Abstract
Sodium (Na) is ubiquitous in soils, and is transported to plant shoots via transpiration through xylem elements in the vascular tissue. However, excess Na is damaging. Accordingly, control of xylem-sap Na concentration is important for maintenance of shoot Na homeostasis, especially under Na stress conditions. Here we report that shoot Na homeostasis of Arabidopsis thaliana plants grown in saline soils is conferred by reactive oxygen species (ROS) regulation of xylem-sap Na concentrations. We show that lack of A. thaliana respiratory burst oxidase protein F (AtrbohF; an NADPH oxidase catalysing ROS production) causes hypersensitivity of shoots to soil salinity. Lack of AtrbohF-dependent salinity-induced vascular ROS accumulation leads to increased Na concentrations in root vasculature cells and in xylem sap, thus causing delivery of damaging amounts of Na to the shoot. We also show that the excess shoot Na delivery caused by lack of AtrbohF is dependent upon transpiration. We conclude that AtrbohF increases ROS levels in wild-type root vasculature in response to raised soil salinity, thereby limiting Na concentrations in xylem sap, and in turn protecting shoot cells from transpiration-dependent delivery of excess Na.
Keywords: Arabidopsis , Na homeostasis, NADPH oxidase, ROS, salt tolerance
Introduction
Soil salinity is one of the major factors limiting global agricultural production, affecting an estimated ∼20 million hectares of cultivated land world-wide (Epstein, 1985; Zhu, 2002; Flowers, 2004; Munns and Tester, 2008). In addition, increased salinisation of cultivated land, especially where production depends on irrigation, is threatening the sustainability of global food production (Greenway and Munns, 1980; Frommer et al, 1999; Tester and Davenport, 2003; Flowers, 2004; Munns and Tester, 2008). Thus, there is an urgent need to advance the understanding of plant soil-salinity tolerance mechanisms, and to increase the salinity tolerance of crops.
Soil salinity is characterized by a high concentration of soluble salts. NaCl is the most soluble and widespread soil salt, and Na+ toxicity is therefore the most prevalent natural salinity stress restricting plant growth (Zhu, 2002; Tester and Davenport, 2003; Munns and Tester, 2008). Many plants have evolved mechanisms to circumvent the effects of high soil Na+ concentrations, including maintenance of shoot and root intracellular Na+ concentrations at non-toxic levels (Zhu, 2002; Munns and Tester, 2008). In Arabidopsis, delivery of Na+ from the soil to shoot is believed to involve the following steps. Initially, Na+ enters root epidermal cells passively via nonselective cation channels (Amtmann and Sanders, 1999; Tester and Davenport, 2003; Munns and Tester, 2008) and likely also via high-affinity K+ (HKT) transporters (Munns and Tester, 2008). Next, Na+ moves across the endodermis, and then enters the cells of the central vascular cylinder (stele). Finally, Na+ is loaded from the stelar cells into the xylem elements specialized for long-distance transport (vessels and tracheids), and is thereby delivered to the shoot in the transpiration stream (Smith, 1991; Davenport et al, 2007; Munns and Tester, 2008). Working against this root-to-shoot Na+ flow, there are mechanisms that restrict the amount of Na+ delivered to the shoot. First, plasma membrane Na+/H+ antiporters (e.g., SOS1) can pump Na+ back into soil solution, reducing the net influx of Na+ into inner cell layers of the root (Shi et al, 2002; Zhu, 2002; Munns and Tester, 2008; Quintero et al, 2011). This reverse pumping of Na+ into the soil is dependent on additional factors (e.g., SOS2, SOS3 and the putative calcium sensor CBL10; Qiu et al, 2002, 2004; Quan et al, 2007). Second, HKT transporters can retrieve Na+ from the transpiration stream in the xylem before it reaches the shoots (Mäser et al, 2002; Ren et al, 2005; Sunarpi et al, 2005; Davenport et al, 2007; Møller et al, 2009; Baxter et al, 2010; Munns et al, 2012). These mechanisms for counteracting root-to-shoot Na+ delivery are essential for maintaining shoot Na+ concentrations at non-toxic levels and thus for soil-salinity tolerance (Zhu, 2002; Munns and Tester, 2008).
Soil-salinity stress causes the in planta accumulation of reactive oxygen species (ROS) (Achard et al, 2008; Miller et al, 2010), which secondarily results in oxidative stress and cell damage (Flowers, 2004; Miller et al, 2010; Xie et al, 2011). In addition, there is evidence suggesting that ROS, as well as being a toxic agent in salinity stress, also acts as a signalling mediator of plant salinity tolerance. For example, Arabidopsis mutants lacking cytosolic and/or chloroplastic H2O2 ascorbate peroxidase removal enzymes were found to be more tolerant of salinity stress, suggesting that increased ROS levels promote tolerance (Miller et al, 2007). In addition, it has recently been reported that the enzyme At5PTase7 regulates ROS production in saline conditions (likely through activation of NAPDH oxidase enzymes), and that mutants lacking At5PTase7 activity are hypersensitive to salinity (Kaye et al, 2011). Finally, treatment of wild-type (WT) Arabidopsis plants with diphenylene iodonium (DPI, an inhibitor of NADPH oxidase activity) blocked salinity-induced ROS production and reduced salinity tolerance by inhibiting gp91phox homologues (also called respiratory burst oxidase; Leshem et al, 2007). ROS therefore plays a dual role in salinity response, potentially causing oxidative damage as well as conferring salinity tolerance (Dat et al, 2000). However, to our knowledge, no ROS-associated genes have previously been identified following forward-genetic screens for mutants with altered salinity sensitivity. In addition, the cellular mechanisms by which in vivo ROS production and ROS signalling promote salinity tolerance remain unknown.
Previous genetic screens for Arabidopsis thaliana mutants displaying altered salinity responses have largely involved in vitro (Petri-dish) environments (Liu et al, 2000; Quesad et al, 2000; Shi et al, 2002), and have enabled the discovery of genes (including SOS1, SOS2 and SOS3) playing major roles in plant salinity response and tolerance (Zhu, 2002). However, the relative artificiality of these in vitro screens (in particular the limitations on transpiration rate) might have resulted in additional genes important to plant soil-salinity response being missed (Møller and Tester, 2007). We therefore devised a novel screen for A. thaliana mutants hypersensitive to soil salinity, with special focus on the possibility that such mutants might be affected in the delivery of Na from root to shoot (see Materials and methods section). We describe here the isolation of a novel soil-salinity hypersensitive mutant from this screen, and show that the mutant phenotype is conferred by a loss-of-function allele of A. thaliana respiratory burst oxidase protein F (AtrbohF), a gene whose product is an NADPH oxidase that catalyses production of ROS. We subsequently show that AtrbohF increases root vascular ROS levels in response to soil salinity, thus reducing xylem-sap Na levels and delivery of Na to the shoot, hence protecting the shoot from the damaging effects of excess Na. In addition, we show that delivery of excess Na to the shoot caused by lack of AtrbohF is dependent upon a functioning transpiration stream. For the first time, we show how tissue-specific ROS production regulates root–shoot Na delivery and consequent Na tolerance in transpiring soil-grown plants typical of natural and agricultural environments.
Results
Identification of an A. thaliana soil-salinity sensitive1-1 (sss1-1) mutant
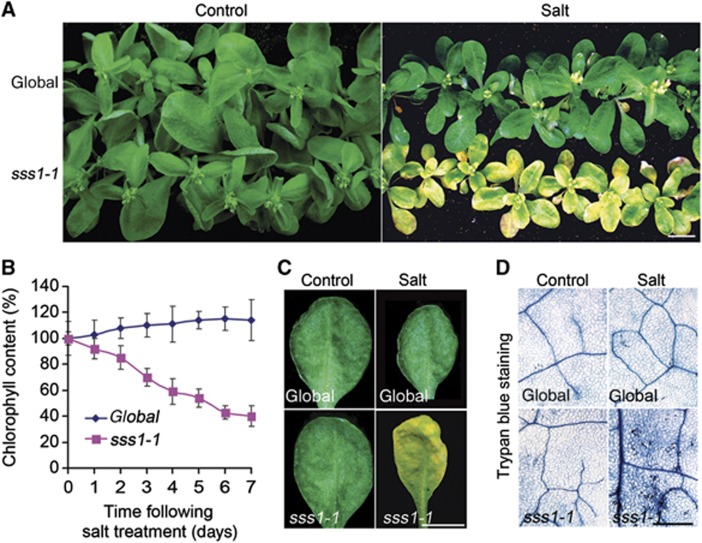
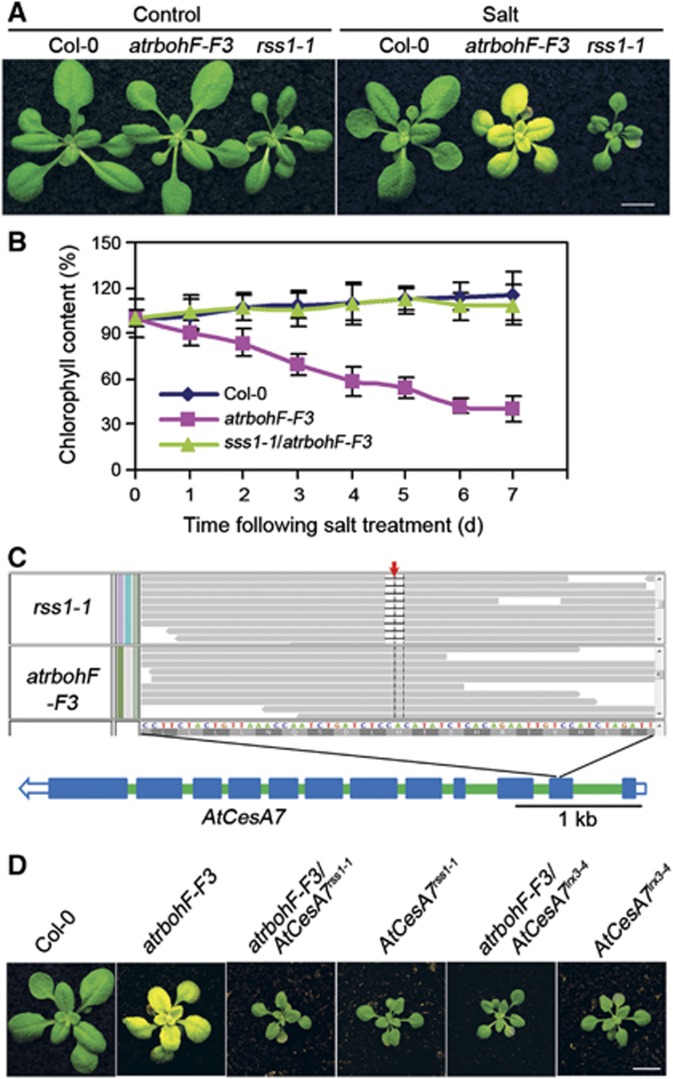
Our previous work had shown that the abscisic acid (ABA) and ethylene signalling pathways can integrate plant salinity responses via the DELLAs, a family of growth-inhibitory proteins particularly associated with gibberellin signalling (Achard et al, 2006). However, DELLA-deficient A. thaliana mutants actually display mild salinity sensitivity (Achard et al, 2006), thus indicating the relative importance of DELLA-independent salinity responses. We therefore screened for mutants displaying soil-salinity hypersensitivity derived from a DELLA-deficient A. thaliana progenitor line (see Materials and methods section). Screening of approximately 50 000 fast neutron-mutagenised DELLA-deficient M2 seedlings (see Materials and methods section; Belfield et al, 2012) yielded 10 mutants displaying hypersensitivity to soil salinity. Among these, the sss1-1 mutant phenotype was shown by backcrossing to be conferred by a single recessive allele. sss1-1 mutants display a characteristic bleaching phenotype 10 days after being watered with 120 mM NaCl, while progenitor controls remain green (Figure 1A). This bleaching is due to a dramatic decrease (∼60%) in sss1-1 chlorophyll content (Figure 1B and C) and associated cell death (Figure 1D). Interestingly, although sss1-1 is hypersensitive to a range of soil NaCl concentrations (Supplementary Figure 1A and B), there was no detectable difference between the salt sensitivities of in vitro grown sss1-1 and control plants (Supplementary Figure 1C). Thus, sss1-1 identifies a gene that regulates plant sensitivity to soil but not to in vitro salinity.
Figure 1.
Initial characterization of the sss1-1 mutant. (A) Mutant (sss1-1) and DELLA-deficient progenitor (global) control plants were grown on soil in standard conditions for 4 weeks, then watered once to soil capacity with either 120 mM NaCl (Salt) or water (Control), and photographed 10 days after treatment. Bar=1 cm. (B) Shoot chlorophyll contents of plants as indicated (salt treatment as in A), expressed as percentage of 0-day controls. Results shown are means±s.e. of three independent replicates. (C) Appearance of the fourth leaves of global and sss1-1 plants grown under control conditions or 10 days after salt treatment. Plant growth and salinity treatments as described in Figure 1A. Bar=1 cm. (D) Close-up images of leaves (as in A) stained with Trypan blue, revealing increased cell death in salt-treated sss-1 leaves. Bar=0.5 mm.
sss1-1 is a novel mutant AtrbohF allele
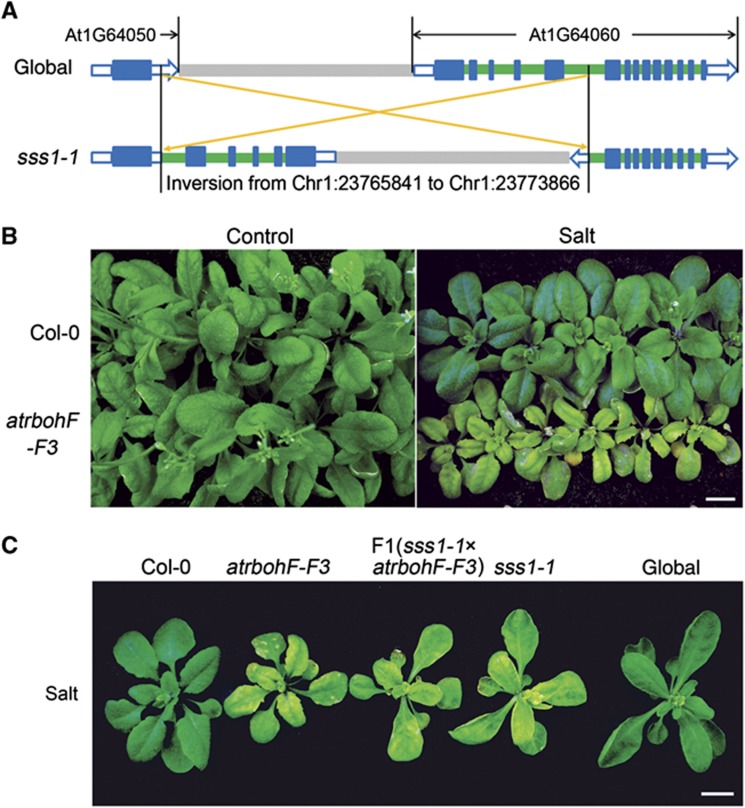
We identified the mutation conferring sss1-1 phenotype via whole-genome sequence analysis (see Materials and methods section; Jiang et al, 2011; Belfield et al, 2012). Of five novel mutations in the sss1-1 genome (versus the progenitor genome; Supplementary Figure 1D), only one (an 8025-bp inversion; Chr1: 23 765 370–23 773 866) was uniformly homozygous in ∼60 soil-salinity hypersensitive F2 plants obtained following a backcross between sss1-1 and the progenitor (data not shown). This inversion interrupts gene At1G64060, a gene encoding a gp91phox homologue and previously designated as AtrbohF (Figure 2A; Torres et al, 2002, 2005).
Figure 2.
sss1-1 is a novel mutant AtrbohF allele. (A) Cartoon showing the interstitial genomic inversion detected in sss1-1 (versus global progenitor genome). The inversion interrupts AtrbohF (At1G64060). Blue boxes represent exons, green boxes introns, and grey boxes intergenic regions. Open blue boxes represent untranslated regions (UTR). (B) WT (Col-0) and atrbohF-F3 plants grown in normal conditions (Control) or 7 days after salt treatment (Salt; salt treatment as in Figure 1A). Bar=1 cm. (C) sss1-1/atrbohF-F3 heterozygote and control homozygote plants (as indicated) 7 days after salt treatment. Plant growth and treatment is as in Figure 1A. Bar=1 cm.
To confirm that loss of AtrbohF function confers the sss1-1 phenotype, we showed that atrbohF-F3 (a loss-of-function AtrbohF allele derived from the A. thaliana Col-0 (WT) genetic background; Torres et al, 2002) homozygotes and sss1-1/atrbohF-F3 heterozygotes both display soil-salinity hypersensitivity (Figure 2B and C). Thus, loss of AtrbohF function confers hypersensitivity to soil salinity. In addition, loss of AtrbohF function confers soil-salinity hypersensitivity irrespective of the presence or absence of DELLA function.
AtrbohF is expressed in root vascular tissue
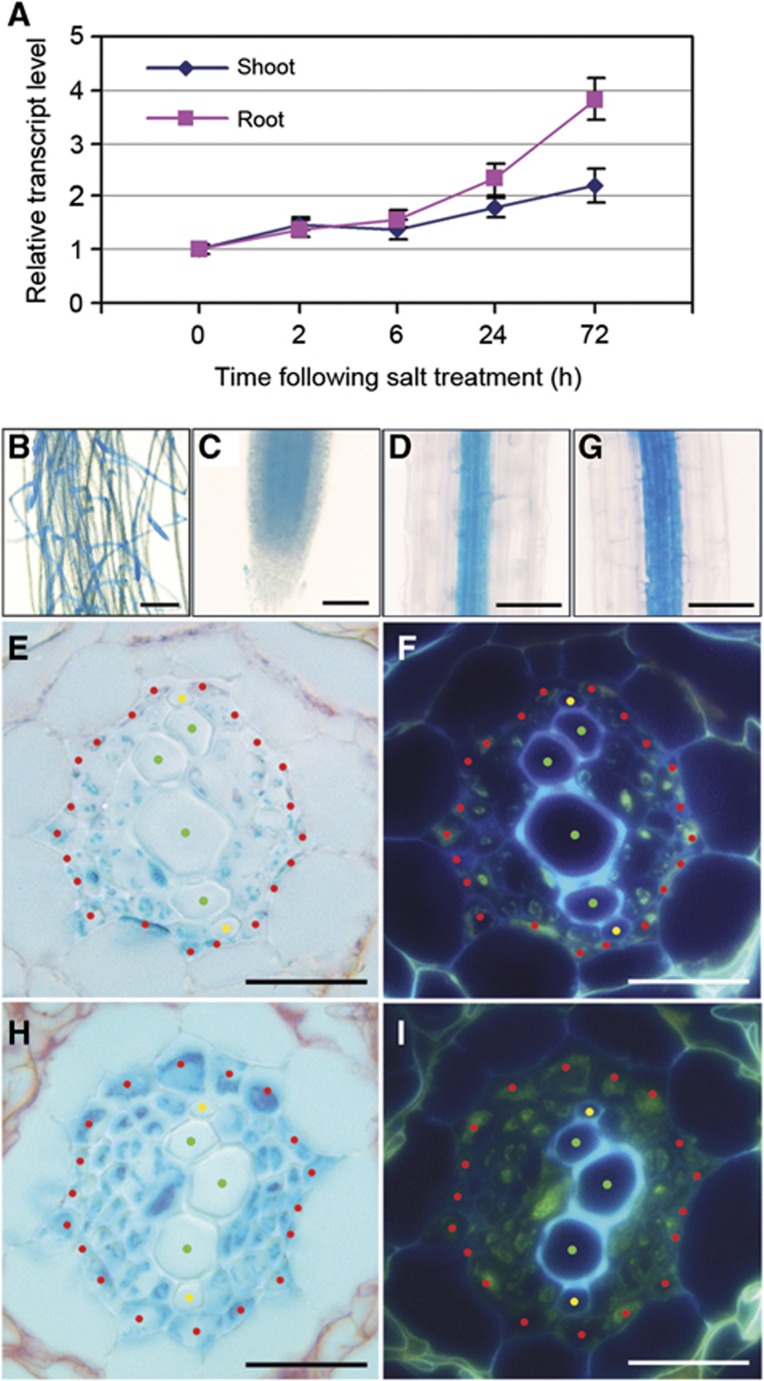
We next studied the effect of salinity on AtrbohF transcripts. Exposure to salinity caused marginal increases in AtrbohF transcript levels 2–6 h following treatment onset, and more substantial increases after 24–72 h (especially in the root; Figure 3A), suggesting that an increase in AtrbohF transcript levels might confer enhanced soil-salinity tolerance. We next investigated the cell/tissue specificity of AtrbohF expression, using pAtrbohF:GUS plants expressing Escherichia coli β-glucuronidase (GUS) under the control of an AtrbohF promoter fragment (Materials and methods section). GUS activity was predominantly detected in roots of pAtrbohF:GUS plants (Figure 3B), mainly in cells comprising the central vascular cylinder or stele (Figure 3C and D). Cross-sectional analysis detected GUS activity primarily within the pericycle cells and associated vascular parenchyma, but not in xylem vessels (Figure 3E and F). In addition, we found that salinity caused a tissue-/cellular-specific increase in GUS staining in root stele cells (Figure 3G–I). These observations indicate that AtrbohF is mainly expressed in the root stele, and that salinity increases stele-specific expression of AtrbohF.
Figure 3.
AtrbohF expression: cell/tissue specificity and salinity induction. (A) Real-time PCR analysis of salt-induced increase in AtrbohF transcript levels. Four-week-old hydroponically grown WT plants (see Materials and methods section) were treated with ¼ MS (control) or ¼ MS plus 100 mM NaCl. Samples were collected at times indicated. Data are expressed as fold increase of experimental samples over 0-h controls, and are means±s.e. of three replicates. (B–I) β-Glucuronidase staining of pAtrbohF:GUS plant tissues. (B) Root system; (C) root tip; (D, G) mature root; (E, H) cross-sections of mature root. (G, H) β-Glucuronidase staining of the mature root of pAtrbohF:GUS plants 3 days following salt (100 mM NaCl) treatment. (F, I) UV fluorescence images of root cross-sections shown in E and H. Bars=1 cm in B; 100 μm in C, D and G; 50 μm in E, F, H and I. Locations of specific cell types are highlighted with red (pericycle), yellow (protoxylem) and green (metaxylem) dots in E, F, H, I; the tissue surrounding the xylem elements interior to the pericycle comprises vascular parenchyma and phloem cells. Results are from 4-week-old plants grown in hydroponic conditions (see Materials and methods section).
AtrbohF confers salinity-induced ROS accumulation and soil-salinity tolerance
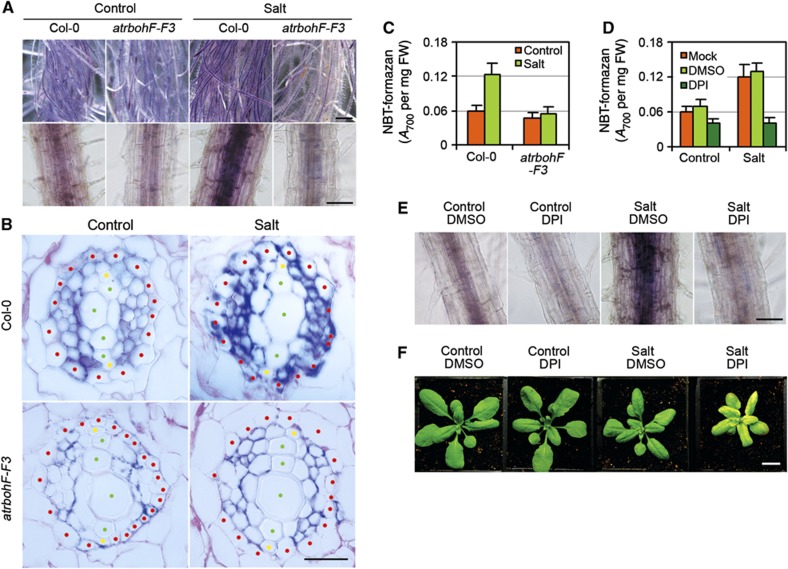
Soil salinity causes ROS accumulation (Zhu, 2002; Munns and Tester, 2008; Miller et al, 2010; Mittler et al, 2011; Suzuki et al, 2011), and we have shown above that lack of AtrbohF (an NADPH oxidase enzyme catalysing ROS production) confers soil-salinity hypersensitivity (Figure 1A). We therefore next studied the relationship between AtrbohF-mediated ROS production and salinity tolerance. We first determined the effect of salinity on in planta ROS levels using nitroblue tetrazolium (NBT; in the presence of superoxide, NBT is reduced to a blue/purple formazan deposit; Carol et al, 2005). Salinity caused an increase in NBT staining in WT root stele cells (Figure 4A). Cross-sectional analysis revealed that this increase is localized primarily within pericycle and vascular parenchyma cells (Figure 4B), a cellular distribution pattern similar to that of AtrbohF transcripts (Figure 3H). In contrast, atrbohF-F3 mutant roots did not display salinity-induced increases in stele-cell NBT staining or formazan deposits (Figure 4A–C). Thus, salinity induces the accumulation of NBT-detectable ROS in root stelar cells, and this accumulation is dependent on AtrbohF function.
Figure 4.
Soil-salinity tolerance conferred by AtrbohF-mediated ROS production in root vasculature. (A) Effect of salt on NBT-visualized ROS levels in WT (Col-0) and atrbohF-F3 roots (see Materials and methods section). Upper panel shows strong NBT staining of the central stele area of salt-treated WT root systems, lower panel single root in close-up. Bar=1 cm in upper panel; 100 μm in lower panels. (B) Root cross-section (as in A). Locations of cell types are highlighted with red (pericycle), yellow (protoxylem) and green (metaxylem) dots; cells surrounding the xylem elements interior to the pericycle represent vascular parenchyma and phloem. Bars=50 μm. (C) Colorimetric quantification of NBT-formazan deposition in indicated root tissue (see Materials and methods section; FM, fresh mass). (D, E) DPI inhibits salt-induced NBT-formazan production in WT roots versus controls (DMSO is carrier; see Materials and methods section). Bar=100 μm. (F) WT (Col-0) plants 7 days following treatments (as indicated). Bar=1 cm. Four-week-old plants were watered with either 120 mM NaCl (Salt) or water (Control), combined with 5 μM of DPI or DPI carrier (DMSO), and photographed 7 days after treatment. Bar=1 cm. Data shown in (C) and (D) are means±s.e. of three replicates.
We next showed that inhibition of NADPH oxidase activity with DPI (Carol et al, 2005) blocked salinity-induced NBT-detectable ROS accumulation in WT root stele (Figure 4D and E). We also found that DPI-treated WT plants phenocopied the soil-salinity hypersensitivity conferred by lack of AtrbohF (Figure 4F). We conclude that AtrbohF NADPH oxidase activity is necessary for salinity-induced ROS production and for consequent salinity tolerance.
Lack of AtrbohF function confers elevated shoot Na levels
Growth of plants on high-salinity soils causes excessive accumulation of Na ions in shoots and resultant cellular damage (due to toxicity and/or osmotic stress). We next determined if the soil-salinity hypersensitivity conferred by lack of AtrbohF function is due to increased shoot Na accumulation. We found that lack of AtrbohF function (atrbohF-F3; sss1-1) caused accumulation of >2-fold more shoot Na than in controls (global progenitor) during a 1- to 7-day period following watering with 120 mM NaCl (Figure 5A). Reduced shoot K content has been reported to cause Na accumulation in saline conditions, and Na accumulation can decrease K uptake (Zhu, 2002; Munns and Tester, 2008). We found that mutants lacking AtrbohF function showed K concentrations similar to WT 1–2 days following salinity treatment, and K concentrations marginally less than WT 4–7 days following treatment (Supplementary Figure 2A). Thus, lack of AtrbohF function confers increased shoot Na accumulation, an accumulation that is not obviously associated with a decrease in K content. The increased shoot Na accumulation caused by lack of AtrbohF function presumably explains the visible shoot soil-salinity hypersensitivity (bleaching and so on, Figure 1A) conferred by AtrbohF loss-of-function mutations.
Figure 5.
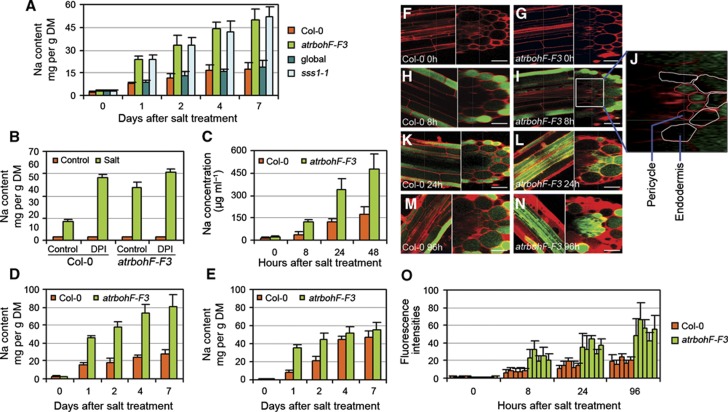
AtrbohF-mediated ROS regulation of root vasculature, xylem-sap and shoot Na levels. (A) Shoot Na contents (genotypes as indicated; global is control for sss1-1; Col-0 for atrbohF-F3; DM, dry mass) 0–7 days following salt treatment (as in Figure 1A). Na content determinations as in Materials and methods section. (B) Effect of DPI treatment on shoot Na content of WT versus atrbohF-F3. Salt and DPI treatment was as in Figure 4D. Shoot Na contents were determined 4 days following treatment. (C) Xylem-sap Na concentrations, measured in root-pressure exudate collected from detopped plants, following salt treatment of soil-grown WT (Col-0) and atrbohF-F3 (see Materials and methods section). (D, E) Shoot (D) and root (E) Na content of hydroponically grown WT (Col-0) and atrbohF-F3 0–7 days following salt treatment. Plant growth and salt treatments as in Figure 2A. Data shown in A–E are means±s.e. of three replicates. (F–N) Cellular localization of Na+ in salt-treated WT and atrbohF-F3 roots. Confocal images of roots stained with CoroNa Green (green) and propidium iodide (red) are shown. (F, G) WT (Col-0; F) and atrbohF-F3 (G) roots without salt treatment; left panel longitudinal section, right panel transverse section. (H–N) Cellular localization of Na+ in WT (H, K, M) and atrbohF-F3 (I, J, L, N roots following 8 h (H–J), 24 h (K, L) and 96 h (M, N) of salt treatment. Salt treatment as in Figure 2A. Bar=50 μm. (O) Quantitative comparison of CoroNa Green fluorescence intensities in root pericycle cells. Five individual plants were measured for each genotype. Values are means±s.e. for 20 pericycle cells from each individual plant.
We next determined the effects of DPI treatment on shoot accumulation of soil Na. Shoots of saline-soil-grown DPI-treated WT plants accumulated higher Na concentrations than controls (Figure 5B), suggesting that NADPH oxidase-generated ROS accumulation counters the accumulation of soil Na in untreated WT shoots. In contrast, DPI treatment of saline-soil-grown atrbohF-F3 mutants caused only a marginal increase in their already elevated shoot Na concentrations (Figure 5B), suggesting that AtrbohF is the predominant NADPH oxidase countering shoot accumulation of soil Na. Consistent with this conclusion, lack of AtrbohF function (versus lack of function of alternative Atrboh genes) uniquely confers both soil-salinity hypersensitivity (Supplementary Figure 2B) and increased shoot Na accumulation (Supplementary Figure 2C). Thus, DPI treatment phenocopies both the salinity hypersensitivity (Figure 4F) and shoot Na accumulation phenotypes (Figure 5B) conferred by lack of AtrbohF function, indicating that ROS generated by AtrbohF has a specific and predominant role in regulating both Na accumulation and soil-salinity tolerance.
Lack of AtrbohF function confers elevation of xylem-sap Na concentration
Root-to-shoot delivery of soil Na is mediated by the xylem-based transpiration stream. AtrbohF has previously been reported to mediate stomatal closure induced by ethylene and ABA (Kwak et al, 2003; Desikan et al, 2006; Joshi-Saha et al, 2011). As stomatal aperture is a major determinant of transpirational flow, we next determined if the increased shoot Na accumulation characteristic of atrbohF-F3 mutants could be due to increased transpiration rate. However, while excess soil-salinity inhibited measured transpiration (Inan et al, 2004) of both WT and atrbohF-F3 plants, there was no detectable difference between the transpiration rates of the two genotypes in either control or high soil-salinity conditions (Supplementary Figure 2D), suggesting that the elevated shoot Na accumulation of atrbohF-F3 is unlikely to be due to effects on transpiration rate.
We next compared Na concentrations in the xylem sap of WT (Col-0) and atrbohF-F3 plants by sampling root-pressure exudate from detopped plants. While the xylem-sap Na concentration of both WT and atrbohF-F3 plants increased progressively over time following initiation of salt treatment, the atrbohF-F3 xylem-sap Na concentration was consistently >2-fold higher than that of WT (Figure 5C; also see Materials and methods section). This elevated xylem-sap Na concentration presumably explains why the shoots of plants lacking AtrbohF function accumulate higher levels of Na than WT plants: increased xylem-sap Na concentrations, in combination with unaltered transpiration rates, will lead to increased delivery of soil Na from root to shoot (Smith, 1991).
Lack of AtrbohF function confers rapid increase in stele cell Na+ levels
We next determined how lack of AtrbohF function causes increased xylem-sap Na concentrations by studying the accumulation and distribution of Na in root tissues of hydroponically cultured plants (see Materials and methods section). Hydroponically cultured atrbohF-F3 mutants displayed a salinity hypersensitivity phenotype similar to that observed when grown in soil (Supplementary Figure 2E), and likewise accumulated higher shoot Na levels than WT plants following onset of salinity treatment (Figure 5D). In addition, while atrbohF-F3 root Na concentrations were higher than those of WT controls for the first 2 days following onset of hydroponic salt treatment (Figure 5E), atrbohF-F3 and WT roots accumulated similar root Na concentrations following longer periods of exposure to salinity (days 4 and 7; Figure 5E).
We next visualized the specific sites of Na+ ion accumulation in roots exposed to salinity using CoroNa Green dye (a green-fluorescent indicator that increases emission intensity upon Na+ binding; Oh et al, 2009; Figure 5F–N). Fluorescence was barely detectable in cells of both WT and atrbohF-F3 control roots (Figure 5F and G), but a strong fluorescence signal was detectable in diverse cell types following exposure to 100 mM NaCl (Figure 5H–N). In salt-treated WT roots, fluorescence was predominantly detected in epidermal cells (Figure 5H, K and M), and was faintly detectable in pericycle and vascular cells. In contrast, fluorescence became clearly detectable in both epidermal and pericycle cells of atrbohF-F3 roots at 8 h after exposure onset (Figure 5I and J), with further increases in pericycle and vascular cells at 24 h (Figure 5L and O) and 96 h (Figure 5N and O). Thus, Na+ accumulates to higher than WT concentrations in the root vasculature of salinity-treated mutants lacking AtrbohF function, presumably explaining the elevated xylem-sap Na concentration characteristic of these mutants.
The saline-soil hypersensitivity of atrbohF-F3 is transpiration dependent
To further understand the soil-salinity hypersensitivity conferred by lack of AtrbohF function, we next screened for novel mutations restoring normal soil-salinity tolerance to atrbohF-F3 (see Materials and methods section). We thereby identified repressor of soil-salinity sensitive 1-1 (rss1-1), a mutation suppressing the effects of atrbohF-F3 on both visible soil-salinity hypersensitivity and chlorophyll levels in saline soils (Figure 6A and B). Whole-genome sequencing identified four novel mutations in the rss1-1 genome (compared to the atrbohF-F3 genome; Supplementary Figure 3A). Only one of the four novel mutations (the 2-bp deletion on Chr5 (−CA); Figure 6C) was uniformly homozygous in all salinity-tolerant F2 plants. This deletion interrupts A. thaliana cellulose synthesis catalytic subunit7 (AtCesA7; Figure 6C).
Figure 6.
rss1-1 is a novel mutant AtCesA7 allele. (A) Soil-grown plants (genotypes as indicated) grown under control conditions (Control) and 7 days after salt treatment (Salt). Bar=1 cm. (B) Chlorophyll content of soil-grown plants. Genotypes as indicated (salt treatments were as described in Figure 1A). (C) The mutation causing rss1-1 phenotype. The top panel shows the 2-bp deletion (−CA; highlighted by the red arrow) that interrupts AtCesA7 in rss1-1. Reads from rss1-1 and atrbohF-F3 are aligned against TAIR9. The bottom panel shows the genomic structure of AtCesA7. Blue boxes represent coding sequence, green boxes introns, open blue boxes untranslated regions (UTR). (D) Demonstration that rss-1 is a mutant AtCesA7 allele. atrbohF-F3/AtCesA7irx3-4 is less sensitive to soil salinity compared to atrbohF-F3 and shows a salt sensitivity similar to that of atrbohF-F3/AtCesA7rss1-1. Bar=1 cm.
To determine if loss of AtCesa7 function was responsible for repressing the salt hypersensitivity of atrbohF-F3 in rss1-1, we generated an independent line lacking AtrbohF and AtCesA7 functions (atrbohF-F3/AtCesA7irx3-4, Turner and Somerville, 1997), studied the salt sensitivity of this line compared to atrbohF-F3, and found that atrbohF-F3/AtCesA7irx3-4 displayed a salt tolerance phenotype (relative to atrbohF-F3) similar to that of atrbohF-F3/AtCesA7rss3-4 (Figure 6D). These observations indicated that rss1-1 is a novel loss-of-function allele of AtCesA7.
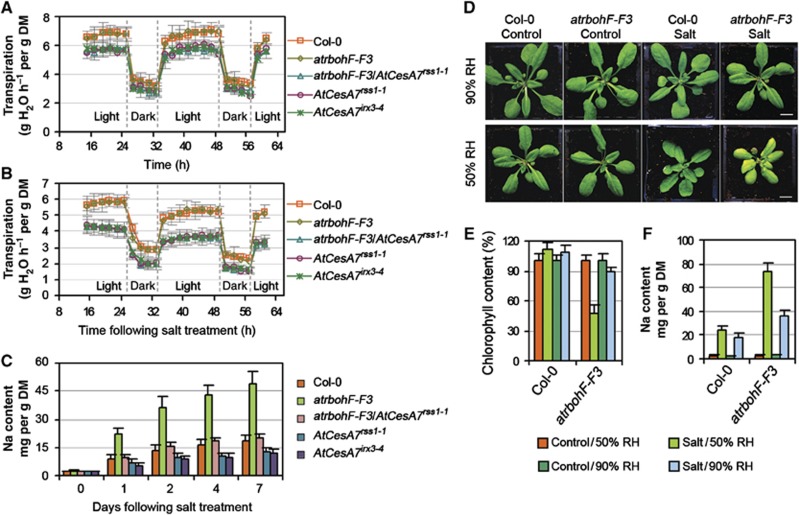
Loss-of-function mutations of AtCesA7 (e.g., irregular xylem3 (irx3)) confer collapsed stem xylem (Turner and Somerville, 1997). Accordingly, our novel AtCesA7rss1-1 allele confers a marked reduction in the number of functional conducting root vasculature xylem elements (Supplementary Figure 3B). Loss of AtCesA7 function has also been reported to confer reduced transpiration rate (Liang et al, 2010). Similarly, AtCesA7rss1-1 confers reduced transpiration rate versus WT and progenitor atrbohF-F3 plants, and suppresses the transpiration of atrbohF-F3 plants in both control and saline-soil growth conditions (Figure 7A and B). In addition, although shoot Na concentrations were relatively similar in atrbohF-F3/AtCesA7rss1-1 and atrbohF-F3 plants grown in control conditions, atrbohF-F3/AtCesA7rss1-1 shoot Na levels were ∼30% those of atrbohF-F3 in saline soil (Figure 7C). As transpiration rate has a major effect on root-to-shoot Na delivery (Zhu, 2002; Møller and Tester, 2007; Munns and Tester, 2008), we conclude that AtCesA7rss1-1 suppresses the soil-salinity hypersensitivity of atrbohF-F3 by reducing the transpiration rate and thus the transpiration-dependent root-to-shoot delivery of Na. This conclusion is strengthened by the observation that transpiration-restricting high atmospheric relative humidity (90%) reduces both the severity of the visible soil-salinity hypersensitivity phenotype and the shoot Na accumulation of atrbohF-F3 mutants grown on saline soil (Figure 7D–F).
Figure 7.
The soil-salinity hypersensitivity of atrbohF-F3 is transpiration-dependent. (A, B) Comparison of transpiration rate of plants (genotypes as indicated) under control conditions (A) or following salt treatment (B) over the experimental period (see Materials and methods section). Values are means±s.e. of at least eight plants. (C) Shoot Na content of plants (genotypes as indicated) 0–7 days following salt treatment (salt treatment as described in Figure 1A). Values are means±s.e. for three replicates. (D–F) The atrbohF-F3 plant displays a mild salt hypersensitivity when grown at 90% relative humidity (RH; versus when grown at 50% RH). Appearance (D), chlorophyll content (E) and shoot Na concentration (F) of Col-0 and atrbohF-F3 plants grown in conditions as indicated. Data are means±s.e. of two replicates.
Discussion
Plant roots supply soil mineral ions to shoots via the transpiration stream (Smith, 1991; Smith et al, 2010). Homeostatic regulation of shoot ion levels is necessary because many ions, such as Na, are toxic in excess. We here describe the discovery of a root-localized mechanism for regulation of root-to-shoot Na delivery. This mechanism contributes to shoot Na homeostasis and protects shoot cells from the damaging effects of excess Na accumulation. In essence, increased soil salinity triggers increased ROS production in root vasculature tissue. Local increases in ROS levels then cause reductions in root xylem-sap Na concentrations and in delivery of Na to the shoot in the transpiration stream.
Soil-salinity stress has long been known to cause the accumulation of ROS in planta (Miller et al, 2010; Mittler et al, 2011). ROS is toxic and causes oxidative stress and cell damage (Miller et al, 2010; Mittler et al, 2011; Suzuki et al, 2011), but is also a potential response signalling mediator (Miller et al, 2007, 2010; Achard et al, 2008; Kaye et al, 2011; Mittler et al, 2011; Suzuki et al, 2011; Ma et al, 2012). We have discovered a cellular signalling mechanism via which soil-salinity-induced ROS regulates shoot Na homeostasis. This discovery rests on our initial observation that AtrbohF (a gene encoding an important NADPH oxidase regulator of ROS production; Sagi and Fluhr, 2006) confers tolerance of high soil salinity (Figures 1, 2, 3). Multiple distinct NADPH oxidases are encoded by the Arabidopsis genome (by genes AtrbohA–AtrbohJ; Sagi and Fluhr, 2006). Previous reports have associated lack of AtrbohJ or AtrbohD function with in vitro salt hypersensitivity (atrbohJ; Kaye et al, 2011) or in vitro salt tolerance (atrbohD; Xie et al, 2011; Ma et al, 2012; Marino et al, 2012), and further studies suggest that AtrbohF acts redundantly with AtrbohD in regulating in vitro salt responses (Xie et al, 2011; Ma et al, 2012). However, these previous and our own studies have shown that lack of AtrbohF function alone had no detectable effect on in vitro salt sensitivity (Supplementary Figure 1C; Xie et al, 2011; Ma et al, 2012). In marked contrast, we find that lack of AtrbohF function alone confers severe soil-salinity hypersensitivity, and that AtrbohF is predominant among NADPH oxidase-encoding loci in the control of soil-salinity tolerance (Supplementary Figure 2B). As transpiration is negligible in in vitro grown plants, we propose that AtrbohF specifically conditions the transpiration-dependent aspects of soil-salinity tolerance. This proposal is strengthened by our observation that a decrease in transpiration rate, either via genetic mutation (e.g., AtCesA7rss1-1; Figure 6A) or via an increase in atmospheric relative humidity (Figure 7D–F), suppresses the soil-salinity hypersensitivity of atrbohF-F3. We conclude that AtrbohF specifically regulates transpiration-dependent soil-salinity response and tolerance.
When plants are grown under Na stress conditions, the accumulation of shoot Na+ ions leads to cellular injury (arising from metabolic toxicity and/or osmotic stress; Zhu, 2002; Munns and Tester, 2008). A previous report indicates that mutants lacking AtrbohF function accumulate similar to WT levels of Na when grown on high salt in vitro (Ma et al, 2012). In contrast, our experiments have shown that AtrbohF-dependent accumulation of ROS in root vasculature actually plays an important role in regulation of vascular Na concentration and consequent shoot Na concentration (Figure 5). This difference is again attributable to large differences in transpiration rates in the two experimental systems (Figure 7). Our observations indicate that, when the transpiration stream is functioning, and under high-salinity conditions, loss of AtrbohF function increases root vasculature Na levels (Figure 5F–N), increases xylem-sap Na concentration (Figure 5C), and in consequence increases delivery of Na to the shoot, resulting in elevated shoot Na accumulation (Figure 5A). This elevated accumulation of Na in the shoot causes the visible damage associated with soil-salinity hypersensitivity.
A previous report has shown that salinity tolerance can be increased by engineered expression of HKT1 in root stele (Møller et al, 2009), indicating that stele-specific regulation of Na concentration underlies salinity tolerance. In addition, while previous reports have suggested that salinity stress induces stele-specific ROS (Dinneny et al, 2008; Dinneny, 2010), the mechanism and biological significance of this induction remained poorly understood. We here show that salinity-induced ROS accumulation in the root stele is substantially attributable to AtrbohF function (Figure 4A–C), and that AtrbohF-dependent accumulation of ROS in the stele confers regulation of shoot Na delivery and tolerance. We propose that, in WT root vasculature, localized salt-induced increases in AtrbohF transcript levels (Figure 3A; and also possible post-translational activation of AtrbohF (Sirichandra et al, 2009; Yun et al, 2011) cause an increase in AtrbohF activity, thereby increasing local ROS levels (Figure 4A–C). The resultant elevated vascular ROS then inhibits net accumulation of Na in the xylem sap and transpiration stream (Figure 5C and F–O), thus protecting cells of the shoot from the elevated Na+ that would otherwise be delivered to them. There is considerable experimental evidence that elevated ROS cause increased cytosolic free Ca2+ concentrations (Foreman et al, 2003), and that these in turn can lead to improved cellular K/Na homeostasis by enhancing Na+ extrusion and maintaining K+ influx (Demidchik et al, 2002; Zhu, 2002; Munns and Tester, 2008). A speculative possibility is that increased ROS might stimulate the activity of HKT transporters in the vasculature, which are known to be important in controlling xylem-sap Na levels and the consequential salt load on the shoot.
In summary, we have discovered a novel salinity-activated, ROS-regulated, root-localized control on root–shoot Na transport via the transpiration stream. This control contributes to essential shoot ionic homeostasis in transpiring plants and identifies a potential target for increasing the salinity tolerance of a wide range of agricultural crop species.
Materials and methods
Plant materials
The DELLA-deficient (gai-t6, rga-t2, rgl1-1, rgl2-1 and rgl3-4; global) progenitor, atrbohF-F3, atrbohD and atrbohF-F3/artbohD mutants were as previously described (Torres et al, 2002; Belfield et al, 2012). atrbohI (N53622), atrbohJ, atrbohH (SALK_558170), atrbohE (SALK_564850), atrbohC (SALK_058191) and AtCesAirx3-4 (SALK_029940C) mutant lines were obtained from the European Arabidopsis Stock Centre (http://Arabidopsis.info).
Soil and hydroponic Arabidopsis culture
Soil: Seeds were sown on Erin Multipurpose compost (Erin Horticulture; http://www.erinhorticulture.com) and grown in controlled environments (16/8-h light/dark cycle, 22°C, 50–60% relative humidity). Hydroponics: Sterilized seeds were sown on sponges (approximately 1 cm thick) saturated with ¼ MS solution. The seeds were first stratified at 4 °C for 3 days in the dark. Subsequently, the sponges were suspended over ¼ MS solution. Plants were grown in controlled environments as described above, and the ¼ MS solution was replaced with new media twice a week.
Mutant screen
Fast neutron-mutagenised M2 global seeds (Belfield et al, 2012) were sown on Erin Multipurpose compost, grown for 4 weeks (conditions as above), then watered once to soil saturation with 120 mM NaCl solution. After 5 days, all plants displaying salinity hypersensitive phenotypes (bleached or pale leaves) were transferred to normal soil and grown to seed set. To screen for atrbohF-F3 suppressors, atrbohF-F3 seeds were fast neutron mutagenised as in Belfield et al (2012). M2 descendants were screened as above, but salt-tolerant individuals (relative to atrbohF-F3) were collected.
Salt treatment under agar plate conditions
Seven-day-old plants grown on MS agar plates (1% (w/v) sucrose); 16/8-h light/dark cycle, 22°C) were transferred to MS agar plates (1% (w/v) sucrose) with a range of NaCl concentrations.
Illumina sequencing and detection of mutations
Genomic re-sequencing of sss1-1 and rss1-1 mutants was performed at the Beijing Genomics Institute, China (using the Illumina Genome Analyzer II platform). We followed previous protocols (Jiang et al, 2011) to detect novel mutations versus progenitor reference genomes (sss1-1 versus the global-reference sequence (Belfield et al, 2012) and rss1-1 versus atrbohF-F3).
Transcription analyses
Real-time PCR was performed as previously described (Jiang et al, 2007). Primer pairs used for PCR amplification of each gene were as follows: AtrbohF: 5 rCAAGCATTGAGCCAAAACCT-3′ and 5anTGCTCCTTTGGCTGTGAGTA-3′; CBP20: 5 PGAAGGAAGACAATGGGGCCG-3′ and 5anCGGCCATAGCGATCATCGTC-3′.
Constructs and plant transformation
Approximately 2.3 kb DNA upstream of AtrbohF was amplified from Col-0 genomic DNA using the following primers: AtrbohF PF, 5PFGGGGACAAGTTTGTACAAAAAAGCAGGCTTCCCCAGTTATGTGTGCATGT-3′, and AtrbohF PR, 5PRGGGGACCACTTTGTACAAGAAAGCTGGGTCCAGATCCAAAGTCGGAATTC-3′. This fragment was cloned into pMDC163 using Gateway Technology (Invitrogen) to generate the pAtrbohF-GUS construct, which was then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into the Col-0 laboratory strain.
Chlorophyll determination
We determined the chlorophyll content as previously described (Jiang et al, 2007).
Cell-death staining and GUS staining
We stained dead cells with Trypan blue as previously described (Carol et al, 2005), and performed GUS staining as described in Jiang et al (2007).
Determination of ROS accumulation
NBT staining and formazan deposit qualification were as previously described (Carol et al, 2005; Myouga et al, 2008).
Determination of transpiration rate
We determined the transpiration rate as previously described (Inan et al, 2004). In brief, 4-week-old plants grown singly in 4-cm pots were watered to soil capacity with water or 120 mM NaCl solution. Pots were then sealed with plastic wrap, leaving shoots outside the wrap (to avoid water loss from soil surface and bottom of the pots), and weighed on an electronic balance every 2 h for 2 days (measurements began 12 h after treatments) to measure transpiration rate.
Determination of ion content
Plant material was harvested, oven dried for at least 24 h at 80 °C and weighed. The material was then digested in concentrated (69%, v/v) HNO3 for at least 12 h for elemental extraction. Concentrations of Na and potassium were determined in appropriately diluted samples by atomic absorption spectrophotometry in an air–acetylene flame (AAnalysis100: Perkin-Elmer; http://www.perkinelmer.com).
Measurement of xylem-sap Na concentration
Five-week-old plants were used for measurement of xylem-sap Na concentration. To collect samples of xylem sap, the rosette leaves and inflorescence stems were excised at the base of the main stem axis. Plants were then kept in a chamber at >95% relative humidity. Sap exuding at the cut surface of the detopped root system was collected and pooled from approximately 20 replicate individual plants. Five microlitres of xylem sap was added to 45 μl of concentrated (69%, v/v) HNO3, incubated for at least 12 h, and then diluted to 5 ml with water for ion content measurement.
Visualization of cellular Na+ concentration
Four-week-old hydroponically grown seedlings were stained and observed by confocal microscopy (Zeiss LSM 510) after salinity treatment. Staining for Na+ was performed as described by Oh et al (2009).
Visualization of xylem
To visualize root xylem cells (Supplementary Figure 3B), we stained transverse root sections (8 μm thick) from 4-week-old plants with toluidine blue.
Supplementary Material
Acknowledgments
We thank Liam Dolan for atrbohI, atrbohJ, atrbohH, atrbohE and atrbohC seeds, Jian-Kang Zhu and Yan Guo for stimulating discussion, and Xiangchao Gan for assistance with genome sequence analysis. This publication is based on the work supported by Award No. KUK-I1-002-03, made by King Abdullah University of Science and Technology, and by Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/F020759/1 and BB/F022697/1. RM and JR are supported by the Wellcome Trust grant 075491/Z/04.
Author contributions: CJ, EB, AV, JACS and NPH conceived the study and wrote the paper. CJ carried out the mutant screens. CJ, AM, JR and RM analysed the genome sequencing data. CJ generated the pAtrbohF:GUS and 35S:AtrbohF/atrbohF-F3 transgenic lines and implemented genetic crosses between mutants. CJ and JACS measured the ion contents. CJ carried out studies of NBT staining, GUS staining, Trypan blue staining, CoroNa staining, chlorophyll measurement and DPI treatment.
Footnotes
The authors declare that they have no conflict of interest.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Amtmann A, Sanders D (1999) Mechanisms of Na+ uptake by plant cells. Adv Bot Res 29: 75–112 [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang Y, Lahner B, Nordborg M, Vitek O, Salt DE (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield EJ, Gan X, Mithani A, Brown C, Jiang C, Franklin K, Alvey E, Wibowo A, Jung M, Bailey K, Kalwani S, Ragoussis J, Mott R, Harberd NP (2012) Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res 22: 1306–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1 controls xylem retrieval of Na+ in Arabidopsis. Plant Cell Environ 30: 497–507 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Davenport RJ, Tester M (2002) Nonselective cation channels in plants. Annu Rev Plant Biol 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Desikan R, Last K, Harrett-Williams R, Tagliavia C, Harter K, Hooley R, Hancock JT, Neill SJ (2006) Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J l47: 907–916 [DOI] [PubMed] [Google Scholar]
- Dinneny JR (2010) Analysis of the salt-stress response at cell-type resolution. Plant Cell Environ 33: 543–551 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Epstein E (1985) Salt-tolerant crops: origins, development, and prospects of the concept. Plant Soil 89: 187–198 [Google Scholar]
- Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Ludewig U, Rentsch D (1999) Taking transgenic plants with a pinch of salt. Science 285: 1222–1223 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, Davies JM, Dolan L (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Greenway H, Munns RA (1980) Mechanisms of salt tolerance in non-halophytes. Annu Rev Plant Physiol 31: 149–190 [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, Shi H, Damsz B, Charbaji T, Gong Q, Ma S, Fredricksen M, Galbraith DW, Jenks MA, Rhodes D, Hasegawa PM et al. (2004) Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Mithani A, Gan X, Belfield EJ, Klingler JP, Zhu J-K, Ragoussis J, Mott R, Harberd NP (2011) Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr Biol 21: 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, Leung J (2011) Brand new START: abscisic acid perception and transduction in the guard cell. Sci Signal 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Kaye Y, Golani Y, Singer Y, Leshem Y, Cohen G, Ercetin M, Gillaspy G, Levine A (2011) Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol 157: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y, Seri L, Levine A (2007) Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J 51: 185–197 [DOI] [PubMed] [Google Scholar]
- Liang Y-K, Xie X, Lindsay SE, Wang YB, Masle J, Williamson L, Leyser O, Hetherington AM (2010) Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana. Plant J 64: 679–686 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F (2012) NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J Exp Bot 63: 305–317 [DOI] [PubMed] [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9–15 [DOI] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, Robertson W, Sussman MR, Schroeder JI (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and a response to abiotic stress. Plant Physiol 144: 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Breusegem FV (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals? Trends Plant Sci 12: 534–540 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotech 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, Jiang X, D'Urzo MP, Lee SY, Zhao Y, Bahk JD, Bressan RA, Yun DJ, Pardo JM, Bohnert HJ (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279: 207–215 [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 9: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, Pardo JM (2011) Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Sagi M, Fluhr R (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu J-K (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Smith AM, Coupland G, Dolan L, Harberd N, Jones J, Martin C, Sablowski R, Amey A (2010) Plant Biology New York, USA: Garland Science, Taylor & Francis Group, LLC, [Google Scholar]
- Smith JAC (1991) Ion transport and the transpiration stream. Bot Acta 104: 416–421 [Google Scholar]
- Sunarpi Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R (2011) Respiratory burst oxidases: the engines of ROS signalling. Curr Opin Plant Biol 14: 691–699 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport RJ (2003) Na+ transport and Na+ tolerance in higher plants. Ann Bot 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defence response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB (2011) Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 66: 280–292 [DOI] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, Pallas JA, Loake GJ (2011) S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268 [DOI] [PubMed] [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.