Abstract
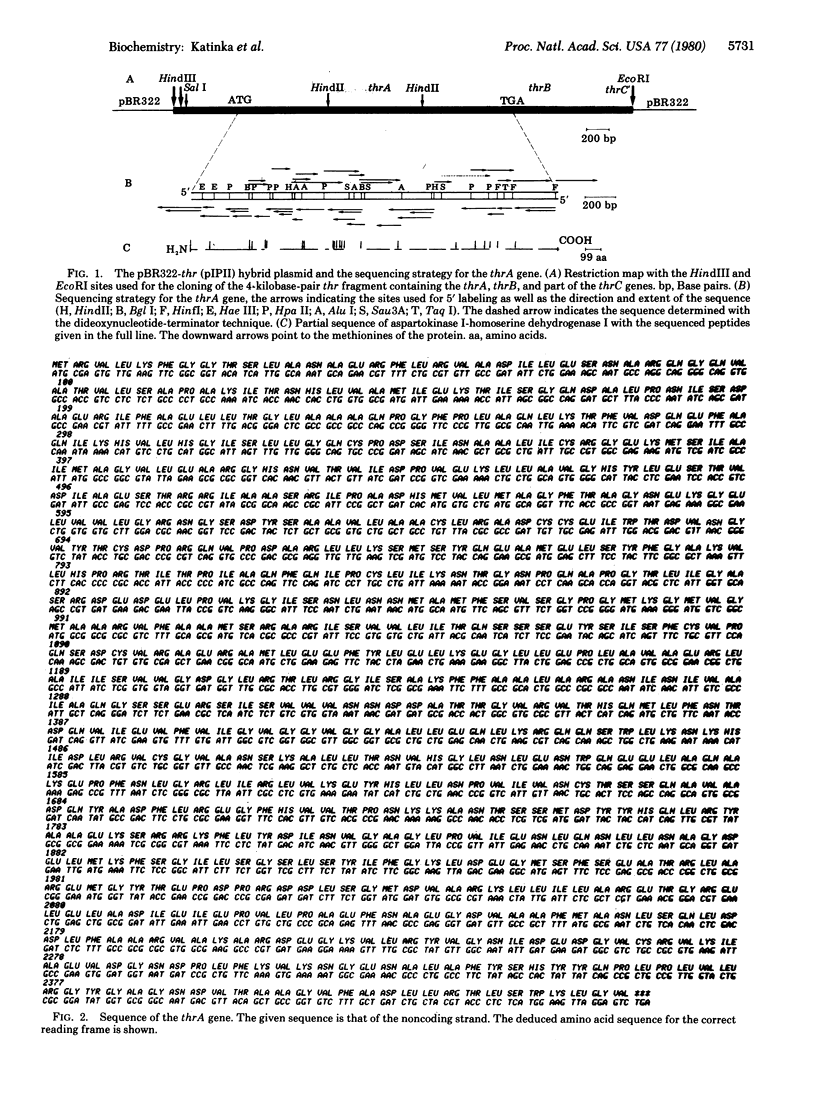
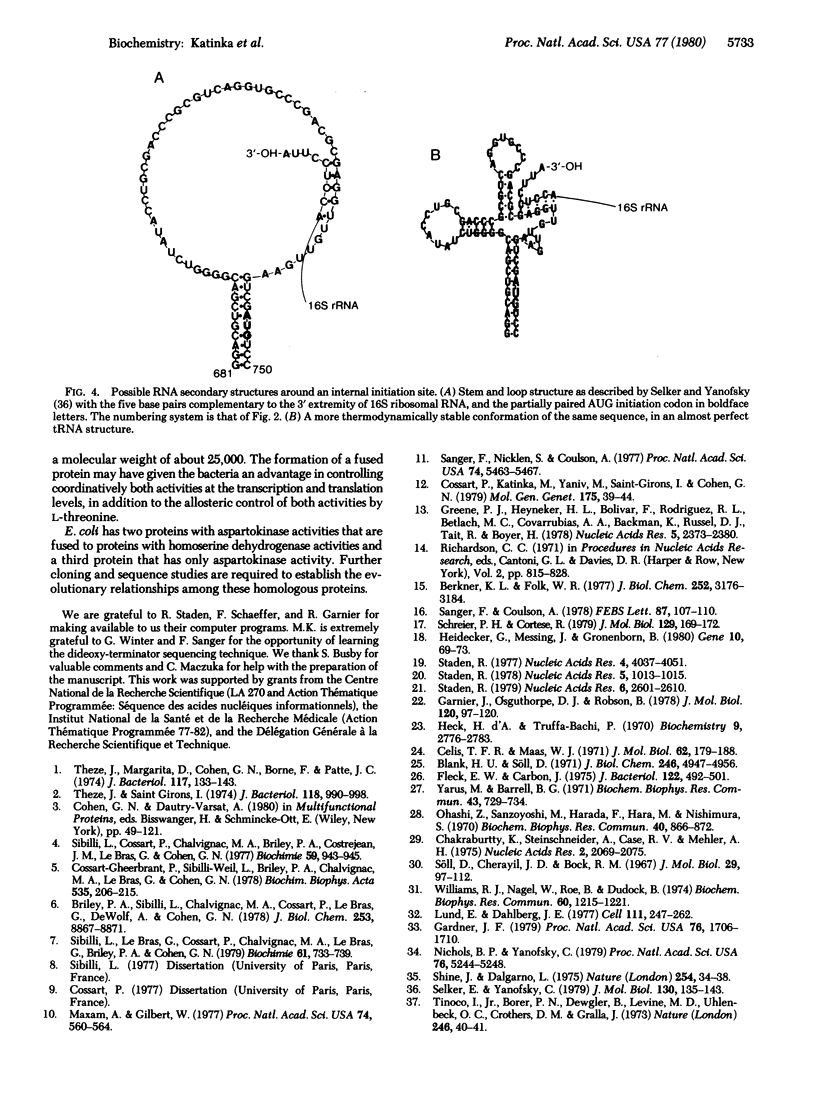
The thrA gene of Escherichia coli codes for a single polypeptide chain having two enzymatic activities required for the biosynthesis of threonine, aspartokinase I and homoserine dehydrogenase I. This gene was cloned in a bacterial plasmid and its complete nucleotide sequence was established. It contains 2460 base pairs that encode for a polypeptide chain of 820 amino acids. The previously determined partial amino acid sequence of this protein is in good agreement with that predicted from the nucleotide sequence. The gene contains an internal sequence that resembles the structure of bacterial ribosome-binding sites, with an AUG preceded by four triplets, each of which can be converted to a nonsense codon by a single mutation. This suggests that the single polypeptide chain was formed by the fusion of two genes and that initiation of translation may occur inside the gene to give a protein fragment having only the homoserine dehydrogenase activity.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Blank H. U., Söll D. Purification of five leucine transfer ribonucleic acid species from Escherichia coli and their acylation by heterologous leucyl-transfer ribonucleic acid synthetase. J Biol Chem. 1971 Aug 25;246(16):4947–4950. [PubMed] [Google Scholar]
- Briley P. A., Sibilli L., Chalvignac M. A., Cossart P., Le Bras G., De Wolf A., Cohen G. N. The primary structure of Escherichia coli K12 aspartokinase I-homoserine dehydrogenase I. Site of limited proteolytic cleavage by subtilisin. J Biol Chem. 1978 Dec 25;253(24):8867–8871. [PubMed] [Google Scholar]
- Celis T. F., Maas W. K. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. IV. Further studies on the role of arginine transfer RNA repression of the enzymes of arginine biosynthesis. J Mol Biol. 1971 Nov 28;62(1):179–188. doi: 10.1016/0022-2836(71)90138-0. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K., Steinschneider A., Case R. V., Mehler A. H. Primary structure of tRNA-Lys of E. coli B. Nucleic Acids Res. 1975 Nov;2(11):2069–2075. doi: 10.1093/nar/2.11.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart-Gheerbrant P., Sibilli-Weill L., Briley P. A., Chalvignac M. A., Le Bras G., Cohen G. N. The primary structure of Escherichia coli K12 aspartokinase I-homoserine dehydrogenase I-Isolation and characterisation of the peptides produced by cyanogen bromide. Biochim Biophys Acta. 1978 Aug 21;535(2):206–215. doi: 10.1016/0005-2795(78)90086-7. [DOI] [PubMed] [Google Scholar]
- Cossart P., Katinka M., Yaniv M., Saint Girons I., Cohen G. N. Construction and expression of a hybrid plasmid containing the Escherichia coli thrA and thrB genes. Mol Gen Genet. 1979 Aug;175(1):39–44. doi: 10.1007/BF00267853. [DOI] [PubMed] [Google Scholar]
- Fleck E. W., Carbon J. Multiple gene loci for a single species of glycine transfer ribonucleic acid. J Bacteriol. 1975 May;122(2):492–501. doi: 10.1128/jb.122.2.492-501.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1706–1710. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck H d'A, Truffa-Bachi P. Circular dichroic and optical rotatory dispersion spectra of the threonine-inhibited aspartokinase-homoserine dehydrogenase of Escherichia coli K 12. Effects of ligand binding and protein denaturation. Biochemistry. 1970 Jul 7;9(14):2776–2783. doi: 10.1021/bi00816a004. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Spacer transfer RNAs in ribosomal RNA transcripts of E. coli: processing of 30S ribosomal RNA in vitro. Cell. 1977 Jun;11(2):247–262. doi: 10.1016/0092-8674(77)90042-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. P., Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oashi Z., Saneyoshi M., Harada F., Hara H., Nishimura S. Presumed anticodon structure of glutamic acid tRNA from E. coli: a possible location of a 2-thiouridine derivative in the first position of the anticodon. Biochem Biophys Res Commun. 1970 Aug 24;40(4):866–872. doi: 10.1016/0006-291x(70)90983-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Selker E., Yanofsky C. Nucleotide sequence of the trpC-trpB intercistronic region from Salmonella typhimurium. J Mol Biol. 1979 May 15;130(2):135–143. doi: 10.1016/0022-2836(79)90422-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Sibilli L., Cossart P., Chalvignac M. A., Briley P. A., Costrejean J. M., Le Bras G., Cohen G. N. The primary structure of Escherichia coli K12 aspartokinase I-homoserine dehydrogenase I. Distribution of the methioninyl residues and of the cysteinyl and tryptophanyl tryptic peptides. Biochimie. 1977;59(11-12):943–946. doi: 10.1016/s0300-9084(78)80712-3. [DOI] [PubMed] [Google Scholar]
- Sibilli L., Le Bras G., Cossart P., Chalvignac M. A., Le Bras G., Briley P. A., Cohen G. N. The primary structure of Escherichia coli K 12 aspartokinase I-homoserine dehydrogenase I : sequence of cyanogen bromide peptide CB 3. Biochimie. 1979;61(5-6):733–739. doi: 10.1016/s0300-9084(79)80174-1. [DOI] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D., Cherayil J. D., Bock R. M. Studies on polynucleotides. LXXV. Specificity of tRNA for codon recognition as studied by the ribosomal binding technique. J Mol Biol. 1967 Oct 14;29(1):97–112. doi: 10.1016/0022-2836(67)90183-0. [DOI] [PubMed] [Google Scholar]
- Thèze J., Margarita D., Cohen G. N., Borne F., Patte J. C. Mapping of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. J Bacteriol. 1974 Jan;117(1):133–143. doi: 10.1128/jb.117.1.133-143.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thèze J., Saint-Girons I. Threonine locus of Escherichia coli K-12: genetic structure and evidence for an operon. J Bacteriol. 1974 Jun;118(3):990–998. doi: 10.1128/jb.118.3.990-998.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Williams R. J., Nagel W., Roe B., Dudock B. Primary structure of E. coli alanine transfer RNA: relation to the yeast phenylalanyl tRNA synthetase recognition site. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1215–1221. doi: 10.1016/0006-291x(74)90328-3. [DOI] [PubMed] [Google Scholar]
- Yarus M., Barrell B. G. The sequence of nucleotides in tRNA Ile from E. coli B. Biochem Biophys Res Commun. 1971 May 21;43(4):729–734. doi: 10.1016/0006-291x(71)90676-0. [DOI] [PubMed] [Google Scholar]


