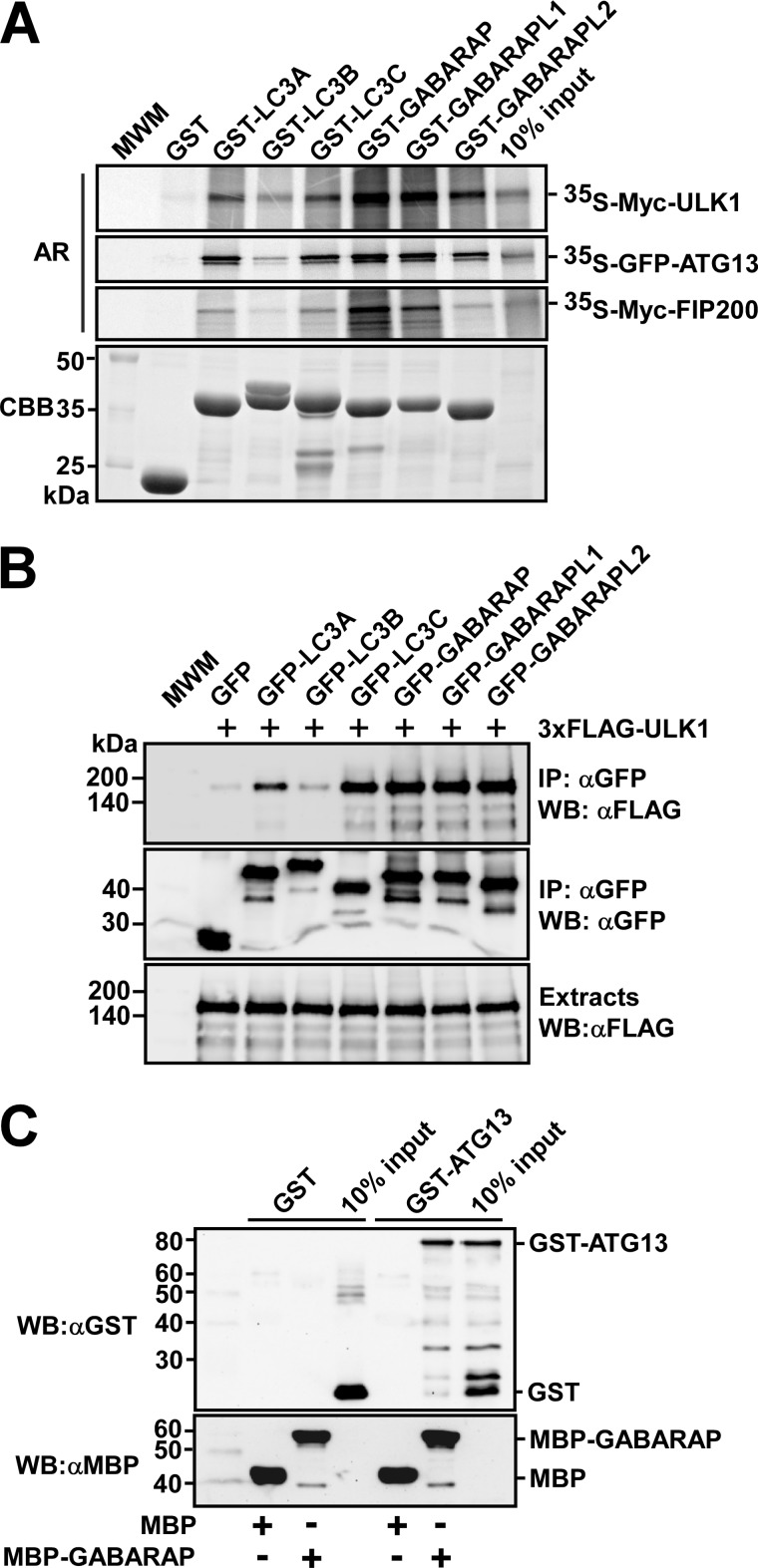
To investigate the interactions between ULK1 and human ATG8 homologues, we expressed different ATG8 homologues as GST fusions in E. coli and tested their ability to interact with in vitro translated ULK1 in GST pulldown assays. ULK1 interacted most strongly with GABARAP and GABARAPL1, but it also interacted with GABARAPL2, LC3A, and LC3C (Fig. 1A). We could also see an interaction between ULK1 and LC3B although it was weak compared with the other interactions (Fig. 1A). Hence, ULK1 seemed to have a preference for GABARAP subfamily proteins. This notion was supported by immunoprecipitation experiments using transiently transfected HEK293 cells (Fig. 1B). In these experiments, ATG8 homologues fused to GFP were co-expressed with ULK1 and immunoprecipitated with anti-GFP antibody. ULK1 co-precipitated with all ATG8 homologues except for LC3B (Fig. 1B).
FIGURE 1.
ULK1, ATG13, and FIP200 interact with human ATG8 family proteins. A, GST pulldown assays showing that ULK1, ATG13, and FIP200 all preferentially bind to the GABARAP subfamily proteins. The indicated proteins were in vitro translated in the presence of [35S]methionine and tested in GST pulldown assays for interaction with the indicated GST-fused recombinant proteins. Autoradiograph (AR, top panels) and Coomassie stained immobilized GST or GST fusion proteins (CBB, bottom panel) are shown. B, co-precipitation of FLAG-ULK1 with GFP-tagged ATG8 homologues from HEK293 cell extracts. Cells were co-transfected with the indicated constructs, and GFP or GFP fusion proteins were immunoprecipitated with GFP antibodies. Precipitated GFP-tagged proteins (middle panel), co-precipitated ULK1 (upper panel), and inputs of ULK1 (lower panel) were analyzed by Western blotting using the indicated antibodies. C, MBP pulldown assay showing direct interaction between ATG13 and GABARAP. All proteins were produced in E. coli and GST and GST-ATG13 were eluted from the beads and tested for interaction with immobilized MBP or MBP-GABARAP. The proteins were detected by Western blotting (WB) using the indicated antibodies. Anti-GST staining (upper panel) and anti-MBP staining (lower panel) are shown.
To test whether other members of the ULK complex can also interact with ATG8 homologues, we performed GST pulldown assays with in vitro translated ATG13 and FIP200. We found ATG13 to have a binding pattern similar to that of ULK1, with strong preference for the GABARAP subfamily and almost none for LC3B (Fig. 1A). Also FIP200 interacted with ATG8 homologues, but the interaction of FIP200 seemed limited to GABARAP and GABARAPL1, and was relatively weaker than observed for both ULK1 and ATG13 (Fig. 1A). We were unable to produce full-length ULK1 or FIP200 as recombinant proteins in E. coli, but GST-ATG13 was efficiently produced. The interaction between ATG13 and GABARAP was therefore verified using recombinant proteins expressed in E. coli. In MBP pulldown assays, recombinant MBP-GABARAP interacted with recombinant GST-ATG13 (Fig. 1C). These results strongly suggest that the interaction between ATG13 and GABARAP is direct, and not mediated by other members of the ULK complex. In conclusion, three different proteins of the ULK complex interact with ATG8 homologues, and they all seem to have a preference for the GABARAP subfamily.
ULK1/ATG1 Kinases Contain a LIR Motif Just C-terminal to Their Kinase Domain
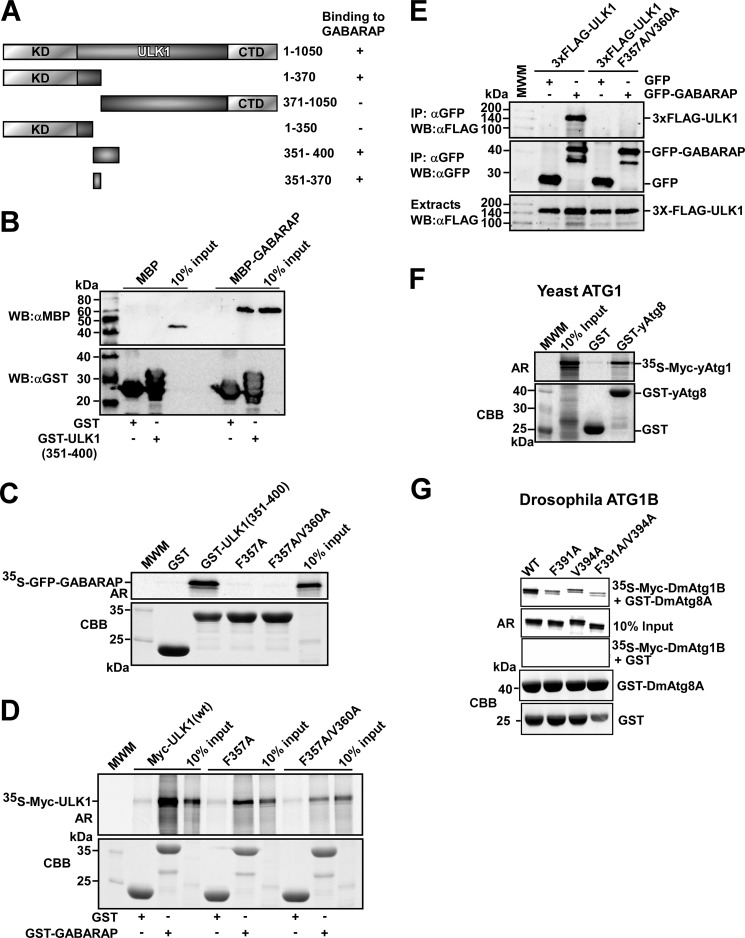
To map the region in ULK1 mediating the binding to ATG8 homologues, deletion constructs of ULK1 were in vitro translated and tested for binding to recombinant GST-GABARAP in GST pulldown assays. The interaction with GABARAP was mapped to amino acids 351–370 of ULK1 (Fig. 2, A-C, and supplemental Fig. S1, A and B). Moreover, a recombinant GST-tagged ULK1 fragment encompassing amino acids 351–400 co-precipitated with MBP-GABARAP in MBP pulldown assays demonstrating that the interaction between amino acids 351 and 370 in ULK1 and GABARAP is direct (Fig. 2B). This region contains a putative core LIR motif (FVMV, amino acids 357–360). Point mutations of the aromatic residue (F357A), and both the aromatic residue and the conserved hydrophobic position (F357A/V360A) of the putative LIR motif abolished the interaction between recombinant GST-ULK1(351–400) and in vitro translated GABARAP (Fig. 2C). These mutations also blocked the interaction between in vitro translated full-length ULK1 and recombinant GST-GABARAP (Fig. 2D). The mapping of the LIR motif was confirmed by immunoprecipitation of ULK1 from HEK293 cells transiently co-expressing FLAG-ULK1 and GFP-GABARAP. Whereas GABARAP was efficiently co-precipitated with wild type ULK1, it was not co-precipitated with ULK1 containing a mutated LIR motif (Fig. 2E). A LIR-mediated interaction between these two proteins was also demonstrated by confocal imaging of transiently transfected HEK293 cells. Overexpressed mCherry-ULK1 has mainly a diffuse cellular distribution, but localization in punctuated structures was seen in some of the cells expressing relatively low levels of ULK1. In contrast, the LIR mutant of ULK1 (F357A/V360A) was completely diffuse (supplemental Fig. S2A). When mCherry-ULK1 and GFP-GABARAP were co-expressed, they co-localized in punctuated structures (supplemental Fig. S2B). Many of these structures had a size of less than 1 μm, suggesting that they are phagophores or autophagosomes. The LIR mutant of ULK1 did not similarly co-localize with GFP-GABARAP in small punctuated structures, although co-localization could be observed in a minority of punctuated structures larger than 1 μm (supplemental Fig. S2C).
FIGURE 2.
Identification of a GABARAP interacting LIR motif in ULK1. A, schematic representation of the domain organization of human ULK1. The ULK1 fragments used to map the GABARAP-interacting region are indicated. (+) and (−) indicate the presence or absence of GABARAP binding, respectively. B, GST pulldown assay showing a direct interaction between GABARAP and a deletion fragment of ULK1 encompassing amino acids 351–400. All proteins were produced in E. coli, and MBP or MBP-GABARAP was eluted from the beads and tested for interaction with immobilized GST or GST-ULK1(351–400). Precipitated GST or GST-ULK1(351–400) and co-precipitated MBP or MBP-GABARAP were analyzed by Western blotting using the indicated antibodies. C, GST pulldown assay demonstrating loss of binding of the F357A and F357A/V360A point mutations of ULK1(351–400) to in vitro translated GFP-GABARAP. D, full-length ULK1 with the F357A and F357A/V360A point mutations that lost binding affinity for GABARAP. The GST pulldown assay was carried out using in vitro translated full-length ULK1 constructs and bacterially expressed and immobilized GST or GST-GABARAP. E, the F357A/V360A point mutations prevent co-precipitation of ULK1 with GFP-GABARAP from HEK293 cell extracts. Cells were co-transfected with the indicated constructs, and GFP or GFP-GABARAP were immunoprecipitated (IP) using GFP antibodies. The inputs of wild type or mutated 3xFLAG-ULK1 (lower panel), precipitated GFP or GFP-GABARAP (middle panel), and co-precipitated FLAG-ULK1 (upper panel) were analyzed by Western blotting (WB) using the indicated antibodies. F, yeast Atg1 interacts with yeast Atg8. In vitro translated yAtg1 was tested in a GST pulldown assay for interaction with GST or GST-yAtg8. Autoradiograph (AR) and Coomassie staining of immobilized GST or GST-yAtg8 (CBB) are shown. G, Drosophila Atg1B interacts with Drosophila Atg8A via a LIR motif. In vitro translated DmAtg1B, wild-type, and LIR mutant, were tested in a GST pulldown assay for interaction with GST or GST-DmAtg8A. Autoradiograph and Coomassie staining of immobilized GST or GST-DmAtg8A (CBB) are shown.
To test whether the presence of a LIR motif in ATG1/ULK1 kinases is an evolutionary conserved feature we looked for binding of yeast Atg1 to yeast Atg8 by pulldown assays and found indeed such an interaction (Fig. 2F). We also spotted an evolutionary conserved LIR motif (DDFVLV) located just C-terminal to the kinase domain of Drosophila Atg1B, almost identical to the DDFVMV found in human ULK1 and similarly located in the protein. Mutation of the conserved aromatic residue and/or hydrophobic residue led to loss of binding in GST pulldown assays confirming the evolutionary conservation of the LIR motif in Drosophila Atg1B (Fig. 2G).
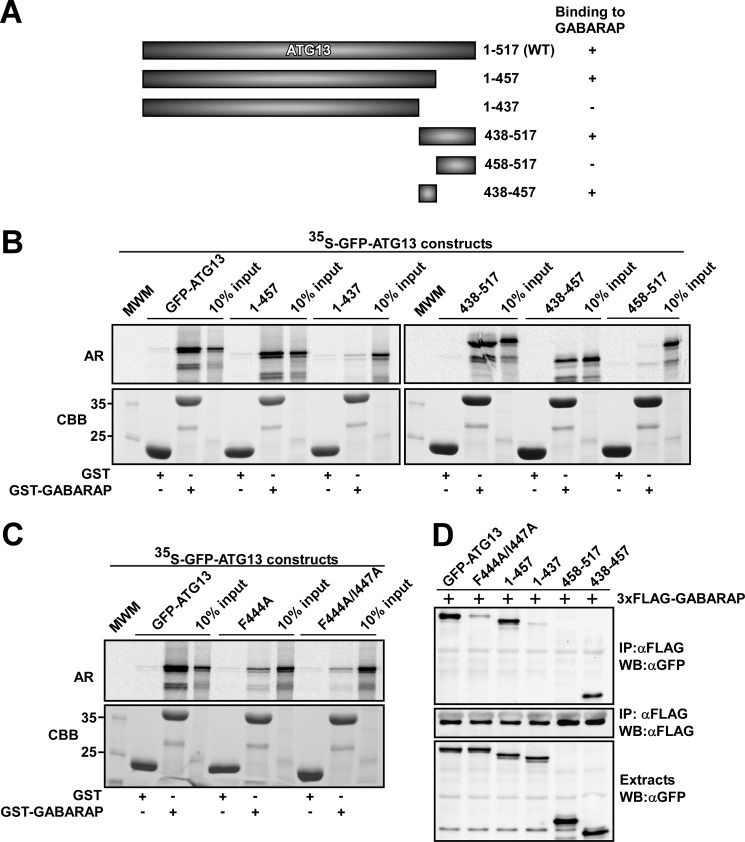
Mapping the LIR Motif in ATG13
Using a series of in vitro translated deletion constructs in GST pulldown experiments we mapped the GABARAP interacting region in ATG13 to amino acids 438–457 (Fig. 3, A and B). Fragments lacking this region displayed no interaction with GABARAP, and this region contains a candidate LIR motif (FVMI; amino acids 444–447). Mutation of either the aromatic residue (F444A) or both conserved residues of the core LIR motif (F444A/I447A) abolished the interaction between ATG13 and GABARAP (Fig. 3C). We verified the importance of the LIR motif for the interaction in vivo by immunoprecipitation following co-expression of FLAG-GABARAP with different ATG13 constructs (wild type and mutants) in HEK293 cells. Immunoprecipitation of FLAG-GABARAP using FLAG antibodies resulted in the co-precipitation of co-expressed wild type ATG13 (Fig. 3D). ATG13 deletion constructs containing amino acids 438–457 were efficiently co-precipitated with GABARAP (Fig. 3D). However, neither deletion constructs of ATG13 lacking amino acids 438–457 nor full-length ATG13 mutated in the LIR motif were co-precipitated with GABARAP (Fig. 3D). Taken together, these data support the conclusion that there is a single LIR motif in ATG13 that interacts with human ATG8 homologues.
FIGURE 3.
Identification of a GABARAP interacting LIR motif in ATG13. A, schematic representation of full-length ATG13 and its deletion constructs used to map the GABARAP-interacting region. (+) and (−) indicate the presence or absence of GABARAP binding, respectively. B, amino acids 438–457 of ATG13 are needed for its binding to GABARAP. The indicated GFP-tagged ATG13 constructs were in vitro translated in the presence of [35S]methionine and tested in GST pulldown assays for binding to recombinant GST or GST-GABARAP. C, the F444A and F444A/I447A point mutations strongly reduce the affinity of ATG13 for GABARAP. The GST pulldown assays were conducted using in vitro translated GFP-ATG13 constructs and bacterially expressed GST or GST-GABARAP. For B and C, autoradiograph (AR, upper panels) and Coomassie staining of immobilized proteins (CBB, lower panels) are shown. D, GFP fusions of wild type or the indicated point or deletion mutants of ATG13 were co-transfected with FLAG-GABARAP in HEK293 cells. FLAG-GABARAP was immunoprecipitated (IP) with FLAG antibodies. The inputs of the various GFP-ATG13 constructs (upper panel), precipitated FLAG-GABARAP (middle panel), and co-precipitated GFP-ATG13 constructs (upper panel) were analyzed by Western blotting (WB) using the indicated antibodies.
The LIR Motifs of ULK1 and ATG13 Interact with both Domains of GABARAP
The LIR motif of p62 binds to the interface between the N-terminal arm and C-terminal ubiquitin-like domain of LC3B, and binding therefore depends on the presence of both LC3B domains (15, 35, 36). Because ULK1 and ATG13 show preference for GABARAP subfamily proteins, we wanted to verify that this is also the case for the interactions of ULK1 and ATG13 with GABARAP. Hence, deletion mutants of GABARAP were tested in GST pulldown assays for their ability to interact with in vitro translated LIR motifs from ULK1 or ATG13 (supplemental Fig. S3A, left panel). As a control, we confirmed that binding of the LIR motif of p62 was strongly affected by a deletion of either the N-terminal arm or C-terminal domain of GABARAP. Next, we tested the LIR motifs of ULK1 and ATG13. As expected, their binding to GABARAP similarly depended on the presence of both domains in GABARAP (supplemental Fig. S3, B and C, left panels). We also tested the interactions of full-length ULK1, ATG13, or FIP200 in the same assay. Whereas both ATG13 and FIP200 lost their binding in the absence of either the N- or C-terminal domain of GABARAP, ULK1 retained some weak affinity for the isolated C-terminal region of GABARAP (supplemental Fig. S3D). However, all interactions were abolished by deletion of the N-terminal arm, which fits well with what has previously been reported for other LIR interactions (19). Moreover, point mutations in the C-terminal domain of GABARAP affecting the interaction with the LIR motif of p62 (supplemental Fig. 3A, right panel) also affected the interactions with the LIR motifs of ULK1 and ATG13 (supplemental Fig. S3, B and C, right panels).
Use of Peptide Arrays to Identify LIR Motifs in ULK1, ATG13 and FIP200
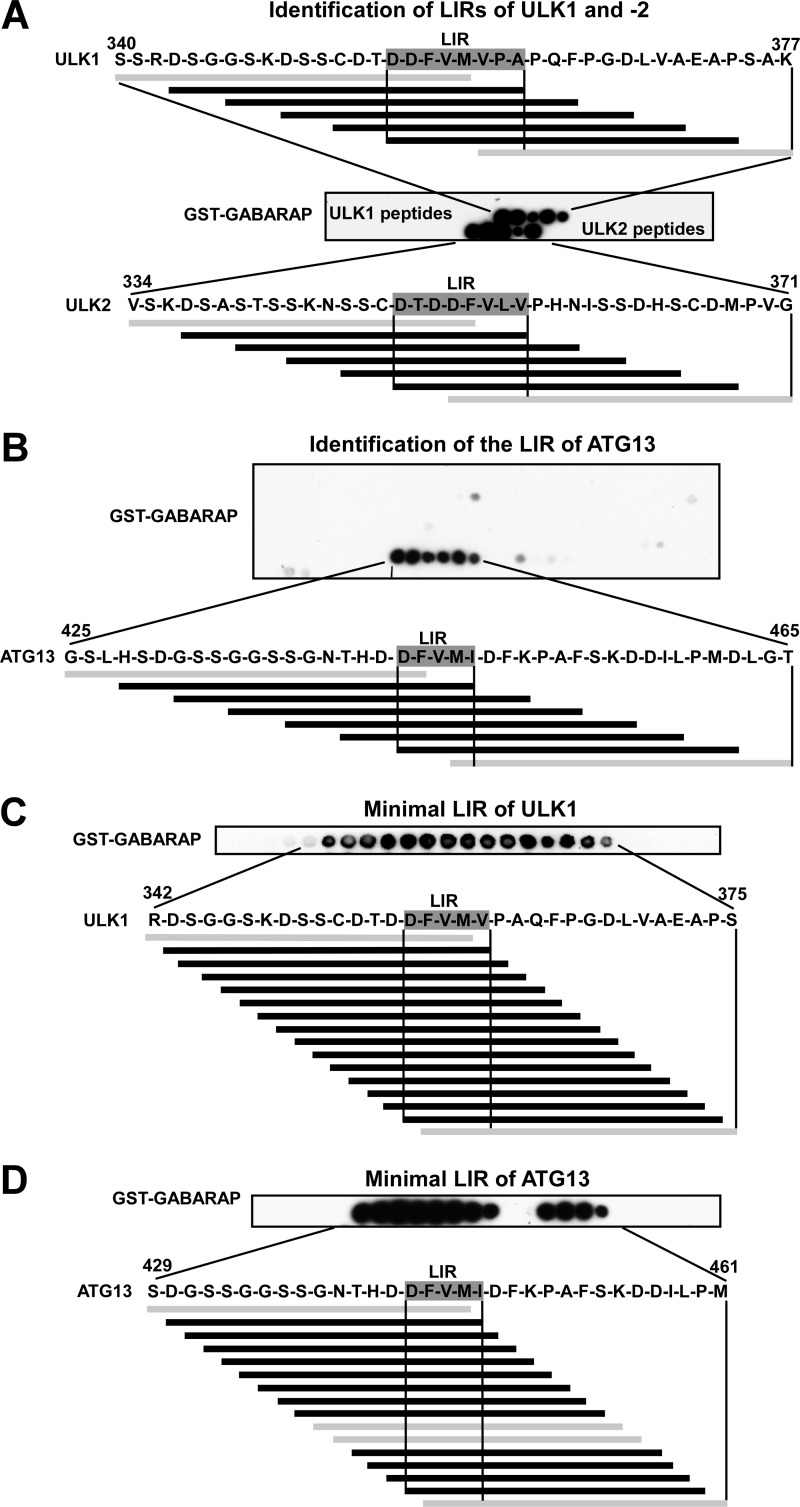
To further investigate the LIR motifs in ULK1 and ATG13, we utilized peptide array analyses. The peptide arrays consisted of short synthetic peptides of ULK1 (the related ULK2 was included) and ATG13 attached to cellulose membranes. For ATG13 we performed a peptide walk covering the complete sequence of the full-length protein using 20-mer peptides with 3-amino acid intervals. For ULK1 and -2 we probed a limited region containing the LIR motif already mapped by deletion mapping and point mutations (Fig. 2 and supplemental Fig. S1). The arrays were probed with recombinant GST or GST-GABARAP and binding was detected with anti-GST antibodies. This way a single LIR was identified in ULK1, ULK2, and ATG13. For ULK1 and ATG13, this region was identical to the LIR motifs identified using GST pulldown assays (Figs. 2, A-E, 3, A-D, and 4, A and B).
FIGURE 4.
Identification of the LIR motifs in ULK1, ULK2, and ATG13 using peptide arrays. A and B, identification of GABARAP-interacting LIR motifs in ULK1, ULK2 (A), and ATG13 (B). Arrays of 20-mer peptides covering the interacting region of ULK1, ULK2, and full-length ATG13 were synthesized on cellulose membranes. Each peptide was shifted three amino acids relative to the previous peptide. C and D, minimal LIR motifs of ULK1 (C) and ATG13 (D). Arrays of 18-mer peptides with single amino acid increments, covering a 42-amino acid region with the LIR motif in the center, were synthesized on cellulose membranes. The arrays were probed with 1 μg/ml of GST or GST-GABARAP for 2 h, and binding to GST (not shown) or GST-GABARAP was detected with anti-GST antibodies. Shown are sequence data for the GABARAP interacting peptides. Interacting peptides are in black, whereas noninteracting peptides are in gray.
The peptide arrays are ideally suited to give information about the minimal size of each identified LIR motif. The 20-mer arrays with a spacing of 3 amino acids indicated a minimal LIR motif for ULK1 of 8 residues with the sequence DDFVMVPA (Fig. 4A; amino acids 355–362). Similarly, in ULK2 the motif was also 8 residues (DTDDFVLV). The peptide array with ATG13 indicated a minimal LIR motif with the sequence DFVMI (Fig. 4B, amino acids 443–447). For high resolution mapping of the borders of the LIR motifs of ULK1 and ATG13 we employed arrays with 18-mer peptides spaced by a single amino acid gap. Because there was no need to screen the full-length proteins, 42-amino acid long fragments with the LIR motif in the center were chosen for this second round of peptide walks. This way we mapped the minimal LIR motif of ULK1 to the 5-amino acid sequence DFVMV (Fig. 4C). A very weak interaction was seen with a fragment lacking the Asp residue, but the much stronger interaction of peptides containing this residue strongly indicates that this residue should be included as part of the minimal LIR motif. The high resolution mapping also confirmed that the 5-mer peptide identified above (DFVMI) constitutes the minimal LIR motif for ATG13 (Fig. 4D). Together, the data support the conclusion that the presence of an acidic residue adjacent to the core motif is crucial for the ability of ULK1 and ATG13 to interact with GABARAP.
For the peptide arrays performed with ATG13 (Fig. 4D), we observed that some of the peptides containing the minimal LIR motif did not interact at all. This is most likely due to neighboring amino acids affecting the presentation of the LIR motifs in these noninteracting peptides. This indicates the importance of using relatively short gaps between peptides when one attempts to identify LIR motif(s) in a given protein. Nevertheless, in most cases, three amino acid gaps between peptides seem good enough to identify all functional LIR motifs within a protein.
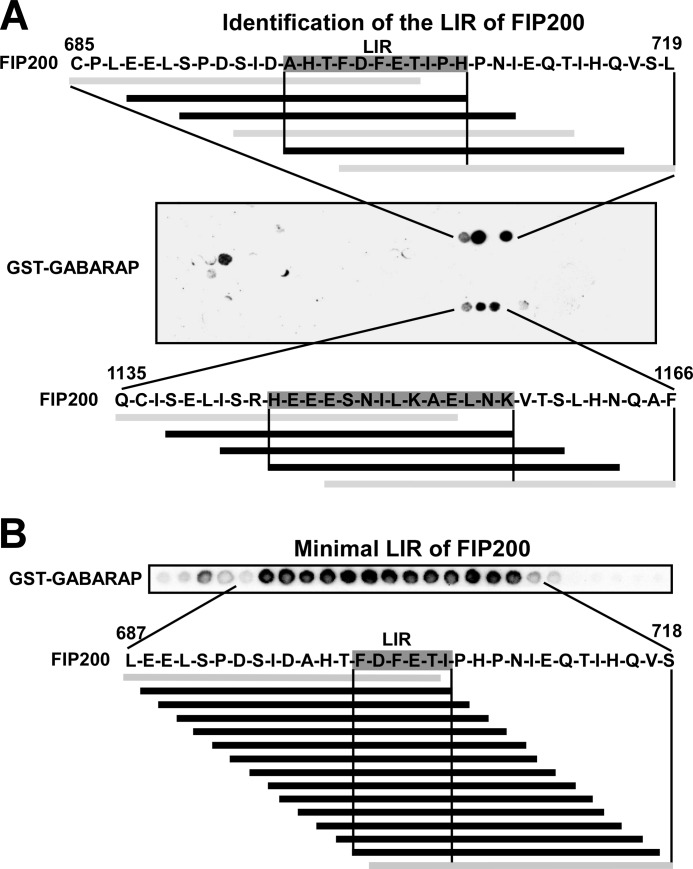
To identify LIR motifs in FIP200 using peptide arrays, full-length FIP200 was screened as 20-mer peptides and two putative GABARAP interacting regions were identified. Only three interacting peptides were identified for each of the sites. This is fewer than for most LIR motifs we have identified using this method, and may reflect that the interaction is less efficient. FIP200 also displayed a weaker interaction with GABARAP than ULK1 and ATG13 in GST pulldown assays (Fig. 1A). Best binding was seen with a series of peptides containing the putative LIR sequence DFETI (Fig. 5A). No typical LIR sequence was seen in the other GABARAP binding peptides (Fig. 5A). Hence, DFETI is the most likely LIR motif of FIP200. In a second peptide array, using 18-mer peptides spaced by one amino acid, the minimal LIR motif of FIP200 was narrowed down to the 6-mer peptide FDFETI (Fig. 5B), indicating the importance of adjacent acidic residues also for the LIR motif of FIP200.
FIGURE 5.
Identification of LIR motifs in FIP200 using peptide arrays. A, an array of 20-mer peptides covering the full-length of FIP200 was performed as described in the legend to Fig. 4. B, an array of 18-mer peptides with single amino acid increments was performed as described for ULK1 and ATG13 (see legend to Fig. 4).
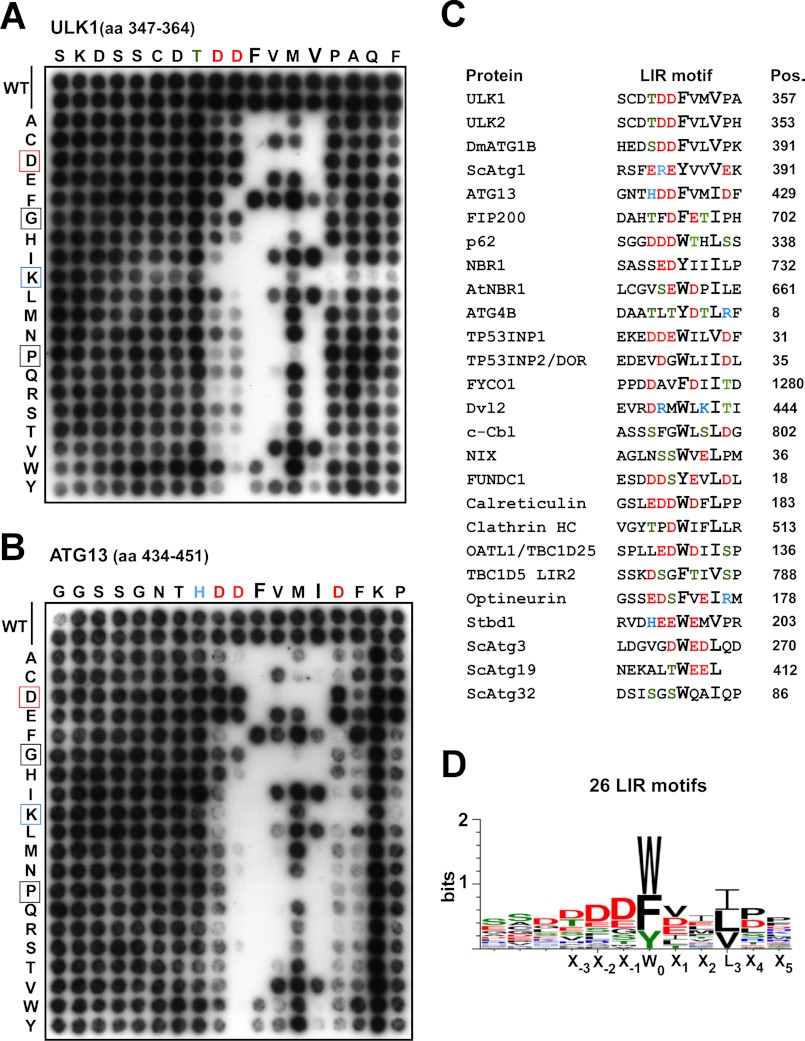
Two-dimensional Mutation Scans Reveal the Sequence Requirements for GABARAP-binding LIR Motifs
To analyze the importance of each position of the LIR motifs of ULK1 and ATG13, peptide arrays were performed with a series of 18-mer peptides containing single point mutations. All 18 positions were mutated to all alternative amino acids, and the effects of the different point mutations on binding to GST-GABARAP were then tested. Based on previously characterized LIR motifs, the consensus is X−3-X−2-X−1-(W/Y/F)-X1-X2-(L/I/V) with (W/Y/F)-X1-X2-(L/I/V) as the core motif (17, 18). In full agreement with the LIR consensus, using the nomenclature from Inagaki and co-workers (17), only the aromatic residues Phe, Trp, and Tyr were accepted in the 0 position of the LIR motif of ULK1 (FVMV) and ATG13 (FVMI)(Fig. 6, A and B). As expected, position 3 of the motifs was also strongly affected by most of the mutations. Clearly, the large hydrophobic residues Val, Ile, or Leu are required in this position although Phe is also tolerated (Fig. 6, A and B). Generally, Gly or Pro residues, affecting the secondary structure, were not tolerated at any of the core positions (0–3). Because the LIR motifs are generally composed of hydrophobic and acidic residues, basic residues (His, Arg, and Lys) are generally not tolerated, except in position 2. Surprisingly, both for the ULK1 and ATG13 core motifs, Asp residues are not tolerated at all in the 1 and 2 positions, whereas Glu works fine. Here, the length of the side chain must be the crucial parameter to explain this selectivity.
FIGURE 6.
Characterization of the LIR motifs of ULK1 and ATG13. A, two-dimensional peptide arrays scans analyzing the effect of single amino acid substitutions at all positions of the indicated 18-mer peptides from ULK1 (amino acids 347–364); B, or ATG13 (amino acids 434–451). Each position of the 18-mer peptides was replaced with all 20 amino acids and GST-GABARAP was detected with anti-GST antibodies. C, alignment of published LIR motifs verified by binding assays. The amino acid position (Pos.) of the conserved aromatic residue in the LIR motifs is indicated for each protein. All sequences are from the human proteins except for the four yeast proteins Atg1, -3, -19, and -32, plant NBR1, and Drosophila ATG1B. The sequences and references are the following: p62 (16), NBR1 (19), AtNBR1 (43), ATG4B (44), TP53INP1 and TP53INP2/DOR (45), FYCO1 (21), Dvl2 (46), NIX (47), FUNDC1 (48), Calreticulin (49), clathrin heavy chain (50), OATL1/TBC1D25 (22), TBC1D5 (23), Optineurin (20), Stbd1 (51), yeast Atg3 (52), yeast Atg19 (35) and Atg32 (53). D, sequence logo of the LIR motifs aligned in C made using WebLogo 3 (54).
A very surprising observation was that position 1 of the core motif was strongly affected by mutations (Fig. 6, A and B). The amino acids that were well tolerated in this position in ULK1 or ATG13 are Val, Ile, Phe, Glu, and Cys, and most of the other substituted amino acids resulted in complete loss of binding. The most striking observation was that, except for glutamate, the remaining tolerated amino acids are all hydrophobic (Fig. 6, A and B). The X2 position in the core LIR motifs of ULK1 and ATG13 tolerates most amino acid substitutions and are clearly less important than the other positions. However, in the LIR of ATG13, in addition to the 4-mer core motif, the negatively charged Asp residue just N-terminal to the core motif seems critically needed (Fig. 6B). Only Asp, Glu, or Gly was tolerated in this position and other substitutions all resulted in a complete loss of interaction with GABARAP. This correlates with the size of the minimal motif in ATG13 as suggested from Fig. 4B. In ULK1, the two Asp residues N-terminal to the 4-mer core motif seem equally important (Fig. 6A). The effect of mutating one of these positions was therefore less severe than for the single important Asp residue in ATG13.
Another notable observation is that the region C-terminal to the core motif is important for the ATG13 motif, but not for the ULK1 motif. Mutations at either of the two positions C-terminal to the core motif of ATG13 strongly reduced its binding to GABARAP. In particular, the presence of an acidic residue adjacent to the core motif appeared to be important for an efficient interaction with GABARAP, and there also seemed to be a preference for a bulky amino acid (Phe or Trp) following the acidic residue (Fig. 6B). In contrast to ATG13, the LIR of ULK1 contains no acidic residues within the region C-terminal to the core motif (Fig. 6A).
Toward a Reliable LIR Motif Consensus
We have compiled 26 published LIR motifs that have been verified by binding assays (Fig. 6C), and calculated a sequence logo (Fig. 6D). These data confirm the requirement for an aromatic amino acid at the 0 position, and that the hydrophobic position 3 is always occupied by Ile, Leu, or Val. Trp and Phe dominate in the aromatic position but three of 26 LIRs have a Tyr at this position. Another important observation is that position −1 and −2 often are occupied by acidic residues, like found for ULK1 and ATG13, for example. Also, eight of the LIRs have a phosphorylatable Ser or Thr at these positions that can contribute strongly to acidity and increase binding in vivo as shown recently for optineurin (20). Another, clear tendency seen both from the sequence compilation and the two-dimensional scans is the preference for either an acidic (Glu or Asp) or hydrophobic residue (Val, Ile, Leu) at position 1 of the core motif. Taken together, our experimental data and the sequence compilations suggest the following LIR consensus (DE)(DEST)(WFY)(DELIV)X(ILV).
Specificity of LIR Interactions
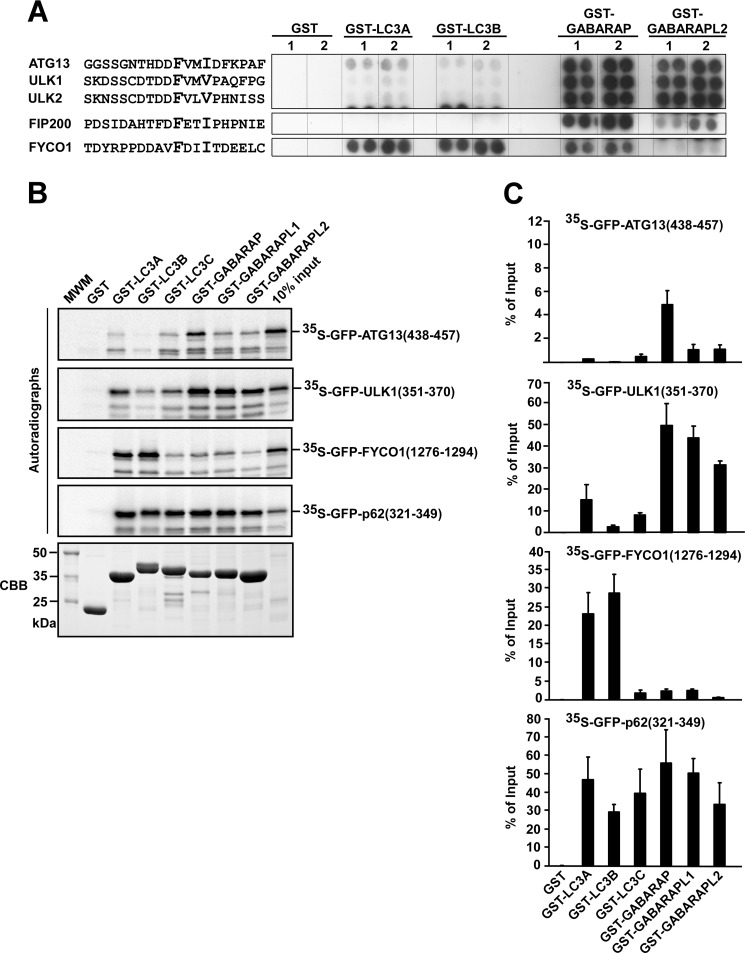
Next, we wanted to study the specificity of selected LIR motifs in peptide arrays. We tested 20-mer LIR peptides from ATG13, ULK1, ULK2, and FIP200 for binding to four different human ATG8 homologues fused to GST; i.e. LC3A, LC3B, GABARAP, and GABARAPL2 (Fig. 7A). We also included the LIR motif of FYCO1 because this protein has a clear preference for LC3B (21, 37), which is a very different specificity compared with ULK1 and ATG13. We have previously mapped the binding of FYCO1 to LC3B to a 19-mer fragment (amino acids 1276–1294 in FYCO1) encompassing a putative core LIR motif (FDII)(21). All the peptides interacted strongly with at least one of the ATG8 homologues, indicating that their LIR motifs were exposed and functional. Of the peptides tested, the only LIR motif that interacted strongly with LC3 subfamily proteins was that of FYCO1 (Fig. 7A). In sharp contrast to the LIR motif of FYCO1, the motifs of ATG13, ULK1, and ULK2 preferentially interacted with GABARAP and GABARAPL2 (Fig. 7A). The predicted LIR motif of FIP200 displayed a very weak affinity for GABARAPL2, but it interacted with GABARAP (Fig. 7A).
FIGURE 7.
Specificity of isolated LIR motifs analyzed by use of peptide arrays and GST pulldown assays. A, the indicated 20-mer peptides from ATG13, ULK1, ULK2, FIP200, and FYCO1 were synthesized on cellulose membranes in parallels (indicated as 1 and 2). The arrays were probed with 1 μg/ml of GST, GST-LC3A, GST-LC3B, GST-GABARAP, or GST-GABARAPL2 for 2 h, as indicated. Binding was detected with anti-GST antibodies. B, GST pulldown assays testing binding of isolated LIR motifs to GST or various ATG8 homologues fused to GST. The indicated fragments of ULK1, ATG13, FYCO1, and p62 were in vitro translated as GFP fusion proteins and tested in GST pulldown assays for binding to GST or the indicated GST fusion proteins. Autoradiographs (upper panels) and Coomassie staining of the immobilized GST or GST fusion proteins used in the assay (CBB, lower panel) are shown. C, quantitative representations of the experiments depicted in B. Quantifications of the mean % binding with S.D. from three independent experiments are shown.
The specificity of the identified LIR motifs was also analyzed by in vitro GST pulldown assays (Fig. 7, B and C). For these assays, 18- or 20-mer LIR peptides fused to GFP were in vitro translated and tested for interaction with the six different GST-tagged human ATG8 homologues. The data we got from these experiments correlated well both with data obtained using full-length proteins (Fig. 1) and data obtained using peptide arrays (Fig. 7A). Again, the LIR motifs of ULK1 and ATG13 displayed a preference for GABARAP subfamily proteins. The LIR motif of ULK1 interacted strongly with all three GABARAP subfamily proteins, whereas it interacted much more weakly with LC3A and LC3C, and did not interact with LC3B (Fig. 7, B and C). The LIR motif of ATG13 interacted strongly with GABARAP and weakly with GABARAPL1 and GABARAPL2, but it did not bind to LC3B (Fig. 7, B and C). As a control, the LIR motif of FYCO1 interacted strongly with LC3A and -B, but not with LC3C or GABARAP subfamily proteins. As yet another control, we tested the LIR motif of p62 in the same assay and it interacted strongly with all the six human ATG8 homologues (Fig. 7, B and C).
The LIR Motif in ULK1 Is Required for the Association of ULK1 with Autophagosomes
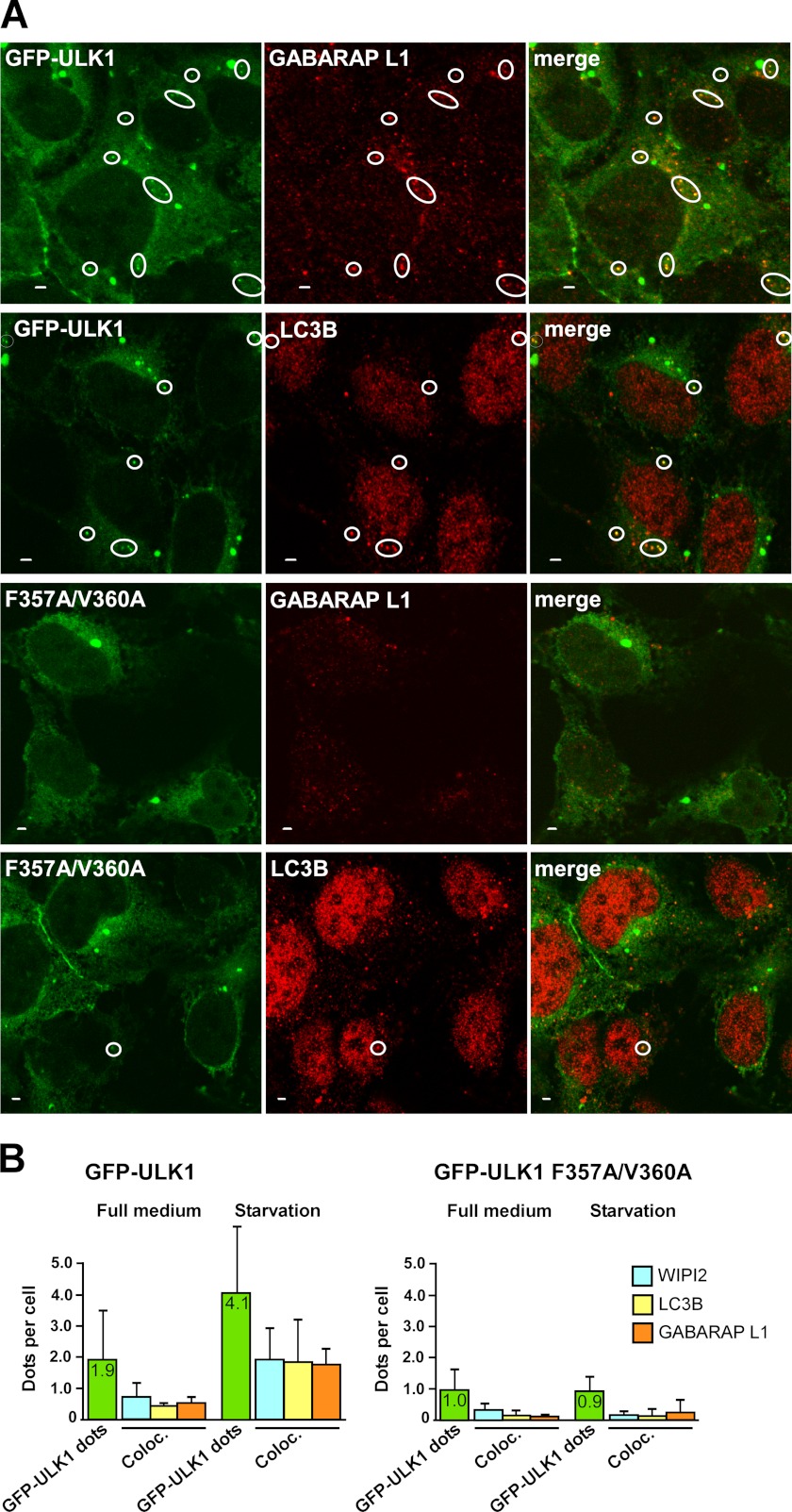
The ULK1 complex is recruited before ATG8 homologues to the site of phagophore formation, but a role of the ULK1-ATG8 interaction may be to keep the ULK1 complex attached to the phagophore. Because a transient overexpression of ULK1 usually has an inhibitory effect on phagophore formation (24), we made HEK293 Flp-In T-Rex cells stably expressing GFP-ULK1 or GFP-ULK1 F357A/V360A from a tetracycline-inducible promoter. Flow cytometric analyses showed a similar expression level of wild type and LIR-mutated GFP-ULK1 in these cells treated with tetracycline for 14 h (supplemental Fig. S4A). We showed above that the LIR motif is required for colocalization of overexpressed ULK1 and GABARAP in small punctuated structures (supplemental Fig. S2). In cells stably expressing wild type and LIR-mutated GFP-ULK1, both constructs displayed a mainly diffuse localization pattern. However, GFP-ULK1 also accumulated in aggresome-like structures of several micromolar and in smaller dots of 100–200 nm (Fig. 8A). Whereas the large aggregate, often a single per cell, was seen with both ULK constructs, the smaller dots were mainly seen with the wild type protein (Fig. 8A). Based on data from experiments where we counted the number of GFP-ULK1-positive dots, the average number of wild type ULK1 dots per cell increased from 1.9 to 4.1 in response to 1 h of amino acid starvation (Fig. 8B). In contrast to this, no increase in the number of dots per cell was observed for LIR-mutated ULK1 in response to amino acid starvation (Fig. 8B).
FIGURE 8.
The LIR motif in ULK1 is needed for amino acid starvation-induced co-localization of ULK1 with endogenous LC3B, GABARAP L1, and WIPI2 on autophagosomes. A, co-localization of wild type GFP-ULK1 with endogenous GABARAP L1 and LC3B in punctuated structures in HEK293 Flp-In T-Rex cells. Expression of GFP-ULK1 (wild type or mutated) was induced for 24 h with tetracycline. Cells were then incubated for 1 h in starvation medium without amino acids, fixed, and stained with anti-LC3B or anti-GABARAP L1 antibodies as indicated. Representative images are shown, the circles indicate dots with co-localization. Bars, 2 μm. B, quantitative analysis of intracellular punctuated structures in HEK293 Flp-In T-Rex cells expressing GFP-ULK1 (wild type or LIR-mutated). Expression of GFP-ULK1 was induced as in A, and cells were then incubated for 1 h in full medium or starvation medium. Cells were fixed and stained with anti-WIPI2, anti-LC3B, or anti-GABARAP L1 antibodies before being analyzed by confocal microscopy. Each bar indicating the number of GFP-ULK1 positive dots per cell (green color) is based on an analysis of 490–570 cells, whereas 100–180 cells were analyzed for co-localization with endogenous WIPI2, LC3B, and GABARAP L1. Under starvation conditions, the average co-localization of WIPI2, LC3B, and GABARAP L1 with wild type ULK1 dots was 47, 45, or 43%, respectively, and for the LIR mutant ULK1 the corresponding numbers were 25, 15, and 22%.
LC3B is the most firmly established autophagosomal marker in mammalian cells. Staining of GFP-ULK1 expressing cells with LC3B antibodies were done to test if endogenous LC3B is recruited to ULK1 positive structures. Analyses of confocal images revealed that in amino acid-starved cells LC3B co-localized with 45% of the wild type ULK1 dots, but only 15% of the dots were formed by LIR-mutated ULK1 (Fig. 8B). A similar co-localization pattern was seen after staining the cells with antibodies specific for GABARAPL1 (38)(Fig. 8B). Neither of the ATG8 homologs co-localized with ULK1 in aggregates larger than 1 μm (Fig. 8A). WIPI2 is recruited to the phagophore before LC3B (39). Staining of GFP-ULK1 expressing cells with WIPI2 antibodies revealed a similar co-localization pattern as with the ATG8 homologues (Fig. 8B). By co-staining the cells with two different antibodies, we were also able to show the co-presence of WIPI2 and LC3B or GABARAPL1 in dots formed by wild type ULK1 (supplemental Fig. S4B). In conclusion, we observed a LIR-dependent formation of punctuated structures containing GFP-ULK1, and these structures co-localized with WIPI2, LC3B, and GABARAPL1. This is consistent with a role for the LIR motif in keeping ULK1 stably attached to the phagophore, whereas the LIR-mutated ULK1 may have a more transient binding.
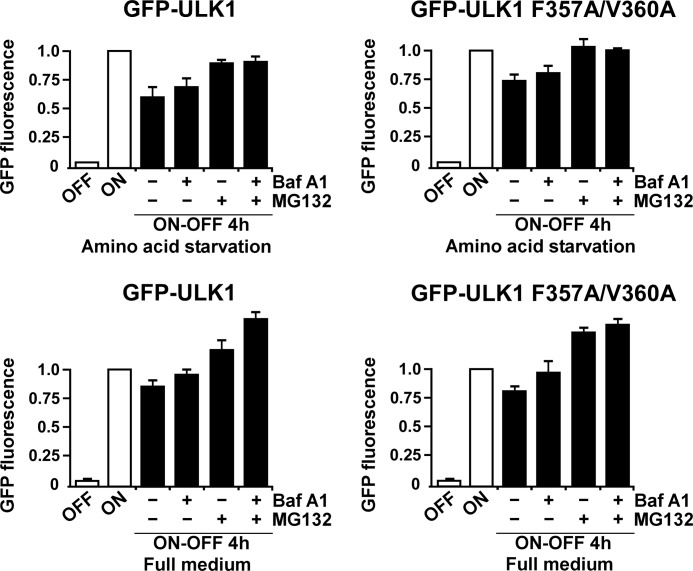
Recent studies have indicated that plant ATG1 is degraded by autophagy (40), suggesting that mammalian ULK1 could similarly be an autophagy substrate. To measure degradation of GFP-ULK1 or GFP-ULK1 F357A/V360A, we used the HEK293 Flp-In T-Rex cells stably expressing these proteins. The cells were grown overnight in rich medium in the presence of tetracycline, resulting in accumulation of GFP-ULK1 or GFP-ULK1 F357A/V360A. The promoter was then shut off by removal of tetracycline and degradation of GFP-ULK1 was measured in live cells by flow cytometry, using loss of green fluorescence as readout. The degradation was measured both in rich medium and in starvation medium lacking amino acids. Starvation medium induces autophagy, but it also helps to inhibit new synthesis of GFP-ULK1, whereas measuring GFP-ULK1 degradation. The starvation medium was supplemented or not with bafilomycin A1 or a proteasomal inhibitor (MG132), to see whether these inhibitors were able to block degradation of the proteins. Both in full medium and in starvation medium, we observed a reduced level of GFP-ULK1 or GFP-ULK1 F357A/V360A 4 h after promoter shut-off (Fig. 9). MG132 inhibited this degradation, suggesting that GFP-ULK1 is mainly degraded by the proteasome (Fig. 9). Bafilomycin A1 had only a minor effect on the degradation of GFP-ULK1, further indicating that GFP-ULK1 is mainly degraded by the proteasome (Fig. 9). However, a minor inhibition of degradation seen with bafilomycin A1, when added alone, suggests that a fraction of GFP-ULK1 is degraded by autophagy. In this assay the LIR motif had no significant effect on autophagy of GFP-ULK1, and the overall rate of degradation of LIR-mutated GFP-ULK1 was similar to the wild type protein (Fig. 9). Hence, these results show that the LIR motif of ULK1 is not mediating significant autophagic degradation of ULK1. Rather, a scaffolding role on the autophagosome, like found for FYCO1 (21), TBCD1D5 (23), and OATL1 (22) is most likely the role played by the GABARAP proteins in interacting with the LIR motif of ULK1.
FIGURE 9.
Only a minor fraction of GFP-ULK1 is degraded by autophagy. HEK293 Flp-In T-Rex cells stably expressing GFP-ULK1 (wild type or LIR-mutated) from an inducible promoter were grown overnight in full medium supplemented with tetracycline, resulting in accumulation of GFP-ULK1 (ON). Tetracycline was removed (promoter shut-off, ON-OFF), and the cells were incubated for 4 h in full medium or starvation medium, both supplemented or not with bafilomycin A1 and/or MG132 as indicated. Degradation of GFP-ULK1 (wild type or LIR-mutated) was measured by flow cytometry, using loss of GFP fluorescence as readout. For each condition, median GFP fluorescence was determined from a population of at least 10,000 cells. Bars show the average GFP fluorescence from three independent experiments, relative to the ON condition.
DISCUSSION
ULK1 has previously been found to interact with GABARAP and GABARAPL2 (33), but whether this interaction is dependent on a LIR motif has not been tested. Here, we show that ULK1 uses a LIR motif to interact with ATG8 homologues. We also identify LIR motifs in ATG13 and FIP200. All these LIR motifs were found to have preference for GABARAP subfamily proteins.
We used peptide array analysis as an alternative method to identify and further characterize LIR motifs. Interestingly, the LIR containing peptides interacted with the ATG8 proteins with the same strength and specificity as observed in other in vitro assays. In addition, for the motifs in ULK1 and ATG13, this method was also used to identify all amino acids that were critical for their interaction with GABARAP. Peptide array analysis works as a specific and efficient method for identifying LIR motifs within full-length proteins. It is highly stringent, and the risk of selecting false positives is low. The method also allows detailed mapping of the extent of the binding motifs and two-dimensional substitution mutagenesis probing each position of the binding motif.
The core LIR sequences of the motifs in ULK1 and ATG13 are FVMV and FVMI, respectively. Interestingly, in addition to the first and last residues of these core sequences, mutation of the second Val residue also strongly negatively affected the ability of these LIR motifs to interact with GABARAP. Hence, the second residue of the core motif is likely more important than previously anticipated. This was also confirmed by our analysis of 26 verified LIR motifs. Moreover, acidic residues adjacent to the core motif are also important for the interactions of ULK1 and ATG13 with GABARAP, completely in line with what has been reported for the LIR motif of p62 (16). Both in ULK1 and ATG13, two Asp residues are located N-terminal to the core motif. In addition to this, the LIR motif in ATG13 contains an essential Asp residue at the C-terminal side of the core motif. This study, therefore, supports the suggestion by Johansen and Lamark (18) that as an extension to the commonly referred (W/Y/F)XX(L/I/V) core motif, the consensus LIR sequence should include acidic residues located N-terminal to the core motif. Our analyses allow us to suggest the following LIR consensus (DE)(DEST)(WFY)(DELIV)X(ILV). Sequence searches using this degenerate motif, and the application of the hand rules about avoidance of Gly, Pro, and basic residues (His, Arg, Lys) in the core motif identified in this study, may greatly aid in the identification of functional LIR motifs in ATG8-interacting proteins.
The LIR motifs of ULK1, ULK2, DmAtg1, ATG13, and FIP200 all contain Phe as the aromatic core residue. These proteins have very similar specificity, preferring the GABARAP subfamily members, and their LIR motifs are also highly similar. Other LIR motifs listed in Fig. 6C with Phe in this position are those of FYCO1, optineurin, and TBC1D5. The LIR of FYCO1 contains two amino acids that are not accepted in the motifs of ULK1 and ATG13, i.e. Val at position −1 and Asp at position 1. This may reflect that FYCO1 has a specificity that is very different from ULK1 and ATG13, as shown in Fig. 7. All residues in the LIR motif of optineurin are accepted in ULK1 and ATG13, with the exception of Ser in position −1. It was recently shown that phosphorylation of this residue by TBK1 increases the binding affinity of the optineurin LIR for LC3B 5-fold (20). ULK1, ULK2, ATG13, FIP200, and DmAtg1 all have a Asp in this position.
Evidence is emerging that assign different roles to the two ATG8 subfamilies in autophagosome biogenesis. In this regard, one particular study showed that the LC3 subfamily is important for phagophore elongation, whereas the GABARAPL2/GATE16 may act at a later stage in the process (14). However, little is currently known about the individual roles of different LC3 and GABARAP subfamily proteins on the phagophore. It is interesting to note that proteins of the ULK complex have preference for the GABARAP subfamily proteins. However, both ULK1 and ATG13 also interacted with LC3A and LC3C. Clearly, more studies are needed to elucidate which ATG8s are the in vivo interaction partners of the endogenous ULK1-ATG13-FIP200-ATG101 complex. As indicated by the large proteomics screen of Behrends et al. (37), the specificity of LIR-mediated interactions is higher in vivo than in binding assays in vitro. It is also possible that the specificity of the complex may differ from the specificity of the individual proteins observed in our in vitro assays.
A recent study in Arabidopsis showed that ATG1 and ATG13 are degraded by autophagy during nutrient starvation (40). It is presently not known if this is selective autophagy mediated by LIR-ATG8 interactions. During revision of this manuscript, two independent studies reported that yeast Atg1 is partially degraded by autophagy, and they identified a LIR/AIM motif in Atg1 that was found to be essential for its autophagic degradation (41, 42). These studies showed that Atg1 is present on autophagosomes, and indicated an important role for Atg1 at later steps during autophagosome formation dependent on the Atg8 interaction. The LIR/AIM motif in Atg1 was not important for the role of Atg1 during initiation of autophagosome formation (42). Kraft et al. (41) also looked at mammalian ULK1. They showed that transiently overexpressed ULK1 is located on WIPI2 positive structures. Their data also indicated that this location of ULK1 depends on a LIR motif in mammalian ULK1, identical to the motif mapped by us in this study.
In our study, we similarly report that mammalian ULK1 stably expressed in mammalian cells is present on WIPI2 and LC3B positive structures. We also show that ULK1 needs the LIR motif to get recruited to these structures. This supports the conclusion that ULK1 is located on phagophores and/or autophagosomes, suggesting a role for ULK1 in later steps of autophagosome formation.
In accordance with recent data obtained by Joo et al. (32), we found that mammalian ULK1 is mainly degraded by the proteasome and with little contribution from autophagy. This suggests that the LIR-GABARAP interaction is not used to capture ULK1 for selective autophagy. We cannot exclude, however, that ULK1 and the entire complex may be degraded by autophagy under certain circumstances. It must be emphasized that it is particularly difficult to study the role of the LIR motif in ULK1 in autophagy because ULK1 overexpression is inhibitory to autophagy and ATG13 and FIP200 also have LIR motifs and can recruit the LIR mutant ULK1 precluding the observation of any phenotype of this mutant in mammalian cells.
Recent studies have revealed that binding to ATG8 family proteins is a feature shared by several of the human core ATG proteins (37). Several of the ATG proteins co-localize with GFP-LC3 positive punctuated structures. Presumably, they are located on the outer surface of the phagophores, and some are also located on autophagosomes. This is also most likely the case for FYCO1 and the TBC1D family proteins found to interact with ATG8 family proteins (21–23). We, therefore, favor the hypothesis that binding of the ULK complex and other ATG proteins to lipidated ATG8 proteins on the surface of the phagophore is required for efficient formation or maturation of an autophagosome. Hence, an important role of ATG8 homologues may be to act as a scaffold for the assembly of autophagy complexes.
Supplementary Material
Acknowledgments
We are grateful to Sharon Tooze for the generous gift of WIPI2 antibody and Ola Rumohr Blingsmo at the Biotechnology Centre, University of Oslo, for synthesizing the peptide arrays. We are indebted to the BioImaging and Proteomics FUGE core facilities at the Institute of Medical Biology, University of Tromsø, for the use of instrumentation and expert assistance.
This work was supported in part by grants from the FUGE and FRIBIO programs of the Norwegian Research Council, the Norwegian Cancer Society, and the Blix Foundation (to T. J.).

This article contains supplemental Table S1 and Figs. S1–S4.
- ULK1
- Unc-51 like kinase 1
- (MAP1)LC3
- microtubule-associated protein 1 light chain 3
- ATG
- AuTophaGy-related
- FIP200
- focal adhesion kinase family interacting protein of 200 kDa
- GABARAP
- γ-aminobutyric acid type A receptor associated-protein
- LIR
- LC3 interacting region
- MBP
- maltose-binding protein.
REFERENCES
- 1. Tomoda T., Bhatt R. S., Kuroyanagi H., Shirasawa T., Hatten M. E. (1999) A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24, 833–846 [DOI] [PubMed] [Google Scholar]
- 2. Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 [DOI] [PubMed] [Google Scholar]
- 3. Chan E. Y. (2012) Regulation and function of uncoordinated-51 like kinase proteins. Antioxidants Redox Signal. 17, 775–785 [DOI] [PubMed] [Google Scholar]
- 4. Weidberg H., Shvets E., Elazar Z. (2011) Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 [DOI] [PubMed] [Google Scholar]
- 5. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms. Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 6. Yang Z., Klionsky D. J. (2010) Mammalian autophagy. Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bodemann B. O., Orvedahl A., Cheng T., Ram R. R., Ou Y. H., Formstecher E., Maiti M., Hazelett C. C., Wauson E. M., Balakireva M., Camonis J. H., Yeaman C., Levine B., White M. A. (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144, 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Longatti A., Tooze S. A. (2009) Vesicular trafficking and autophagosome formation. Cell Death Differ. 16, 956–965 [DOI] [PubMed] [Google Scholar]
- 10. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 11. Shpilka T., Weidberg H., Pietrokovski S., Elazar Z. (2011) Atg8. An autophagy-related ubiquitin-like protein family. Genome Biol. 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) LC3, GABARAP, and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 14. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 16. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 17. Noda N. N., Ohsumi Y., Inagaki F. (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 18. Johansen T., Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 20. Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pankiv S., Alemu E. A., Brech A., Bruun J. A., Lamark T., Overvatn A., Bjørkøy G., Johansen T. (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itoh T., Kanno E., Uemura T., Waguri S., Fukuda M. (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 192, 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Popovic D., Akutsu M., Novak I., Harper J. W., Behrends C., Dikic I. (2012) Rab GTPase-activating proteins in autophagy. Regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan E. Y., Kir S., Tooze S. A. (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 [DOI] [PubMed] [Google Scholar]
- 25. Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. (2009) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979 [DOI] [PubMed] [Google Scholar]
- 28. Mercer C. A., Kaliappan A., Dennis P. B. (2009) A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5, 649–662 [DOI] [PubMed] [Google Scholar]
- 29. Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kundu M., Lindsten T., Yang C. Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A., Thompson C. B. (2008) Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joo J. H., Dorsey F. C., Joshi A., Hennessy-Walters K. M., Rose K. L., McCastlain K., Zhang J., Iyengar R., Jung C. H., Suen D. F., Steeves M. A., Yang C. Y., Prater S. M., Kim D. H., Thompson C. B., Youle R. J., Ney P. A., Cleveland J. L., Kundu M. (2011) Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell 43, 572–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okazaki N., Yan J., Yuasa S., Ueno T., Kominami E., Masuho Y., Koga H., Muramatsu M. (2000) Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain. Possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 85, 1–12 [DOI] [PubMed] [Google Scholar]
- 34. Larsen K. B., Lamark T., Øvervatn A., Harneshaug I., Johansen T., Bjørkøy G. (2010) A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 6, 784–793 [DOI] [PubMed] [Google Scholar]
- 35. Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 36. Shvets E., Fass E., Scherz-Shouval R., Elazar Z. (2008) The N terminus and Phe-52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 121, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 37. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Le Grand J. N., Chakrama F. Z., Seguin-Py S., Fraichard A., Delage-Mourroux R., Jouvenot M., Risold P. Y., Boyer-Guittaut M. (2011) GABARAPL1 antibodies. Target one protein, get one free!. Autophagy 7, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 39. Polson H. E., de Lartigue J., Rigden D. J., Reedijk M., Urbe S., Clague M. J., Tooze S. A. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 [DOI] [PubMed] [Google Scholar]
- 40. Suttangkakul A., Li F., Chung T., Vierstra R. D. (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. The Plant cell 23, 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kraft C., Kijanska M., Kalie E., Siergiejuk E., Lee S. S., Semplicio G., Stoffel I., Brezovich A., Verma M., Hansmann I., Ammerer G., Hofmann K., Tooze S., Peter M. (2012) Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31, 3691–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakatogawa H., Ohbayashi S., Sakoh-Nakatogawa M., Kakuta S., Suzuki S. W., Kirisako H., Kondo-Kakuta C., Noda N. N., Yamamoto H., Ohsumi Y. (2012) The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 287, 28503–28507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Svenning S., Lamark T., Krause K., Johansen T. (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sancho A., Duran J., García-España A., Mauvezin C., Alemu E. A., Lamark T., Macias M. J., DeSalle R., Royo M., Sala D., Chicote J. U., Palacín M., Johansen T., Zorzano A. (2012) DOR/Tp53inp2 and Tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PloS One 7, e34034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X., Chen Y. G. (2010) Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12, 781–790 [DOI] [PubMed] [Google Scholar]
- 47. Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dötsch V., Ney P. A., Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185 [DOI] [PubMed] [Google Scholar]
- 49. Mohrlüder J., Stangler T., Hoffmann Y., Wiesehan K., Mataruga A., Willbold D. (2007) Identification of calreticulin as a ligand of GABARAP by phage display screening of a peptide library. FEBS J 274, 5543–5555 [DOI] [PubMed] [Google Scholar]
- 50. Mohrlüder J., Hoffmann Y., Stangler T., Hänel K., Willbold D. (2007) Identification of clathrin heavy chain as a direct interaction partner for the γ-aminobutyric acid type A receptor associated protein. Biochemistry 46, 14537–14543 [DOI] [PubMed] [Google Scholar]
- 51. Jiang S., Wells C. D., Roach P. J. (2011) Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism. Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 413, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamaguchi M., Noda N. N., Nakatogawa H., Kumeta H., Ohsumi Y., Inagaki F. (2010) Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 54. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kramer R. M., Roberts E. F., Um S. L., Borsch-Haubold A. G., Watson S. P., Fisher M. J., Jakubowski J. A. (1996) p38 mitogen-activated protein kinase phosphorylates cytosolic phospholipase A2 (cPLA2) in thrombin-stimulated platelets: Evidence that proline-directed phosphorylation is not required for mobilization of arachidonic acid by PLA2. J. Biol. Chem. 271, 27723–27729 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.