Summary
Pseudomonas aeruginosa responds to growth on agar surfaces to produce cyclic-di-GMP, which stimulates biofilm formation. This is mediated by an alternative cellular function chemotaxis-like system called Wsp. The receptor protein WspA, is bioinformatically indistinguishable from methyl-accepting chemotaxis proteins. However, unlike standard chemoreceptors, WspA does not form stable clusters at cell poles. Rather, it forms dynamic clusters at both polar and lateral subcellular locations. To begin to study the mechanism of Wsp signal transduction in response to surfaces, we carried out a structure-function study of WspA and found that its C-terminus is important for its lateral subcellular localization and function. When this region was replaced with that of a chemoreceptor for amino acids, WspA became polarly localized. In addition, introduction of mutations in the C-terminal region of WspA that rendered this protein able to form more stable receptor-receptor interactions, also resulted in a WspA protein that was less capable of activating signal transduction. Receptor chimeras with a WspA C-terminus and N-terminal periplasmic domains from chemoreceptors that sense amino acids or malate responded to surfaces to produce c-di-GMP. Thus the amino acid sequence of the WspA periplasmic region did not need to be conserved for the Wsp system to respond to surfaces.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that is also a model organism for studies of biofilms, defined as communities of bacteria encased in an extracellular matrix. P. aeruginosa can cause acute infections, but it is also known for its ability to cause chronic biofilm infections in the lungs of cystic fibrosis patients (Hoiby, 2002, Singh et al., 2000) and on indwelling medical devices. Biofilm infections are problematic because they are resistant to antibiotic treatment and tend to escape immune surveillance (Mah & O’Toole, 2001). The ability of P. aeruginosa to adopt a biofilm lifestyle is determined by the intracellular signaling molecule, cyclic-di-GMP (c-di-GMP). Diguanylate cyclases (DGCs) with GGDEF domains synthesize c-di-GMP from two monomers of GTP, and phosphodiesterases (PDEs) with EAL or HD-GYP domains degrade c-di-GMP (Christen et al., 2005, Paul et al., 2004, Ryan et al., 2006, Schmidt et al., 2005, Tamayo et al., 2005). P. aeruginosa has genes for over forty of these enzymes, many of which have additional protein domains that are likely to sense environmental stimuli to control intracellular levels of c-di-GMP (Kulasakara et al., 2006). In many bacteria, relatively high levels of intracellular c-di-GMP are associated with elevated biofilm formation and reduced motility (Hengge, 2009, Simm et al., 2004).
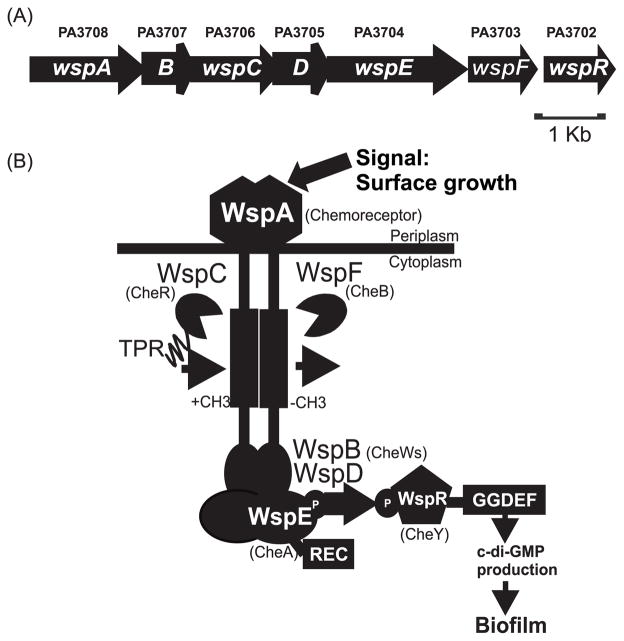
Probably the best studied DGC from P. aeruginosa is WspR. wspR mutants are defective in some, but not all c-di-GMP associated traits. They are partially defective in synthesis of Pel polysaccharide, a component of the biofilm matrix, but they have normal swarming, swimming and twitching motility (Huangyutitham and Harwood, unpublished), (Kulasakara et al., 2006). WspR is part of the Wsp complex, a membrane bound chemotaxis-like system that belongs to the “alternative cellular function” (ACF) chemotaxis family (Wuichet et al., 2007, Guvener & Harwood, 2007, Hickman et al., 2005) (Fig. 1). Wsp consists of six proteins, including a predicted methyltransferase (CheR homolog), a methylesterase (CheB homolog), a CheA-like histidine kinase and two CheW-like adaptor proteins. In addition, the CheA-like protein (WspE in the Wsp system) has a predicted response regulator receiver domain (REC) at its C-terminus and the CheR homologue (WspC in the Wsp system) has a tetratricopeptide repeat (TPR) domain at its C-terminus. These proteins are predicted to form a complex with WspA, a membrane-bound methyl-accepting chemotaxis protein (MCP) or chemoreceptor protein, of domain class H40 (Alexander & Zhulin, 2007) (Fig 1). The most distinguishing feature of ACF systems is that they have response regulator receiver proteins that differ from the chemotaxis response regulator CheY (Guvener & Harwood, 2007, Hickman et al., 2005). In the case of WspR this is a composite REC-GGDEF protein.
Fig. 1. Schematic model of the Wsp system.
(A) Genetic organization of the wsp locus in PAO1. (B) Model of the Wsp protein complex. The chemotaxis counterparts of the Wsp proteins are indicated in parentheses. WspA, the receptor, detects a signal associated with growth on a surface. WspB and WspD are CheW-like proteins. WspE is a CheA-like histidine kinase with an additional receiver domain (REC) at the C-terminus. WspR is a REC-GGDEF domain protein that has diguanylate cyclase activity. WspC is a predicted CheR-like methyltransferase with a C-terminal tetratricopeptide repeat (TPR) domain and WspF is a predicted CheB-like methylesterase. In our model, detection of surface growth by WspA results in phosphorylation of WspR. Phosphorylated WspR synthesizes c-di-GMP, which triggers biofilm formation.
Wsp has two major phenotypic features in addition to c-di-GMP synthesis that distinguish it from flagella-based chemotaxis (Che) systems. First, instead of responding to small soluble compounds, like amino acids, the Wsp system senses a signal associated with a physical condition, growth on a surface (Guvener & Harwood, 2007). Another difference between the Che and Wsp systems is their subcellular localization properties (Guvener & Harwood, 2007). Bacterial Che proteins, including P. aeruginosa Che proteins, localize at the cell poles (Guvener et al., 2006, Bardy & Maddock, 2005). It is thought that polar localization is important for promoting the integration of signals from different chemicals and for increasing the sensitivity of the chemotaxis machinery to sensory inputs (Sourjik & Berg, 2004). By contrast, qualitative observations of Wsp protein-YFP fusions suggested that the membrane bound Wsp complex is laterally localized in P. aeruginosa around the cell periphery including at cell poles (Guvener & Harwood, 2007). The differences in subcellular localization between the Wsp and Che proteins may reflect functional differences between a system that controls biofilm formation in response to surfaces and a system that controls flagella motility in response to gradients of chemicals. We wanted to probe this hypothesis in more detail for two reasons. The first is to learn more about the mechanism of action of a complex bacterial signal transduction system that influences biofilm formation. The second is to characterize in more detail a member of the ACF family of signal transduction systems.
In this study we quantitated the subcellular localization properties of WspA, the membrane-bound receptor that anchors associated Wsp proteins, and examined effects of various Wsp system mutations on WspA localization. Using standard wide-field fluorescence microscopy, we can observe Wsp signal transduction visually and in single cells. When WspR-YFP is unphosphorylated, it is diffuse and no protein localization is observed. However upon phosphorylation, WspR-YFP forms visible subcellular clusters (Guvener & Harwood, 2007). We used this tool to look at the ability of chimeric WspA receptors to respond to the signal of surface growth. Here we found that several of the Wsp proteins and also the C-terminal region of WspA are important for its lateral localization and function. Analysis of P. aeruginosa chemoreceptor-WspA chimeras indicates that surface sensing by WspA does not require a specific type of periplasmic sensing domain.
RESULTS
Quantitative and real time analysis of WspA distribution
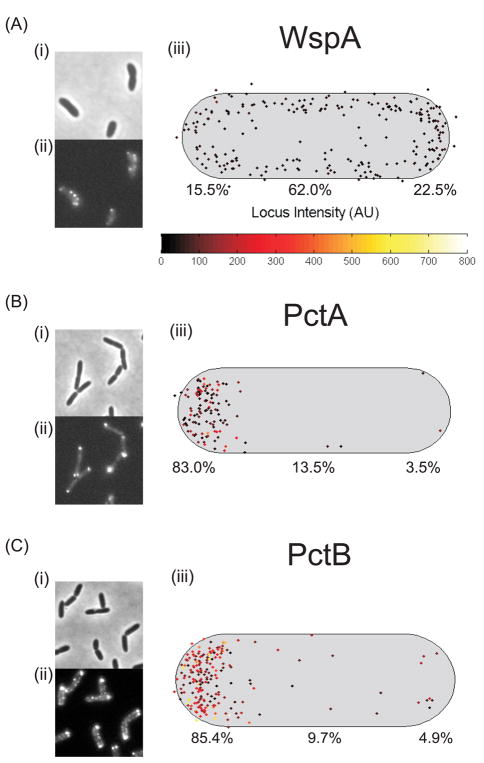
We examined the subcellular localization and intensity of the receptor WspA tagged with YFP by fluorescence microscopy. For a given experiment about 100 cells were analyzed. The brightest cluster in each cell was identified and the data for the population of cells were combined to generate a single composite image that shows many spots of varying intensity. Cells that had recently divided and were still touching were used for analysis. Daughter cells have a “new” cell pole at the recent site of division and an “old” cell pole that is retained from the predivisional cell. We found that WspA was localized to polar and lateral regions of wild-type strain PAO1 cells (Fig 2A), with up to five visible clusters of WspA-YFP per cell. The WspA-YFP fusion is functional; the YFP fusion leads to hyperactivation of the Wsp system, resulting in overproduction of c-di-GMP and wrinkly colony morphology (Guvener & Harwood, 2007). We analyzed the subcellular distribution of two P. aeruginosa chemoreceptors, PctA and PctB, which mediate chemotaxis to amino acids (Taguchi et al., 1997) and found that they were mostly present at the old cell pole. The polar clusters of these YFP fusion proteins had high fluorescence intensity, likely reflecting their relatively high abundance in cells (Fig. 2B and C). We also analyzed the localization of WspA in pairs of optically sectioned cells (touching at the new cell pole) (Movie 1) and found that WspA-YFP clusters are present along the length of the cells. In time lapse studies, we observed the distribution of WspA-YFP in cells at intervals of 5 s over periods of 2 min before the YFP protein bleached. Lateral WspA-YFP clusters appeared to form and dissolve (Movie 2 and 3). Polar clusters of WspA-YFP appeared larger and brighter than lateral clusters.
Fig. 2. Subcellular localization of YFP-tagged (A) WspA and the chemoreceptors (B) PctA and (C) PctB.
(i) Phase contrast image, (ii) YFP image, (iii) Composite data showing the subcellular localization of clusters in a population of cells. The cell length and width represents the average cell size, the cluster intensity is represented by the locus intensity heat map, the brightest cluster in each cell was plotted. The proportion (%) of clusters found in the old (left) or new (right) cell pole or laterally localized is indicated.
Previous studies have shown that MreB is a dynamic, cytoplasmic-membrane-associated protein that may act as a scaffold for peptidoglycan synthesis (Dominguez-Escobar et al., 2011, Garner et al., 2011, Salje et al., 2011). Since MreB is important for localization of other bacterial proteins (Cowles & Gitai, 2010, White et al., 2010, Shaevitz & Gitai, 2010, Gitai et al., 2004), we tested the possible involvement of MreB in mediating WspA localization. When we treated cells with the MreB-depolymerizing drug A22 (Bean et al., 2009), MreB localization was disrupted but the localization of WspA remained unaffected (Fig. S1).
Effect of other Wsp proteins on WspA subcellular location and Wsp activity
To assay signal transduction we count the percentage of cells with visible WspR-YFP clusters, a measure of WspR phosphorylation (Guvener & Harwood, 2007). When cells are grown in liquid culture very few cells have WspR-YFP clusters. However, growth on an agar surface stimulates an increase in the number of cells with WspR-YFP foci. We have previously shown that WspA and WspE are required for Wsp signal transduction (Guvener & Harwood, 2007). In addition, deletion of the wspF methylesterase gene causes overstimulation of the Wsp system as revealed by a high percentage of cells with subcellular clusters of WspR-YFP (Guvener & Harwood, 2007). WspC encodes a predicted methyltransferase that is expected to act in opposition to the activity of the WspF methylesterase. Indeed, we found that signal transduction was abolished in a wspC deletion mutant and in a wspC-TPR domain deletion mutant, regardless of the presence of absence of wspF (Table 1). A wspE-REC domain deletion mutant retained about half of wild-type signaling activity. ΔwspB and ΔwspD mutants were inactive (Table 1). The subcellular distribution of WspA-YFP was altered in both wspD (59% of the WspA fluorescent clusters were located at the old cell pole, compared to 24% in a wild-type strain background) and the wspE deletion mutants (52% of the WspA clusters were polar), suggesting that the lateral localization of WspA in cells is correlated with a functional Wsp system (Fig. 3). However WspA can retain its lateral location and be part of a non-functional Wsp system. Such is the case for WspA-YFP in a wspB deletion background (Table 1 and Fig. 3)
Table 1.
Effect of various wsp mutations on Wsp function, as measured by WspR-YFP clusteringa.
| Percentage of population with WspR-YFP clusters | ||
|---|---|---|
| Strain name | Liquid | Surface |
| WT | 1 | 31 |
| ΔwspC | 0 | 0 |
| wspCΔTPR | 1 | 1 |
| wspEΔREC | 2 | 15 |
| ΔwspB | 0 | 0 |
| ΔwspD | 0 | 0 |
| ΔwspF | 30 | 64 |
| ΔwspF ΔwspC | 0 | 0 |
| ΔwspF wspCΔTPR | 4 | 1 |
| ΔwspFwspEΔREC | 15 | 25 |
| ΔwspF ΔwspB | 3 | 1 |
| ΔwspF ΔwspD | 0 | 0 |
At least 100 cells were counted in each of two different experiments
Fig. 3. Subcellular localization of WspA-YFP in wild-type (A) and mutant backgrounds (B–D).
(i) Phase contrast image, (ii) YFP image, (iii) Composite data showing the subcellular localization of clusters in a population of cells. The cell length and width represents the average cell size, the cluster intensity is represented by the locus intensity heat map, the brightest cluster in each cell was plotted. The proportion (%) of clusters found in the old (left) or new (right) cell pole or laterally localized is shown.
Analysis of a WspA-PctA chimera indicates that the C-terminus of WspA is important for its lateral localization
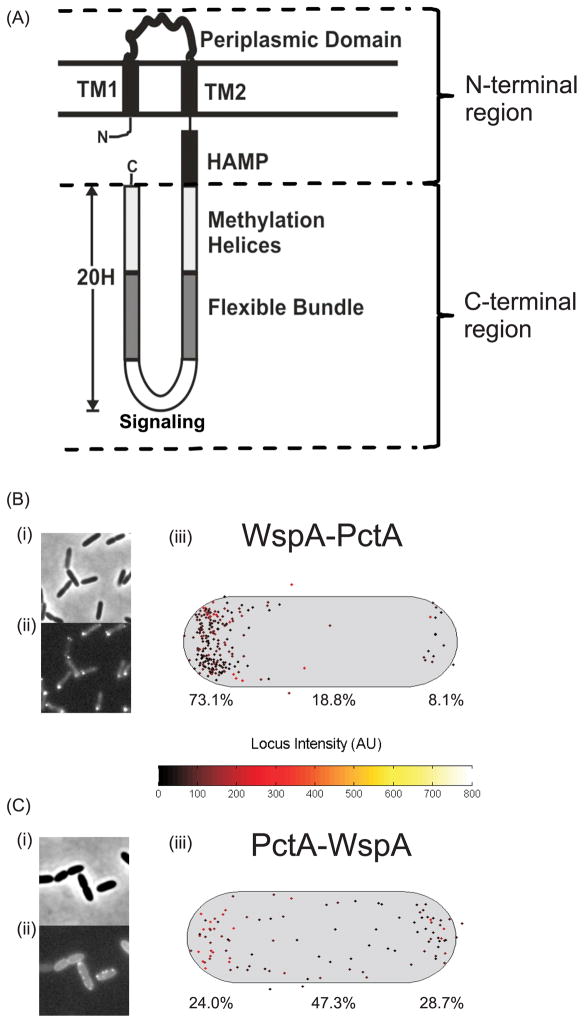
WspA has a typical chemoreceptor domain organization with an N-terminal region consisted of two transmembrane domains that flank a periplasmic domain, a HAMP domain and a highly conserved signaling domain (Fig 4A). The periplasmic domains of the other studied PAO1 chemoreceptors (PctA, PctB, PctC and PA2652) share only 4–14% amino acid identity with WspA. However, the HAMP and signaling domains of the chemoreceptors share 33–39% amino acid identity with the WspA HAMP domain and 58–62% identity with the WspA signaling domain. Each of these chemoreceptors and WspA has 40 heptad repeats in the C-terminal region (Alexander & Zhulin, 2007). To test possible effects of the various chemoreceptor domains on WspA localization we constructed chimeras where either the N-terminus or the C-terminus of WspA was replaced with that of another chemoreceptor and fused this chimera to YFP (Fig. 4A). A WspA-PctA chimera had a polar localization, similar to that of wild type PctA (compare Figs. 2B and 4B). However a PctA-WspA chimera was localized around the cell periphery, similar to the wild-type WspA (compare Figs. 2A and 4C).
Fig. 4. Construction and subcellular localization of chemoreceptor – WspA chimeras.
(A) Schematic of the topology of the P. aeruginosa chemoreceptors PctA, PctB, PctC, PA2652 and WspA. (B) Subcellular localization of WspA-PctA and (C) PctA-WspA chimeras. (i) Phase contrast image, (ii) fluorescent image, (iii) Composite data showing the subcellular localization of clusters in a population of cells. The cell length and width represents the average cell size, the cluster intensity is represented by the locus intensity heat map, the brightest cluster in each cell was plotted. The proportion (%) of clusters found in the old (left) or new (right) cell pole or laterally localized is shown.
WspA responds to surfaces when its periplasmic domain is replaced with that of another chemoreceptor
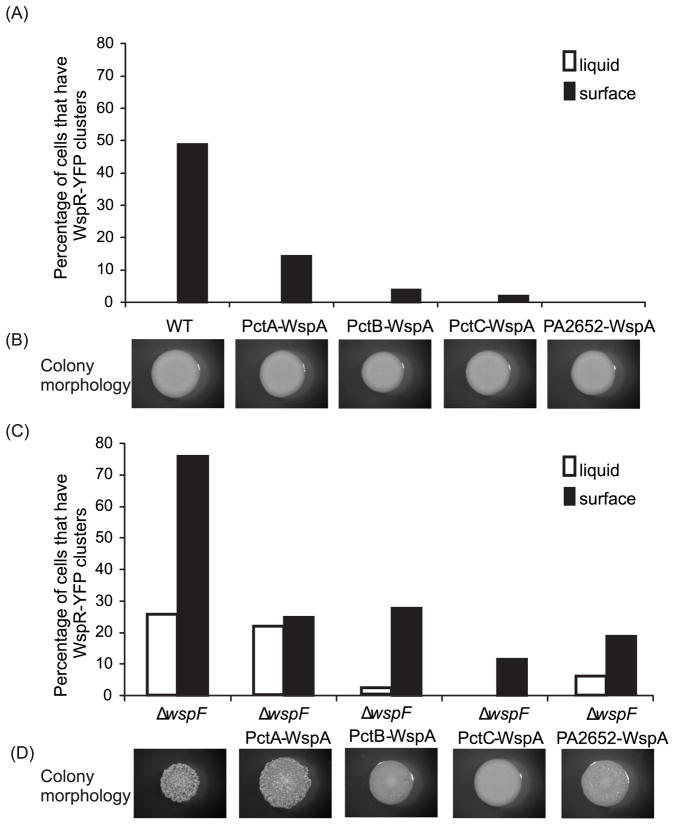
P. aeruginosa chemoreceptors PctA, PctB and PctC each detect a subset of amino acids and PA2652, detects malate (Kuroda et al., 1995, Taguchi et al., 1997, Alvarez-Ortega & Harwood, 2007). Fusion of the N-terminal region of PctA, PctB, PctC or PA2652 to the C-terminal region of WspA (tagged with CFP) resulted in chimeras that localized laterally in cells, similar to the wild-type WspA (Fig 4C, Fig S2). Further, when we analyzed chemoreceptor-WspA chimeras that were not tagged with CFP, each was active in surface sensing although they were less functional than WspA (Fig. 5). PctA-WspA was the most functional, followed by PctB-WspA, PA2652-WspA and PctC-WspA. This result was observed both in a wild-type background and in a ΔwspF background (Fig. 5). The frequency of WspR-YFP clustering was not affected by exposure of cells expressing PctA-WspA to leucine or serine, compounds sensed as chemoattractants by PctA (Taguchi et al., 1997),
Fig. 5. Functional analysis of chemoreceptor-WspA chimeras.
(A) and (C) ability of each chemoreceptor-WspA chimera to phosphorylate WspR during liquid (white bars) and surface (black bars) growth, measured as percentage of cells in the population that have at least one WspR-YFP cluster. (B) and (D) colony morphology of each strain. (A) and (B) wild-type background encoding WspA or chemoreceptor-WspA variants. (C) and (D) ΔwspF background encoding WspA or chemoreceptor-WspA variants.
Identification of amino acids important for WspA function and self-assembly
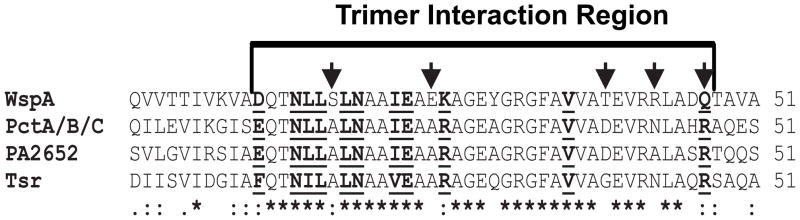
Work with E. coli suggests that a stochastic self-nucleation mechanism accounts for the formation of chemoreceptor clusters. Newly synthesized receptors are inserted into lateral membranes, but receptor-receptor interactions lead to the assembly of relatively large complexes that are pushed to the cell poles by insertion of new cell membrane components and cell division processes. Possibly this is influenced by membrane curvature (Alley, 2001, Greenfield et al., 2009). Previous work identified conserved residues in the signaling domain of the E. coli chemoreceptor Tsr that are important for stable receptor-receptor interactions and formation of trimers-of-dimers of Tsr (Ames & Parkinson, 2006, Ames et al., 2002, Mowery et al., 2008). We hypothesized that WspA monomers may have a reduced ability to form stable trimers-of-dimers compared to polar chemoreceptors, and thus a reduced ability to form larger arrays and localize to the cell pole. This is consistent with our observation that the C-terminus of WspA is important for its lateral subcellular localization. To identify residues that might influence WspA self-assembly, we aligned the signaling domain sequences of WspA, PctA, PctB, PctC and PA2652 with that of the E. coli Tsr receptor (Fig. 6). We found that the trimer interaction region of all five PAO1 receptors is highly conserved. However, WspA has five non-conservative amino acid differences from the chemoreceptors (Fig. 6). We mutated these WspA residues to match those of PctA and examined effects on WspA-YFP localization in E. coli and P. aeruginosa.
Fig. 6. Identification of WspA amino acids that may participate in stabilization of trimer interactions.
ClustalW alignment of the trimer interaction region. Likely trimer interaction residues (bold and underlined) were identified by comparison to those identified in Tsr (Ames et al., 2002). * identical,: highly conserved,. weakly conserved. Arrows show amino acids of WspA that have non-conservative changes compared to PctA.
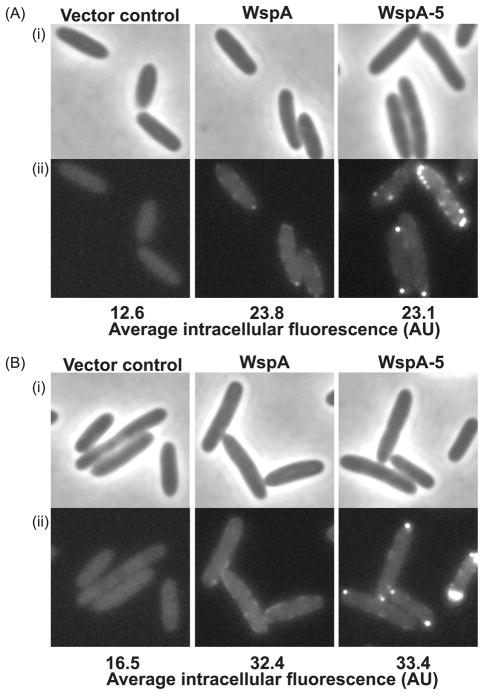
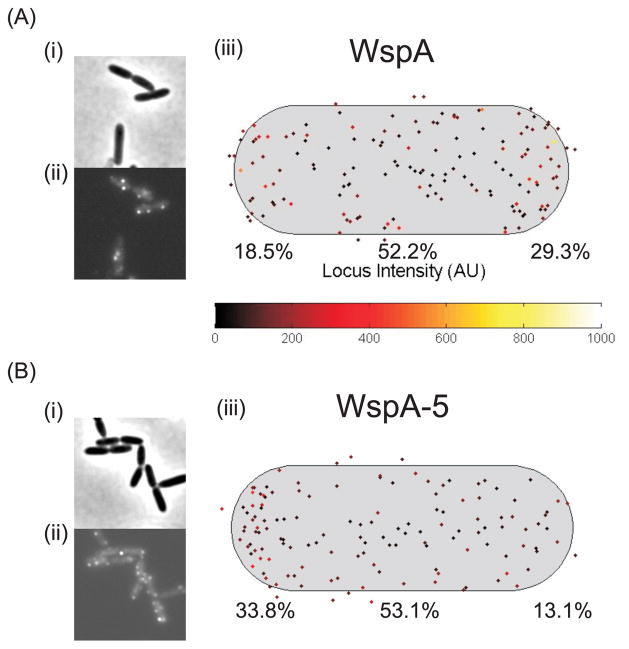
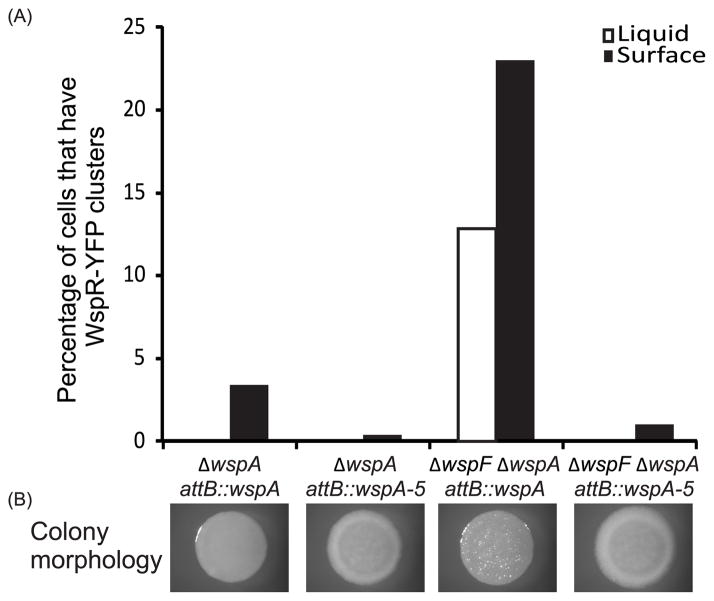
When expressed in E. coli, the mutated WspA (called WspA-5) formed a greater number of visible clusters than the wild-type protein and this was not dependent on the E. coli chemotaxis machinery being present (Fig. 7). This suggests that the changes that we made in WspA stabilized predicted trimer-of-dimer interactions. When WspA-5 variant protein was expressed in P. aeruginosa quantitative analysis showed that WspA-5 had an increased tendency to be located at the old cell pole (Fig. 8). The WspA-5 variant was less functional than the wild-type protein as evidenced by a substantially reduced ability to activate WspR phosphorylation in a ΔwspF background (Fig. 9). Furthermore, a ΔwspF mutant has a wrinkly colony morphology, caused by overexpression of EPS, but colonies formed by the wspA-5ΔwspF strain were smooth (Fig. 9B). These data suggest that WspA receptor interactions are less stable than those of the PctA chemoreceptor and that receptor-interaction residues are important for Wsp signal transduction.
Fig. 7. Localization of WspA-YFP and WspA- 5-YFP.
(A) E. coli RP437 (chemotaxis proficient) strain background, (B) HCB437 (chemotaxis genes deleted) strain background. (i) phase contrast, (ii) YFP.
Fig. 8. Subcellular localization of (A) WspA-YFP and (B) WspA-5-YFP in PAO1.
(i) Phase contrast, (ii) YFP (iii) Composite data showing the subcellular localization of clusters in a population of cells. The cell length and width represents the average cell size, the cluster intensity is represented by the locus intensity heat map, the brightest cluster in each cell was plotted. The proportion (%) of clusters found in the old (left) or new (right) cell pole or laterally localized is shown.
Fig. 9. Functional analysis of WspA and WspA-5.
(A) Ability of WspA/WspA-5 to phosphorylate WspR, measured by percentage of cells in the population that have at least one WspR-YFP cluster. Liquid grown cells: white bars; surface grown cells: black bars. (B) Colony morphology of corresponding strains. Vector control strains were ΔwspA attB::vector control and ΔwspFΔwspA attB::vector control, these strains did not have any WspR-YFP clusters and had smooth colony morphology (data not shown).
DISCUSSION
Our data show that the Wsp system has several features that contribute to its lateral subcellular localization and to its ability to function in sensing surfaces. One of these features is the C-terminal domain of WspA. A protein chimera with an N-terminal WspA periplasmic domain and a C-terminal chemoreceptor domain localized to the cell pole (Fig. 4B), whereas chimeras with a chemoreceptor periplasmic domain and a WspA C-terminus localized laterally in cells (Fig. 4C and S2) and responded to surfaces to stimulate the WspR response-regulator diguanylate cyclase to produce c-di-GMP (Fig. 5). The periplasmic domains of the four P. aeruginosa chemoreceptors that were tested, PctA, PctB, PctC and PA2652, share very little amino acid sequence identity with the periplasmic domain of WspA. This suggests that the specific amino acid sequence of the periplasmic domain of WspA is not a critical determinant of surface sensing.
Another important feature of a functional Wsp system is the interaction of the C-terminal WspA signaling domain with the CheW-like protein, WspD and the CheAY-like protein, WspE. In the absence of WspD or WspE, WspA becomes mostly polar and is inactive in signal transduction (Fig. 3, Table 1). It could be that WspD interacts with WspA such that trimer-of dimer formation is unstable, and formation of higher order arrays is prevented. Consistent with this we found that the sequence of the WspA trimer interaction region is important for function (Fig. 9). A version of WspA (WspA-5-YFP) that was mutated to have the same timer interaction sequence as the P. aeruginosa chemoreceptor PctA formed bright, defined clusters when expressed in E. coli (Fig 7), whereas wild-type WspA-YFP, expressed at equivalent levels, was barely visible presumably because these receptors interacted less efficiently and were diffuse (Fig. 7). When WspA-5-YFP was expressed in P. aeruginosa it was more often located at the old cell pole than WspA and was less functional (Fig. 8 and 9). This suggests that relatively weak timer of dimer interactions of WspA are an important feature of its activity. Another potential consequence of the relatively weak trimer of dimer interactions could be that they render WspA less likely to associate with chemoreceptors, thus reducing the chance of the Wsp and chemotaxis systems interfering with each other.
Previous work showed that chemotaxis signal transduction can be activated by changes in osmotic pressure due to effects on the stability of chemoreceptor clusters (Vaknin & Berg, 2006). It is tempting to speculate that the instability of WspA clusters allows for more sensitive detection of mechanical perturbations of the cell membrane. In support of this we found no instance where versions of WspA that were polarly-localized or more stable were active in signal transduction. However, firm demonstration that instability of WspA clusters is important for sensing will require a better understanding of the specific nature of the surface associated signal that stimulates the system. It is also possible that the subcellular location of the Wsp complex is important for delivery of c-di-GMP to a specific receptor. However, c-di-GMP is a small molecule that can diffuse rapidly. The differential effects that c-di-GMP has on cellular functions could be explained by differences in affinities of receptor proteins for c-di-GMP without the necessity to evoke subcellular compartmentalization of c-di-GMP synthesis and signaling.
Although this study sheds some light on the functioning of the Wsp system, there is still much we don’t understand. A hallmark feature of bacterial chemotaxis systems is that they sense chemical gradients by a temporal mechanism where environments are sampled over intervals of time (Berg & Tedesco, 1975, Dahlquist et al., 1976, Block et al., 1983). Since the Wsp system includes methylation and demethylation enzymes that are important for temporal sensing in homologous chemotaxis systems, we assume that it also temporally senses and adapts to the signal associated with surface growth. The exact nature of the signal remains elusive. We speculate that WspA is stimulated by changes in the physical environment of the periplasm, either by mechanical stress associated with surface or cell-cell contact, or possibly by physiological changes associated with switching to a biofilm lifestyle.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table S1. Pseudomonas aeruginosa strain PAO1 was the wild type used. Unless otherwise stated E. coli and P. aeruginosa strains were grown in Luria–Bertani (LB) medium (Sambrook et al., 1989) at 37°C with shaking. For microscopy, overnight cultures were diluted 60-fold in a final volume of 3 ml, and grown at 28 –30°C with aeration to an OD600 of 0.2–0.4 to produce liquid-grown cells. These strains were then diluted to an OD600 of 0.005, 100 μl aliquots were spread inoculated onto LB 2.5% agar plates and incubated at room temperature for about 20 hours to produce surface-grown cells. To visualize colony morphology, 2 μl of broth-grown cells that had been diluted to an OD600 of 0.005 were spot inoculated onto Tryptone-Congo Red plates (Friedman & Kolter, 2004) and incubated at room temperature for 2–3 days. Colonies were photographed using a digital camera mounted on a dissection microscope (Olympus SZX-ILLK100). Antibiotic concentrations were 100 μg ml−1 ampicillin, 10 μg ml−1 gentamicin (Gm) and 10 μg ml−1 tetracycline (Tc) for E. coli and 50 μg ml−1 Gm and 50–100 μg ml−1 Tc for P. aeruginosa.
DNA manipulations
DNA manipulations were carried out according to standard protocols (Sambrook et al., 1989). Ligation-independent cloning was achieved using an In Fusion Cloning kit, according to the manufacturer’s instructions (ClonTech). Phusion Hot Start DNA polymerase (Finnzymes) was used to PCR amplify DNA, strain PAO1 gDNA was used as the template unless otherwise stated. DNA was isolated with Qiagen DNA purification kits. Oligonucleotides were synthesized by Integrated DNA Technologies. DNA sequencing was performed by Genewiz Inc.
Genetic manipulation of P. aeruginosa
yfp or cfp translational fusion and in-frame deletion/replacement suicide plasmids were based on plasmid pEX18Gm or pEX19Gm and generated as described in Tables S2 (plasmids) and S3 (primers). Suicide plasmids were transferred to P. aeruginosa by conjugation with E. coli S17-1 and plasmid inserts were integrated into the chromosome by homologous recombination (Ferrandez et al., 2002). Potential mutants were screened by PCR. For each mutant, the region of the chromosome that had been manipulated was PCR amplified and sequenced to confirm the strain had the correct genetic arrangement.
Construction of MINICTX2-based plasmids was achieved as described in Table S2. The resulting vectors were introduced into P. aeruginosa by conjugation from E. coli S17-1 and integrated into the attB site on the chromosome by site-specific recombination, as described (Hoang et al., 2000). Transconjugants were screened by PCR. The wspA genes that were inserted into the attB site were PCR amplified and sequenced to confirm the correct genetic arrangement of the strains. The pUC57-3′wspA-4 mutations plasmid was purchased from Genewiz Inc., see Fig. S3 for a description of the mutations introduced into wspA. SOE PCR was used to introduce a fifth mutation into wspA using pUC57-3′wspA-4 mutations and PAO1 gDNA as templates (Table S2, Table S3, Fig S3).
Fluorescence microscopy techniques
Samples were prepared and static images captured from at least two biological replicates as previously described (Guvener & Harwood, 2007). Movies of WspA-YFP cluster dynamic behavior were captured with the same filters and microscope, equipped with a Nikon Z-motor focus unit, a FN-MT Magnification Changer Turret and a Photometrics Evolve EM CCD Camera. NIS-Elements Advanced Research Complete Duo Software Package was used for capturing the movies. WspA-YFP movies were captured using an exposure time of 400 ms (movie 2) or 300 ms (movie 3) with the mercury lamp set at 75%, at a readout speed of 5 MHz and 1X conversion gain. The 100X oil immersion objective was used with the magnification changer turret set to 1.5X. A total of 12 exposures were captured before the YFP photobleached.
Z-stack analysis of WspA-YFP localization was conducted with a Nikon A1RSi confocal microscope equipped with a 50 mW multiline argon laser unit (Spectra Physics). A 60X oil DIC objective was used with a 514 nm laser at 3% power, with a pinhole size of 76 um. The Z-stack was sampled in 13 steps of 0.175 um each. Data was processed with the NIS-Elements Advanced Research Software Package.
Measurement of average intracellular fluorescence and WspR-YFP clustering
Phase contrast images were used to select candidate cells for analysis. Metamorph software was used to assess WspR-YFP clustering frequency, as previously described (Guvener & Harwood, 2007), except that the threshold value to determine the presence of a WspR-YFP cluster was set to >1.78. WspR-YFP clustering frequency data represents an average of at least two biological replicates, carried out on different days. Around 100 cells were counted per experiment.
Measurement of subcellular localization of WspA and other chemoreceptors
Standard phase-contrast microscopy images were segmented using custom MATLAB software. A watershed-based algorithm was used to transform phase-contrast images into segmented regions with well defined boundaries. The region boundaries were then optimized using a user-trained scoring algorithm dependent on cell type and growth and microscopy conditions. Aligned fluorescent images were overlaid over the segmented images and fluorescent foci were fit using a Gaussian point-spread function. The optimized regions were marked as cells, and relevant physical characteristics, e.g. area, long and short-axis length, foci position, were recorded. Cells used for analysis were gated based on physical characteristics relevant to specific analysis goals and the results were visualized using the same suite of custom MATLAB software. Only cells that had recently divided and were still touching at the new cell pole were used for analysis. This method was used to identify the old and new cell pole. The old and new cell pole were designated as 15% of total cell length at each end.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grant GM56665 from the National Institute of General Medical Sciences to CSH, as well as grants NSF PHY-084845 and NSF MCB-1151043 to PAW. PAW also received support from the Sloan Foundation and the University of Washington Research Royalty Fund. We thank Howard C. Berg for providing strains and John S. Parkinson for providing strains and for helpful discussions. We also thank Kimberly Cowles and Zemer Gitai for protocols for the MreB experiments.
References
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR. The highly conserved domain of the Caulobacter McpA chemoreceptor is required for its polar localization. Mol Microbiol. 2001;40:1335–1343. doi: 10.1046/j.1365-2958.2001.02476.x. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ortega C, Harwood CS. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol. 2007;73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc Natl Acad Sci U S A. 2006;103:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy SL, Maddock JR. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J Bacteriol. 2005;187:7840–7844. doi: 10.1128/JB.187.22.7840-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Tedesco PM. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Segall JE, Berg HC. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Cowles KN, Gitai Z. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol Microbiol. 2010;76:1411–1426. doi: 10.1111/j.1365-2958.2010.07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist FW, Elwell RA, Lovely PS. Studies of bacterial chemotaxis in defined concentration gradients. A model for chemotaxis toward L-serine. J Supramol Struct. 1976;4:329–342. doi: 10.1002/jss.400040304. [DOI] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Hoiby N. Understanding bacterial biofilms in patients with cystic fibrosis: current and innovative approaches to potential therapies. Journal of Cystic Fibrosis. 2002;1:249–254. doi: 10.1016/s1569-1993(02)00104-2. [DOI] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Kumano T, Taguchi K, Nikata T, Kato J, Ohtake H. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J Bacteriol. 1995;177:7019–7025. doi: 10.1128/jb.177.24.7019-7025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Salje J, van den Ent F, de Boer P, Lowe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143(Pt 10):3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:592–596. doi: 10.1073/pnas.0510047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.