Abstract
Purpose
To investigate the association of two reported regions on chromosome 15 with moderate to high myopia in two Chinese cohorts from southern China.
Methods
Two candidate regions on 15q14 and 15q25 were selected based on reported association with refractive error in the literature. Five single nucleotide polymorphisms (SNPs) were genotyped in 300 university students with high myopia at Guangzhou and 308 without refractive error, and 96 university students of Chaoshan Chinese origin with moderate to high myopia and 96 without refractive error. Genotypes were evaluated using direct sequencing and analyzed with chi-square, Armitage trend, and Mantel-Haenszel tests, and regression analysis.
Results
Of the five SNPs screened, alleles of rs634990 and rs524952 in the 15q14 region showed evidence of allelic association with moderate to high myopia (p<8.81×10−7 and p<1.57×10−6, respectively) in the Guangzhou group, but not in the Chaoshan group. The SNPs at 15q25 did not show significant association in any group. Association of rs634990 and rs524952 were still significant when both groups were combined into a single analysis (p<1.66×10−6 and p<2.72×10−6, respectively), and for genotypic, additive, and dominant models.
Conclusions
This study confirms the significant association of rs634990 and rs524952 on chromosome 15q14 previously reported in European and Japanese populations with high myopia in the Guangzhou but not the Chaoshan Chinese populations, suggesting that genetic contributors to high myopia in the Chaoshan population might be different from other Chinese populations.
Introduction
Myopia is the leading cause of visual impairment worldwide, contributing to an overall prevalence of 30% across the globe, although the prevalence reached as high as 50%–70% in some urban East Asian populations [1-7]. High myopia, characterized as a refractive error greater giving a spherical equivalent less than –6D, can be associated with myopic retinopathy, retinal detachment, glaucoma, and cataracts [8]. Currently, although there appear to be some common risk factors, the precise relationship between high and low or medium myopia remains unclear. Although the molecular mechanisms of myopia have yet to be delineated, environmental and genetic factors contribute to its pathogenesis [9-13]. This is well documented in several epidemiological studies. In recent years, the prevalence of high myopia has appeared to be increasing, especially in East Asia [4,5,7], as education levels rise with concomitant increases in time spent in near work such as reading and writing, and decreases in time spent outdoors [14].
Although epidemiological and twin studies provide the most compelling evidence for an environmental contribution to myopia [7,12], several linkage studies have identified Mendelian loci contributing to myopia susceptibility, termed MYP1–3 and MYP 5–19 [15-29]. Most often, myopia is multifactorial with a complex inheritance pattern. Genetic loci contributing to myopia occurring in the general population have been identified in several genome-wide association studies [30-35], although for most of these loci identification of the causative sequence changes and their related genes has not yet been possible.
Two studies have shown an association of markers on chromosome 15 with refractive error. One was performed in a Dutch population from the Rotterdam study and replicated in four independent sample groups of European ancestry [32]. A second study was performed in the TwinsUK cohort and replicated in six independent sample groups of European ancestry [31]. These two loci have been examined for association with high myopia in a Japanese population [36]. In the latter study, the support for the 15q14 locus was unambiguous, while that for markers in the 15q25 region was less robust. In addition, this suggested that these loci confer susceptibility not only to general refractive error but also to high myopia, at least in the Japanese population. Recently, data from these three studies and from the Singapore Malay Eye Study (SIMES), Singapore Indian Eye Study (SINDI), Singapore Prospective Study Program (SP2), and Singapore Cohort study Of the Risk factors for Myopia (SCORM) studies of Singaporean populations were combined in a meta-analysis confirming association with the chromosome 15q14 locus [37]. However, the results for Asian population groups were mixed in this study, which included Japanese as well as individuals of Chinese and Malaysian descent from Singapore. The present study aims to evaluate single nucleotide polymorphisms (SNPs) in these two candidate regions on chromosome 15q14 and 15q25, previously reported to be associated with refractive error and high myopia in European and Japanese populations, in two Chinese university student populations, one of Chaoshan origin in Guangzhou and the second of Han Chinese but not Chaoshan origin in Guangzhou.
Methods
This study protocol was approved by the Institutional Review Board and Ethics Committee of the Zhongshan Ophthalmic Center, Sun Yat-Sen University, Guangzhou, China, and the CNS Institutional Review Board, NIH, Bethesda, Maryland. Patients were recruited from the Zhongshan Ophthalmic Center and its clinics, and the laboratory part of the study was performed at the National Eye Institution (NEI) after written informed consent was obtained from all the participants in accordance with the tenets of Declaration of Helsinki.
Enrollment of cases and controls
Individuals with moderate to high or high myopia or without myopia were enrolled for this study. Criteria for diagnosis of moderate to high myopia included a spherical equivalent ≤ −4.0 D and exclusion of other known ocular or systemic diseases. Criteria for diagnosis of high myopia included a spherical equivalent ≤ −6.0 D and exclusion of other known ocular or systemic diseases. Individuals must have received at least 12 years of education and have a best aided visual acuity of 0.8 or better without other known eye or systemic diseases. Control individuals all had bilateral refraction between −0.50 D and +1.0 D spherical equivalent without a family history of high myopia. They also must have received at least 12 years of education and have a best unaided visual acuity of 1.0 or better without other known eye or systemic diseases. Individuals with or without myopia were recruited through the Zhongshan Ophthalmic Center from two population groups. The first group consisted of 300 university students in Guangzhou with high myopia and 308 control individuals, university students in Guangzhou of Han Chinese ethnicity (individuals from Chaoshan were excluded). The second group of 96 patients with moderate to high myopia and 96 control individuals was also recruited from university students in Guangzhou but of Chaoshan origin, a region in the eastern part of Guangdong province with an independent dialect and culturally distinct population.
All individuals underwent a clinical evaluation comprising a fundus examination with a direct ophthalmoscope and a slit lamp biomicroscopic examination. In addition, all patients underwent visual acuity (unaided, near, and best acuity) and color vision examinations. Refractive errors were measured with a Topcon KR-80000 (Paramus, NJ) auto refractometer after cycloplegia induced with Mydrin-P (Santen Pharmaceutical C. Ltd., Osaka, Japan). Ocular biometric axial length was measured using an IOL Master V5 (Carl Zeiss Meditec AG, Jena, Germany). Selected individuals also underwent electroretinography and fundus photography. Comparison of the refraction between the left and right eyes showed no statistical difference, so the phenotypic analysis was based on refraction of the right eyes.
Single nucleotide polymorphism selection
A total of five SNPs in two candidate regions were selected for the present study. The selected SNPs are in chromosome 15q14 (rs634990 and rs524952) and chromosome 15q25 (rs8027411, rs17175798, and rs939658). Primers were designed for each SNP using Primer3 v. 0.4.0. Primer sequences were listed in Table 1.
Table 1. The primer pairs used for PCR.
| SNP | Direction | Primer (5′-3′) |
|---|---|---|
|
rs634990 |
F |
CCCTCTGCTCCATCTGCTA |
| |
R |
TGATGGGCCATTATCTGTGA |
|
rs524952 |
F |
TGCTCATGACATTTGTGAACC |
| |
R |
ACCAGGAAAAGGGCTTCAAT |
|
rs8027411 |
F |
CTCTTCATGGGGGAAGCAG |
| |
R |
CAGCCTAGCAGACAGAGCAA |
|
rs17175798 |
F |
TAGCTCCCTTGGGGAAAGAT |
| |
R |
CAACCAGAGAACAGGCTTTCA |
|
rs939658 |
F |
ACAGAAATTGATCGCCCACA |
| R | AAAACTTAGTGGTCAATGTGATGG |
Genotyping
Total genomic DNA was extracted from peripheral blood leukocytes using a standard phenol/chloroform method as described previously [38]. The SNPs were amplified with PCR using a 9700 Thermo Cycler (Applied Biosystems, Inc. [ABI], Foster City, CA). PCR reactions were performed in 10 µl reaction volumes containing 40 ng genomic DNA, 10 µM primer pairs, 1 µl 10X PCR Buffer II (GeneAmp; ABI), 0.6 µl 10 mM dNTP mix (GeneAmp; ABI), 2.5 mM MgCl2, and 1 µl Taq DNA polymerase. Initial denaturation was performed for 5 min at 95 °C, followed by 35 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 40 s, with a final elongation of 10 min at 72 °C followed by a hold at 4 °C. Genotyping was performed with directional sequencing using Big Dye Terminator Ready reaction mix according to the manufacturer’s instructions (Applied Biosystems). Genotyping was performed by sequencing to minimize cost and repetition. Sequencing was performed on an ABI PRISM 3130×l Genetic Analyzer (Applied Biosystems). Sequence traces were analyzed using Mutation Surveyor (Soft Genetics Inc., State College, PA) and the SeqMan program of DNASTAR Software (DNASTAR Inc., Madison, WI).
Statistical analysis
Chi-square and Fisher’s exact tests were used to test the allelic, genotypic, and model-based associations of all the SNPs. The Hardy–Weinberg equilibrium of each SNP in the control and affected individuals was also examined using a Χ2 test, all as implemented in the Golden Helix SVS software suite 7.5.2 (Golden Helix, Bozeman, MT). Odds ratio (OR), relative risk, and call rate were also calculated using the same program. Because this is a directed search of candidate genes with a presumed high a priori probability of being associated, a p value <0.05 was considered statistically significant. Corrections for multiple testing were done with the Bonferroni method, where indicated, giving a p<0.00066 for 76 tests (three genotypic, one allelic, and one additive model for each of the five markers in the Guangzhou and Chaoshanese groups, six additional tests for the two markers showing association, and haplotype analysis in each group). Haplotypes were estimated using the Golden Helix SVS software suite version 7.5.2 and the Haploview program using the expectation-maximization (EM) algorithm. Mantel-Haenszel tests were performed using a continuity correction. Power calculations were done for chi-square statistics for allelic associations using the power for association with error (PAWE) program for data assuming no errors and an invariant control allele frequency and varying case allele frequency [39,40].
Results
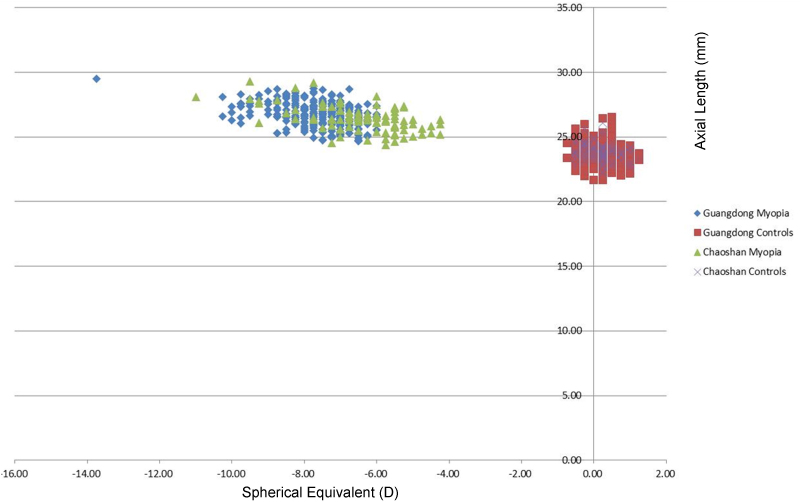
The clinical characteristics of the study subjects are shown in Table 2 and Figure 1. Although there was a slight preponderance of men (about 65%) in the myopia and control groups from Chaoshan, only about 49% of the Han Chinese patients with myopia were male as opposed to 64% of the control group. The ages of the patients and controls in both groups were similar, averaging about 21.6–22.2 years old (Table 2). Although the diagnosis of high myopia in this study was based on refraction, the axial lengths of patients with myopia in the Chaoshan and Guangzhou groups were similar, about 26 mm, while those of the controls in both groups averaged between 23 and 24 mm. The refraction in diopters was also similar between patients in both groups, although the mean was slightly higher (−6.52 OD, −6.37 OS) in the Chaoshan group than in the Guangzhou group (−7.28 OD, −7.20 OS); the values for the controls from both groups were close to 0. All markers were in Hardy–Weinberg equilibrium (p>0.05) in the Chaoshan, Guangdong, and combined groups. Comparison of the affected and control individuals between the Chaoshan and Guangdong population groups did not show a statistically significant difference for either rs634990 or rs524952.
Table 2. Clinical Characteristics of affected and unaffected individuals from the Chaoshan and Guangzhou population groups.
| Parameters | Chaoshan |
Guangzhou |
Combined |
|||
|---|---|---|---|---|---|---|
| Myopia (n=96) | Controls (n=96) | Myopia (n=300) | Controls n=308) | Myopia (n=396) | Controls (n=404) | |
| Males (%) |
65.63 |
65.63 |
49.17 |
63.64 |
53.15 |
64.11 |
| Age (mean±SD) | 21.80±1.27 | 21.68±1.30 | 22.19±1.67 | 21.66±1.54 | 22.10±4.77 | 21.67±1.48 |
| Axial length (mm) | ||||||
|---|---|---|---|---|---|---|
| O.D. |
26.28±0.96 |
23.78±0.72 |
26.60±0.89 |
23.66±0.80 |
26.53±0.91 |
23.69±0.78 |
| O.S. | 25.22±1.02 | 23.72±0.68 | 26.60±.092 | 23.62±0.79 | 26.52±0.95 | 23.64±0.77 |
| Refraction (diopters) | ||||||
|---|---|---|---|---|---|---|
| OD |
−6.54±1.31 |
0.27±0.51 |
−7.82±0.87 |
0.19±0.48 |
−7.51±1.14 |
0.21±0.49 |
| OS | −6.28±1.84 | 0.33±0.44 | −7.79±1.12 | 0.31±0.44 | −7.42±1.48 | 0.31±0.44 |
Figure 1.
Distribution of refraction and axial length among high myopic and control individuals in the Guangzhou and Chaoshan myopia and unaffected groups. The spherical equivalent of each individual is shown on the abscissa while the axial length is shown on the ordinate axis. Although the control samples are similarly clustered around 0 D: and 23 mm, the Chaoshan myopic group includes individuals with spherical equivalents < −4.
Among the five SNPs studied, the three on chromosome 15q25 (rs8027411, rs17175798, and rs93958) showed no association with moderate or high myopia in either the Guangzhou or Chaoshan group (Table 3). In contrast, the two SNPs on chromosome 15q14, rs634990 and rs5249952, showed association with high myopia in the Guangzhou group but not with high to moderate myopia in the Chaoshan group. The highest levels of association were seen with the G (minor) allele of rs634990 (p<1.57×10−6, OR=1.75, 95% confidence interval [CI]=1.39–2.20) and the T (minor) allele of rs5249952 (p<8.8×10−7, OR=1.78, 95% CI=1.41–2.23). Genotypic association followed a similar pattern with the odds ratios increasing from the homozygous major allele to about 2 for heterozygotes for either marker, to slightly over 3 for homozygous minor alleles of either marker (Table 3). In addition, tests of specific models were also highly significant; the genotypic test for rs634990 gave p<6.59×10−6, the additive model gave p<1.45×10−6, the dominant model showed increased risk with a p<1.91×10−5 (OR=2.12, 95% CI=1.50–3.01), and a comparison of the homozygous GG and AA genotypes gave p<3.09×10−6 (OR=3.12, 95% CI=1.91–5.07). For rs524952, the genotypic test gives p<8.53×10−6, the additive model gave p<7.01×10−7, the dominant model gave p<1.91×10−5 (OR=2.28, 95% CI=1.61–3.25), and a comparison of the TT and AA homozygous genotypes gave p<2.35×10−6 (OR=3.17, 95% CI=1.94–5.16).
Table 3. Association of high myopia in the Chaoshan and Guangzhou population groups.
| Guangzhou | ||||||
|---|---|---|---|---|---|---|
| Region | SNP | Test | Myopia (300 total) | Control (308 total) | Odds ratio* | P** |
| 15q14 |
rs634990 |
AA |
73 |
125 |
1 |
1.91×10−5 |
| |
|
GA |
156 |
144 |
1.86 (1.29–2.68) |
0.2 |
| |
|
GG |
71 |
39 |
3.12 (1.92–5.07) |
4.25×10−4 |
| |
|
G allele % |
0.5 |
0.36 |
1.75 (1.39–2.20) |
1.57×1−6 |
| |
|
Additive |
|
|
|
1.45×10−6 |
| |
rs634990 |
AA |
69 |
125 |
1 |
3.31×10−6 |
| |
|
TA |
161 |
143 |
2.04 (1.41–2.95) |
0.07 |
| |
|
TT |
70 |
40 |
3.17 (1.95–5.16) |
9.22×10−4 |
| |
|
T allele % |
0.5 |
0.36 |
1.78 (1.41–2.23) |
8.8×10−7 |
| |
|
Additive |
|
|
|
7.01×10−7 |
| 15q25 |
rs8027411 |
TT |
113 |
108 |
1 |
0.5 |
| |
|
GT |
146 |
151 |
0.92 (0.65–1.31) |
0.92 |
| |
|
GG |
41 |
49 |
0.80 (0.49–1.31) |
0.44 |
| |
|
G allele % |
0.38 |
0.4 |
0.90 (0.72–1.14) |
0.39 |
| |
|
Additive |
|
|
|
0.38 |
| |
rs17175798 |
CC |
112 |
107 |
1 |
0.51 |
| |
|
TC |
148 |
152 |
0.93 (0.66–1.32) |
0.99 |
| |
|
TT |
40 |
49 |
0.78 (9.48–1.28) |
0.37 |
| |
|
T allele % |
0.38 |
0.4 |
0.90 (0.71–1.30) |
0.36 |
| |
|
Additive |
|
|
|
0.35 |
| |
rs939658 |
CC |
121 |
116 |
1 |
0.5 |
| |
|
TC |
139 |
147 |
0.93 (0.66–1.32) |
0.73 |
| |
|
TT |
40 |
45 |
0.91 (0.69–1.30) |
0.65 |
| |
|
T allele % |
0.37 |
0.39 |
0.92 (0.73–1.16) |
0.36 |
| Additive | 0.48 | |||||
| Chaoshan | ||||||
|---|---|---|---|---|---|---|
| Region | SNP | Genotype | Myopia (96 total) | Control (96 total) | Odds Ratio | P** |
| 15q14 |
rs634990 |
AA |
28 |
34 |
1 |
0.35 |
| |
|
GA |
48 |
46 |
1.26 (0.67–2.41) |
0.77 |
| |
|
GG |
20 |
16 |
1.51 (0.66–3.47) |
0.46 |
| |
|
G allele % |
0.46 |
0.41 |
1.23 (0.83–1.85) |
0.3 |
| |
|
Additive |
|
|
|
0.3 |
| |
rs524952 |
AA |
28 |
34 |
1 |
0.35 |
| |
|
TA |
48 |
46 |
1.26 (0.67–2.41) |
0.77 |
| |
|
TT |
20 |
16 |
1.52 (0.66–3.47) |
0.46 |
| |
|
T allele % |
0.46 |
0.41 |
1.24 (0.83–1.35) |
0.3 |
| |
|
Additive |
|
|
|
0.3 |
| 15q25 |
rs8027411 |
TT |
37 |
26 |
1 |
0.09 |
| |
|
GT |
44 |
54 |
0.57 (0.30–1.08) |
0.15 |
| |
|
GG |
14 |
15 |
0.66 (0.27–1.59) |
0.84 |
| |
|
G allele % |
0.38 |
0.44 |
0.79 (0.52–1.19) |
0.21 |
| |
|
Additive |
|
|
|
0.2 |
| |
rs17175798 |
CC |
37 |
27 |
1 |
0.13 |
| |
|
TC |
44 |
53 |
0.61 (0.32–1.15) |
0.19 |
| |
|
TT |
14 |
15 |
0.68 (0.28–1.64) |
0.84 |
| |
|
T allele % |
0.41 |
0.38 |
0.79 (0.52–1.18) |
0.25 |
| |
Additive |
|
|
|
0.24 |
|
| |
rs939658 |
CC |
35 |
31 |
1 |
0.68 |
| |
|
TC |
48 |
50 |
0.93 (0.66–1.32) |
0.77 |
| |
|
TT |
13 |
15 |
0.91 (0.69–1.30) |
0.54 |
| |
|
T allele % |
0.37 |
0.39 |
0.92 (0.73–1.16) |
0.53 |
| Additive | 0.52 | |||||
| Combined | ||||||
|---|---|---|---|---|---|---|
| Region | SNP | Genotype | Myopia (396 total) | Control (404 total) | Odds Ratio* | P** |
| 15q14 |
rs634990 |
AA |
97 |
159 |
1 |
6.63×10−6 |
| |
|
GA |
209 |
189 |
1.81 (1.32–2.50) |
0.09 |
| |
|
GG |
90 |
56 |
2.63 (1.73–4.00) |
0.001 |
| |
|
G allele % |
0.49 |
0.37 |
1.75 (1.39–2.20) |
1.66×10−6 |
| |
|
Additive |
|
|
|
1.42×10−6 |
| |
rs524952 |
AA |
101 |
159 |
1 |
2.89×10−5 |
| |
|
TA |
204 |
190 |
1.69 (1.23–2.32) |
0.2 |
| |
|
TT |
91 |
55 |
2.60 (1.72–3.95) |
6.10×10−4 |
| |
|
T allele % |
0.49 |
0.37 |
1.78 (1.41–2.23) |
2.72×10−6 |
| |
|
Additive |
|
|
|
2.59×10−6 |
| 15q25 |
rs8027411 |
TT |
150 |
134 |
1 |
0.16 |
| |
|
GT |
190 |
205 |
0.83 (0.61–1.12) |
0.43 |
| |
|
GG |
55 |
64 |
0.77 (0.50–1.18) |
0.44 |
| |
|
G allele % |
0.38 |
0.41 |
0.90 (0.72–1.14) |
0.17 |
| |
|
Additive |
|
|
|
0.17 |
| |
rs17175798 |
CC |
149 |
134 |
1 |
0.38 |
| |
|
TC |
192 |
205 |
0.84 (0.62–1.14) |
0.52 |
| |
|
TT |
54 |
64 |
0.75 (0.49–1.17) |
0.19 |
| |
|
T allale % |
0.38 |
0.41 |
0.90 (0.71–1.13) |
0.17 |
| |
|
Additive |
|
|
|
0.16 |
| |
rs939658 |
CC |
156 |
147 |
1 |
0.55 |
| |
|
TC |
187 |
197 |
0.93 (0.66–1.32) |
0.66 |
| |
|
TT |
53 |
60 |
0.83 (0.54–1.28) |
0.38 |
| |
|
T allele % |
0.37 |
0.39 |
0.91 (0.74–1.11) |
0.37 |
| Additive | 0.35 | |||||
* Relative to major allele, or major allele homozygote **p<0.00066 is significant, corresponding to a p<0.-05 after Bonferroni correction for 76 tests.
The proportion of men in the Guangzhou myopia group was significantly lower than the other groups (Table 2). To examine the effect on association with alleles of rs634990 and rs524952, the analyses were performed separately in men and women. Men alone showed an allelic p=8.0×10−6 and 5.1×10−5 for rs634990 and rs5249952, respectively, and women showed corresponding values of 0.04 and 0.02. In addition, the ORs for men were 2.0 (95% CI: 1.48, 2.75) and 1.89 (95% CI: 1.39, 2.58) and for women, the ORs were 1.48 (95% CI: 1.0, 2.03) and 1.5 (95% CI: 1.1, 2.1) for rs634990 and rs5249952, respectively—slightly less but overlapping. When association in the Guangzhou group was tested for men and women as two separate groups using the Mantel-Haenszel test, association was confirmed with p=4.1×10−6 and 7.5×10−6 for rs634990 and rs5249952, respectively. Finally, logistic regression of the affection status against the number of copies of the minor allele correcting for the covariant effect of gender gives p=1.91×10−6 and p=4.13×10−6 for rs634990 and rs5249952, respectively, confirming that the genetic association is not merely a reflection of gender differences in the Guangzhou sample set.
When the Guangzhou and Chaoshan groups were combined and analyzed, the results remained highly significant (Table 2). Specific models also remained highly significant for rs634990; the genotypic test gave p<6.59×10−6, the additive model gave p<1.42×10−6, the dominant model showed increased risk with a p<6.63×10−6 (OR=2.00, 95% CI=1.47–2.71), and comparison of the GG and AA homozygous genotypes gave p<4.39×10−6 (OR=2.63, 95% CI=1.73–4.00). For rs524952, tests of specific models also remained highly significant; the genotypic test gave p<1.49×10−5, the additive model gave p<2.59×10−6, the dominant model gave p<2.89×10−5 (OR=1.90, 95% CI=1.40 – 2.56), and a comparison of the GG and AA homozygous genotypes gave p<5.42×10−6 (OR=2.60, 95% CI=1.72–3.95). To account for possible differences in the Guangzhou and Chaoshanese populations (see Discussion), association of rs634990 and rs524952 alleles with myopia in the combined groups was also assessed using the Mantel-Haenszel test, yielding p=3.54×10−6 and 2.17×10−6 for rs634990 and rs524952, respectively. In addition, logistic regression correcting for Guangzhou or Chaoshan origin as a covariate gave p=2.31×10−6 and 4.18×10−6 for rs634990 and rs524952, respectively. Although the odds ratios for these markers were somewhat lower in the Chaoshan group (Table 3), the two sample sets were not differentiated significantly on this basis.
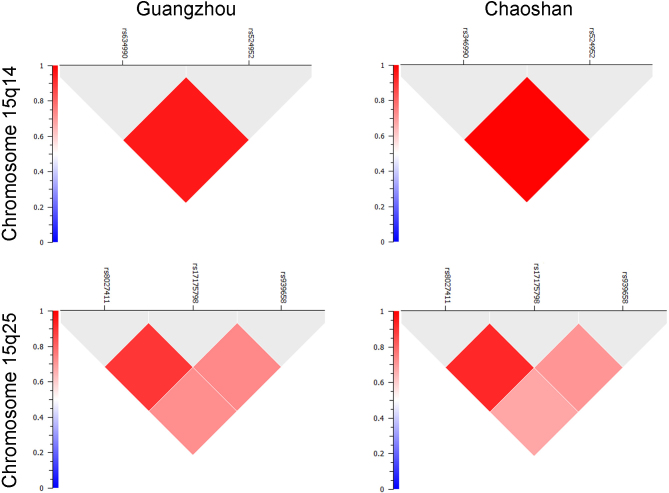
These results suggest that rs634990 and rs524952 might be in strong linkage disequilibrium, and this is the case (Table 4). In the Chaoshan population group, rs634990 and rs524952 showed an r2 value of 1 with frequencies for the AA haplotype of 0.54 in cases and 0.59 in controls, and frequencies for the GT haplotype of 0.46 in cases and 0.41 in controls. Association of neither haplotype with myopia reached significance in this group. In the Guangzhou group, the markers showed an r2 value of 0.95 with frequencies of 0.49 in cases and 0.64 in controls for the AA haplotype and 0.49 in cases and 0.36 in controls for the GT haplotype. Both haplotypes were associated with myopia with p<1.20×10−6; the AA haplotype was protective with an OR=0.57 (95% CI=0.45–0.71), and the GT haplotype increased the risk with an OR=1.76 (95% CI=1.40–2.23). When the two groups were combined, the association held, with r2=0.96 and frequencies of 0.50 and 0.63 for the AA haplotype and 0.48 and 0.37 for the GT haplotypes in cases and controls, respectively. Both haplotypes were associated with myopia with p<2.20×10−6; the AA haplotype was protective with an OR=0.62 (95% CI=0.51–0.75), and the GT haplotype increased the risk with an OR=1.62 (95% CI=1.33–1.98).
Table 4. Haplotype analysis of high myopia in the Chaoshan and Guangzhou population groups.
| Guangzhou | |||||||
|---|---|---|---|---|---|---|---|
| SNPs | r2 | D' | Haplotype | Myopia (300 total) | Control (308 total) | Odds ratio | p** |
|
rs634990 |
0.95 |
0.98 (0.96 - 1) |
AA |
0.49 |
0.64 |
0.57 (0.45–0.71) |
1.20×10−6 |
| rs524952 | GT | 0.49 | 0.36 | 1.76 (1.40–2.23) | |||
| Chaoshan | |||||||
|---|---|---|---|---|---|---|---|
| SNPs | r2 | D' | Haplotype | Myopia (96 total) | Control (96 total) | Odds ratio | p** |
|
rs634990 |
1 |
1 (0.98 - 1) |
AA |
0.54 |
0.59 |
0.81 (0.54–1.21) |
0.3 |
| rs524952 | GT | 0.46 | 0.41 | 1.24 (0.82–1.85) | |||
| Combined |
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|
| SNPs | r2 | D' | Haplotype | Myopia (396 total) | Control (404 total) | Odds Ratio | p** |
|
rs634990 |
0.96 |
0.99 (0.97 - 1) |
AA |
0.5 |
0.63 |
0.62 (0.51–0.75) |
2.20×10−6 |
| rs524952 | GT | 0.48 | 0.37 | 1.62 (1.33–1.98) |
**p<0.00066 is significant, corresponding to a p<0.-05 after Bonferroni correction for 76 tests
Discussion
We evaluated the potential association of five polymorphisms in two previously identified candidate regions on chromosome 15 in patients with moderate to high myopia in an unselected Han Chinese population and a Chaoshanese population; both groups were university students in Guangzhou, China. Chaoshan is a specific area in eastern Guangdong province, in which the people have unique cultural and origin features as we have previously described [38]. However, principal component analysis of Chaoshanese compared to other Chinese populations shows that although the Chaoshanese population appears to be more homogeneous than mixed Han Chinese samples, this population is more similar to the Guangzhou population than populations of other major cities such as Shanghai and Hong Kong, which are Han Chinese [38]. In addition, neither the affected, control, nor combined Chaoshanese and Guangzhou sample sets were statistically significantly differentiated based on any of the markers studied here (data not shown), although the Chaoshan group was small. In addition, the haplotype structures SNPs tested in the 15q14 and 15q25 regions is similar in the two groups (Figure 2). Taken together with the small size of the Chaoshan sample set, these provided a rationale for combining the two groups into a single analysis. The SNPs in the chromosome 15q14 region, rs634990 and rs524952, but none of the SNPs in the 15q25 region, rs8027411, rs17175798, and rs939658, were significantly associated with high myopia. These results are consistent across a broad variety of tests and models, with the similar results for rs634990 and rs524952 probably resulting from the strong linkage disequilibrium between them. These results are also consistent with Hayashi et al.’s [36] results showing an association of markers in the 15q14 but not the 15q25 region with high myopia.
Figure 2.
Linkage disequilibrium of markers at chromosome 15q14 and chromosome 15q25 in the Guangzhou and Chaoshan populations. Linkage disequilibrium is shown as r2. The chromosomal positions are rs634990 (35,006,073), rs524952 (35,005,886), rs8027411 (79,461,029), rs17175798 (79,463,960), and rs939658 (79,451,869) from GRCh37.p5, build 37.3. For reference, the 5′ end of RASGRF1 is at 79,383,215, the 5′ end of GOLGA8B is at 34,875,771, the 3′ end of ACTC1 is at 35,080,297, and the 3′ end of GJD2 is at 35,044,679; the loci are not included in the regions shown here.
This study was designed to confirm the results of two genome-wide association studies of refractive error and myopia in European populations showing association with markers on chromosome 15q14 and 15q25 [31,32], and a more recent confirmatory study in a Japanese population [36] as well as a meta-analysis including European and Asian population groups [37], Table 5. The chromosome 15q14 region includes three genes, the Golgi autoantigen golgin-67 (GOLGA8B), cardiac muscle cardiac 1 (ACTC1), and Connexin36 (GJD2), of which exons of GJD2 were sequenced without showing any associated sequence changes. Several common high myopia loci [26,34,35,41], as well as Mendelian loci [23,28,42,43], have been identified in Han Chinese. SNPs in genes including PAX6 [44-46], LAMA1 [47], LUM [48-50], TGFB1 [51,52], and CTNND2 [33,41], and mutations in ZNF44 [53] also have been identified in Chinese families with Mendelian high myopia. However, some regions have not been consistently identified as risk loci [54,55]. The results presented here provide strong support for a risk locus for high myopia in the Han Chinese population on chromosome 15q14 even though the original studies tested refractive errors including low or medium myopia.
Table 5. Summary of association of myopia with markers on chromosome 15q14 and 15q25 from the literature and the present study.
| Locus | best SNPs* | Gene(s) | Study | Population | best P | best OR |
|---|---|---|---|---|---|---|
| 15q14 |
rs634990
rs524952 |
GOLGA8B ACTC1 GJD2 |
Solouki et al. [57] |
European |
2.21×10−14 |
1.81 (1.42–2.36)# |
| |
rs634990
rs524952 |
Hayashi et al. [36] |
Japanese |
8.781×10−7 |
1.32 (1.11–1.56) |
|
| |
rs634990 |
|
Verhoeven et al. [37]** |
European Japanese Singapore |
9.2×10−23^ |
1.88 (1.64–2.16)# |
| |
rs634990 |
|
|
European |
3.87×10−12^ |
na |
| |
rs8032019 |
|
Asian |
9.65×10−4^ |
na |
|
| |
rs634990 |
|
present study |
Han Chinese |
1.66×10−6+ |
1.75 (1.39–2.20) |
| 15q25 |
rs8027411
rs939658 |
RASGRF1 |
Hysi et al. [31] |
European |
2.07×10−9 |
1.16 (1.02–1.28)# |
| |
rs8027411
rs939658 |
Hayashi et al. [36] |
Japanese |
0.031 |
1.17 (1.03–1.33) |
|
| |
rs939661 |
|
Verhoeven et al. [37]** |
European Japanese Singapore |
1.22×10−4& |
na |
| rs8027411 | present study | Han Chinese | 0.17+ | 0.9 (0.72–1.14) |
* not exhaustive; **meta-analysis including results of Hysi et al. and Soluki et al. with others; ^ p<3.56×10–3 considered significant after Bonferroni correction; & p<1.92×10–3 considered significant after Bonferroni correction; # OR for homozygous carriers not available; na not available; +p<0.00066 is considered significant after Bonferroni correction
Although this study easily has sufficient power to confirm the 15q14 association, the relatively small number of patients and controls analyzed cannot exclude the possibility of alleles of the three SNPs in the chromosome 15q25 region showing some association with myopia. The combined data set has about 80% power to detect an OR=1.3 but only 50% to detect an OR=1.2 with p<0.05. However, it is unlikely that the level of association in the 15q25 markers is greater than the 95% confidence limits of the odds ratios shown in Table 3 [56], which are between 0.71 and 1.41 for allelic association, and between 0.57 and 1.44 for the genotype odds ratios. The size of the Chaoshan test group does not allow exclusion of low levels of association to the 15q14 locus in that population, requiring odds ratios of 1.8 and 1.5 to provide 80% and 50% power for p<0.05, respectively. However, the Guangzhou group alone does somewhat better with odds ratios of 1.4 and 1.26 providing 80% and 50% power for p<0.05, respectively. Although the odds ratios are less extreme in the Chaoshan than in the Guangzhou group, the trends are in the same direction in both groups, and the confidence intervals overlap. This is complicated by the lower threshold of refractive error for recruiting cases in the Chaoshan population, which might decrease the genetic contribution to myopia in that group as well as shifting the contributions of different candidate genes, depending on their relationship to high and moderate myopia. Finally, associations in the chromosome 15q25 region seen in other populations might not be present in Chinese. This could be because of different population histories, which could alter the haplotype block structure and result in a lack of allelic association. In this regard, the haplotype structure at the 15q14 and 15q25 loci is identical in the Guangzhou and Chaoshan groups (Figure 2). In addition, differing genetic and environmental contributions to the disease risk in various ethnic and geographic groups could also result in a lack of association.
In conclusion, we confirmed the association of alleles of rs634990 and rs524952 on chromosome 15q14 with high myopia in a Han Chinese population of university students in Guangzhou. In contrast, the results in the Chaoshan population group and with markers rs8027411, and rs939658 on chromosome 15q25 were inconclusive. This study also confirms that the 15q14 locus, previously shown to affect refraction in mild or moderate myopia, is important for high myopia, implying at least some overlap in the genetic determinants of these traits. Sequencing of genes in the region, including ACTC1, GOLGA8B, and GJD2, might show sequence changes not seen in the European population, perhaps providing a clue to the specific gene responsible for association at this locus.
Acknowledgments
The authors like to thank all the participants for their kind cooperation in this study. Grant information: This study was supported by National Science Fund for Distinguished Young Scholars (30,725,044 to Q.Z.) and the Fundamental Research Funds of State Key Lab of Ophthalmology, Sun Yat-sen University.
References
- 1.Vitale S, Ellwein L, Cotch MF, Ferris FL, 3rd, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–9. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, Taylor HR, Ikram MK, Congdon NG, O'Colmain BJ. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 3.Young TL. Molecular genetics of human myopia: an update. Optom Vis Sci. 2009;86:E8–22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, Cheung EY, Lai RY, Chew SJ. Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci. 2004;45:1071–5. doi: 10.1167/iovs.03-1151. [DOI] [PubMed] [Google Scholar]
- 5.He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci. 2004;45:793–9. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- 6.Sawada A, Tomidokoro A, Araie M, Iwase A, Yamamoto T, Tajimi Study Group Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–70. doi: 10.1016/j.ophtha.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 7.Wong TY, Foster PJ, Hee J, Ng TP, Tielsch JM, Chew SJ, Johnson GJ, Seah SK. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000;41:2486–94. [PubMed] [Google Scholar]
- 8.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25:381–91. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 9.Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet. 2011;79:301–20. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saw SM, Chua WH, Wu HM, Yap E, Chia KS, Stone RA. Myopia: gene-environment interaction. Ann Acad Med Singapore. 2000;29:290–7. [PubMed] [Google Scholar]
- 11.Feldkämper M, Schaeffel F. Interactions of genes and environment in myopia. Dev Ophthalmol. 2003;37:34–49. doi: 10.1159/000072037. [DOI] [PubMed] [Google Scholar]
- 12.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–6. [PubMed] [Google Scholar]
- 13.Pacella R, McLellan J, Grice K, Del Bono EA, Wiggs JL, Gwiazda JE. Role of genetic factors in the etiology of juvenile-onset myopia based on a longitudinal study of refractive error. Optom Vis Sci. 1999;76:381–6. doi: 10.1097/00006324-199906000-00017. [DOI] [PubMed] [Google Scholar]
- 14.Kleinstein RN, Jones LA, Hullett S, Kwon S, Lee RJ, Friedman NE, Manny RE, Mutti DO, Yu JA, Zadnik K. Refractive error and ethnicity in children. Arch Ophthalmol. 2003;121:1141–7. doi: 10.1001/archopht.121.8.1141. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]
- 16.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–19. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–6. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 19.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide Linkage Scan for Myopia Susceptibility Loci among Ashkenazi Jewish Families Shows Evidence of Linkage on Chromosome 22q12. Am J Hum Genet. 2004;75:448–59. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 22.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–36. [PMC free article] [PubMed] [Google Scholar]
- 26.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 27.Paget S, Julia S, Vitezica ZG, Soler V, Malecaze F, Calvas P. Linkage analysis of high myopia susceptibility locus in 26 families. Mol Vis. 2008;14:2566–74. [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Xiao X, Li S, Zhang Q. Clinical and linkage study on a consanguineous Chinese family with autosomal recessive high myopia. Mol Vis. 2009;15:312–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Ma JH, Shen SH, Zhang GW, Zhao DS, Xu C, Pan CM, Jiang H, Wang ZQ, Song HD. Identification of a locus for autosomal dominant high myopia on chromosome 5p13.3-p15.1 in a Chinese family. Mol Vis. 2010;16:2043–54. [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YJ, Goh L, Khor CC, Fan Q, Yu M, Han S, Sim X, Ong RT, Wong TY, Vithana EN, Yap E, Nakanishi H, Matsuda F, Ohno-Matsui K, Yoshimura N, Seielstad M, Tai ES, Young TL, Saw SM. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–75. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Qu J, Xu X, Zhou X, Zou H, Wang N, Li T, Hu X, Zhao Q, Chen P, Li W, Huang K, Yang J, He Z, Ji J, Wang T, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20:2861–8. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Qu J, Zhang D, Zhao P, Zhang Q, Tam PO, Sun L, Zuo X, Zhou X, Xiao X, Hu J, Li Y, Cai L, Liu X, Lu F, Liao S, Chen B, He F, Gong B, Lin H, Ma S, Cheng J, Zhang J, Chen Y, Zhao F, Yang X, Yang C, Lam DS, Li X, Shi F, Wu Z, Lin Y, Yang J, Li S, Ren Y, Xue A, Fan Y, Li D, Pang CP, Zhang X, Yang Z. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011;88:805–13. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi H, Yamashiro K, Nakanishi H, Nakata I, Kurashige Y, Tsujikawa A, Moriyama M, Ohno-Matsui K, Mochizuki M, Ozaki M, Yamada R, Matsuda F, Yoshimura N. Association of 15q14 and 15q25 with High Myopia in Japanese. Invest Ophthalmol Vis Sci. 2011;52:4853–8. doi: 10.1167/iovs.11-7311. [DOI] [PubMed] [Google Scholar]
- 37.Verhoeven VJ, Hysi PG, Saw SM, Vitart V, Mirshahi A, Guggenheim JA, Cotch MF, Yamashiro K, Baird PN, Mackey DA, Wojciechowski R, Ikram MK, Hewitt AW, Duggal P, Janmahasatian S, Khor CC, Fan Q, Zhou X, Young TL, Tai ES, Goh LK, Li YJ, Aung T, Vithana E, Teo YY, Tay W, Sim X, Rudan I, Hayward C, Wright AF, Polasek O, Campbell H, Wilson JF, Fleck BW, Nakata I, Yoshimura N, Yamada R, Matsuda F, Ohno-Matsui K, Nag A, McMahon G, Pourcain BS, Lu Y, Rahi JS, Cumberland PM, Bhattacharya S, Simpson CL, Atwood LD, Li X, Raffel LJ, Murgia F, Portas L, Despriet DD, van Koolwijk LM, Wolfram C, Lackner KJ, Tonjes A, Magi R, Lehtimaki T, Kahonen M, Esko T, Metspalu A, Rantanen T, Parssinen O, Klein BE, Meitinger T, Spector TD, Oostra BA, Smith AV, de Jong PT, Hofman A, Amin N, Karssen LC, Rivadeneira F, Vingerling JR, Eiriksdottir G, Gudnason V, Doring A, Bettecken T, Uitterlinden AG, Williams C, Zeller T, Castagne R, Oexle K, van Duijn CM, Iyengar SK, Mitchell P, Wang JJ, Hohn R, Pfeiffer N, Bailey-Wilson JE, Stambolian D, Wong TY, Hammond CJ, Klaver CC. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131:1467–80. doi: 10.1007/s00439-012-1176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Wang P, Li S, Xiao X, Jia X, Guo X, Kong QP, Yao YG, Zhang Q. Mitochondrial DNA haplogroup distribution in Chaoshanese with and without myopia. Mol Vis. 2010;16:303–9. [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 40.Edwards BJ, Haynes C, Levenstien MA, Finch SJ, Gordon D. Power and sample size calculations in the presence of phenotype errors for case/control genetic association studies. BMC Genet. 2005;6:18. doi: 10.1186/1471-2156-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu B, Jiang D, Wang P, Gao Y, Sun W, Xiao X, Li S, Jia X, Guo X, Zhang Q. Replication study support CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci. 2011;52:8258–61. doi: 10.1167/iovs.11-7914. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Xiao X, Li S, Jia X, Yang Z, Huang S, Caruso RC, Guan T, Sergeev Y, Guo X, Hejtmancik JF. Mutations in NYX of individuals with high myopia, but without night blindness. Mol Vis. 2007;13:330–6. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Guo X, Xiao X, Yi J, Jia X, Hejtmancik JF. Clinical description and genome wide linkage study of Y-sutural cataract and myopia in a Chinese family. Mol Vis. 2004;10:890–900. [PubMed] [Google Scholar]
- 44.Han W, Leung KH, Fung WY, Mak JY, Li YM, Yap MK, Yip SP. Association of PAX6 polymorphisms with high myopia in Han Chinese nuclear families. Invest Ophthalmol Vis Sci. 2009;50:47–56. doi: 10.1167/iovs.07-0813. [DOI] [PubMed] [Google Scholar]
- 45.Ng TK, Lam CY, Lam DS, Chiang SW, Tam PO, Wang DY, Fan BJ, Yam GH, Fan DS, Pang CP. AC and AG dinucleotide repeats in the PAX6 P1 promoter are associated with high myopia. Mol Vis. 2009;15:2239–48. [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang B, Yap MK, Leung KH, Ng PW, Fung WY, Lam WW, Gu YS, Yip SP. PAX6 haplotypes are associated with high myopia in Han chinese. PLoS ONE. 2011;6:e19587. doi: 10.1371/journal.pone.0019587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao YY, Zhang FJ, Zhu SQ, Duan H, Li Y, Zhou ZJ, Ma WX, Li Wang N. The association of a single nucleotide polymorphism in the promoter region of the LAMA1 gene with susceptibility to Chinese high myopia. Mol Vis. 2011;17:1003–10. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F, Zhu T, Zhou Z, Wu Y, Li Y. Association of lumican gene with susceptibility to pathological myopia in the northern han ethnic chinese. J Ophthalmol. 2009;2009:514306. doi: 10.1155/2009/514306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen ZT, Wang IJ, Shih YF, Lin LL. The association of haplotype at the lumican gene with high myopia susceptibility in Taiwanese patients. Ophthalmology. 2009;116:1920–7. doi: 10.1016/j.ophtha.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 50.Lin HJ, Kung YJ, Lin YJ, Sheu JJ, Chen BH, Lan YC, Lai CH, Hsu YA, Wan L, Tsai FJ. Association of the lumican gene functional 3′-UTR polymorphism with high myopia. Invest Ophthalmol Vis Sci. 2010;51:96–102. doi: 10.1167/iovs.09-3612. [DOI] [PubMed] [Google Scholar]
- 51.Lin HJ, Wan L, Tsai Y, Tsai YY, Fan SS, Tsai CH, Tsai FJ. The TGFbeta1 gene codon 10 polymorphism contributes to the genetic predisposition to high myopia. Mol Vis. 2006;12:698–703. [PubMed] [Google Scholar]
- 52.Zha Y, Leung KH, Lo KK, Fung WY, Ng PW, Shi MG, Yap MK, Yip SP. TGFB1 as a susceptibility gene for high myopia: a replication study with new findings. Arch Ophthalmol. 2009;127:541–8. doi: 10.1001/archophthalmol.2008.623. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Li Y, Zhang D, Zhang H, Lu F, Liu X, He F, Gong B, Cai L, Li R, Liao S, Ma S, Lin H, Cheng J, Zheng H, Shan Y, Chen B, Hu J, Jin X, Zhao P, Chen Y, Zhang Y, Lin Y, Li X, Fan Y, Yang H, Wang J, Yang Z. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Wang P, Gao Y, Li S, Xiao X, Zhang Q. High myopia is not associated with single nucleotide polymorphisms in the COL2A1 gene in the Chinese population. Mol Med Report. 2012;5:133–7. doi: 10.3892/mmr.2011.626. [DOI] [PubMed] [Google Scholar]
- 55.Wang P, Li S, Xiao X, Jia X, Jiao X, Guo X, Zhang Q. High myopia is not associated with the SNPs in the TGIF, lumican, TGFB1, and HGF genes. Invest Ophthalmol Vis Sci. 2009;50:1546–51. doi: 10.1167/iovs.08-2537. [DOI] [PubMed] [Google Scholar]
- 56.Smith AH, Bates MN. Confidence limit analyses should replace power calculations in the interpretation of epidemiologic studies. Epidemiology. 1992;3:449–52. doi: 10.1097/00001648-199209000-00011. [DOI] [PubMed] [Google Scholar]
- 57.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS. van RG, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]