Abstract
Cell signaling networks propagate information from extracellular cues via dynamic modulation of protein–protein interactions in a context-dependent manner. Networks based on receptor tyrosine kinases (RTKs), for example, phosphorylate intracellular proteins in response to extracellular ligands, resulting in dynamic protein–protein interactions that drive phenotypic changes. Most commonly used methods for discovering these protein–protein interactions, however, are optimized for detecting stable, longer-lived complexes, rather than the type of transient interactions that are essential components of dynamic signaling networks such as those mediated by RTKs. Substrate phosphorylation downstream of RTK activation modifies substrate activity and induces phospho-specific binding interactions, resulting in the formation of large transient macromolecular signaling complexes. Since protein complex formation should follow the trajectory of events that drive it, we reasoned that mining phosphoproteomic datasets for highly similar dynamic behavior of measured phosphorylation sites on different proteins could be used to predict novel, transient protein–protein interactions that had not been previously identified. We applied this method to explore signaling events downstream of EGFR stimulation. Our computational analysis of robustly co-regulated phosphorylation sites, based on multiple clustering analysis of quantitative time-resolved mass-spectrometry phosphoproteomic data, not only identified known sitewise-specific recruitment of proteins to EGFR, but also predicted novel, a priori interactions. A particularly intriguing prediction of EGFR interaction with the cytoskeleton-associated protein PDLIM1 was verified within cells using co-immunoprecipitation and in situ proximity ligation assays. Our approach thus offers a new way to discover protein–protein interactions in a dynamic context- and phosphorylation site-specific manner.
Introduction
In receptor tyrosine kinase networks, such as the epidermal growth factor receptor (EGFR) network, phosphorylation is the dominant mechanism for transducing extracellular signals into intracellular and phenotypic changes.1 These phosphorylation events are temporally and spatially controlled through the actions of EGFR, and the downstream kinases and phosphatases that become recruited to the vicinity of the activated receptor. Ultimately, the extracellular signal is propagated through transient, phosphorylation-driven protein–protein interactions. Unfortunately, current methods are not well suited for predicting or discovering these transient phosphorylation-specific protein–protein interactions, particularly if the half-life of the complex is short. Many phosphotyrosine-dependent protein–protein interactions are mediated through modular binding domains that directly recognize phosphorylated tyrosine residues within specific sequence contexts, such as SH2 and PTB domains2,3 These domains are found on a large number of signaling proteins, including several key protein kinases and phosphatases that are themselves recruited to activated RTKs. The activities of these receptor-recruited kinases and phosphatases are subsequently modulated by auto- and trans-phosphorylation events, resulting in complex patterns of dynamic phosphorylation and the assembly of multi-protein complexes, ultimately resulting in stimulus-dependent feed-back and feed-forward loops. Consequently, the dynamic trajectory of a single phosphorylation site on a protein is shaped by the responsible kinase and phosphatase activities, as well as by secondary events such as phospho-dependent sequestration away from these enzymes or recognition by a phosphorylation-specific binding domain. Thus, similar dynamic behavior at different phosphorylation sites within one protein, or at sites within two different proteins could indicate shared regulation and/or localization, and thus could indicate that the two proteins interact as part of a multi-protein complex, either directly, or via a protein scaffold.
Given this mechanism for phosphorylation-mediated protein complex formation, it should be possible to predict transient protein interactions by finding evidence of strong phosphorylation site co-regulation over time across two or more proteins. How can such co-regulated sites on different proteins be accurately identified?
The ability to measure phosphorylation sites within cellular signaling networks has rapidly expanded through the use of quantitative mass spectrometry. In the application of these methods to RTK signaling, phosphotyrosine-containing peptides are typically enriched for using metal ion affinity chromatography or immunoprecipitation with anti-phosphotyrosine antibodies.4 When combined with sample mass-labeling by iTRAQ or SILAC, these methods allow relative quantitation of phosphorylation events across multiple conditions, or as a function of time.5 Quantitative phosphoproteomics approaches have allowed a single investigator to measure an extensive number of phosphorylation events, on the order of hundreds to thousands, producing dynamic, high-dimensionality phosphoproteomic datasets. Making sense of the resulting data, however, has proven to be a bottleneck in relating these phosphorylation events to the observed cell phenotypes.6
As a general method, clustering techniques can be enormously helpful at finding co-regulated events within high-dimensionality datasets, and can be useful in identifying common functionality and enzymatic control.7,8 Clustering algorithms create clusters of vectors such that vectors within a cluster are more similar to each other than they are to vectors in the remaining clusters. However, one complication of clustering techniques is that perturbations in the parameters used to perform the clustering, such as varying the method of data transformation, the distance metric, the algorithm, or the number of clusters sought, greatly influences the clustering solution.9 Since all of these perturbations are technically correct mathematical solutions to the clustering problem, it is difficult to determine, a priori, a particular parameter set to apply, or which of the clustering solutions obtained using different clustering metrics, are the most biologically meaningful. Therefore, applying clustering methods to search for co-regulated phosphorylation sites, in an attempt to infer protein macromolecular complex formation, could yield a variety of disparate solutions.
In this work we combine quantitative measurements of tyrosine phosphorylation dynamics with a high-throughput computational analysis technique called Multiple Clustering Analysis Methodology (MCAM)9 to explore whether robust co-regulation of phosphorylation dynamics, based on considering a very large set of different clustering solutions, can help to infer protein–protein interactions. The use of MCAM, which applies a variety of clustering algorithms with a variety of clustering parameters to create an ensemble of clustering solutions, allows robust co-regulation of different phosphorylation sites to be recognized by virtue of their consistent capture into the same cluster across the ensemble. We show here that phosphorylation sites on proteins that show such robust co-clustering likely reflect common co-regulatory phosphorylation events, and therefore likely indicate protein–protein interactions. We explicitly identify just such an interaction between EGFR and the cytoskeletal protein PDLIM1.
Results
General methodology
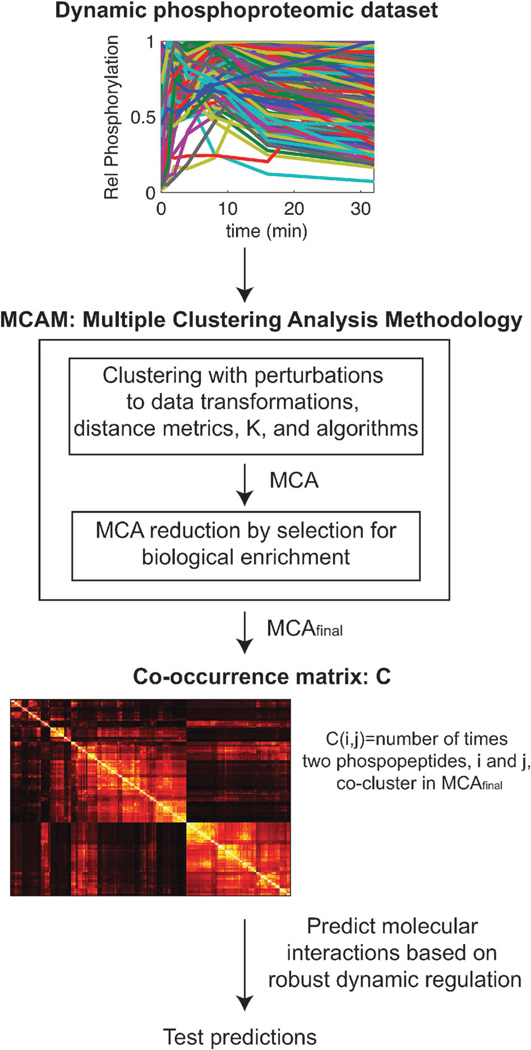
The general methodology for prediction of protein macromolecular complex formation from dynamic phosphoproteomic data is shown in Fig. 1. A quantitative, mass spectrometry (MS) experiment using human mammary epithelial cells (HMECs) stimulated with 100 ng mL−1 EGF was used as the primary data source.10 Following stimulation, the cells were lysed, proteolyzed, and labeled with one of an isobaric series of tags and subjected to anti-phosphotyrosine immunoprecipitation with discovery, monitoring and quantitation following liquid chromatography tandem MS. This dataset represents one of the largest dynamic, phosphotyrosine coverage of the EGFR network to date with 222 species measured across 7-time points (0, 1, 2, 4, 8, 16, and 32 minutes). The web-based database of post-translational modification experiments, PTMScout (http://ptmscout.mit.edu),11 was utilized to access, modify, and explore this dataset. We found that eighteen phosphopeptides in the dataset were redundantly sampled due to a missed proteolytic cleavage event. To prevent obscuring the biological enrichment results between non-redundant species, the mis-cleaved form of each redundantly captured phosphopeptide sequence was removed, leaving 204 unique phosphopeptides derived from 141 unique proteins.
Fig. 1.
Methodology for predicting protein macromolecular interactions through the robust co-regulation of tyrosine phosphorylation dynamics. A quantitative, dynamic, global dataset of phosphorylation is subjected to MCAM, which combinatorially applies a variety of transforms, distance metrics, algorithms, and cluster numbers (K) to produce an ensemble of clustering solutions, termed MCA for Multiple Clustering Analysis. The initial MCA is then pruned to remove those sets that yield little meaningful biological insight according to statistical significance of enrichment for biological terms such as Gene Ontology annotations, protein domains, and predictions of the responsible kinase. This pruned set, MCAfinal, is then analyzed for robust dynamic regulation by considering the number of times any pair of phosphopeptides co-cluster in the full set. Robust and interesting results can then be tested for accuracy regarding dynamic molecular interaction in the original conditions the dataset was collected under.
We subjected this 204 phosphopeptide × 7 time point dataset to multiple clustering analysis methodology, a methodology for analyzing high-dimensionality biological datasets that utilizes ensemble clustering and feature selection.9 MCAM employs clustering to discover co-regulated biological events and biological feature enrichment to understand how those events are tied to biological processes. Since the result of a clustering solution relies heavily on any transformations made to the data, as well as the choice of distance metric, clustering algorithm, and the number of partitions (K), MCAM applies a variety of these in a combinatorial fashion to produce a large number of clustering set solutions. The initial parameters of clustering were chosen in order to explore a large variety of possible clusters and this combination produced 2015 individual clustering solutions, as shown in Table 1.
Table 1.
Parameters of clustering in the first round of MCAM and those pruned to produce the final MCA. NormMax transform means all dynamics were normalized to their maximum value, normMax_log10 indicates taking the normMax followed by the base-10 log transform, and zscore_log10 indicates mean-centered and univariate scaled data that is then transformed by the base-10 log transform. SOM stands for self-organizing maps
| Original MCA | Removed in MCAfinal |
Remains in MCAfinal | |
|---|---|---|---|
| Transforms | center, zscore, normMax, rangeScale, log10, power^(1/2), pareto, FFT, differential, normMax_log10, zscore_log10 | center, pareto, zscore_log10 | zscore, normMax, rangeScale, log10, power^(1/2), FFT, differential, normMax_log10, |
| Distance metrics | Euclidean, correlation, cityblock, cosine, chebychev | correlation | Euclidean, cityblock, cosine, chebychev |
| K | 3–30 in increments of 3 | 3, 6, 9 | 12–30 in increments of 3 |
| Algorithms | Hierarchical, Kmeans, Ncut, SOM, Affinity Propagation | Hierarchical, Kmeans, Ncut, SOM, Affinity Propagation | |
| Resulting number of cluster sets | 2015 | 793 |
Biological feature enrichment was then used to select for those combinations of algorithmic parameters that produced clusters with enrichment in meaningful biological information including Gene Ontology terms, kinase and binding protein predictions, linear amino acid sequence motifs, and protein domains. Following feature selection, 793 individual clustering solutions remained and this set of clustering solutions is used throughout the paper and referred to as MCAfinal. Feature selection for biological information indicated that cluster sets with nine clusters or fewer were relatively uninformative. Additionally, the correlation distance metric and three different transformations were removed from consideration because the overall feature enrichment resulting from their removal was greatly improved, indicating they also contributed very little to the possibility for biological inference and may instead add noise to the ensemble result, Table 1.
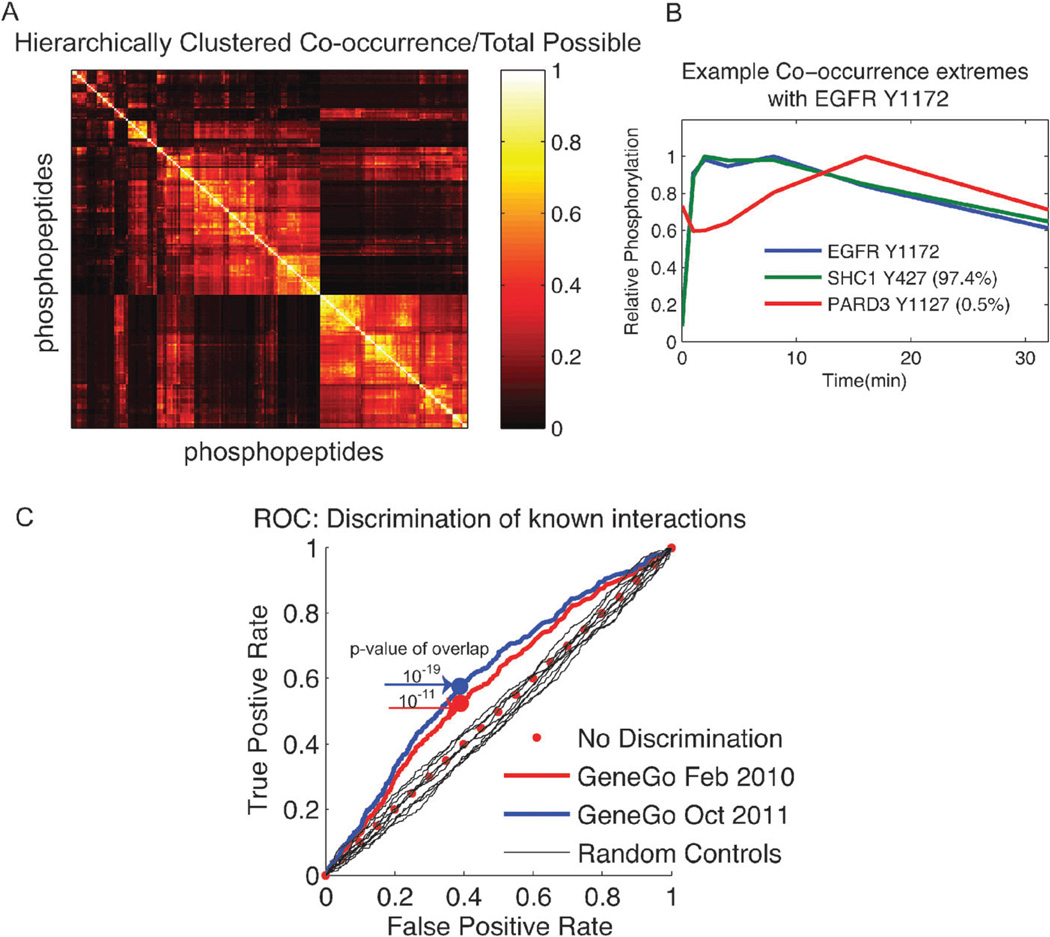
MCAfinal has high dimensionality and analyzing the information contained within this set of clustering solutions therefore becomes difficult, since there is no easy way to visualize all of the solutions simultaneously. One way to condense the results of the MCAM framework is to consider the co-occurrence matrix – a single matrix view of the potential molecular complexes. The co-occurrence matrix represents the number of times any two phosphopeptides cluster together across all sets within MCAfinal, for all phosphopeptide pairs in the dataset. The resulting hierarchically clustered co-occurrence matrix, C, obtained from our dataset is shown in Fig. 2A. According to the co-occurrence matrix results in MCAfinal, there is a significant amount of variability across clustering solutions, with only 0.04% of all possible phosphopeptide pairs never clustering together within at least one clustering solution within MCAfinal, indicating that almost all possible phosphopeptides cluster with each other at least once. This observation highlights the need to consider more than a single clustering solution to find co-regulation amongst dynamic phosphotyrosine events in large phosphoprotein datasets. Two extremes of phosphorylation site co-occurrence values based on this analysis are depicted for the time-dependent phosphorylation of sites on SHC, PARD3, and EGFR shown in Fig. 2B. SHC1 Y427 phosphorylation co-clusters with EGFR Y1172 phosphorylation 97.4% of the time in MCAfinal, whereas PARD3 Y1127 phosphorylation is drastically different and co-clusters only 0.5% of the time. The fact that clustering solutions exist where such robust results as EGFR Y1172:SHC1 Y427 are not seen and solutions where disparate phosphorylation dynamics like EGFR Y1172:PARD3 Y1127 do co-cluster indicates that all solutions should be considered when trying to understand the relationship between two dynamic measurements, and that our methodology, based on examining multiple clustering solutions is likely to be more informative than any single clustering solution.
Fig. 2.
Robust co-clustering predicts known protein–protein interactions. (A) The hierarchically clustered MCAM co-occurrence matrix C. C(i,j) indicates the fraction of times two phosphopeptides, i and j, co-cluster out of the total possible cluster sets (793 in this case). (B) An example of two extreme C(i,j) for phosphopeptides co-clustering with EGFR Y1172 phosphorylation. EGFR Y1172 co-clusters with SHC1 Y42 97.4% of the time, whereas EGFR Y1172 and PARD3 Y1127 co-cluster less than 0.5% of the time. (C) The resulting receiver operator curve from the overlap between all C′ and A was made to demonstrate the ability to discriminate known protein–protein interactions from information in the matrix C. Subjecting randomized data matrixes to the MCAM framework generated ten random controls. The discrimination ability of random controls is with regard to October 2011 GeneGo data. The p-values, according to the hypergeometric test, for the point of best classification on the ROC values are given for the indicated two years of GeneGo data.
Robustness in the co-occurrence matrix overlaps with known protein–protein interactions
Since tyrosine phosphorylation is often involved in directing protein–protein interactions, we first examined whether tight co-regulation of tyrosine phosphorylation events identified by robust co-clustering overlapped with known protein– protein interactions. To do this we quantified the extent of overlap between the co-occurrence matrix, C, and handcurated protein–protein interactions in the database GeneGo (http://www.genego.com) from February 2010 and October 2011 for the 141 unique proteins contained in our dataset. To facilitate quantitative comparison, an unweighted, undirected, adjacency matrix (A) was created representing whether two phosphopeptides came from two proteins that were known to interact directly based on that GeneGo version. Next, the co-occurrence matrix was converted into a binary matrix (C′) by setting all values in C to the value one if they were above a given threshold and to the value zero otherwise. The threshold used to assign a value of one was progressively increased from zero values of co-occurrence up to complete co-occurrence, creating 794 different C′ matrixes. The ability to discriminate between known protein interactions in GeneGo was then calculated for all C′ matrixes. At each threshold value the significance of the overlap between C′ and A was calculated using a hypergeometric test.
If a value of one in a thresholded C′ matrix is considered to be a predicted protein interaction, then a true positive occurs when a predicted interaction overlaps with a known protein–protein interaction. A false positive interaction is defined as a predicted protein–protein interaction that has not been validated. Fig. 2C shows the resulting ROC curve for the co-occurrence values of MCAfinal at various thresholds. As a test for statistical significance, we also generated 10 controls by randomly re-ordering the data matrix before subjecting it to clustering using the final MCA parameters to produce a co-occurrence matrix. In marked contrast to these ten random controls, the co-occurrence values of MCAfinal, over a range of different thresholds, were able to discriminate mapped from unmapped protein–protein interactions. At the point of best classification along the ROC curve in Fig. 2C, the statistical significance of the overlap between the C′ matrix and the GeneGo matrix showed a p-value on the order of 10−19 for 2011 GeneGo data and 10−11 for 2010 GeneGo data, based on a hypergeometric test. Since all phosphopeptides co-cluster with themselves 100% of the time, the co-occurrence matrix is unable to discriminate self-interactions, and these were therefore ignored in the calculation of significance and discrimination. In 2010, there were 570 known peptide interactions between the 141 unique proteins in our dataset, in the GeneGo knowledgebase and 566 interactions in the 2011 GeneGo revision. These data resulted in a total of 45 changes between the adjacency matrixes of 2010 and 2011, and a corresponding increase in the ability of our method to predict known protein–protein interactions. The increase in the statistical significance of the overlap between true and predicted interactions as the database of known protein– protein interactions is expanded and refined over time, as revealed by the change in discrimination curves in Fig. 2C, suggests that our method may be capturing true protein–protein interactions that have not yet been independently validated.
That is, in addition to predicting a significant portion of known protein–protein interactions, robust co-clustering also predicts a large number of protein–protein interactions for which there is no current evidence in GeneGo. At the peak of significance, where the p-value is on the order of 10−19, the number of predicted interactions in the matrix is 8159; compared to 566 GeneGo annotated predictions (a total of 382 GeneGo interactions are captured by the 8159 predictions). This peak occurs at a co-occurrence ratio of 0.15 and greater, which indicates that if one were to consider individual clustering solutions, two phosphopeptides might only co-cluster in 15% of the solutions. To understand what is gained from a robust co-clustering approach compared to individual clustering solutions, we examined the overlap between the adjacency matrix A and every single clustering solution’s adjacency matrix. Only 21 of the 793 sets have a better overlap than the peak overlap of C′ at a threshold of 0.15, based on the p-value resulting from the significance test. However, if these 21 individual adjacency matrixes are combined in such a way that all interactions across all 21 sets are considered significant, then it represents capturing 74% of the total possible number of pairwise interactions (20 706 total possible interactions exist among the 204 phosphopeptide combinations). In contrast, at even the low cutoff threshold of 15%, robust clustering identifies only 40% of the possible relationships that would be considered significant, resulting in a smaller and more believable number of possible resulting interactions to explore. These findings indicate the utility of considering only the robust results from a combination of many mathematical solutions to the clustering problem when deploying limited experimental resources to expand current knowledge.
Robust co-clustering indicates EGFR phosphorylation site-specific protein interactions
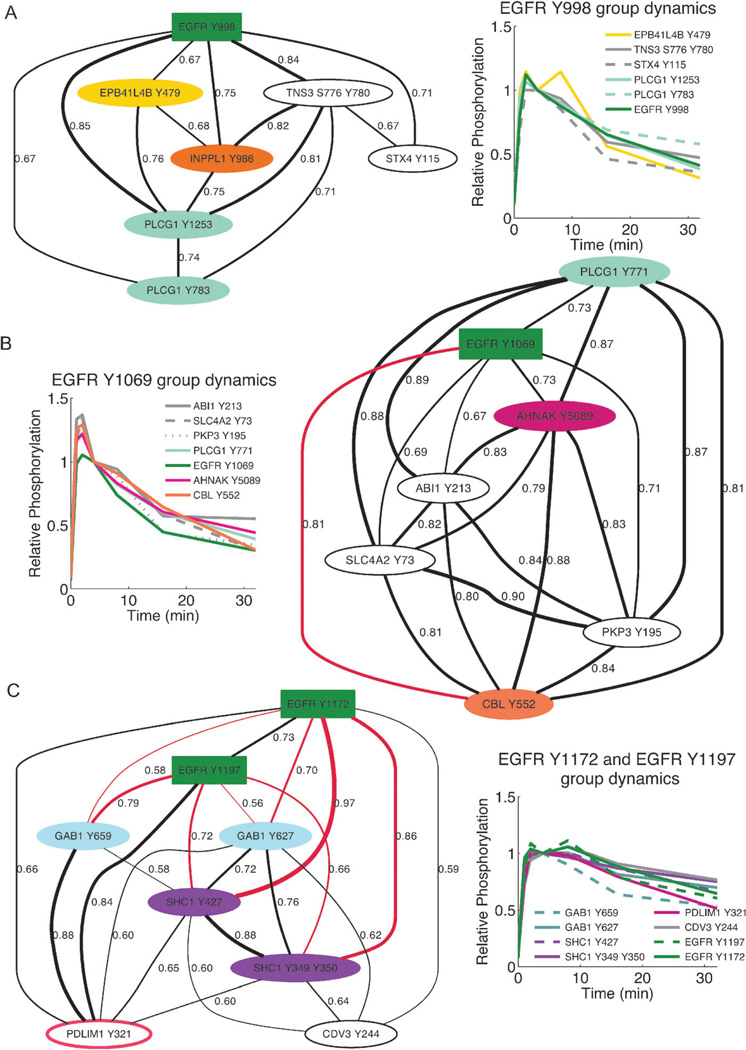
We next investigated whether the protein–protein interactions predicted by robust co-clustering of tyrosine phosphorylation dynamics could be used to reveal phospho-site-specific relationships between these proteins. To accomplish this, the co-occurrence matrix was treated as a weighted adjacency matrix between phosphopeptides, i.e. the fraction of co-occurrence between phosphopeptides was used as a weighted edge to create a connected graph, Fig. 3. For the EGFR phosphorylation sites, which likely represent the most well studied nodes in this dataset, the generated graphs from the co-occurrence matrix were compared with current knowledge. Fig. 3 shows the resulting weighted graphs generated for four phosphorylation sites on the EGF receptor (Swissprot identifier P00533), Y998, Y1069, Y1172 and Y1197. The edge weights, appearing next to the line, are also depicted by the thickness of the line and represent the fractional co-occurrence between the two nodes that edge connects. The edges are colored if there is direct evidence that the indicated protein interacts specifically with the EGFR phosphorylation site the edge is connecting. The nodes themselves are filled in with color if there is evidence of known protein interactions between the colored node and EGFR, based on knowledge in iRefWeb.12 At a cutoff value of 0.15, where the most significant overlap with known protein–protein interactions occurs, the number of nodes connected to each site is extensive (ESI†). Therefore, the graphs in Fig. 3 were generated by finding the co-occurrence cutoff value such that no more than six nodes were directly connected to the site of interest on EGFR and only edges having value greater than the cutoff are indicated. Fig. 3 demonstrates that in addition to recapitulating known protein–protein interactions related to EGFR, potentially novel interactions may be captured by this method.
Fig. 3.
EGFR graphs built from the co-occurrence matrix indicate robust co-clustering yields site-specific insight. Graphs were built around the four singly phosphorylated EGFR peptides measured in the dataset using a fraction of co-occurrence threshold such that six direct nodes were attached to the EGFR tyrosine site of interest. In the graphs, colored nodes indicate a protein known to interact directly with EGFR based on experimental evidence, as indicated in iRefWeb.12 Edges are weighted according to their fraction of co-occurrence and colored red if that protein is known to interact with the indicated EGFR phosphorylation site directly. Edge weights (i.e. fraction of co-occurrence) are indicated by the thickness of the line and the exact edge weight value. (A) The EGFR Y998 group includes proteins known to interact with EGFR including multiple sites on phospholipase-c gamma-1, PLCG1, inositol polyphosphate phosphatase-like protein 1, INPPL1/SHIP2, and erythrocyte membrane protein band 4.1 like 4B isoform 1, EPB41L4B. The fractional threshold used to determine the graph structure from the matrix C was 0.66. (B) The EGFR Y1069 group includes a third site on PLCG1 as well as sites on AHNAK and CBL, which are also known to interact with EGFR. It has been shown that CBL, an E3 ubiquitin ligase, is recruited directly to Y1069 on EGFR. The fractional threshold used was 0.66. (C) EGFR Y1197 and Y1172 are robustly connected together and to known EGFR protein interactions of the adaptor proteins SHC1 and GAB1. SHC1, and subsequently, GAB1 are recruited to the receptor through Y1172 and Y1197. The minimum threshold value to connect six nodes to both sites was 0.55.
All of the EGFR tyrosine phosphorylation sites, and their robustly co-clustered partners, undergo a large and rapid increase in phosphorylation at early time points. It appears that the major distinction being made by clustering algorithms likely lies in the profile of down-regulation. The sites associated with EGFR Y1069 phosphorylation experience the fastest decrease in phosphorylation. The EGFR Y1172 and Y1197 sites robustly co-cluster with each other and that group experiences the slowest decrease in phosphorylation amongst the singly-phosphorylated tyrosine peptides in the dataset. It appears that these rather minor distinctions in temporal profiles are enough to distinguish site-specific interactions. For example, EGFR Y1069 is a known recruitment site for the E3 ubiquitin ligase CBL, Fig. 3B.13 EGFR Y1172 recruits the adaptor protein SHC through SHC’s PTB domain; whereas Y1197 recruits SHC through a SHC’s SH2 domain,14 Fig. 3C. Subsequently, SHC phosphorylation recruits GAB1, via GRB2 binding, to the receptor.14 This knowledge is recapitulated in the module in Fig. 3C that contains both the sites on EGFR known to recruit SHC, and multiple phosphorylation events on SHC and GAB1. Not only are known interactions recapitulated, but the robust groups across EGFR sites are significantly different from each other, indicating that this approach can distinguish functional roles of the different sites on EGFR via the phosphopeptides they are most robustly co-regulated with.
Prediction of the interaction between EGFR and PDLIM1 is validated
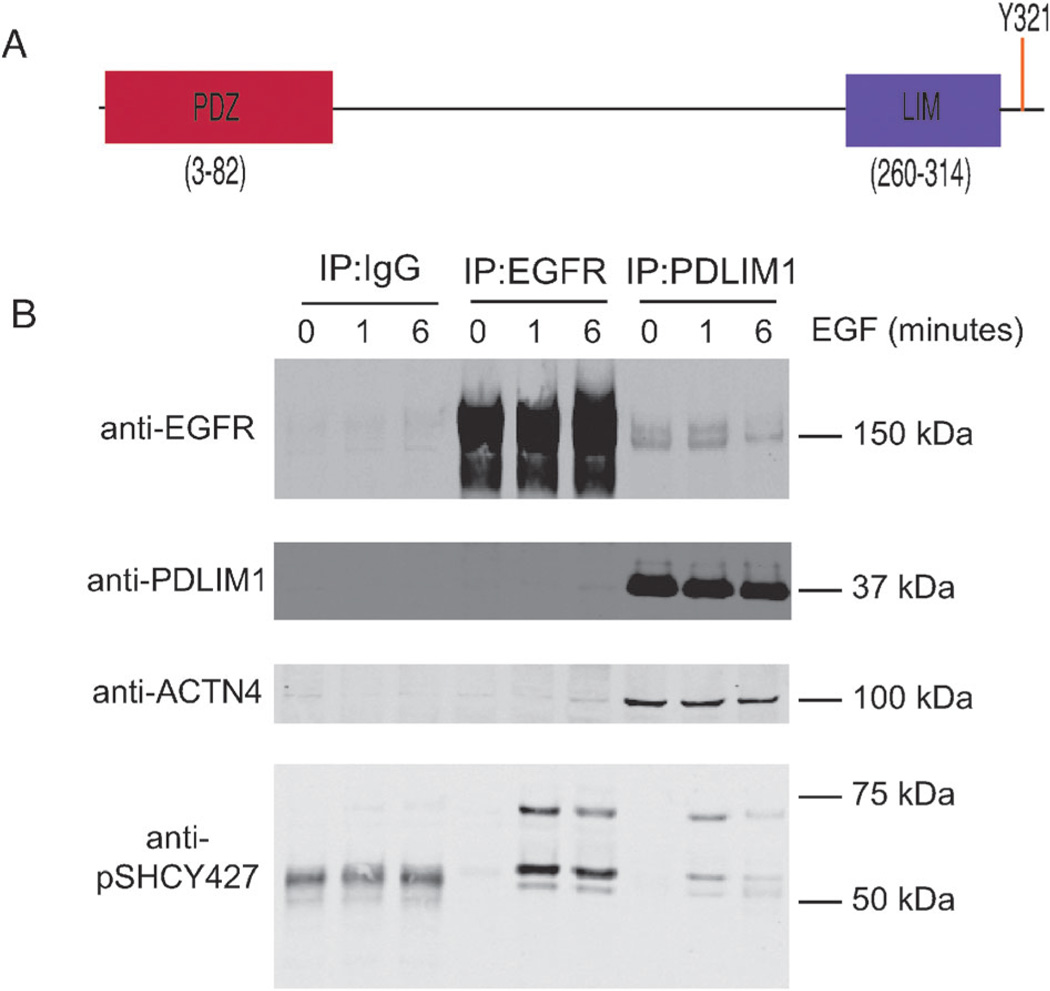
As a final explicit experimental test of the method, we asked whether robust co-clustering of phosphorylation-site dynamics could be used to identify context-dependent protein–protein interactions that had not yet been experimentally observed. One of the more surprising and robust relationships that emerged from our analysis was the temporal co-occurrence of PDLIM1 Y321 phosphorylation (Swissprot O00151) with phosphorylation on the sites in EGFR known to recruit SHC, Y1197 and Y1172 (Fig. 3C). PDLIM1, also known as CLP-36 and hCLIM1, contains an N-terminal PDZ domain and a C-terminal LIM domain (Fig. 4A) and is known to associate with alpha-actinin-1 and alpha-actinin-4. Interactions between these proteins and the PDLIM1 PDZ domain are believed to localize PDLIM1 to actin stress fibers in non-muscle cells.15 It was later shown that PDLIM1 recruits a kinase, Clik1, to actin stress fibers through its LIM domain.16 We found that the time course of PDLIM1 phosphorylation on Y321, a site located C-terminal to the LIM domain and very near the extreme C-terminus, was the top-most co-clustering trajectory with that of EGFR Y1197 phosphorylation, as well as being robustly co-regulated with the rest of the EGFR-SHC module (see Fig. 3C). To date, however, there is no evidence for a connection between PDLIM1 and the EGFR network.
Fig. 4.
Co-immunoprecipitation of EGFR and PDLIM1. (A) The structure of PDLIM1, indicating the N-terminal PDZ domain and the C-terminal LIM domain. The phosphorylation site of interest, Y321, lays eight residues upstream of the C-terminus. (B) Confluent plates of HMECs were treated with either a media-control (0 minutes) or media supplemented with EGF (total concentration 100 ng mL−1) for 1 or 6 minutes, as indicated. Lysates were then immunoprecipitated with agarose beads conjugated to an IgG control, EGFR antibody, or PDLIM1 antibody, as indicated. Eluted proteins from the IP were than blotted for the presence of EGFR or PDLIM1, the known PDLIM1-interactor alpha-actinin 4, ACTN4, or the known EGFR-interactor SHC using a phospho-SHC Y427 antibody.
We therefore examined potential interactions between EGFR, SHC, and PDLIM1 using co-immunoprecipitation (Fig. 4B). Confluent plates of HMECs were serum starved, stimulated with EGF, and lysates immunoprecipitated using antibodies against EGFR or PDLIM1. The immunoprecipitated proteins were then immunoblotted using a different set of EGFR and PDLIM1 antibodies, in addition to antibodies against alpha-actinin-4 and phospho-SHC Y427. We were unable to identify an antibody against total SHC protein that was sensitive enough to detect endogenous SHC in crude HMEC lysates. However, excellent results were obtained using a phospho-SHC antibody directed against the Y427 site, which was the most robustly co-regulated site with both PDLIM1 Y321 and EGFR Y1197 (Fig. 3C). As shown in Fig. 4B, alpha-actinin-4 (ACTN4) was found to interact with PDLIM1 in HMEC cells in an EGF-independent manner. In contrast, Tyr-427 phosphorylated forms of SHC were detected in both the EGFR and PDLIM1 immunoprecipitates only following EGF stimulation. Findings suggestive of an interaction between EGFR and PDLIM1 were observed in the PDLIM1 IPs, though the signal was quite weak and appeared to be independent of EGF stimulation. This interaction could not be confirmed in the EGFR IPs; however, this could perhaps be a consequence of low interaction stoichiometry.
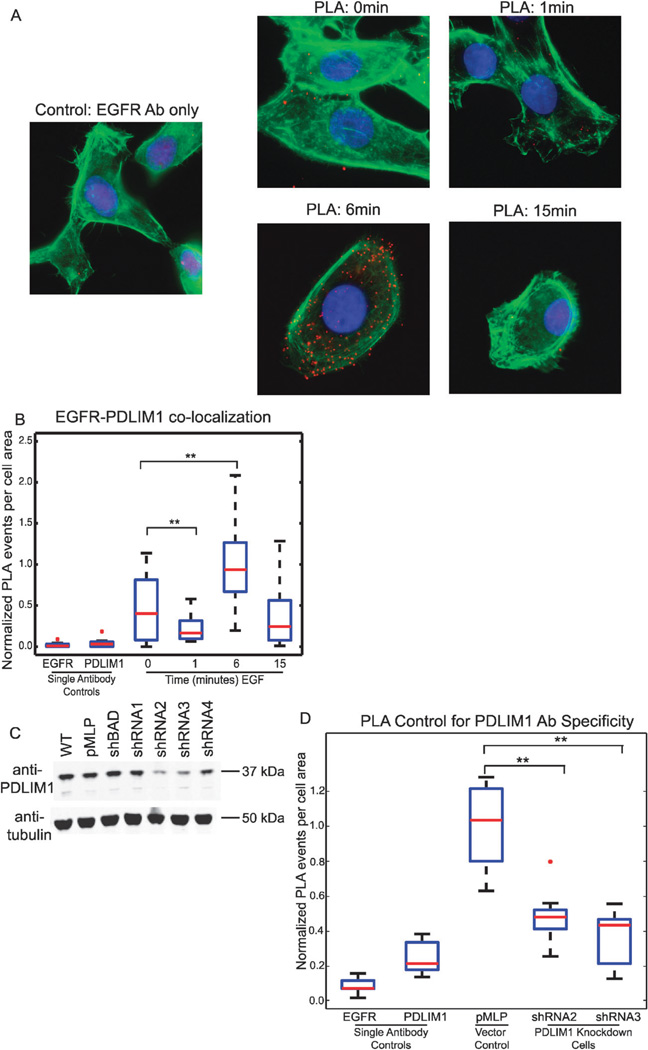
In order to provide further confirmation for an interaction between EGFR and PDLIM1 the proximity ligation assay (PLA17), was used to examine in situ interaction between EGFR and PDLIM1, and the effect of EGF stimulation, using immunofluorescence microscopy. PLA is an extension of co-immunofluorescence, where the secondary antibodies are conjugated to short DNA sequences, which can then be ligated by the addition of two oligonucleotides when they are within 40 nm of each other. This proximal protein event can then be amplified and detected by labeled, complementary DNA. In situ PLA has been used to detect protein interactions and protein modifications. In these experiments, cells seeded on collagen were serum starved and then stimulated with EGF for 1, 6 or 15 minutes before being fixed and assayed for the proximity of EGFR and PDLIM1. Three replicates were performed and five images captured for each condition, which included, on average, 1.5 cells per image resulting in around 22 cells imaged for every condition. A positive PLA signal indicates the EGFR antibody epitope and the PDLIM1 antibody epitope are within 30–40 nm of each other. As shown in Fig. 5A, we observed significant EGFR-PDLIM1 proximity events under all conditions examined, as compared to single antibody background controls, corroborating our co-immunoprecipitation results (full quantitation available in ESI†). Furthermore, these results showed a heavy dynamic dependence of the EGFR-PDLIM1 co-association upon EGF stimulation, with a peak interaction detected by PLA at 6 minutes post-stimulation and returning to a basal state of interaction by 15 minutes. There was a small, but statistically significant decrease in PLA interactions at 1 minute, compared to basal interactions, Fig. 5B. Thus, both the PLA data and co-immunoprecipitation results demonstrated a macromolecular complex involving PDLIM1 and EGFR. The additional spatial resolution of the in situ proximity ligation assay, which includes cell fixation, may account for our increased ability to detect the EGF-dependent component of the interaction.
Fig. 5.
Proximity ligation reveals an EGF-dependent dynamic, in vivo interaction between EGFR and PDLIM1. (A) Merged images of DAPI, phalloidin and PLA staining of HMEC cells treated with control media or EGF-containing media for 1, 6, or 15 minutes before being fixed. All images are from the same biological replicate and are representative of that replicate. (B) The combined quantitation of three biological replicates of PLA stained HMEC cells (fifteen frames counted per condition in total). The number of PLA events per cell area was measured and normalized to that observed following a 6 minute stimulation. Asterisks indicate statistically significant differences, which were observed both between the single antibody controls and all PLA experiments, and between the 0 and 1 minute samples, and the 0 and 6 minute samples. (C) Representative blot of a biological and technical triplicate measurement of PDLIM1 knockdown in wild type cells, vector control cells, cells infected with a scrambled hairpin control against the gene for BAD and four cell lines expressing hairpins designed against the UTR regions of PDLIM1. Knockdown measurements for all replicates indicate shRNA2 knocks PDLIM1 down by 69%and shRNA3 knocks down PDLIM1 by 74%. (D) Demonstration of the specificity of the PDLIM1 antibody recognition in the PLA events. Analysis of the two knockdown HMEC lines revealed a significant decrease in the number of PLA events observed between EGFR and PLDIM1. Two biological replicates were combined in this quantitation.
To further ensure that the PLA events measured in HMECs are a direct result of antibody-mediated recognition of PDLIM1, stable PDLIM1 knockdown cells were created. Two of four hairpins that we designed were successful at knocking down PDLIM1; shRNA2 knocks down PDLIM1 protein levels by 69% while shRNA3 knocks them down by 74% (Fig. 5C). As shown in Fig. 5D, there was a significant decrease in the number of PLA events detected in PDLIM1 knock-down HMECs following stimulation with EGF for 6 minutes, as compared to knock-down vector control cells. Two control cell types were included, an empty vector control and a vector with a scrambled hairpin to the gene BAD, which should control for any changes resulting from non-specific shRNA processing. These data indicate that the co-immunoprecipitation and PLA results are specific for the correct recognition of PDLIM1 by the antibody used in the experiment, and provide further validation that EGFR interacts specifically with PDLIM1 in a dynamic, EGF-dependent manner.
Discussion
The prediction, discovery and documentation of protein interactions remain extremely active areas of research. However, most currently available experimental and computational methods have limited ability to detect transient phosphodriven interactions. Yeast two-hybrid (Y2H) methods, for example, are performed within an organism that has very few, if any, dedicated Tyr kinases, making the detection of pTyr-dependent PPIs virtually impossible. However, a recent study from Vinayagam et al. did show that directed-PPI networks generated from Y2H data can be computationally annotated for information flow and verified by combining the data with time-resolved global phosphoproteomic data.18 Yeast 3-hybrid approaches have been proposed to address this problem of phosho-dependent PPIs,19 but few successful such screens have been reported to date. An alternative approach in mammalian cells is the use of tandem affinity purification (TAP) experiments.20,21 However, the degree of purification and washing steps that used in typical TAP-tagging methodologies is not ideal for discovering transient protein–protein interactions,22 although protein crosslinking prior to cell lysis and TAP purification methods may improve the ability to detect transient interactions.23
In the computational realm, a growing number of databases attempt to capture current knowledge of protein–protein interactions, including BIND,24 BioGRID,25 MINT,26 and DIP.27 Since many of these databases capture incomplete but overlapping subsets of the information, other resources, such as iRefWeb,12 have emerged to consolidate these resources into a single database view for analyzing the veracity of a particular protein–protein interaction. In addition, there are handcurated pathway databases that capture protein–protein interactions, such as GeneGo (http://www.genego.com), and tools such as STRING that attempt to predict protein interactions by combining both experimental information and text-based searches in the literature to support potential for protein interactions.28 However, much of this computational/informatics information has been gathered in the absence of a particular context of interest, such as the transient phospho-dependent protein–protein interactions induced by RTK activation. Additionally, nearly all protein–protein interaction databases and resources lack specific information regarding the mechanism (i.e. the particular phosphorylation site) involved in driving phosphorylation-specific interactions.
Two key pieces of evidence support the notion that robust co-regulation of tyrosine phosphorylation provides evidence for protein–protein interactions; first that it overlaps with current protein–protein interaction knowledge and second that it recapitulates known interactions at the site-specific level. However, the prediction of current protein interaction knowledge, although clearly better than by random chance, could be improved. One major contributor to the limited ability of our method to capture protein–protein interactions is the fact that many protein–protein interactions currently captured in GeneGo are not driven by phosphorylation, or not by the phosphorylation events captured in the dataset that we analyzed. In addition, we believe that other protein– protein interactions that are predicted by robust phospho-site co-clustering prediction have not yet been mapped due to limited state of current experimental knowledge. This is supported by the fact that there is an improvement in the ability to predict protein–protein interactions based on co-regulated phosphorylation dynamics as the protein interaction networks have evolved in time, i.e. predictions have better overlap with knowledge in October of 2011 than they do with knowledge in February of 2010. Additionally, this method is based on phosphorylation dynamics, which may then be capable of predicting transient events under network stimulation, whereas the majority of current knowledge comes from static interactions or systems with different dynamic trajectories. Despite the lack of perfect overlap with currently known protein–protein interactions, the method presented here is able to accurately predict protein–protein interactions that were then confirmed experimentally, and produce a large number of new hypotheses regarding other, currently unknown interactions.
In addition to being able to predict protein–protein interactions, which may be dynamic in nature, this method, based on particular phosphorylation residue dynamics, is also able to recapitulate specific residue phosphorylation interactions. This indicates that robust co-regulation of tyrosine phosphorylation can be used to tease apart the role of specific phosphorylation sites on a protein. For example, the role of Y1069 as a CBL recruitment site is separated from that of Y1172 and Y1197, which recruit SHC to the receptor. Given these findings, we would therefore predict that EGFR Y998 phosphorylation drives recruitment of lipid signaling molecules, specifically PLCG1 and SHIP2 (INPPL1) to the receptor (see Fig. 3A). Further, we see that individual sites on these proteins are co-regulated with specific, individual sites on the receptor. EGFR Y1069 is robustly co-regulated with PLCG1 Y771 phosphorylation, which is not the PLCG1 phosphorylation site that is robustly co-regulated with EGFR Y998. Although these latter hypotheses have not been tested experimentally, the EGFR site-specific groupings suggest that this method holds promise for distinguishing the function and interactions amongst multiple phosphorylation sites on the same protein.
Perhaps the most telling evidence that suggests novel protein–protein interactions can be predicted using robust co-regulation of tyrosine phosphorylation dynamics was the ability to predict a novel interaction between an extremely well studied protein, EGFR, and one that had not been previously studied in the context of EGF-signaling, PDLIM1. Both co-IP and PLA experiments indicate a macromolecular complex exists between these two proteins. PLA experiments further demonstrate the transient nature of this complex. Additionally, the co-IP experiments demonstrate that both PDLIM1 and EGFR interact with phospho-SHC Y427, which is also predicted based on robust co-regulation of phosphorylation of SHC at Y427 with that of EGFR Y1172 and Y1197 and PDLIM1 Y321. Based on linear amino acid sequence alone, Y321 of PDLIM1 matches the known consensus sequence for EGFR kinase recognition and possible recognition by SHC’s SH2 domain.29 It might also furnish a recruitment site for GRB2, based on linear amino acid sequence alone,29 and may explain the reason that the most robustly co-regulated site with PDLIM1 is GAB1 Y659, which could be recruited secondarily to PDLIM1 via GRB2. Given PDLIM1’s structure and its interaction with SHC, robust co-regulation with GAB1, and its interaction with alpha-actinin proteins, it is tempting to speculate that PDLIM1’s role in the EGFR network may be to help integrate the cytoskeletal elements of signaling with proliferation, although we have not been able to confirm this experimentally due to technical issues surrounding the inability to overexpress PDLIM1 constructs and their variants within the human mammary epithelial cell line.
The method proposed in this work may help to expand our current knowledge of RTK signaling networks, or other networks regulated by protein phosphorylation such as the DNA damage response, in a manner that differs from other currently existing experimental technologies or computational techniques. Time- and stimulus-dependent phosphoproteomic experiments are increasingly being performed in a manner that captures signaling dynamics. The method presented here harnesses that type of information to predict dynamically-modulated macromolecular complex formation in a highly context-specific manner, and implicitly integrates the activation (or inhibition) of the specific network of interest, the cells or tissue-specific proteins that were measured, and the dynamical properties of the network itself.
Methods
Dataset preparation and MCAM implementation
The PTMScout11 interface was used to download the quantitative mass spectrometry measurements of human mammary epithelial cells stimulated with saturating concentrations of EGF, at 100 ng mL−1, for 0, 1, 2, 4, 8, 16, or 32 minutes.10 There were 18 phosphopeptides represented by two forms of a peptide cleavage, a perfectly trypsinized peptide and a mistrypsinized peptide. To avoid skewing the enrichment results by the presence of multiple forms of the same phosphorylation sites, the miscleaved forms of redundant peptides were removed from the dataset, leaving 204 phosphopeptides measured across seven time points. Default protein assignments were then made for ambiguous peptide assignments, through the PTMScout interface. The final form of this dataset “Default Assignments, Miscleaveages Removed” was then exported for clustering for MCAM analysis in Matlab. The parameters of clustering used in the initial MCAM round and those pruned from the final round are given in Table 1. Set pruning was determined by removing those sets whose removal improved the overall biological enrichment by 10% or greater, while not decreasing the impact of any one biological category by more than 3%. To produce a random control, the original data matrix was randomly reordered and subjected to the same parameters of clustering as used in MCAfinal. Ten random controls were generated.
Statistical significance of biological enrichment was calculated using the PTMScout MCAM interface for Gene Ontology terms, PFAM domains, Scansite binding and kinase predictions, linear amino acid sequence motifs, and enrichment for the domain a phosphorylation site falls in. This analysis used the default PTMScout parameters of 5 × 10−2 for an alpha corrected value, corrected by Benjamini and Hochberg FDR procedure,30 1 × 10−5 for significant PFAM domain consideration, 1 × 10−2 cutoff for motif search analysis, and a Scansite cutoff of 3. At the time statistical significance of enrichment was calculated, GO terms on PTMScout were at version 1.2 and downloaded October 11, 2011 from GO (http://www.geneontology.org). Matlab code for enrichment analysis and pruning was downloaded from http://ptmscout.mit.edu/MCAM_July_2011.zip. Further details regarding the process of MCAM including enrichment analysis and pruning can be found in ref. 9.
Protein interaction network overlap and co-occurrence graphs
Refseq identifiers for proteins in the dataset were imported into a network in GeneGo’s pathway analysis tool (http://www.genego.com). Direct interactions between these proteins provided by GeneGo were than exported and turned into a binary adjacency matrix, A, that aligned with the co-occurrence matrix, C. C(i,j) indicates the fractional number of times phosphopeptides i and j co-cluster in MCAfinal. When multiple peptides were measured from the same protein, A has multiple entries for that protein to match the entries in C. Therefore, A(i,j), indicates whether the proteins that phosphopeptides i and j arise from are known to interact (value 1) or not known to interact (value 0), according to GeneGo. The protein interaction network from GeneGo was captured for this experiment in October 2011 and in February 2010. C′ is created by setting all values in C to binary value 1 if C(i,j) >=threshold and value 0 otherwise. For every threshold value between 0 and 1, the overlap between C′ and A is tested for significance according to the hypergeometric distribution (see eqn (1)). Since A, C, and therefore C′, are all symmetric matrices, i.e. A is a non-directional adjacency matrix, and the diagonal of C′ is meaningless since it will always be 1 (all phosphopeptides co-cluster with themselves the maximum number of times), the number of interactions in A and C′ are only considered for the upper triangular portion of the matrix.
Eqn (1): Hypergeometric test for the significance of overlap between A and C′
| (1) |
N is the total number of possible phosphopeptide interactions (20 706 in this case), where m represents the number of phosphopeptides, n is the number of “significant” interactions in C′, i.e. those values above the threshold, k represents the total intersection of C′ = 1 and A = 1, and K is the total number of interactions in A.
To build the ROC curve we took all C′ matrixes and calculated the corresponding true positive and false positive rates. True positives are defined as when a value in C′ is 1 and the same value in A is 1, i.e. a predicted interaction according to C′ is a known interaction according to A. A false positive is when C′ had a value of 1 and A had a value of 0, indicating a prediction is not a known interaction.
Non-directional, weighted graphs were created from C′ using GraphViz, (http://www.graphviz.org/). A dot file output was created from Matlab from the adjacency matrix C′, where the threshold for C was set such that the phosphopeptide of interest had six directly connected nodes. The edge weights for C were left in C′ for those nodes and all other values were set to zero. The number of nodes for a map was set based on the readability of a graph.
Cell culture, media, and serum starvation
Human mammary epithelial cells, the 184A1 cell line used in the original phosphoproteomic study,10 were purchased from ATCC. Cells were cultured in DFCI-1 media and passaged weekly and maintained at 37 °C in 5% CO2. Serum starvation involved washing twice with PBS and leaving overnight in DFCI-1 lacking EGF, bovine pituitary extract, and serum components. All experiments were performed on cells passaged less than 15 times.
Co-immunoprecipitation
Antibodies for EGFR, PDLIM1, and an IgG control were conjugated to agarose for co-immunoprecipitation (co-IP). Pre-conjugated EGFR and IgG beads were purchased from Santa Cruz, SC-03AC and SC-2345, respectively. PDLIM1 antibody (Abcam ab64971, Rabbit pAb) was conjugated one to two days before the co-IP experiment using a Pierce Co-Immunoprecipitation kit at a concentration of 100 µg of antibody per 100 µL of agarose beads, to match the same conjugation density as the EGFR and IgG beads. For each of the biological replicate co-IP assays, nine 15 cm plates were seeded with 5 × 106 cells on day 1. On day 2 they were serum starved and on the morning of day 3 they were stimulated with 100 ng ml−1 of EGF for the indicated times. Following stimulation, plates were placed on ice, rapidly washed twice with ice cold PBS and lysed in 750 µL of lysis buffer (50 mM Hepes, 10% glycerol, 1% NP-40 and 150 mM NaCl) supplemented with protease and phosphatase inhibitors (Roche Complete Mini Protease Inhibitor Cocktail, 0.1 M phenylmethanesulfonyl fluoride, and Boston BioProducts Phosphatase Inhibitor Cocktail-I) for 30 minutes at 4 °C. Lysates were microfuged for 15 minutes and the supernatant from the three plates for each condition combined. Samples were pre-cleared at 4 °C for 30 to 45 minutes with 20 µL of protein A beads that had been previously blocked with BSA. 900 µL of pre-cleared lysate was combined with 30 µL of EGFR, PDLIM1, or IgG control conjugated agarose beads and incubated at 4 °C for 4 hours. Following incubation, the recovered beads were washed 3 times with 300 µL of lysis buffer and eluted at room temperature for 30 minutes, in 50 µL of 6Murea with agitation.
Eluted protein samples were analyzed by SDS polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and blotted, for the presence of phospho-SHC Y427 (Cell Signaling Technologies, CST, 2431 Rabbit pAb), EGFR (CST 4267, Rabbit mAb), PDLIM1 (Abcam ab17022, Goat pAb), and alpha-actinin4, ACTN4 (Epitomics 2885-1, Rabbit pAb). Western blots were imaged using a LICOR Odyssey Infrared Imaging System (Licor Biosciences, Lincoln, NE, USA).
Proximity ligation assay
Collagen coated MatTek dishes (10 mm, 1.5 glass) were seeded with 5 × 104 cells in 2 mL of media. 6–8 hours later the media was removed and replaced with serum-free media overnight. In the AM cells were stimulated with 100 ng mL−1 EGF for 0, 1, 6, and 15 min. Cells were rapidly washed twice with ice cold PBS, then fixed at room temperature for 20 minutes in 4% paraformaldehyde in PHEM fixation buffer (pH 7.0), permeabilized for 10 minutes in 0.5% BSA, 0.2% TX-100 in PBS and then blocked for 30 minutes in 10% BSA in PBS. After washing once in 0.5% BSA, cells were incubated for 1 hour at room temperature with Mouse anti-EGFR (Invitrogen clone 31G7) at 1 : 200 and Goat anti-PDLIM1 (Abcam) at 1 : 200 using 100 µL volumes. Duolink Anti-Goat MINUS and anti-Mouse PLUS secondary antibodies and Far-Red Detection Reagents were purchased from Olink Bioscience (Uppsala Sweden). Secondary incubation, ligation, and amplification steps for PLA were performed as suggested by Olink using 40 µL volumes. Following amplification a first wash was performed for 10 min in Olink Buffer B, pH 7.5, followed by a 10 min wash in 0.5% BSA. Slides were then incubated in Alexa Fluor 568 phalloidin (Invitrogen 1 : 200) for 45 minutes at room temperature, washed 3 × 5 minutes in 0.5% BSA and mounted using Duolink Mounting Media with DAPI. Slides were stored at −20 °C and imaged using a DeltaVision deconvolution microscope (Applied Precision) within one week. Five images per condition were captured for analysis. On average, each image contained 1.5 cells.
Images were analyzed using ImageJ software. To calculate cell area, cells were outlined manually based on phalloidin staining. PLA events were automatically quantified by first applying a Yen threshold to create a binary image and then automatic particle counting in ImageJ was done, assuming particle sizes from 2–50 pixels in diameter. When PLA events merged to create particles larger than 50 pixels, the area was measured and number of events was assumed to be particle area divided by 10, since 10 pixels was roughly the median size of most PLA events. Five frames per condition in each replicate were collected. The number of PLA events per cell area for each condition was normalized to the 6 minute condition for that experiment. In the PDLIM1 knockdown experiment, the experiments were normalized to the wild type condition before being combined (duplicate experiment). Statistical significance was assessed using a two-tailed Student’s t-test. All quantitation for PLA experiments is available in ESI.†
PDLIM1 knockdown cell lines
Four hairpins designed against the 3′ UTR sequence of human PDLIM1 were cloned into LTR-driven MiR30 Puro-IRES-GFP (pMLP) vectors.31 The hairpin targeting sequences were: shRNA1: 5′-ACGTTCTTCTCTGCTGCATTTG-3′, 5′-CTCAGCCAAATTTGAACCCAAT-3′, and shRNA4: 5′-CTCTCTCAGTGTTCTGGCCCTC-3′. Control cell lines included a vector control (pMLP with no hairpin insert) and a pMLP vector with target shRNA of a scrambled sequence designed against BAD (shBAD). The scrambled hairpin provides a control for any non-specific effects of shRNA processing. After retroviral infection, puromycin selection was used to purify shRNA-containing HMECs. PDLIM1 knockdown efficiency was then validated by western blot using both the Rabbit pAb and Goat pAb from Abcam, ab64971 and ab17022, which were raised against different PDLIM1 protein sequences. Membranes were imaged using the LICOR Odyssey Infrared Imaging System and densitometry was used to measure signal. PDLIM1 amounts were normalized to alpha-tubulin loading control detected by Mouse mAb clone DM1A. Quantitation of all replicates are available in ESI.†
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants U54-CA112967, R01-CA96504, P50-GM68762 and R01 ES-015339. The authors would like to thank members of the Lauffenburger and Yaffe labs for useful discussions, manuscript editing and assistance in assay development, particularly Shannon Hughes, Justin Pritchard, Michael Lee, Alexandra Gardino, Melody Morris, Brian Joughin, and Douglas Jones. We would also like to thank the Koch Institute microscopy core facility and Eliza Vasile for their services.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mb25200g
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278(5346):2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 2002;3(3):177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21(3):255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 5.Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics. 2008;8(21):4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaffe MB, White FM. Signaling networks get the global treatment. Genome Biol. 2007;8(1):202. doi: 10.1186/gb-2007-8-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Naegle KM, et al. MCAM: multiple clustering analysis methodology for deriving hypotheses and insights from high-throughput proteomic datasets. PLoS Comput. Biol. 2011;7(7) doi: 10.1371/journal.pcbi.1002119. e1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf-Yadlin A, et al. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U. S. A. 2007;104(14):5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naegle KM, et al. PTMScout, a Web resource for analysis of high throughput post-translational proteomics studies. Mol. Cell. Proteomics. 2010;9(11):2558–2570. doi: 10.1074/mcp.M110.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner B, et al. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database. 2010;2010 doi: 10.1093/database/baq023. baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oksvold MP, et al. Serine mutations that abrogate ligand-induced ubiquitination and internalization of the EGF receptor do not affect c-Cbl association with the receptor. Oncogene. 2003;22(52):8509–8518. doi: 10.1038/sj.onc.1207117. [DOI] [PubMed] [Google Scholar]
- 14.Batzer AG, et al. The phosphotyrosine interaction domain of Shc binds an LXNPXY motif on the epidermal growth factor receptor. Mol. Cell. Biol. 1995;15(8):4403–4409. doi: 10.1128/mcb.15.8.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallenius T, Luukko K, Makela TP. CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J. Biol. Chem. 2000;275(15):11100–11105. doi: 10.1074/jbc.275.15.11100. [DOI] [PubMed] [Google Scholar]
- 16.Vallenius T, Makela TP. Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J. Cell Sci. 2002;115(Pt 10):2067–2073. doi: 10.1242/jcs.115.10.2067. [DOI] [PubMed] [Google Scholar]
- 17.Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 18.Vinayagam A, et al. A directed protein interaction network for investigating intracellular signal transduction. Sci. Signal. 2011;4(189) doi: 10.1126/scisignal.2001699. rs8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Lautar S. A yeast three-hybrid method to clone ternary protein complex components. Anal. Biochem. 1996;242(1):68–72. doi: 10.1006/abio.1996.0429. [DOI] [PubMed] [Google Scholar]
- 20.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 21.Williamson MP, Sutcliffe MJ. Protein–protein interactions. Biochem. Soc. Trans. 2010;38(4):875–878. doi: 10.1042/BST0380875. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, et al. The tandem affinity purification method: an efficient system for protein complex purification and protein interaction identification. Protein Expression Purif. 2010;72(2):149–156. doi: 10.1016/j.pep.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Guerrero C, et al. An integratedmass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics. 2006;5(2):366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Bader GD, et al. BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29(1):242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35(Database issue):D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xenarios I, et al. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28(1):289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snel B, et al. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obenauer JC, Cantley LC, Yaffe MB, et al. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 31.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 2005;37(11):1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.