Abstract
MAGI-1, a member of the MAGUK family of proteins, is shown to be rapidly cleaved during Fas-induced apoptosis in mouse 3T3 A31 cells, and in UV irradiation- and staurosporine-induced apoptosis in HaCaT cells. This generates a 97 kDa N-terminal fragment that dissociates from the cell membrane; a process that is largely prevented in the presence of the caspase inhibitor Z-VAD-fmk. In addition, we show that in vitro translated radiolabelled MAGI-1 is efficiently cleaved into 97 kDa and 68 kDa fragments by caspases-3 and -7 at physiological concentrations and mutating the MAGI-1 Asp761 to Ala completely abolished the caspase-induced cleavage. Moreover, in HaCaT cells over-expressing the MAGI-1 Asp761Ala mutant the disruption of cell-cell contacts was delayed during apoptosis, whereas other caspase-dependent processes such as nuclear condensation were not affected, suggesting that cell detachment is parallel to them. Thus, MAGI-1 cleavage appears to be an important step in the disassembly of cell-cell contacts during apoptosis.
Keywords: Apoptosis, Caspase, MAGI-1, PDZ, Tight junction
Introduction
Apoptosis or programmed cell death is a general mechanism used by all multicellular organisms to remove excessive, infected, damaged or potentially dangerous cells during development or homeostasis and also in a number of diseases [1, 2]. Cysteine proteases from the caspase family play a major role in apoptosis. They cleave a set of cellular proteins exclusively after an Asp residue, being thus unique among proteases [3–5], and they contribute to the typical morphological changes seen during apoptosis.
A major step in apoptosis is the dismantling of cell-cell contacts; thus proteins that are involved in the formation of these contacts represent potential targets for caspases. Several of them have already been shown to be cleaved by caspases, among them: β-catenin, plakoglobin [6–8], α-fodrin [9], actin [10], gelsolin [11], E-cadherin [12], focal adhesion kinase [13], desmosomal plaque proteins [14], the tight junction proteins ZO-1, ZO-2 and occludin, [15] and also hDLG, a constituent of adherens junctions in epithelial and endothelial cells [16].
The PDZ domain-containing proteins are thought to play a major role in the organization of protein-protein complexes at plasma membranes and they are typically associated with cell junctions, including synapses of the central nervous system [17, 18]. The best known and most abundant are the MAGUK (membrane associated guanylate kinase) proteins which are characterized by modular arrangements of structural domains, and consist of one or more PDZ domains, an SH3 or WW domain and an inactive guanylate kinase (GuK) homology domain, all of which can act as protein-protein interaction domains. MAGUKs are localised at regions of cell-cell contact, such as tight junctions in epithelial cells and synaptic junctions in neurons, where they are believed to function as molecular scaffolds in the formation of multimolecular (for reviews see [19–22]).
Our goal was to elucidate the role of MAGI-1 protein (membrane associated guanylate kinase inverted), a member of the MAGUK family with a wide tissue distribution [23, 24], in dismantling cell-cell contacts during apoptosis. MAGI-1 has been shown to colocalise with ZO-1 at cell tight junctions [25] where it interacts with β-catenin [26], Rap GEP [27], synaptopodin [27], alpha-actinin-4 [28] and JAM4 [28]. We show here that the cleavage of MAGI-1 is an important and widely found step in the dismantling of cell-cell contacts during the early stages of apoptosis.
Materials and methods
Plasmids
For in vitro expression the MAGI-1 gene was cloned into the HindIII/EcoRI sites of pCDNA3 [30]. The NT-MAGI-1 was cloned by excising a HindIII/ApaI fragment from HA tagged MAGI-1 in pGWI [30] and ligating it into pCDNA3(+). The CT-MAGI-1 was constructed by similarly excising an ApaI/EcoRI fragment from pGWI-HA-MAGI-1 and ligating it into pCDNA3(−). The first methionine residue is at position 780. The mutant MAGI-1 Asp761Ala was constructed using overlap polymerase chain reaction on NT-MAGI-1 cDNA as template using 5′-TACATGCTCCTGGACTGG-3′ primer and 5′-CGACTCACTATAGGGAGA-3′ and 5′-GACACTATAGAATAGGGC-3′ as forward and reverse primers, respectively. The HindIII/ApaI N-terminal fragment was ligated to the ApaI/EcoRI C-terminal fragment and then cloned into HindIII/EcoRI sites of pCDNA3. The sequences were verified by DNA sequencing using an Abi Prism 310 automated DNA sequencer (Perkin Elmer, USA). For GFP fusions wild type and mutant MAGI-1 cDNA were amplified using 5′-CTGCAAGCTTATGTCGAAAGT-3′ and 5′-ACTGGAATTCGATGCTGAGG-3′ as forward and reverse primers, respectively, and cloned into the HindIII/EcoRI sites of pEGFP-N2 vector (Clonetech, USA).
Caspase expression
Recombinant caspases-3, -6 and -7 were expressed in E. coli and purified as described previously [31]. Protein concentrations were determined by absorbance measurements according to Pace et al. [32]. Prior to other experiments, all three caspases were active site titrated using Z-VAD-fmk (Bachem, Bubbendorf, Switzerland) as described [32].
Tissue culture and apoptosis induction
Mouse 3T3 A31 and human HaCaT cells were grown in DMEM plus 10% Foetal Calf Serum (FCS). The day prior to induction of apoptosis, the cells were plated onto 90 mm dishes so as to be approximately 50% confluent. The Fas apoptotic pathway was induced by treating the cells with 10 μg/ml hamster anti-mouse Fas antibody (Pharmingen, San Diego, USA). UV irradiation-induced apoptosis was initiated by removing the growth medium, exposing the cells to 200 J/s of UVC light and then incubating them in growth medium containing 1% FCS. Staurosporine-induced apoptosis was initiated by treating the cells with 140 nM staurosporine (Sigma, Germany). Inhibition of caspases in vivo was performed by including Z-VAD-fmk (Calbiochem) at 50μM at the same time as the induction of apoptosis.
Western blotting
Cells were harvested as described previously [33] at various times post induction of apoptosis, as indicated in the text. Levels of MAGI-1 protein, β-catenin protein and caspase cleavage products were then determined by western blotting and development with ECL according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Uppsala, Sweden). MAGI-1 was detected using a rabbit polyclonal antibody raised against the highly conserved WW region of MAGI-1 (amino acid residues 300–380) fused to GST, β-catenin was detected using a mouse monoclonal antibody from Santa Cruz (USA, clone E-5), whereas E-cadherin was detected using a mouse monoclonal antibody from Becton Dickinson (USA).
Fluorescence microscopy
3T3 A31 cells were grown overnight on coverslips before incubation with anti-Fas antibody (10 μg/ml) and protein G (2 μg/ml; Sigma, Germany) for 0, 1 and 2 h. The cells were then washed with PBS and fixed in 4% paraformaldehyde in PBS for 10 min, followed by permeabilisation with 0.2% (v/v) Triton X-100 in PBS for 5 min. After extensive washing with PBS the cells were incubated with anti MAGI-1 antibody diluted 1/50 in PBS and anti β-catenin antibody diluted 1/100 in PBS for 90 min at 37°C. Following an additional round of extensive washing with PBS the cells were incubated either with goat anti-rabbit rhodamine-conjugated antibody or with goat anti-mouse fluorescein-conjugated antibody (Molecular Probes, USA) for 60 min at 37°C. After this time the cells were washed in PBS, mounted on slides and visualised by confocal microscopy using a Zeiss Axiovert 100 M microscope and LSM510 software (Zeiss, Germany).
HaCaT cells were transfected with C-terminally GFP tagged MAGI-1 constructs, using the Nucleofector™ device (Amaxa, USA) according to manufacturer’s instructions, and seeded on coverslips. Apoptosis was induced 24 h post transfection with UV irradiation. Three hours later, cells were collected and treated as described above, except that the monoclonal anti E-cadherin antibodies (Pharmingen) and Alexa546 conjugated anti-mouse antibodies (Molecular Probes) were used. In addition, DAPI (Sigma, Germany) was used to stain the nuclei. Morphological changes were monitored using fluorescence microscopy (Olympus IX71, Japan).
FACS analysis
The mouse 3T3 A31 cells were harvested 5 h after treatment with anti-Fas antibody, and the human HaCaT cells were harvested at times indicated after UV irradiation. The cells were washed and trypsinised, as for normal passaging, but culture medium supernatant and PBS washes were also retained to ensure that both floating and adherent cells were analysed. After being washed once in DMEM/10% foetal calf serum and once in PBS the cells were incubated for 15 min, with PE-conjugated Annexin V (Becton Dickinson, USA) according to the manufacturer’s instructions. The cells were then subjected to FACS analysis using a FACScalibur flow cytometer (Becton Dickinson, USA) and CellQuest software.
In vitro translation of proteins
For in vitro expression the genes of MAGI-1 and its mutants were cloned in pCDNA3 (Invitrogen, USA). Proteins were expressed using the Promega TNT coupled transcription-translation system, and were radiolabelled with [35S]-cysteine (Amersham Pharmacia Biotech, Uppsala, Sweden). Efficiency of translation was monitored by analysing 1 μl of the translate by SDS-PAGE and phosphorimaging.
Caspase cleavage assays
Caspase cleavage assays were performed in 20 mM Hepes buffer, pH 7.2, containing 100 mM NaCl, 10 mM dithiothreitol, 1 mM EDTA, 0.1 (w/v) CHAPS and 10% (w/v) sucrose at 30°C [27]. Briefly, the caspases were incubated for 5 min in the reaction buffer at 30°C prior to the addition of 4 μl of MAGI-1 translation product to the final volume of 25 μl. After an appropriate time, the reaction was terminated by the addition of 10 μl of SDS-loading buffer and boiling. The reaction mixtures were analyzed by SDS-PAGE and autoradiography. Final concentration of the caspases was 1 μM throughout if not otherwise stated.
Results
MAGI-1 is cleaved by caspases during Fas-induced apoptosis
Fas-induced apoptosis is one of the best characterised of the apoptotic pathways; therefore 3T3 A31 cells were chosen, since they have readily detectable levels of MAGI-1 protein and are also susceptible to apoptosis induced by commercially available anti-Fas antibody. The cells were harvested at different times after the addition of 10 μg/ml anti-Fas antibody, and the level of MAGI-1 protein was followed by western blot analysis. The results obtained are shown in Fig. 1(A). An increase in an apparent cleavage product of MAGI-1, migrating at approximately 97 kDa, can be clearly seen within 1 h of adding the anti-Fas antibody. This cleavage is completely abolished in the presence of 50 μM Z-VAD-fmk, which indicates that MAGI-1 is most probably cleaved by caspases. Since the anti-MAGI-1 antibody is raised against the highly conserved WW domain of MAGI-1 (amino acid residues 300–380), this suggests that the 97 kDa cleavage product comprises the amino terminal half of the MAGI-1 protein (see below). For comparison, β-catenin, which is a MAGI-1 ligand [26] and a known caspase target [6] was also followed during apoptosis. The first cleavage products were also observed an hour after induction of apoptosis (Fig. 1(B)), suggesting that β-catenin cleavage occurs at approximately the same time as MAGI-1 cleavage.
Fig. 1. Fas-induced apoptosis in 3T3 A31 cells: time course of MAGI-1 and β-catenin cleavage and apoptosis quantification.
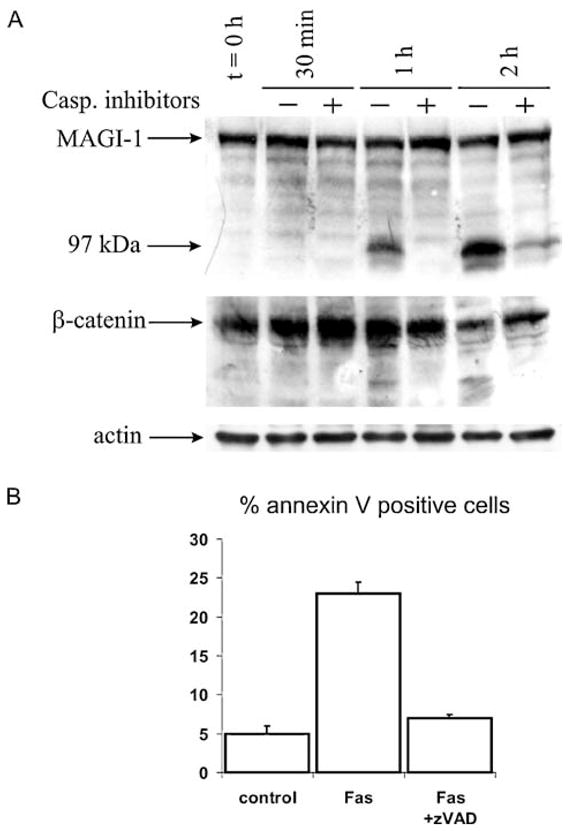
(A) Experiments were performed in the absence (−) or presence (+) of 50 μM Z-VAD-fmk. Cells were harvested at indicated time points post induction of apoptosis by anti-Fas antibody. Following SDS polyacrylamide gel electrophoresis, samples were analyzed by Western blot using an antibody raised against the WW domains of MAGI-1 or an antibody against β-catenin (Santa Cruz). The full length MAGI-1c isoform is arrowed together with the 97 kDa cleavage product. Actin was used as a loading control. (B) Quantification of apoptosis by FACS analysis 1 and 2 h post induction of apoptosis. Experimental details were as described under Materials and methods
In a further experiment, 3T3 A31 cells were tested for exposure of phosphatidylserine, which is another marker of apoptosis [34, 35], by flow cytometry. Annexin V staining (Fig. 1(B)) shows a significant number of cells entering apoptosis 2 hrs after the addition of the anti-Fas antibody, and this is greatly decreased in the presence of Z-VAD-fmk. Caspase activity was measured as described previously [36] in order to verify that cells were indeed apoptotic. A significant caspase activation (~20-fold), based on DEVD-ase activity, which could be completely abolished by z-VAD-fmk was observed (data not shown).
Changes in MAGI-1 localisation during Fas-induced apoptosis
Next, potential changes in MAGI-1 localisation during this process were investigated. Previous studies had demonstrated that MAGI-1 localises to regions of cell-cell contact, and it was reasoned that MAGI-1 cleavage could be a means of releasing cell contact during apoptosis. To investigate this, immunofluorescence analyses of MAGI-1 and, for comparison, β-catenin were performed at 1 h and 2 h after the addition of anti-Fas antibody to the 3T3 A31 cells. The results of these analyses are shown in Fig. 2. It can be seen that both MAGI-1 and β-catenin exhibit a similar pattern of expression at the zero time point, being localised to the cell membrane, particularly at regions of cell-cell contact, consistent with previous studies [6, 26]. It can also be seen that at the 2 h time point the β-catenin remains largely localised on the cell membrane. In contrast, the MAGI-1 localisation alters dramatically, with much more of the protein being detected within the cytoplasm of the apoptotic cells. Since the anti-MAGI-1 antibody is raised against the amino terminal portion of the protein and can detect the 97 kDa cleavage product, these results suggest that the 97 kDa N-terminal product is released from the membrane upon caspase cleavage.
Fig. 2. Immunolocalisation of MAGI-1 and β-catenin during Fas induced apoptosis in 3T3 A31 cells.
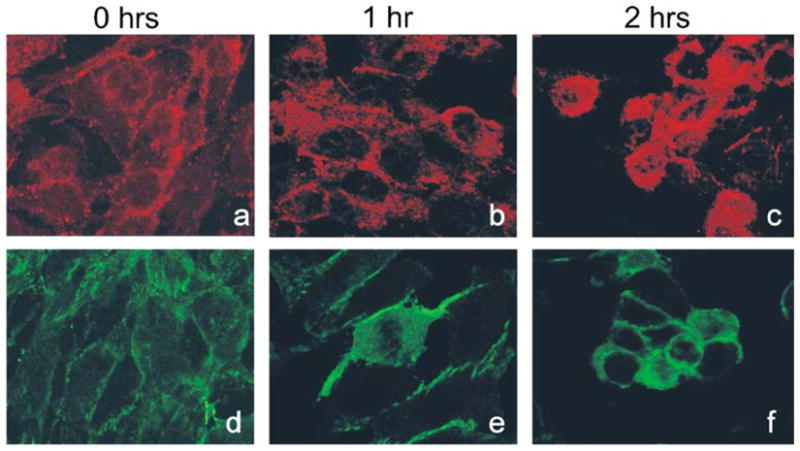
Experimental conditions were as described under Materials and methods. The panels show MAGI-1 (upper panel) at 0 h (a), 1 h (b) and 2 h (c) post addition of anti Fas antibody and β-catenin (lower panel) at 0 h (d), 1 h (e) and 2 h (f) post addition of anti Fas antibody
MAGI-1 is cleaved by caspases during UV irradiation-and staurosporine-induced apoptosis
To test whether MAGI-1 cleavage by caspases is a conserved feature of apoptosis, found in different cells and in response to different stimuli, we investigated UV-induced apoptosis in HaCaT epithelial cells. The cells were subjected to UV irradiation and then harvested at different time points; to investigate caspase involvement, some samples were pre-treated with 50 μM caspase inhibitor. The MAGI-1 protein was then detected by western blot analysis and the results obtained are shown in Fig. 3(A). It can be seen that the MAGI-1 protein was again cleaved to the 97 kDa product following UV irradiation, with readily detectable levels being visible 4 hrs post treatment. In addition, the MAGI-1 cleavage was prevented in the presence of caspase inhibitor. FACS analysis was used to confirm that apoptosis had indeed been induced by the UV irradiation and the results obtained are shown in Fig. 3(B). As can be seen, the number of annexin V positive cells increased and reached 41% after 24 h, while addition of Z-VAD-fmk rescued cells from apoptosis. Similarly, caspase activity (DEVD-ase activity) was largely increased (~ 10-fold after 16 h; data not shown). Cleavage of E-cadherin, which is an important player in establishing and maintaining cell-cell contacts (for review [37]), and which is a known caspase target [12], was also followed during UV irradiation-induced apoptosis. The first cleavage products were also observed 4 h post induction of apoptosis (Fig. 3(C)), suggesting that E-cadherin cleavage occurs at about the same time as MAGI-1 cleavage.
Fig. 3. MAGI-1 and E-cadherin are cleaved during UV irradiation-induced apoptosis in HaCaT cells.
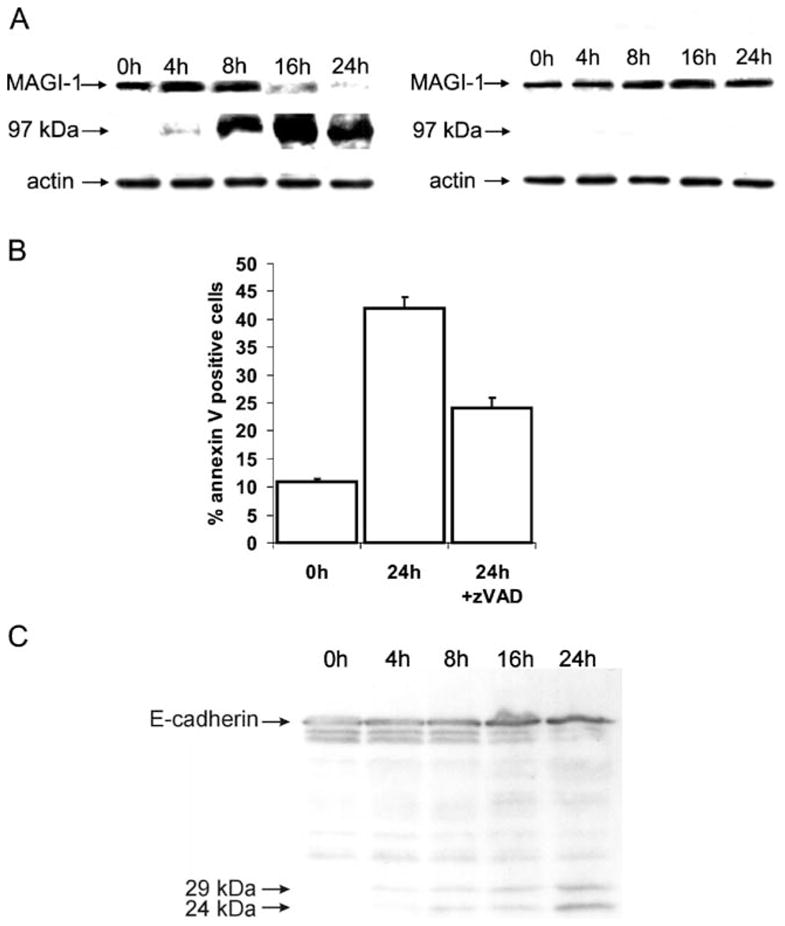
(A) Cells were harvested at indicated time points and Western blot analysis of MAGI-1 protein was performed as described in Material and methods. Experiments were performed in the absence (left panel) or presence (right panel) of 50 μM caspase inhibitor Z-VAD-fmk. The position of MAGI-1 and the 97 kDa cleavage product are indicated by the arrows. (B) Quantification of apoptosis by FACS analysis based on Annexin V staining. Analysis was done using the CellQuest software. Samples were taken at times indicated. (C) Western blot analysis of E-cadherin cleavage during UV irradiation-induced apoptosis. Cells were harvested at times indicated. The position of E-cadherin and its cleavage products are indicated by arrows
Since it was possible that UV irradiation-induced apoptosis might also induce the extrinsic cell death pathway, we chose to investigate MAGI-1 cleavage in HaCat cells following treatment with the protein kinase C inhibitor staurosporine, which primarily triggers the intrinsic caspase activation pathway, leading to mitochondrial cytochrome c release and subsequent caspase-9 and caspase-3 activation. Cells were treated with 140 nM staurosporine, both in the presence and absence of Z-VAD-fmk, and then harvested at different time points. The MAGI-1 protein was detected by western blot analysis and the results obtained are shown in Fig. 4(A). It is clear that MAGI-1 is again cleaved to the 97 kDa product following staurosporine treatment, with readily detectable levels being visible 6 h post addition of staurosporine. In addition, the MAGI-1 cleavage is greatly reduced in the presence of Z-VAD-fmk, even at the later time-points of the assay. FACS analysis was also performed on the cells at the 18 h time point and the results obtained are shown in Fig. 4(B). As can be seen, there is a large number of cells entering apoptosis, as determined by Annexin V staining, and this number is only slightly reduced following treatment with caspase inhibitor. However, caspase activity, which was significantly increased after 18 h (~ 15-fold), was completely ablated in the presence of Z-VAD-fmk (not shown). Taken together these results demonstrate that MAGI-1 is also cleaved by caspases during staurosporine-induced apoptosis in HaCat cells.
Fig. 4. MAGI-1 cleavage during staurosporine-induced apoptosis in HaCat cells.
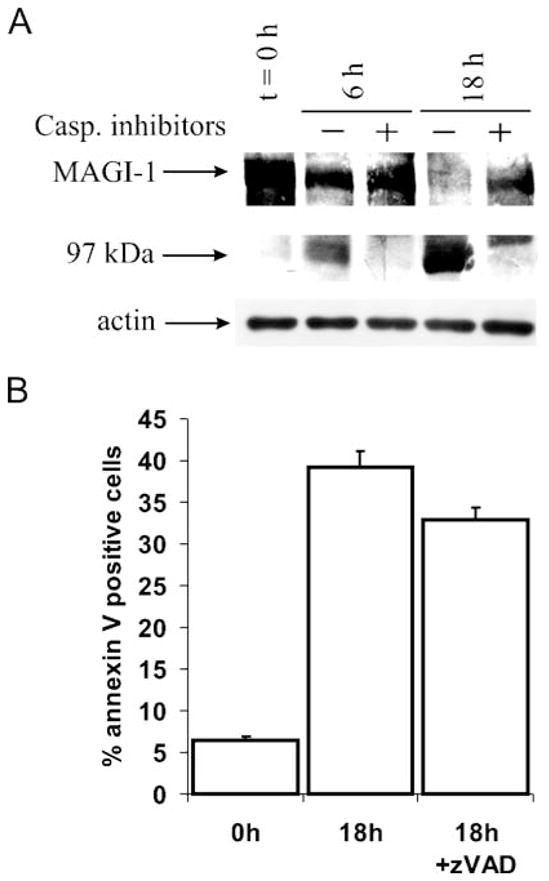
(A) Cells were harvested at indicated time points and Western blot analysis of MAGI-1 protein was performed as described in Fig. 1. Experiments were performed in the absence (−) or presence(+) of 50 μM concentration of Z-VAD-fmk. The position of MAGI-1c and the 97 kDa cleavage product are indicated by the arrows. (B) Quantification of apoptosis by FACS analysis 18 h post induction of apoptosis. Analysis was done using the CellQuest software
In vitro analysis and identification of the caspases responsible for MAGI-1 cleavage
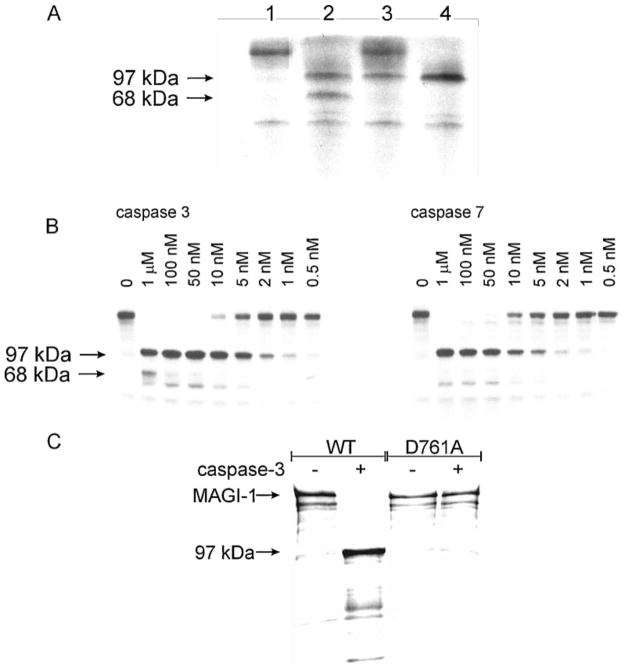
To investigate which of the caspases is responsible for the MAGI-1 cleavage, in vitro translated MAGI-1 protein, radiolabelled with [S35]-Cys, was incubated for 60 min in the presence of the executioner caspases-3, -6 and -7, and then analysed by SDS-PAGE on 15% polyacrylamide gel to allow detection of any smaller degradation products. As can be seen in Fig. 5(A), caspases-3 and -7 efficiently cleaved MAGI-1, whereas the cleavage by caspase-6 was incomplete. A major band, migrating at ~ 97 kDa was observed with all three caspases, consistent with the in vivo data on 3T3 A31 and HaCaT cells, probably representing the N-terminal fragment (see above). In addition, another band, migrating at ~ 68 kDa, was observed in the presence of caspase-3, indicating the presence of an additional cleavage site for caspase-3 in MAGI-1. In additional experiments, various concentrations of caspase-3 and -7 were used to determine whether these cleavage fragments could be generated under physiological conditions. As can be seen from Fig. 5(B), both caspase-3 and -7 can efficiently cleave MAGI-1 at concentrations as low as 5 nM. It can also been seen that the 68 kDa fragment is generated by a subsequent cleavage of the 97 kDa fragment and is not present if the concentration of caspase-3 is below 10 nM, thus raising the question of whether this product would be generated under physiological conditions (Fig. 5(B)). These results demonstrate that caspase-3 and caspase-7 at physiological concentrations can cleave MAGI-1 in vitro to give rise to the 97 kDa product, equivalent to that seen in vivo.
Fig. 5. Cleavage of MAGI-1 by caspases-3, -6 and -7 in vitro.
(A) Radiolabelled full-length MAGI-1 protein was incubated for 1 h with 1μM caspase-3 (lane 2), caspase-6 (lane 3) or caspase-7 (lane 4). Lane 1, control experiment in the absence of caspases. (B) Radiola-belled MAGI-1 was incubated for 1 h with decreasing concentrations of caspase-3 (right panel) or caspase-7 (left panel) and protein bands were visualized by autoradiography. In control experiments caspase was omitted. (C) Radiolabelled WT and MAGI-1 Asp761Ala protein were incubated for 1 h with (+) or without (−) 1 μM caspase-3 and protein bands visualized by autoradiography
The major caspase cleavage site in MAGI-1 is D758QTD761 ↓ S
A detailed PDZ deletion analysis of MAGI-1 revealed that the major caspase cleavage site is located between the second and the third PDZ domain (not shown), pointing to D758QTD761 S as the likely candidate for the cleavage site. To confirm that this is indeed the case, a MAGI-1 mutant was constructed in which Asp761 was replaced by an alanine. Both the wild type MAGI-1 and MAGI-1 Asp761Ala were incubated with caspase-3, and, as can be seen in Fig. 5(C), no cleavage of the MAGI-1 Asp761Ala mutant was observed, indicating that Asp761 Ser762 is the true caspase cleavage site.
Cleavage of MAGI-1 is required for normal cell-cell detachment
To evaluate the role of MAGI-1 cleavage during apoptosis we tried to generate HaCaT and 3T3 A31 cell lines stably overexpressing wild-type and MAGI-1 Asp761Ala proteins, either alone or C-terminally fused to EGFP. However, none of the numerous attempts was successful. Use of the nucleofector technology (Amaxa) allowed up to ~ 70% efficiency of transient transfection in HaCaT cells, as judged by transfection of GFP (data not shown). However, most of the cells died within 24 h of transfection with either wild-type or mutant MAGI-1, suggesting that high levels of MAGI-1 are not tolerated by either of the two cell lines used (not shown). Therefore, the transfection procedure was optimized to result in ≤ 10% MAGI-1-transfected HaCaT cells, which was the highest level tolerated. Using these transfected cells we first confirmed the membrane localisation of wild type MAGI-1 and MAGI-1 Asp761Ala, both C-terminally fused to EGFP (Fig. 6(A)). Next, the effect of the mutation on the progression of UV-induced apoptosis was evaluated. Within 4 h of the induction of apoptosis wt-MAGI-1 was seen to be cleaved and translocated to the cytosol in the transfected cells (white and yellow arrowheads). However, E-cadherin was only cleaved in cells that also exhibited nuclear condensation and fragmentation (Fig. 6(B), upper panel, white arrowheads), suggesting that MAGI-1 cleavage is an earlier event in apoptosis than E-cadherin cleavage. In contrast, in cells overexpressing MAGI-1 D761A, both the MAGI-1 and E-cadherin proteins retained their submembraneous localisation even after the first signs of nuclear condensation became visible (Fig. 6(B), lower panel, white arrow).
Fig. 6. Fluorescence imaging of HaCaT cells overexpressing WT and MAGI-1 Asp761Ala/EGFP fusion proteins.
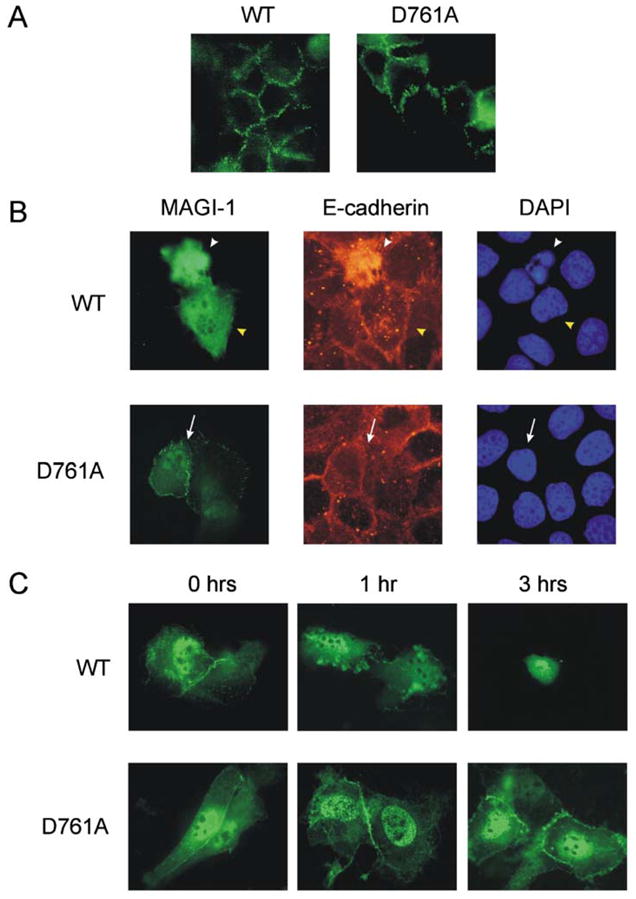
(A) HaCaT cells were transfected with C-terminally GFP tagged WT or D761A mutant MAGI-1 and examined by fluorescent microscopy for their localisation. Green fluorescence is visible at the sites of cell-cell contacts. (B) HaCaT cells were transfected with C-terminally GFP tagged WT or D761A mutant MAGI-1 and seeded on glass coverslips. Apoptosis was induced 24 h post transfection with UV irradiation. Pictures were taken four hours after apoptosis induction. (C) HaCaT cells were transfected with C-terminally GFP tagged WT or D761A mutant MAGI-1 and seeded on glass coverslips. Apoptosis was induced with 500 nM staurosporine and pictures were taken at times indicated
Owing to the different times of apoptosis onset after UV irradiation, and its quick progression thereafter, it was difficult to find cells that were at a comparable stage of apoptosis to observe and quantify any differences between wild-type-transfected and mutant MAGI-1-transfected cells. Typically, some 500 transfected cells at a similar stage of apoptosis could be observed on one slide (i.e. approximately 50,000 cells per slide, about 10% of which were successfully transfected). By morphological criteria, about 10% of the transfected cells (500 ± 50) were estimated to be apoptotic, which agreed well with the number of annexin V positive cells determined by flow cytometry.
In cells transfected with MAGI-1 D761A there were approximately 50 cells per slide (51 ± 3 from a triplicate experiment) in which the membrane localisation of MAGI-1 and E-cadherin was preserved, and nuclear condensation was readily detectable. In contrast, in cells overexpressing GFP alone or MAGI-1 WT there were similar numbers of cells (49 ± 5 from a triplicate experiment) in which membrane localisation of MAGI-1 was lost and the protein could only be detected in the cytosol prior to any detectable nuclear condensation. However, even in the case of the MAGI-1 D761A-expressing cells, considerable numbers of shrunken cells with fragmented or condensed nuclei and cytosolic MAGI-1 were present, suggesting that the prevention of MAGI-1 cleavage cannot block cell-detachment downstream of caspase activation. These results further suggest that cell-cell detachment is independent of nuclear fragmentation in apoptosis. This was corroborated by observation of staurosporine-induced apoptosis in which cell-cell contacts in HaCaT cells overexpressing the D761A mutant were preserved substantially longer than in cells overexpressing wt-MAGI-1 (Fig. 6(C)). From this we concluded that the cleavage of MAGI-1 is required for proper detachment of the cells.
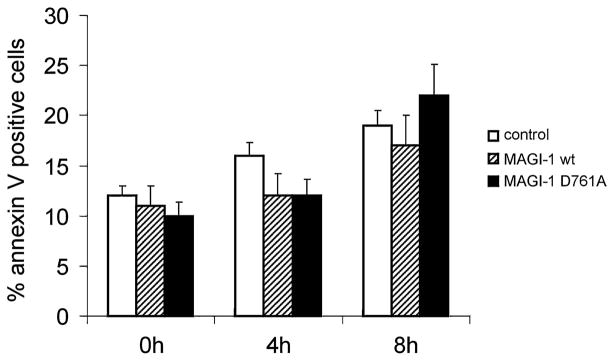
In order to investigate whether PS exposure was also independent of cell-cell detachment, the same experiment in HaCaT cells (see above) was repeated. PS exposure was then analysed by flow cytometry, using the green fluorescence of EGFP as the detection threshold to ensure that only transfected cells were analysed. No difference was observed in the levels of PS exposure between the cells overexpressing wt MAGI-1 or MAGI-1 D761A (Fig. 7), suggesting that PS exposure is also independent of cell-cell detachment.
Fig. 7. Quantification of apoptotic cells after apoptosis induction by UV.
PS exposure was measured after apoptosis induction at times indicated in nontransfected and in WT or D761A MAGI-1 transfected HaCaT cells (open, striped and closed bars, respectively). All other experimental details are described in Materials and methods
Discussion
Tight junctions represent the most apical intercellular junctions of epithelial and endothelial cells. They are known to form semi-permeable diffusion barriers between individual cells, and to act as a fence to prevent mixing of apical and basolateral cellular components, thus maintaining cell polarity. Recently their importance in cell-cell adhesion has also begun to emerge [38, 39]. Since a major feature of apoptosis is the gross structural changes which take place in the cell, including the loss of cell-cell contact, we reasoned that this could be brought about by disruption of the cell’s tight junctions during the apoptotic process. This is supported by an earlier study demonstrating that interruption of tight junctions between the neighbouring superficial cells in developing mouse urinary bladder was the first step of urothelial cell detachment during embryonic development and first urine accumulation in bladder lumen [40]. Similar results were also obtained by Bojarski and coworkers, who also observed cleavage of tight junction proteins ZO-1, ZO-2 and occludin, presumably by caspases, during apoptosis [15]. In addition, the adherens junction protein hDLG was also found to be cleaved during UV irradiation and staurosporine—induced apoptosis [16].
Several MAGUKs, such as ZO-1 [41], ZO-2 [42], ZO-3 [43], hDlg [44], MAGI-1 [23, 26], MAGI-2/S-SCAM [45] and MAGI-3 [46] are localised to cellular tight junctions. However MAGI-1 was chosen as the model MAGUK in this study because of its wide tissue distribution [23, 24]. We found that MAGI-1 protein is cleaved to a 97 kDa product by caspases in 3T3 A31 cells in the Fas model of apoptosis, and in HaCaT cells in UV irradiation- and staurosporine-induced apoptosis. Although the results from these apoptosis models cannot be directly compared, it is clear that MAGI-1 is a general caspase target, supporting the notion that MAGI-1 cleavage is a conserved process found both in different cells and in response to different apoptotic stimuli.
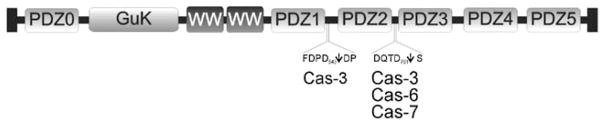
The caspases, like most highly specific proteases, generally cleave their substrates at only a limited number of sites [47, 48]. This is consistent with the generation of the large stable 97 kDa N-terminal MAGI-1 fragment. However, the C-terminal MAGI-1 fragment could not be followed during apoptosis in cell extracts due to the lack of specific antibodies. In vitro analyses were therefore performed in order to identify both the caspase(s) responsible for the cleavage and the cleavage site(s). All three caspases examined (-3, -6 and -7) were shown to cleave MAGI-1 at D758QTD761 S, generating the N-terminal fragment including the GuK domain, both WW domains and PDZ domains 0, 1 and 2, and the shorter C-terminal fragment including PDZ domains 3–5 (Fig. 8). This cleavage is, at least in the case of caspase-3, followed by further separation of the PDZ domain 2 from the remaining N-terminal fragment. It seems, however, that the first cleavage is necessary for the second one to take place, as the MAGI-1 Asp761Ala mutant is not cleaved by caspase-3 at all (Fig. 5(C)). Cleavage by caspase-6 was only partial in these assays, despite the very high concentration of the enzyme used in the experiment (1 μM), thus tending to eliminate its physiological relevance. This is consistent with other studies, since the only protein exclusively cleaved by caspase-6 in vivo found thus far is lamin A, suggesting that caspase-6 has a role in cleaving a very specific set of proteins [49]. The vast majority of caspase targets during apoptosis are, however, cleaved by caspase-3 (for review [4, 47, 48]). At first glance our data also point to caspase-3, and not caspase-7, as the primary executioner. There are several observations that support this idea: (i) caspase-3 activity is required for caspase-7 activation [50] and (ii) the generally higher concentrations of caspase-3 than caspase-7 in vivo. Although the last is not well defined, it has been shown, at least for the 293 human embryonic kidney cells, that intracellular caspase-3 concentration can be as high as 100–200 nM [51] and that of caspase-7 around 65 nM (H. Stennicke and G. Salvesen, personal communication). In addition, MAGI-2, a protein closely related to MAGI-1, was shown to be cleaved by caspase-3 in vitro, which supports our data [52]. Nevertheless, the possible involvement of caspase-7 cannot be formally excluded.
Fig. 8. Schematic representation of MAGI-1 and caspase cleavage site(s).
Diagram of MAGI-1 (grey bar) showing the positions of the guanylate kinase domain, two WW domains and 6 PDZ domains, and the positions and sequences of caspase cleavage sites
It is clear that a considerable amount of MAGI-1 protein is cleaved within two hours of Fas-induced apoptosis in 3T3 A31 cells. This parallels the appearance of apoptosis-related morphological changes, observed by FACS analysis of PS exposure where ~ 25% of the cells were entering apoptosis at the same time (Fig. 1(B)). Similar conclusions were also reached for staurosporine- and UV irradiation-induced apoptosis in HaCaT cells.
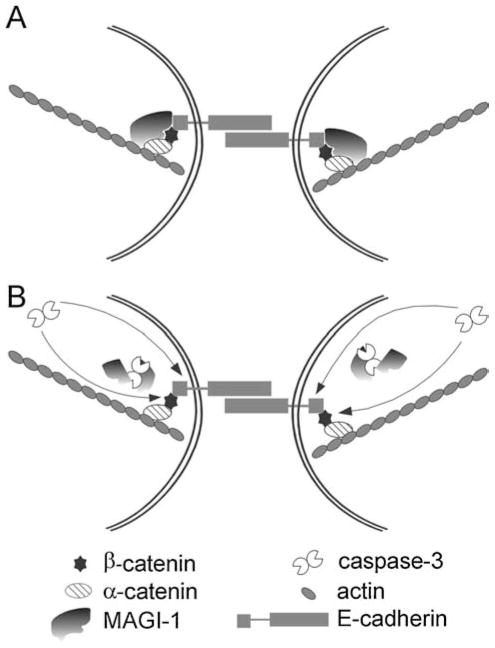
To understand the physiological consequences of MAGUK proteolysis one should consider the mechanisms of cell-cell adhesion (Fig. 9). Cells are linked together by organizing actin filaments to the plasma membrane. Transmembrane Ca2+-dependent adhesion receptors, cadherins, in particular E-cadherin, play an important role in this process by forming complexes with the actin cytoskeleton with the help of two cytoplasmic plaque proteins, α- and β-catenin. Whereas α-catenin links E-cadherin to the actin cytoskeleton, β-catenin links α-catenin with E-cadherin (for review [37]). The whole process is dynamic and involves sequential changes in E-cadherin and actin distribution [53]. In this zipper model, the driving force is directed actin polymerization, which seals the cell borders. However, a number of other proteins have been shown to be recruited to the cadherin-actin complexes [54]. They probably strengthen the adhesion by interacting with actin filaments and other proteins. This is a function for which MAGUKs are extremely well adapted: mediating such protein-protein interactions [17]. Indeed, several MAGUKs have been found to bind to components of the cadherin-actin system [55–59] and it has also been suggested that they link the tight junction constituents, occludin and claudins, with actin filaments [39]. MAGI-1 itself has been shown to localise to tight and adherens junctions [28], where it interacts with the β-catenin-E-cadherin complex [26], with the synaptopodin and alpha-actinin-4 actin binding proteins [28], and with the JAM4 cell adhesion protein [29]. β-catenin was the first protein involved in regulating cell contact that was shown to be cleaved by caspases during apoptosis [6] and E-cadherin is also known to be cleaved by caspases during apoptosis [12]. The data presented here demonstrates that MAGI-1 cleavage occurs at a similar time as β-catenin and E-cadherin cleavage during apoptosis (Figs. 1(A), 3(C)). However, a considerable fraction of MAGI-1, but not β-catenin or E-cadherin, was found in the cytosol following apoptosis induction (Figs. 2, 5(B)), suggesting that MAGI-1 cleavage is the initial event. The cleavage of MAGI-1 implies that many, if not all, of the MAGI-1-mediated interactions between various proteins at the plasma membrane, are lost in apoptosis. Thus, upon the initial cleavage of MAGI-1, and probably of other MAGUKs, the various interactions mediated by these proteins are lost, thereby weakening protein interactions at the cell-cell interface and allowing easier cell-cell detachment. At the same time the release of proteins and cleaved protein fragments, generated by the caspases, from the membrane allows the proteolytic attack of caspases into the core of the adhesion complexes, i.e. the cadherin-catenin complexes, which are probably less accessible normally. These sequential, zipper-like events appear to be important for the progress of apoptosis since our data indicate that the Asp761Ala mutation, in protecting MAGI-1 from caspase cleavage, considerably delays cell-cell detachment. This leads to the suggestion that the interactions formed last in the process of cell-cell adhesion may need to be eliminated first in the opposite process (Fig. 9), i.e. cell-cell detachment. This could be extremely important in processes such as wound healing or in the removal of damaged cells from epithelial tissues, where it has been suggested that apoptotic cells are extruded from epithelial monolayers rather than being engulfed by the neighbouring cells, thus preserving the epithelial integrity. The extrusion was shown to be driven by actin ring formation and contraction in the neighbouring cells, in response to the signal for this contraction coming from the dying cell [60]. This further suggests that the cleavage of MAGI-1, and possibly of other MAGUKs, is a vital point in cell-cell detachment during apoptosis.
Fig. 9. Sequence of events during cell-cell detachment in apoptosis.
(A) Schematic representation of cell-cell adhesion complex. E-cadherin is connected to actin cytoskeleton via α- and β-catenin. MAGI-1 strengthens the complex. (B) After caspase activation, MAGI-1 is the first protein cleaved in this complex thus allowing access to other constituents, such as β-catenin and E-cadherin
Conclusions
This study has identified MAGI-1, a scaffold protein of the MAGUK family and a component of the cellular actin network, as a caspase target during Fas-, staurosporine- and UV irradiation-induced apoptosis. The study has further shown that such cleavage of MAGI-1, and possibly of other MAGUKs, is an important step in cell-cell dissociation during apoptosis. In addition, various steps in the apoptosis program were shown to occur independently of others, suggesting that their parallel occurrence may accelerate the apoptotic process.
Acknowledgments
The Caspase-3, -6 and -7 clones in pET expression vectors were a generous gift of Guy S. Salvesen. This work was supported in part by research grants from Slovene Research Agency, ICGEB CRP/SLO02-01 and a short term EMBO Fellowship to B.T., a research grant from the Associazione Italiana per la Ricerca sul Cancro to L. B. and NIH R01 CA58541 grant to R.J.
Contributor Information
Uros Gregorc, Department of Biochemistry and Molecular Biology, J. Stefan Institute, Ljubljana, Slovenia.
Saska Ivanova, Department of Biochemistry and Molecular Biology, J. Stefan Institute, Ljubljana, Slovenia.
Miranda Thomas, International Centre for Genetic Engineering and Biotechnology, Padriciano 99, I-34012 Trieste, Italy.
Ernesto Guccione, International Centre for Genetic Engineering and Biotechnology, Padriciano 99, I-34012 Trieste, Italy.
Britt Glaunsinger, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA.
Ron Javier, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA.
Vito Turk, Department of Biochemistry and Molecular Biology, J. Stefan Institute, Ljubljana, Slovenia.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology, Padriciano 99, I-34012 Trieste, Italy.
Boris Turk, Email: boris.turk@ijs.si, Department of Biochemistry and Molecular Biology, J. Stefan Institute, Ljubljana, Slovenia, International Centre for Genetic Engineering and Biotechnology, Padriciano 99, I-34012 Trieste, Italy.
References
- 1.Steller H. Mechanism and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 4.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 5.Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 6.Brancolini C, Lazarevic D, Rodriguez J, Schneider C. Dismantling cell-cell contacts during apoptosis is coupled to a caspase-dependent proteolytic cleavage of β-catenin. J Cell Biol. 1997;139:759–771. doi: 10.1083/jcb.139.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancolini C, Sgorbissa A, Schneider C. Proteolytic processing of the adherens junctions components β-catenin and γ-catenin/plakoglobin during apoptosis. Cell Death Diff. 1998;5:1042–1050. doi: 10.1038/sj.cdd.4400443. [DOI] [PubMed] [Google Scholar]
- 8.Van de Craen M, Berx G, Van Den Brande I, Fiers W, Declercq W, Vandenabeele P. Proteolytic cleavage of beta-catenin by caspases: an in vitro analysis. FEBS Lett. 1999;458:157–160. doi: 10.1016/s0014-5793(99)01153-9. [DOI] [PubMed] [Google Scholar]
- 9.Cryns VL, Bergeron L, Zhu H, Li H, Yuan Y. Specific cleavage of alpha-fodrin during Fas- and tumor necrosis factor-induced apoptosis is mediated by an interleukin-1 beta-converting enzyme/Ced 3 protease distinct from the poly(ADP-ribose)polymerase protease. J Biol Chem. 1996;271:31277–31282. doi: 10.1074/jbc.271.49.31277. [DOI] [PubMed] [Google Scholar]
- 10.Mashima T, Naito M, Noguchi K, Miller DK, Nicholson DW, Tsuruo T. Actin cleavage by CPP-32/apopain during the development of apoptosis. Oncogene. 1997;14:1007–1012. doi: 10.1038/sj.onc.1200919. [DOI] [PubMed] [Google Scholar]
- 11.Kothakota S, Azuma T, Reinhard C, et al. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 12.Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 13.Levkau B, Herren B, Koyama H, Ross R, Raines EW. Caspase-mediated cleavage of focal adhesion kinase pp125FAK and disassembly of focal adhesions in human endothelial cell apoptosis. J Exp Med. 1998;187:579–586. doi: 10.1084/jem.187.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiske J, Schoneberg T, Schroder W, Hatzfeld M, Tauber R, Huber O. The fate of desmosomal proteins in apoptotic cells. J Biol Chem. 2001;276:41175–41181. doi: 10.1074/jbc.M105769200. [DOI] [PubMed] [Google Scholar]
- 15.Bojarski C, Weiske J, Schoneberg T, Schroder W, Mankertz J, Schulzke JD, Florian P, Fromm M, Tauber R, Huber O. The specific fates of tight junction proteins in apoptotic epithelial cells. J Cell Sci. 2004;117:2097–2107. doi: 10.1242/jcs.01071. [DOI] [PubMed] [Google Scholar]
- 16.Gregorc U, Ivanova S, Thomas M, Turk V, Banks L, Turk B. hDLG/SAP97, a member of the MAGUK protein family, is a novel caspase target during cell-cell detachment in apoptosis. Biol Chem. 2005;386:705–710. doi: 10.1515/BC.2005.082. [DOI] [PubMed] [Google Scholar]
- 17.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner CG, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 19.Anderson JM. Cell signalling: MAGUK magic. Curr Biol. 1996;6:382–384. doi: 10.1016/s0960-9822(02)00501-8. [DOI] [PubMed] [Google Scholar]
- 20.Gomperts SN. Clustering membrane proteins: it’s all coming together with the PSD-95/SAP90 protein family. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 21.Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- 22.Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 23.Dobrosotskaya I, Guy RK, James GL. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J Biol Chem. 1997;272:31589–31597. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- 24.Wood JD, Yuan J, Margolis RL, et al. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11:149–160. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]
- 25.Laura RP, Ross S, Koeppen H, Lasky LA. MAGI-1: a widely expressed, alternatively spliced tight junction protein. Exp Cell Res. 2002;275:155–170. doi: 10.1006/excr.2002.5475. [DOI] [PubMed] [Google Scholar]
- 26.Dobrosotskaya IY, James GL. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem Biophys Res Commun. 2000;270:903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- 27.Mino A, Ohtsuka T, Inoue E, Takai Y. Membrane-associated guanylate kinase with inverted orientation (MAGI)-1/brain angiogenesis inhibitor 1-associated protein (BAP1) as a scaffolding molecule for Rap small G protein GDP/GTP exchange protein at tight junctions. Genes Cells. 2000;5:1009–1016. doi: 10.1046/j.1365-2443.2000.00385.x. [DOI] [PubMed] [Google Scholar]
- 28.Patrie KM, Drescher AJ, Welihinda A, Mundel P, Margolis B. Interaction of two actin-binding proteins, synaptopodin and alpha-actinin-4, with the tight junction protein MAGI-1. J Biol Chem. 2002;277:30183–30190. doi: 10.1074/jbc.M203072200. [DOI] [PubMed] [Google Scholar]
- 29.Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol. 2003;23:4267–4282. doi: 10.1128/MCB.23.12.4267-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glaunsinger BA, Lee SS, Thomas M, Banks L, Javier R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene. 2000;19:5270–5280. doi: 10.1038/sj.onc.1203906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 32.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardiol D, Kühne C, Glaunsinger B, Lee SS, Javier R, Banks L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 34.Thiagarajan P, Tait JF. Binding of annexin V/placental anticoagulant protein I to platelets. Evidence for phosphatidylserine exposure in the procoagulant response of activated platelets. J Biol Chem. 1990;265:17420–17423. [PubMed] [Google Scholar]
- 35.Martin SJ, Reutelingsperger CP, McGahon AJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirman T, Oresic K, Droga-Mazovec G, Turk V, Reed JC, Myers RM, Salvesen GS, Turk B. Selective disruption of lysosomes in HeLa cells triggers apoptosis, mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279:3578–3587. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 37.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 38.Balda MS, Matter K. Tight junctions. J Cell Sci. 1998;111:541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- 39.Tsukita S, Furuse M, Itoh M. Structural and signalling molecules come together at tight junctions. Curr Opin Cell Biol. 1999;11:628–633. doi: 10.1016/s0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 40.Jezernik K, Sterle M, Batista U. The distinct steps of cell detachment during development of mouse uroepithelial cells in the bladder. Cell Biol Int. 1997;21:1–6. doi: 10.1006/cbir.1996.0109. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypep-tide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;107:2401–2408. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and Drosophila tumor suppressor gene dlg-A. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumine A, Ogai A, Senda T, et al. Binding of APC to the human homolog of the Drosophila disc large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Hepner K, Castelino-Prabhu S, et al. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci USA. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, Dowbenko D, Spencer S, et al. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J Biol Chem. 2000;275:21477–21485. doi: 10.1074/jbc.M909741199. [DOI] [PubMed] [Google Scholar]
- 47.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Diff. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 49.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 50.Denault JB, Salvesen GS. Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem. 2003;278:34042–34050. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- 51.Stennicke HR, Jürgensmeier JM, Shin H, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 52.Torres J, Rodriguez J, Myers MP, et al. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. J Biol Chem. 2003;278:30652–30660. doi: 10.1074/jbc.M212610200. [DOI] [PubMed] [Google Scholar]
- 53.Adams CL, Chen Y-T, Smith SJ, Nelson WJ. Mechanism of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 55.Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reuver SM, Garner CG. E-cadherin mediated cell adhesion recruits SAP97 into the cortical cytoskeleton. J Cell Sci. 1998;111:1071–1080. doi: 10.1242/jcs.111.8.1071. [DOI] [PubMed] [Google Scholar]
- 57.Fanning AS, Jameson B, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 58.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to a-catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itoh M, Morita K, Tsukita S. Characterization of ZO-2 as a MAGUK family member associated with tight and adherens junctions with a binding affinity to occludin and a-catenin. J Biol Chem. 1999;274:5981–5986. doi: 10.1074/jbc.274.9.5981. [DOI] [PubMed] [Google Scholar]
- 60.Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr Biol. 2001;11:1847–1857. doi: 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]