Abstract
To identify quantitative trait loci (QTLs) associated with the primary target traits for selection in practical rice breeding programs, backcross inbred lines (BILs) derived from crosses between temperate japonica rice cultivars Nipponbare and Koshihikari were evaluated for 50 agronomic traits at six experimental fields located throughout Japan. Thirty-three of the 50 traits were significantly correlated with heading date. Using a linkage map including 647 single-nucleotide polymorphisms (SNPs), a total of 122 QTLs for 38 traits were mapped on all rice chromosomes except chromosomes 5 and 9. Fifty-eight of the 122 QTLs were detected near the heading date QTLs Hd16 and Hd17 and the remaining 64 QTLs were found in other chromosome regions. QTL analysis of 51 BILs having homozygous for the Koshihikari chromosome segments around Hd16 and Hd17 allowed us to detect 40 QTLs associated with 27 traits; 23 of these QTLs had not been detected in the original analysis. Among the 97 QTLs for the 30 traits measured in multiple environments, the genotype-by-environment interaction was significant for 44 QTLs and not significant for 53 QTLs. These results led us to propose a new selection strategy to improve agronomic performance in temperate japonica rice cultivars.
Keywords: Oryza sativa L., japonica rice cultivars, quantitative trait loci (QTLs), agronomic traits
Introduction
Understanding the genetic control of agronomic traits is essential for the improvement of cultivars in rice (Oryza sativa L.). Many agronomically and economically important traits, including heading date, grain productivity, disease resistance and stress tolerance, are primary targets for selection in rice breeding programs. Rice breeders evaluate a large number of breeding lines for various kinds of traits throughout the cultivation period, i.e., from the germination stage to the maturity stage and after harvest. Most of the agronomic traits are quantitatively inherited and are largely affected by environmental factors (Hallauer and Miranda 1988). Therefore, stable and reproducible selection is a challenge in breeding programs.
Temperate japonica rice cultivars are predominantly grown in temperate regions in Japan; Korea; some regions of China; California, USA and so on (Mackill 1995). In the past, the extremely low frequency of DNA polymorphism has prevented molecular genetic analysis for agronomic trait variation among temperate japonica rice cultivars. The International Rice Genome Sequencing Project (IRGSP) released the genome sequence of the temperate japonica rice cultivar Nipponbare (IRGSP 2005). This sequence information has been used for the design of numerous simple sequence repeat (SSR) markers (IRGSP 2005, McCouch et al. 2002). By using these markers, it has been possible to detect polymorphism even among temperate japonica cultivars and to identify quantitative trait loci (QTLs) controlling natural variation for agronomic traits. Several QTLs have been found for heading date (Fujino and Sekiguchi 2005, Fujino et al. 2008a, Fujino and Iwata 2011, Matsubara et al. 2008), grain size (Kato et al. 2011), grain quality (Kobayashi et al. 2007, Tabata et al. 2007), culm length (Hori et al. 2009, Kwon et al. 2008, Monden et al. 2009), panicle architecture (Guo and Hong 2010), pre-harvest sprouting resistance (Fujino et al. 2004, Hori et al. 2010), and eating quality (Kobayashi et al. 2008, Kobayashi and Tomita 2008, Kwon et al. 2011, Wada et al. 2008) in progenies derived from crosses among temperate japonica rice cultivars. Recently, whole-genome sequencing of other temperate japonica rice cultivars, including Koshihikari (Yamamoto et al. 2010), Rikuu 132 and Eiko (Nagasaki et al. 2010) and Omachi (Arai-Kichise et al. 2011), was conducted using next-generation sequencing techniques. Many single-nucleotide polymorphisms (SNPs) were detected from comparisons of genome sequences between Nipponbare and the three recently sequenced cultivars (i.e., Koshihikari, Rikuu 132 and Eiko), and some of those SNPs have been successfully used in further genetic analysis (Nagasaki et al. 2010, Yamamoto et al. 2010). These SNPs will enhance genetic dissection of phenotypic differences among temperate japonica cultivars by facilitating QTL analysis (Shibaya et al. 2011).
A set of reciprocal backcross inbred lines (BILs) have been developed from a cross between the temperate japonica rice cultivars Nipponbare and Koshihikari (Matsubara et al. 2008). Both Nipponbare and Koshihikari have been extensively cultivated in Japan. Nipponbare was a leading cultivar until 1978; at that time, Koshihikari became a top cultivar and has continued to be so until the present (Yokoo et al. 2005). Koshihikari has an earlier heading date, better eating quality, stronger cold tolerance at the booting stage, and stronger pre-harvest sprouting resistance than Nipponbare, but weaker resistance to leaf blast and greater susceptibility to lodging. QTLs analyses in the BILs revealed QTLs for heading date (Matsubara et al. 2008), eating quality (Takeuchi et al. 2008), culm length (Hori et al. 2009) and pre-harvest sprouting resistance (Hori et al. 2010). However, many other primary target traits of breeding selection, such as yield-related traits, grain quality, lodging resistance, disease resistance and others, had not been genetically analyzed in these populations.
The objective of the present study was to detect QTLs for the primary target traits in breeding selection in the BILs at six experimental fields located throughout Japan. A high level of phenotypic correlation between heading date and other traits was observed. A large number of QTLs were found, many of which were localized near previously identified heading date QTLs. Based on these results, we discuss the potential limitations of conventional phenotypic selection that are caused by variation in heading date and we propose a strategy of pyramiding large numbers of QTLs using high-density SNP genotyping.
Materials and Methods
Plant materials
A total of 127 BILs produced by self pollinations after a single backcross of the Nipponbare/Koshihikari F1 to Koshihikari were developed by Matsubara et al. (2008); in the present study, we used 100 of these lines (BIL601 to BIL700). Seeds of all 127 BILs are available at the Rice Genome Resource Center in National Institute of Agrobiological Sciences (NIAS) (http://www.rgrc.dna.affrc.go.jp/index.html.en).
Evaluation of primary target traits for selection in rice breeding
Forty-seven agronomic traits and three disease resistance traits (resistance to leaf blast, panicle blast and bacterial leaf blight) were investigated at six experimental fields in Japan in 2009 (Table 1). Agronomic traits were evaluated based on conventional breeding methods in accordance with the UPOV test guidelines (http://www.upov.int/en/publications/tg_rom/) and the Rice Breeding Manual (Yamamoto et al. 1996) at three fields: the National Agriculture and Food Research Organization (NARO) Kyushu Okinawa Agricultural Research Center (KARC; Izumi, Chikugo, Fukuoka, N33.12, E130.29), the NARO Institute of Crop Science (NICS; Nikkawa, Tsukubamirai, Ibaraki, N36.01, E140.02) and the NARO Agricultural Research Center, Hokuriku Research Center (NARCHRC; Inada, Joetsu, Niigata, N37.60, E138.16). In the field at KARC, seeds were sown on 29 May and seedlings were transplanted into the paddy field on 26 June at a planting density of 18.5 individuals per m2. In the field at NICS, seeds were sown on 24 April and seedlings were transplanted into the paddy field on 21 May at a planting density of 23.8 individuals per m2. In the field at NARCHRC, seeds were sown on 27 April and seedlings were transplanted into the paddy field on 27 May at a planting density of 20.6 individuals per m2. Cultivation management followed the standard procedures used at each experimental station.
Table 1.
List of 50 agronomic traits investigated in Nipponbare/Koshihikari backcross inbred lines (BILs) at six research centers throughout Japan
| Trait namea | Unit (score) | Fieldb | Trait scores | ||
|---|---|---|---|---|---|
|
| |||||
| Nipponbarec | Koshihikarid | Range in BILs | |||
| Seedling height at transplanting stage (SHT) | (1–9; short–long) | NICS | 5.0 | 6.5 | 4.0–7.5 |
| Leaf color at transplanting stage (LCT) | (1–9; light–dark) | NICS | 5.0 | 3.5 | 3.0–6.0 |
| Plant height after transplanting in paddy field (PHP) | (1–9; short–long) | NICS | 5.0 | 6.3 | 5.0–4.2 |
| Tiller number after transplanting in paddy field (TNP) | (1–9; low–high) | NICS | 5.8 | 6.0 | 4.5–6.0 |
| Degree of plant growth after transplanting in paddy field (DPP) | (1–9; low–high) | NICS | 5.5 | 6.3 | 5.0–7.0 |
| Leaf color after transplanting in paddy field (LCP) | (1–9; light–dark) | NICS | 5.0 | 3.5 | 3.0–6.0 |
| Plant height at maximum tiller number stage (PHM) | (1–9; low–high) | KARC | 5.0 | 5.3 | 4.0–6.0 |
| NARCHRC | 5.0 | 6.0 | 5.0–7.0 | ||
| Leaf color at maximum tiller number stage (LCM) | (1–9; light–dark) | KARC | 4.7 | 4.7 | 4.0–6.0 |
| NARCHRC | 5.0 | 5.0 | 4.0–6.0 | ||
| Leaf width at maximum tiller number stage (LWM) | (1–9; narrow–broad) | KARC | 5.0 | 5.0 | 4.0–6.0 |
| NARCHRC | 5.0 | 5.0 | 4.0–6.0 | ||
| Leaf color at heading date (LCH) | (1–9; light–dark) day | NICS | 5.5 | 4.3 | 4.0–6.0 |
| Days to heading (DTH) | MARI | 72.0 | 70.0 | 68.0–81.0 | |
| KARC | 82.7 | 79.0 | 71.0–92.0 | ||
| NICS | 110.5 | 100.5 | 93.0–120.0 | ||
| NARCHRC | 116.3 | 105.7 | 99.0–126.0 | ||
| Culm length (CL) | cm | KARC | 59.3 | 65.7 | 50.0–74.0 |
| NICS | 87.9 | 96.7 | 80.5–103.1 | ||
| NARCHRC | 78.5 | 86.6 | 64.6–98.1 | ||
| Culm diameter (CDI) | (2–8; thin–thick) | KARC | 5.0 | 4.7 | 3.0–7.0 |
| NICS | 5.0 | 5.0 | 4.0–5.0 | ||
| NARCHRC | 5.3 | 4.8 | 4.0–6.0 | ||
| Culm stiffness (CSF) | (2–8; stiff–weak) | KARC | 5.3 | 5.7 | 3.0–7.0 |
| NICS | 5.0 | 6.8 | 5.0–7.0 | ||
| NARCHRC | 4.0 | 6.3 | 3.0–7.0 | ||
| Lodging degree (LD) | (0–9; low–high) | KARC | 0.0 | 1.0 | 0.0–2.0 |
| NICS | 0.5 | 2.8 | 0.0–5.0 | ||
| NARCHRC | 0.8 | 9.0 | 0.0–9.0 | ||
| Tiller angle (TA) | (3–7; erect–spreading) | KARC | 4.0 | 4.0 | 4.0–5.0 |
| Panicle length (PL) | cm | KARC | 19.9 | 19.8 | 16.6–21.2 |
| NICS | 20.3 | 19.9 | 17.9–23.0 | ||
| NARCHRC | 18.7 | 19.0 | 16.2–21.3 | ||
| Number of panicles (NOP) | number/plant | KARC | 7.9 | 7.8 | 6.2–10.6 |
| NICS | 11.9 | 12.5 | 10.3–13.9 | ||
| NARCHRC | 14.1 | 14.2 | 10.1–17.9 | ||
| Glume density per panicle (GDP) | (2–8; low–high) | KARC | 4.0 | 5.0 | 4.0–6.0 |
| NICS | 5.0 | 6.0 | 5.0–6.5 | ||
| NARCHRC | 5.0 | 5.0 | 4.0–6.0 | ||
| Flag leaf length (FLL) | (1–9; short–long) | KARC | 3.3 | 3.7 | 3.0–5.0 |
| NICS | 7.0 | 7.0 | 5.0–7.0 | ||
| NARCHRC | 4.0 | 4.5 | 3.0–8.0 | ||
| Flag leaf: angle of leaf blade (FLA) | (1–9; erect–recurved) | KARC | 6.0 | 6.7 | 1.0–8.0 |
| NICS | 5.3 | 4.8 | 3.0–7.0 | ||
| NARCHRC | 3.0 | 4.0 | 1.0–6.0 | ||
| Panicle exsertion length (PEL) | (1–9; enclosed–well-exserted) | KARC | 5.7 | 6.0 | 2.0–7.0 |
| NICS | 5.3 | 6.0 | 3.0–6.0 | ||
| NARCHRC | 7.5 | 8.3 | 6.0–9.0 | ||
| Presence of awns (POA) | (0–9; absent–present) | KARC | 0.0 | 0.0 | 0.0–0.0 |
| NICS | 3.0 | 1.0 | 1.0–4.0 | ||
| NARCHRC | 2.8 | 1.7 | 0.0–7.0 | ||
| Awn length (AL) | (2–8; short–long) | NICS | 3.5 | 2.0 | 2.0–6.0 |
| NARCHRC | 4.0 | 2.0 | 1.0–5.0 | ||
| Apiculus color of awn (ACA) | (1–9; weak–strong) | KARC | 1.0 | 1.0 | 1.0–1.0 |
| NICS | 1.0 | 1.0 | 1.0–1.0 | ||
| NARCHRC | 1.0 | 1.0 | 1.0–1.0 | ||
| Lemma and palea color (LPC) | (1–9; white–black) | KARC | 1.0 | 1.0 | 1.0–1.0 |
| NICS | 1.0 | 1.0 | 1.0–1.0 | ||
| NARCHRC | 1.0 | 1.0 | 1.0–1.0 | ||
| Days to maturity (DTM) | day | KARC | 36.7 | 35.3 | 31.0–45.0 |
| NICS | 43.8 | 38.8 | 35.5–48.5 | ||
| NARCHRC | 57.3 | 46.0 | 41.0–70.0 | ||
| Plant appearance at maturity stage (AMS) | (3–7; superior–inferior) | KARC | 7.0 | 7.0 | 5.0–7.0 |
| NICS | 6.0 | 3.0 | 3.0–7.0 | ||
| Seed fertility (SF) | (1–10; low–high) | NICS | 10.0 | 10.0 | 10.0–10.0 |
| Seed shattering (SH) | (2–8; easy–hard) | KARC | 3.0 | 3.0 | 3.0–3.0 |
| NICS | 3.0 | 3.0 | 3.0–3.0 | ||
| NARCHRC | 2.0 | 2.0 | 2.0–2.0 | ||
| Senescence (SNC) | (0–9; late–early) | KARC | 1.0 | 3.3 | 1.0–4.0 |
| NICS | 5.0 | 5.0 | 4.0–7.0 | ||
| NARCHRC | 4.8 | 5.0 | 3.0–7.0 | ||
| Pre-harvest sprouting resistance (PHS) | (2–8; strong–weak) | NARCHRC | 6.7 | 5.5 | 3.0–7.0 |
| Aboveground dry weight (ADW) | g/plant | NICS | 79.3 | 77.5 | 61.0–105.7 |
| Gross brown rice weight (GBW) | g/plant | KARC | 12.4 | 14.7 | 6.4–16.8 |
| Head brown rice weight (HBW) | g/plant | KARC | 10.1 | 10.7 | 6.3–16.4 |
| NICS | 23.5 | 26.5 | 20.8–34.9 | ||
| Weight of aborted rice (ARW) | g/plant | KARC | 2.3 | 4.0 | 0.1–4.0 |
| 1000-grain weight (TGW) | g | NICS | 22.5 | 22.9 | 20.2–24.6 |
| Protein content of brown rice (PCB) | % | NICS | 6.7 | 6.2 | 5.5–7.2 |
| Grain appearance (GA) | (1–9; good–poor) | KARC | 3.5 | 3.5 | 2.0–7.0 |
| NARCHRC | 2.9 | 5.0 | 2.1–6.0 | ||
| Normal kernel ratio (NKR) | % | NICS | 86.3 | 83.1 | 74.7–92.1 |
| NARCHRC | 85.0 | 72.9 | 56.5–90.7 | ||
| Degree of occurrence of white-belly kernels (DWB) | (0–9; low–high) | KARC | 0.7 | 0.3 | 0.0–2.0 |
| White-belly kernel ratio (WBR) | % | NICS | 1.5 | 0.9 | 0.1–3.5 |
| NARCHRC | 1.0 | 2.2 | 0.1–8.3 | ||
| Degree of occurrence of white-back and white-based kernels (DWK) | (0–9; low–high) | KARC | 1.7 | 2.0 | 0.0–7.0 |
| White-backed and white-based kernel ratio (WKR) | % | NICS | 0.2 | 1.1 | 0.1–4.1 |
| NARCHRC | 1.0 | 4.0 | 0.2–9.4 | ||
| Degree of occurrence of white-center kernels (DWC) | (0–9; low–high) | KARC | 2.7 | 0.7 | 0.0–6.0 |
| Degree of occurrence of milky-white kernels (DMW) | (0–9; low–high) | KARC | 0.0 | 0.3 | 0.0–2.0 |
| Milky-white kernel ratio (MWR) | % | NICS | 1.1 | 1.8 | 0.2–4.3 |
| NARCHRC | 1.7 | 4.2 | 0.3–9.7 | ||
| Leaf blast resistance: lesion area index (LBR) | (0–10; none–dead) | KARC | 0.0 | 0.0 | 0.0–0.0 |
| AARCMARI | 7.7 | 8.8 | 6.0–10.0 | ||
| NARCHRC | 0.2 | 0.5 | 0.0–4.0 | ||
| TARC | 4.0 | 4.2 | 4.1–7.8 | ||
| Panicle blast resistance: degree of occurrence of diseased panicles (PBR) | (0–10; none–almost all) | KARC | 0.0 | 0.0 | 0.0–0.0 |
| NICS | 3.1 | 5.5 | 3.4–7.4 | ||
| NARCHRC | 1.2 | 1.7 | 0.0–5.0 | ||
| Bacterial leaf blight resistance: lesion length (BLB) | (cm) | MARI | 0.6 | 4.0 | 0.6–5.3 |
Agronomic traits were evaluated based on conventional breeding methods in accordance with the UPOV test guidelines (http://www.upov.int/en/publications/tg_rom/) and the Rice Breeding Manual (Yamamoto et al. 1996).
KARC, National Agriculture and Food Research Organization (NARO) Kyushu Okinawa Agricultural Research Center; NICS, NARO Institute of Crop Science; NARCHRC, NARO Agricultural Research Center Hokuriku Research Center; MARI, Miyazaki Agricultural Research Institute; TARC, NARO Tohoku Agricultural Research Center; AARCMARI, Aichi Agricultural Research Center Mountainous Region Agricultural Research Institute.
Donor parent
Recurrent parent
Resistance to leaf blast was evaluated in 2009 in the experimental fields at KARC, NARCHRC, the NARO Tohoku Agricultural Research Center (TARC; Shimo-furumichi, Daisen, Akita, N39.29, E140.29) and the Aichi Agricultural Research Center, Mountainous Region Agricultural Research Institute (AARCMARI; Inabu, Toyota, Aichi, N35.12, E137.30). Resistance to panicle blast was evaluated in the experimental fields at KARC, NICS and NARCHRC. Disease resistance scores for leaf blast and panicle blast at KARC and NARCHRC were evaluated in fields where cultivation practices are studied. Evaluation of panicle blast resistance at NICS was conducted in the experimental field (Gozenyama, Hitachi-omiya, Ibaraki, N36.34, E140.20). Seeds were sown on 23 April and seedlings were transplanted into the paddy field on 14 May at a planting density of 22.2 individuals per m2 in double row plots with two replications per BIL. The disease severity on 30 to 35 days after heading was scored as the degree of blast infection, and the score of BILs which headed after 16 August was excluded because the disease severity of them was apparently lower than the earlier ones. At TARC and AARCMARI, seeds of each BIL were sown in the upland fields on 3 June and 28 May, respectively. The BILs were cultivated in single-row plots 40 cm long with two to three replications per BIL. Nitrogen fertilizer was applied at 20 kg/ha at both TARC and AARCMARI. The disease severity on 30- to 70-day-old plants was scored as the degree of blast infection.
Bacterial leaf blight resistance was scored in the field at the Miyazaki Agricultural Research Institute (MARI; Sadowara, Miyazaki, Miyazaki, N32.00, E131.28) in 2009. Seeds of each BIL were sown on 15 June, and seedlings were transplanted into the paddy field on 10 July in single-row plots with a distance of 18 cm between plants and 30 cm between rows, with two replications per BIL. Nitrogen fertilizer was applied at 10 kg/ha. The leaf blades were inoculated by scissor clipping, and the length of lesion expansion was scored after 15 days.
DNA marker analysis
Total DNA was extracted from the leaves of each plant in the BIL plots by the CTAB method (Murray and Thompson 1980). Two types of DNA markers, SSRs and SNPs, were used for linkage map construction. A total of 150 SSRs reported by IRGSP (2005) and McCouch et al. (2002) and found to be polymorphic between Nipponbare and Koshihikari were used for the analysis. Polymerase chain reaction (PCR) of the SSR markers was performed according to the method of Matsubara et al. (2008). A total of 647 SNPs were selected from genome-wide SNPs data from crosses between Nipponbare and Koshihikari and used for genotyping (Yamamoto et al. 2010). The SNPs were detected using the BeadsStation 500G system (Illumina, San Diego, USA), according to the manufacturer’s instructions. The genotypes of the BILs for the 150 SSR markers and 647 SNP markers used here are available at the Rice Genome Resource Center at NIAS (http://www.rgrc.dna.affrc.go.jp/index.html.en).
QTL detection
We performed linkage mapping using version 3.0 of MAPMAKER/EXP (Lander et al. 1987) and used the Kosambi function to calculate genetic distances. We performed QTL analyses using composite interval mapping, as implemented by the Zmapqtl program (model 6) provided by version 2.5 of the QTL Cartographer software (Basten et al. 2005). We used genome-wide threshold values (α = 0.05) to detect putative QTLs based on the results of 1000 permutations. Genotype-by-environment interaction effects were tested for the agronomic traits that were evaluated at multiple experimental fields. Analysis was performed by two-way ANOVA with the software package JMP 6.0 (SAS Institute, Cary, USA). Significant levels for correlation coefficients and interaction effects were calculated by the statistical method of Benjamini and Hochberg (1995) to correct familywise error rates.
Results
Evaluation of primary target traits for selection in rice breeding
Fifty morphological, physiological and disease resistance traits were evaluated at the six experimental stations: 32 traits at KARC, 36 traits at NICS, 29 traits at NARCHRC, 2 traits at MARI and 1 trait each at TARC and AARCMARI. A total of 101 trait–location combinations were represented in the present study, and the data are summarized in Table 1. Relative to Koshihikari, Nipponbare showed later heading; a longer maturity period; shorter seedling height; stronger lodging resistance, attributed to its short length and thick culm diameter; more panicles with awns; lower gross brown rice weight and head brown rice weight; a smaller percentage of white immature grains; weaker pre-harvest sprouting resistance and stronger resistance to blast and bacterial leaf blight. A wide range of variation was found in the BILs for almost all of the traits (Table 1 and Supplemental Fig. 1). The distributions for most of the traits were continuous, and transgressive segregation was observed in the BILs, indicating the involvement of many genes with alleles showing both positive and negative effects being contributed by both Nipponbare and Koshihikari.
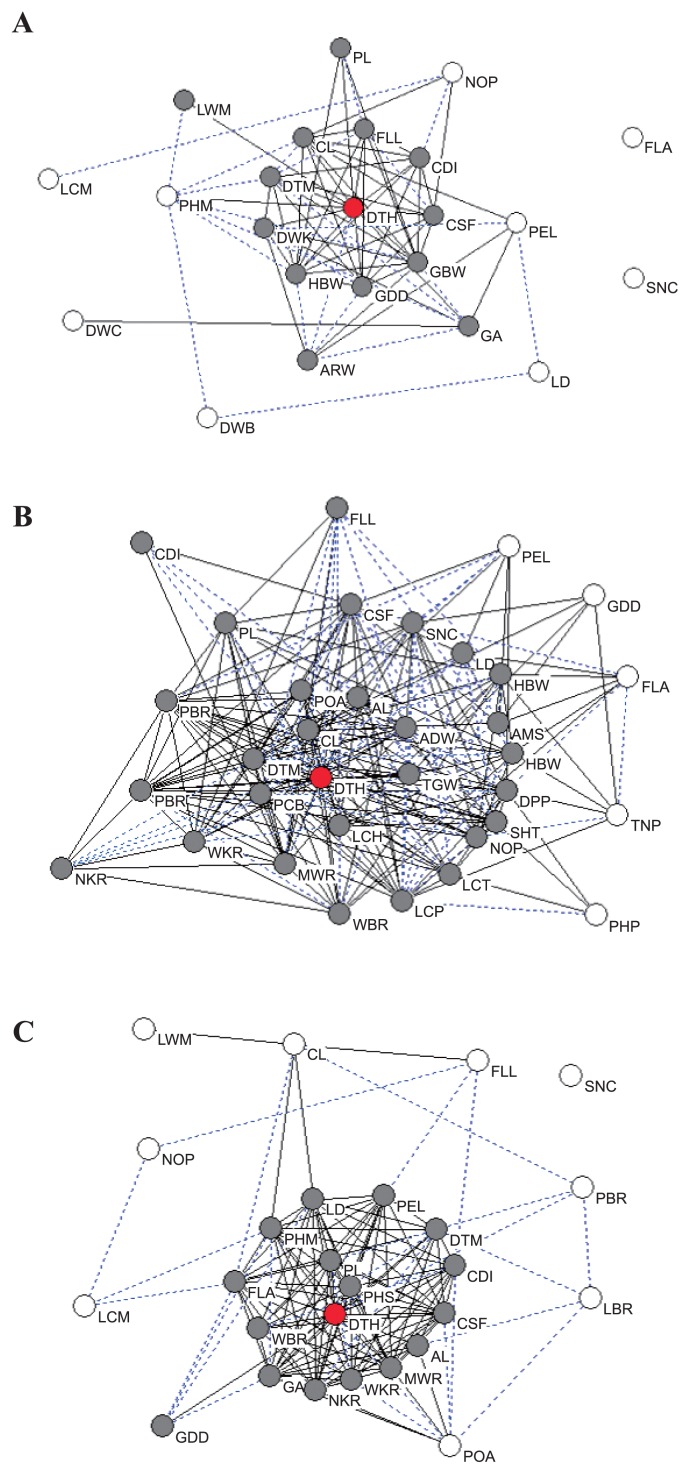
Out of the 50 traits measured in this study, 33 were significantly correlated with days to heading (DTH) at the 5% level in at least one location (Fig. 1 and Table 2). By site, this was 14 of the 31 traits measured at KARC, 21 of 36 traits measured at NICS and 18 of the 28 traits measured at NARCHRC. For example, positive correlations with days to heading (DTH) were found for days to maturity (DTM), culm diameter (CDI), gross brown rice weight (GBW) and head brown rice weight (HBW), whereas negative correlations were found for grain appearance (GA), white immature grains of white-belly kernel ratio (WBR) and white-backed and -based kernel ratio (WKR). Phenotypes with early heading showed a tendency to also have short maturity periods, thin culms, lower gross and head brown rice weights and high numbers of white immature grains. The BILs with late heading date showed similar phenotypes to Nipponbare for most traits. On the other hand, leaf color at maximum tiller number stage (LCM), leaf width at maximum tiller number stage (LWM), number of panicles (NOP), and senescence (SNC) showed relatively low correlations with the other traits at multiple experimental fields. The correlation coefficients among all of the traits investigated in the present study are shown in Supplemental Table 1.
Fig. 1.
Diagrams of correlation coefficients among 50 agronomic traits in the Nipponbare/Koshihikari backcross inbred lines (BILs) measured in the fields at KARC (A), NICS (B) and NARCHRC (C). Circles indicate traits evaluated in the present study. Trait abbreviations are as shown in Table 1. Gray solid lines and blue dotted lines indicate significant correlations between traits at the 1% and 5% levels, respectively. Red circles indicate days to heading (DTH), gray circles indicate traits showing significant correlations with DTH at the 5% level, and open circles represent traits without significant correlations to DTH.
Table 2.
Correlation of agronomic traits with days to heading (DTH) and the number of QTLs for each agronomic trait in the Nipponbare/Koshihikari BILs
| Trait name | KARCa | NICSa | NARCHRCa | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Correlation with DTHb | No. of QTLsc | Correlation with DTHb | No. of QTLsc | Correlation with DTHb | No. of QTLsc | |
| Seedling height at transplanting stage (SHT) | — | — | −0.49 *** | 2(1) | — | — |
| Leaf color at transplanting stage (LCT) | — | — | 0.33 ** | 1(1) | — | — |
| Plant height after transplanting in paddy field (PHP) | — | — | −0.10 | 0 | — | — |
| Tiller number after transplanting in paddy field (TNP) | — | — | −0.15 | 0 | — | — |
| Degree of plant growth after transplanting in paddy field (DPP) | — | — | −0.53 *** | 1(1) | — | — |
| Leaf color after transplanting in paddy field (LCP) | — | — | 0.48 *** | 1(1) | — | — |
| Plant height at maximum tiller number stage (PHM) | −0.27 * | 0 | — | — | −0.58 *** | 2(2) [2] |
| Leaf color at maximum tiller number stage (LCM) | −0.11 | 0 | — | — | 0.19 | 0 |
| Leaf width at maximum tiller number stage (LWM) | 0.28 * | 1(1) [1] | — | — | −0.16 | 0 |
| Leaf color at heading date (LCH) | — | — | 0.08 | 0 | — | — |
| Days to heading (DTH) | 1.00 | 2(2) [2] | 1.00 | 2(2) [2] | 1.00 | 2(2) [2] |
| Culm length (CL) | 0.38 *** | 1 | 0.21 | 4(1) | −0.01 | 2 |
| Culm diameter (CDI) | 0.60 *** | 2(2) [1] | 0.22 * | 0 | 0.49 *** | 3(1) [2] |
| Culm stiffness (CSF) | −0.44 *** | 1(1) [1] | −0.56 *** | 1(1) [1] | −0.58 *** | 4(1) [4] |
| Lodging degree (LD) | −0.13 | 0 | −0.23 * | 1(1) [1] | −0.54 *** | 0 |
| Tiller angle (TA) | 0.18 | 0 | — | — | — | — |
| Panicle length (PL) | 0.28 * | 1(1) [1] | 0.52 *** | 2(1) [1] | −0.41 *** | 0 |
| Number of panicles (NOP) | 0.00 | 0 | 0.31 ** | 0 | 0.00 | 1[1] |
| Glume density per panicle (GDP) | 0.58 *** | 2(1) [1] | 0.10 | 0 | −0.31 ** | 1(1) [1] |
| Flag leaf length (FLL) | 0.43 *** | 0 | 0.20 | 1 | 0.14 | 2 [2] |
| Flag leaf: angle of leaf blade (FLA) | −0.07 | 0 | −0.13 | 3 | −0.63 *** | 2(2) [2] |
| Panicle exsertion length (PEL) | 0.04 | 2 [1] | −0.19 | 3[1] | −0.53 *** | 3(1) [1] |
| Presence of awns (POA) | — | — | 0.55 *** | 1(1) | 0.27 * | 0 |
| Awn length (AL) | — | — | 0.61 *** | 2(1) | 0.24 * | 1 |
| Days to maturity (DTM) | 0.59 *** | 1(1) [1] | 0.80 *** | 1(1) [1] | 0.67 *** | 1(1) [1] |
| Appearance at maturity stage (AMS) | −0.72 *** | 1(1) [1] | 0.13 | 0 | — | — |
| Senescence (SNC) | 0.09 | 2[1] | 0.29 ** | 2(2) [2] | 0.10 | 1[1] |
| Pre-harvest sprouting resistance (PHS) | — | — | — | — | −0.58 *** | 3(2) |
| Aboveground dry weight (ADW) | — | — | 0.79 *** | 2(2) | — | — |
| Gross brown rice weight (GBW) | 0.66 *** | 1(1) | — | — | — | — |
| Head brown rice weight (HBW) | 0.66 *** | 1(1) | 0.00 | 0 | — | — |
| Weight of aborted rice (ARW) | 0.20 | 3 | — | — | — | — |
| 1000-grain weight (TGW) | — | — | −0.29 ** | 3(1) | — | — |
| Protein content of brown rice (PCB) | — | — | −0.62 *** | 3(1) | — | — |
| Grain appearance (GA) | −0.42 *** | 2(1) [1] | — | — | −0.54 *** | 2(1) [1] |
| Normal kernel ratio (NKR) | — | — | 0.23 * | 4(1) [1] | 0.56 *** | 1(1) [1] |
| Degree of occurrence of white-belly kernels (DWB) | 0.13 | 0 | — | — | — | — |
| White-belly kernel ratio (WBR) | — | — | −0.31 ** | 1(1) | −0.43 *** | 3[3] |
| Degree of occurrence of white-back and white-based kernels (DWK) | −0.47 *** | 3(1) | — | — | — | — |
| White-backed and white-based kernel ratio (WKR) | — | — | −0.50 *** | 1(1) [1] | −0.65 *** | 2(2) [1] |
| Degree of occurrence of white-center kernels (DWC) | 0.01 | 0 | — | — | — | — |
| Degree of occurrence of milky-white kernels (DMW) | 0.16 | 0 | — | — | — | — |
| Milky-white kernel ratio (MWR) | — | — | −0.51 *** | 0 | −0.64 *** | 2(2) [2] |
| Leaf blast resistance: lesion area index (LBR) | — | — | — | — | −0.11 | 1 |
| Panicle blast resistance: degree of occurrence of diseased panicles (PBR) | — | — | −0.45 *** | 6(1) | −0.24 * | 2 |
| Bacterial leaf blight resistance: lesion length (BLB) | — | — | — | — | — | — |
| Trait name | MARI | TARC | AARCMARI | |
|---|---|---|---|---|
|
|
|
|
||
| Correlation with DTHb | No. of QTLsc | No. of QTLsc | No. of QTLsc | |
| Days to heading (DTH) | 1.00 | 3(3) [2] | — | — |
| Leaf blast resistance: lesion area index (LBR) | — | — | 1[1] | 1[1] |
| Bacterial leaf blight resistance: lesion length (BLB) | −0.07 | 2 | — | — |
KARC, National Agriculture and Food Research Organization (NARO) Kyushu Okinawa Agricultural Research Center; NICS, NARO Institute of Crop Science; NARCHRC, NARO Agricultural Research Center Hokuriku Research Center; MARI, Miyazaki Agricultural Research Institute; TARC, NARO Tohoku Agricultural Research Center; AARCMARI, Aichi Agricultural Research Center Mountainous Region Agricultural Research Institute.
*, **, *** indicate significant correlations at P < 0.05, P < 0.01 and P < 0.001, respectively.
Figures in parentheses indicate the number of QTLs located near the two main QTLs for days to heading (DTH), and figures in square brackets indicate the number of QTLs showing genotype-by-environment interaction effects.
QTL detection for primary target traits in rice breeding
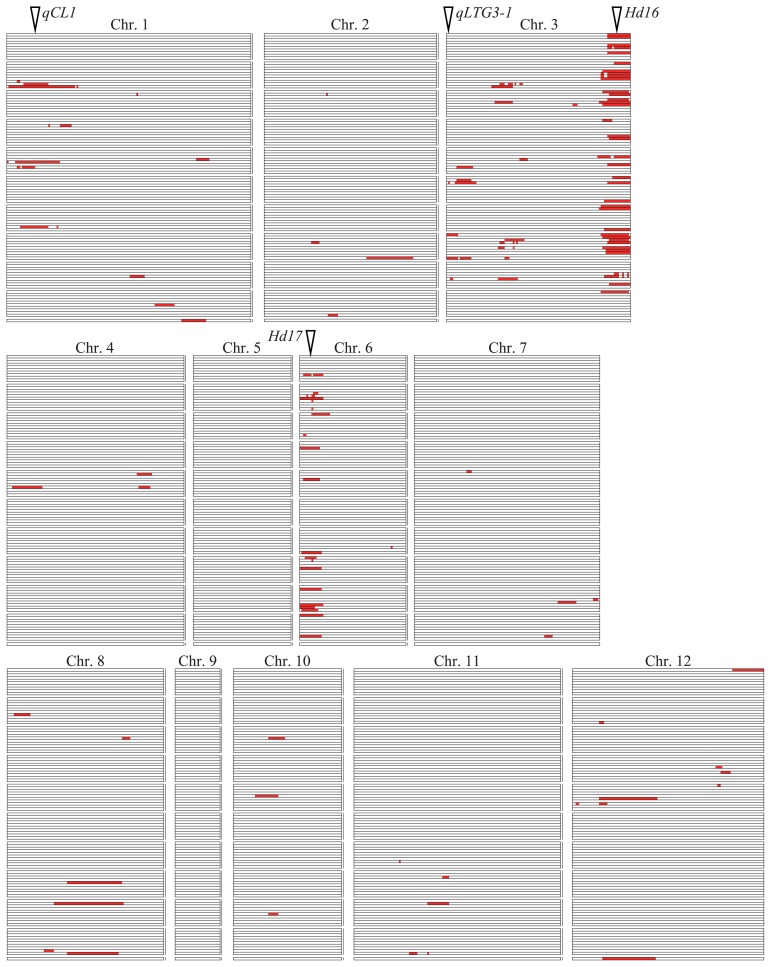
Using a linkage map consisting of 150 SSRs and 647 SNPs, a total of 122 QTLs were detected for 38 of the 50 traits at the six experimental fields (Table 2). The marker interval, LOD score and percentage of variance explained by each QTL is listed in Supplemental Table 2. The QTLs were distributed on all of the rice chromosomes except chromosomes 5 and 9 (Fig. 2). QTL clusters were observed on the short arm of chromosome 1, the short and long arms of chromosome 3 and the short arm of chromosome 6. Fifty-seven of the 122 QTLs explained more than 15% of the phenotypic variance for the trait, whereas 65 of the 122 QTLs explained less than 15% of the phenotypic variance. QTLs with relatively large effects were located within the QTL clusters; thus, it appears that the agronomic traits in this population are controlled mainly by the genes in these clusters. QTLs for days to heading (DTH) were detected at three chromosome regions: the long arm of chromosome 3, the short arm of chromosome 6 and the short arm of chromosome 8. The days to heading QTLs on chromosomes 3 and 6 were detected in the same marker intervals as heading date QTLs Hd16 and Hd17, respectively (Matsubara et al. 2008). In the present study, two of the QTLs for culm length (CL) were detected on chromosomes 1 and 3 and one of the QTLs for resistance to pre-harvest sprouting (PHS) was detected on the short arm of chromosome 3. The culm length QTLs were located near the culm length QTLs qCL1 and qCL3-1 detected by Hori et al. (2009) and the pre-harvest sprouting QTL was located near the low-temperature germinability gene qLTG3-1 detected by Hori et al. (2010).
Fig. 2.
Chromosomal locations of 122 QTLs for 38 agronomic traits detected in the Nipponbare/Koshihikari BILs. Marker intervals showing LOD scores exceeding the threshold for significance are indicated by red boxes. Each trait is shown in a vertical row in the same order as in Table 1. The positions of qCL1 (Hori et al. 2009), qLTG3-1 (Fujino et al. 2008b, Hori et al. 2010) and Hd16 and Hd17 (Matsubara et al. 2008) were defined by the positions of flanking markers and are shown here as white triangles.
QTLs for disease resistance were detected for leaf blast resistance (LBR) on chromosomes 1 and 7; for panicle blast resistance (PBR) on chromosomes 2, 3, 6, 7, 8 and 11 and for bacterial leaf blight resistance (BLB) on chromosomes 1 and 12. QTLs were found on the short arm of chromosome 3 for weight of aborted rice (ARW) and grain quality traits including grain appearance (GA) and white-belly kernel ratio (WBR). No QTLs were found for 12 traits: plant height after transplanting in paddy field (PHP), tiller number after transplanting in paddy field (TNP), leaf color at maximum tiller number stage (LCM), leaf color at heading date (LCH), tiller angle (TA), apiculus color of the awn (ACA), lemma and palea color (LPC), seed fertility (SF), seed shattering (SH), degree of occurrence of white-belly kernel (DWB), degree of occurrence of white-center kernel (DWC) and degree of occurrence of milky-white kernel (DMW), because of small differences in the phenotypes of the BILs (Table 1 and Supplemental Fig. 1).
Among the 122 QTLs, 58 QTLs for 33 traits were found near the heading date QTLs Hd16 and Hd17 (Supplemental Table 2 and Fig. 1). All of these traits showed significant correlations with heading date. The remaining 64 QTLs, representing 23 traits, were located elsewhere in the rice genome. To detect QTLs independent of the effects of the heading date QTLs Hd16 and Hd17, we selected 51 BILs homozygous for Koshihikari segments in these two chromosome regions. QTL analysis in the 51 selected BILs detected 40 QTLs for 27 traits (Supplemental Table 3). The 40 QTLs included 17 of the 122 QTLs detected in the full set of 100 BILs and an additional 23 QTLs that had not been previously detected. Sixteen of the QTLs detected in the 51 selected BILs were in a cluster on the short arm of chromosome 3.
Genotype-by-environment interaction effects
We scored 11 traits at two experimental fields, 17 traits at three experimental fields and 2 traits at four experimental fields (Table 1). The ranges of scores for these 30 traits in the BILs differed among the fields (environments). The BILs at KARC showed early heading, a short maturity period, a short culm length, a small number of panicles, absent awns and a short flag leaf compared with the BILs at NICS and NARCHRC. The BILs at NARCHRC showed greater panicle number and longer maturity period, whereas the BILs tested at NICS showed a long culm, panicle and flag leaf. Since the latitudinal positions, cultivation conditions of day length and temperature and cultivation managements of applied fertilizer and planting density were different among the experimental stations, genotype-by-environment interaction effects were investigated in the present study.
ANOVA revealed that 44 QTLs for 19 traits showed significant effects for genotype-by-environment interactions at the 5% level (Table 2 and Supplemental Table 2). No significant interaction effects were detected for 53 QTLs for 20 traits. The 44 significant QTLs included the heading date QTLs on chromosomes 3 and 6 and other QTLs in these regions. For three traits (leaf width at maximum tiller number stage [LWM], panicle length [PL] and glume density per panicle [GDP]), the additive effects of QTLs for these traits were positive at certain experimental fields, but negative at others. For example, QTLs for glume density per panicle (GDP) were detected near the heading date QTL Hd16 on chromosome 3 in two experimental fields: the additive effects of the QTLs were positive at KARC, but negative at NARCHRC. No QTLs were detected at NICS. The correlations between days to heading (DTH) and glume density per panicle (GDP) were positive at KARC, but negative at NARCHRC and low level of correlation at NICS. Quantitative differences in genotype-by-environment interactions were observed for QTLs for the 12 traits having significant interactions in at least one location. The additive effects of these QTLs were significant at certain experimental fields but not significant at other experimental fields. The directions of additive effects of these QTLs were the same at all experimental fields. For the remaining five traits tested at multiple locations (culm length [CL], presence of awns [POA], awn length [AL], head brown rice weight [HBW] and panicle blast resistance [PBR]), none of the QTLs had significant genotype-by-environment interaction effects.
Discussion
Significant correlations with heading date have been reported for a number of agronomic traits in previous studies (Kwon et al. 2008, Mei et al. 2003, Takeuchi et al. 2008). In this study, significant correlations were also observed between heading date and other traits (Table 2 and Fig. 1). In addition, 58 of the 122 QTLs detected in this study were located near two previously identified heading date QTLs on chromosomes 3 and 6 (Fig. 2 and Supplemental Table 2). There are at least two possible explanations for these observations. One is that some traits are greatly influenced by heading date, and the QTL clusters represent the pleiotropic effects of heading date genes. Another is that genes involved in other agronomic traits are located near these heading date genes. In the present study, many traits seem to be affected by heading date, which corresponds to the former explanation. For examples, late heading would prolong vegetative growth period, increase plant biomass, and result in phenotype changes of culm length (CL), panicle length (PL), flag leaf length (FLL), aboveground dry weight (ADW), gross brown rice weight (GBW) and head brown rice weight (HBW). Days to maturity (DTM), pre-harvest sprouting resistance (PHS), panicle blast resistance (PBR), grain appearance (GA) and chalky grain ratios are mainly influenced by difference of accumulated temperature and amount of insolation at maturing stage, which are caused by heading date alteration. Lodging degree (LD) is one of important agronomic traits and is affected by culm length (CL), culm diameter (CDI) and culm stiffness (CSF). Eight QTLs for these four traits were detected near the two heading date QTLs of Hd16 and Hd17. But, the QTLs were not detected in the QTL analysis using the 51 BILs without large variation in heading date. These results suggest that variations in culm length (CL), culm diameter (CDI) and culm stiffness (CSF) are resulted from the alteration of heading date, and their contributions to lodging degree (LD) are explained by pleiotropic effects of the heading date QTLs. However, it is very hard to conclude with certainty the genetic basis of co-localization of QTLs for heading date and other traits based solely on the data obtained in this study. Further analysis such as fine-mapping using a large population will be required to make more definite conclusions about the reasons for co-localization of these QTLs. It will also be interesting to conduct the same type of QTL analysis using plant materials derived from different cross combinations.
The Nipponbare/Koshihikari BILs that were used in this study have previously been used in other QTL analyses for several traits. QTL analyses have been reported for days to heading (DTH) (Matsubara et al. 2008; Hd16 and Hd17), culm length (CL) (Hori et al. 2009; qCL1 and qCL3-1) and resistance to pre-harvest sprouting (PHS) (Hori et al. 2010; qLTG3-1). Here, we detected QTLs for these traits in the same chromosome regions as in these previous studies. We also compared the QTL positions for other traits with previous reports by using the QTL Annotation Rice Online database (Q-TARO), which is collecting data from QTL studies in rice, including the genomic locations and genetic parameters of each QTL (Yonemaru et al. 2010). QTLs for culm diameter (CDI) on the long arm of chromosome 1 and for 1000-grain weight (TGW) on the long arm of chromosome 11 were detected on similar chromosome regions to those reported by Mao et al. (2003) and Sasahara et al. (1999), respectively. The QTLs detected here for white-belly kernel ratio (WBR; a measure of grain chalkiness) on the short arm of chromosome 3 might be the same as the QTL for chalkiness detected by Kobayashi et al. (2007). Novel QTLs were detected for leaf blast resistance (LBR), panicle blast resistance (PBL) and bacterial leaf blight resistance (BLB). The 9 of 13 QTLs would associate with the strong resistance to leaf blast, panicle blast and bacterial leaf blight in Nipponbare.
In the present study, 30 traits were scored at multiple experimental fields. This allowed us to compare the additive effects of those QTLs in different environments. The 44 QTLs for 19 traits showed significant effects for genotype-by-environment interactions (Supplemental Table 2). These traits should be influenced by climate conditions, such as temperature and cultivation managements at each experimental field. For example, on head brown rice weight (HBW), effects of heading date were different among environmental conditions. Significant correlation with heading date was observed at KARC, but not at the other experimental field of NICS (Table 2). And, one QTL was detected at the Hd16 region at KARC, but no QTL was detected at NICS (Supplemental Table 2). The significant effect of heading date at KARC may be due to short-day length condition and resultant short period of vegetative growth. These particular genotype-by-environment interactions should be confirmed by using near-isogenic lines for the target QTLs. For application of the QTL information to breeding at different cultivation areas, it would be necessary to assess presence and extent of genotype-by-environmental interactions. The traits showing significant genotype-by-environmental interactions should be evaluated at experimental fields near the possible cultivation areas of the varieties under development.
A number of Japanese rice cultivars and elite breeding lines have been developed through crosses between temperate japonica cultivars by using conventional breeding approaches; for example, Nipponbare and Koshihikari originated from temperate japonica rice landraces and their progenies during the 1950s in Japan. The present breeding efforts might have been achieved by accumulating some of the desirable alleles at the QTLs associated with the target traits in breeding programs. For further agronomic improvement of rice cultivars, it will be necessary to identify additional QTLs and generate new combinations of desirable alleles. Based on the results obtained in the present study, we should reconsider the effectiveness of current strategies for selection of progeny from closely related cultivars of temperate japonica rice. A conventional phenotype-based selection strategy cannot be always applied effectively for the improvement of these primary target traits, because of the high level of correlation with heading date and small additive effect of some QTLs for other traits. Breeders have evaluated agronomic traits within only breeding materials showing the same heading date in the conventional breeding programs, to avoid pleiotropic effects of heading date. However, in cases of small population size and large variation for heading date in breeding populations, it is difficult to select the most preferable lines having all of the desirable QTL alleles due to linkages between the heading date QTLs and the other traits QTLs. Similar problems would be occurred in case of selecting individuals with preferred phenotypes on target traits to backcross breeding lines with different heading date. In the present study, 64 QTLs were detected in locations unlinked to heading date QTLs when the full set of BILs was used (Table 2 and Supplemental Table 2) and an additional 23 QTLs were found by QTL analysis of the 51 BILs each having the same chromosome segments around the two heading date QTLs (Supplemental Table 3). Therefore, it is important to accumulate a greater number of desirable alleles at QTLs that are independent from heading date to improve other target traits. Disease resistance and grain chalkiness traits for which a large number of QTLs have been detected will become good targets to accumulate the QTL alleles (Supplemental Tables 2, 3). Effective marker-assisted selection (MAS) and QTL pyramiding will be required to obtain novel cultivars of temperate japonica rice with high agronomic performance.
In general, it is difficult to combine desirable alleles at many loci simultaneously, because the expected probability of the desirable homozygous genotype in an F2 population is 0.25 for one target gene and 0.0625 for two target genes. A two- or three-step pyramiding strategy can be applied to solve this problem. For example, in this study, we identified one BIL (BIL645) based on the genotypes of DNA markers that showed the same genotype for heading date QTLs as Koshihikari and a preferable genotypic constitution (combination of QTL alleles) for many other traits. This type of line can be crossed with Koshihikari or other BILs to make a secondary segregating population to combine additional desirable QTL alleles. Additional individual selection must be done in the secondary segregating population to create the most desirable genotype. Multiple cycles of population development and selections of desirable individuals based on genotype would be one method to facilitate allele pyramiding at many loci. The success of this type of selection largely depends on the reliability of QTL analysis in the primary population, such as the BILs in the present study.
SNP genotyping provides detailed information about the genomic compositions of each breeding line and can distinguish even among temperate japonica rice cultivars (Arai-Kichise et al. 2011, Ebana et al. 2010, Nagasaki et al. 2010, Yamamoto et al. 2010). Previous studies detected a total of 81,449 SNPs from comparisons of genome sequences among temperate japonica rice cultivars (Nagasaki et al. 2010, Yamamoto et al. 2010). The large number of SNPs distributed throughout the rice genome can overcome the limitations once caused by an extremely low frequency of DNA markers in temperate japonica rice cultivars. High-density SNP genotyping would be a powerful tool for MAS and QTL pyramiding, allowing breeders to effectively select superior breeding lines containing all of the desirable QTL alleles which were detected in independent positions from heading date QTLs and showing high agronomic performance at multiple environmental conditions in temperate japonica rice breeding populations. Combination of new selection strategies and high-throughput genotyping technology will improve breeding success in rice.
Supplementary Materials
Acknowledgements
We thank all of the staff members in the rice breeding sections at the National Agriculture and Food Research Organization (NARO) Kyushu Okinawa Agricultural Research Center, the NARO Institute of Crop Science, the NARO Agricultural Research Center Hokuriku Research Center, the NARO Tohoku Agricultural Research Center, the Aichi Agricultural Research Center Mountainous Region Agricultural Research Institute, the Miyazaki Agricultural Research Institute and the National Institute of Agro-biological Sciences for their excellent technical support. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation NVR-0003).
Literature Cited
- Arai-Kichise Y., Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, Wakasa K. (2011) Discovery of genome-wide DNA polymorphisms in a landrace cultivar of Japonica rice by whole-genome sequencing. Plant Cell Physiol. 52: 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57: 289–300 [Google Scholar]
- Basten C.J., Weir B.S., Zeng Z.B. (2005) QTL Cartographer, version 1.17. Department of Statistics, North Carolina State University, Raleigh, p. 189 [Google Scholar]
- Ebana K., Yonemaru J, Fukuoka S, Iwata H, Kanamori H, Namiki N, Nagasaki H, Yano M. (2010) Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza sativa L.). Breed. Sci. 60: 390–397 [Google Scholar]
- Fujino K., Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin S.Y., Yano M. (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 108: 794–749 [DOI] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H. (2005) Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.). Theor. Appl. Genet. 111: 393–398 [DOI] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H. (2008a) Mapping of quantitative trait loci controlling heading date among rice cultivars in the northernmost region of Japan. Breed. Sci. 58: 367–373 [Google Scholar]
- Fujino K., Sekiguchi H., Matsuda Y., Sugimoto K., Ono K., Yano M. (2008b) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 105: 12623–12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Iwata N. (2011) Identification of QTLs controlling heading date on the short arm of chromosome 3 in rice (Oryza sativa L.). Breed. Sci. 62: 294–300 [Google Scholar]
- Guo Y., Hong D. (2010) Novel pleiotropic loci controlling panicle architecture across environments in japonica rice (Oryza sativa L.). J. Genet. Genomics 37: 533–544 [DOI] [PubMed] [Google Scholar]
- Hallauer A.R., Miranda J.B. (1988) Quantitative genetics in maize breeding, 2nd edn Iowa State University Press, Ames, p. 205 [Google Scholar]
- Hori K., Yamamoto T, Ebana K, Takeuchi Y, Yano M. (2009) A novel quantitative trait locus, qCL1, involved in semi-dwarfism derived from Japanese rice cultivar Nipponbare. Breed. Sci. 59: 285–295 [Google Scholar]
- Hori K., Sugimoto K, Nonoue Y, Ono N, Matsubara K, Yamanouchi U, Abe A, Takeuchi Y, Yano M. (2010) Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor. Appl. Genet. 120: 1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (IRGSP) (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Genliang B, Shenghai Y, Tomita K. (2007) Detection of quantitative trait loci for white-back and basal-white kernels under high temperature stress in japonica rice varieties. Breed. Sci. 57: 107–116 [Google Scholar]
- Kobayashi A., Tomita K, Yu F, Takeuchi Y, Yano M. (2008) Verification of quantitative trait locus for stickiness of cooked rice and amylose content by developing near-isogenic lines. Breed. Sci. 58: 235–242 [Google Scholar]
- Kobayashi A, Tomita K. (2008) QTL detection for stickiness of cooked rice using recombinant inbred lines derived from crosses between japonica rice cultivars. Breed. Sci. 58: 419–426 [Google Scholar]
- Kato T., Segami S, Toriyama M, Kono I, Ando T, Yano M, Kitano H, Miura K, Iwasaki Y. (2011) Detection of QTLs for grain length from large grain rice (Oryza sativa L.). Breed. Sci. 61: 269–274 [Google Scholar]
- Kwon S.J., Cho Y.C., Kwon S.W., Oh C.S., Suh J.P., Shin Y.S., Kim Y.G., Holligan D, Wessler S.R., Hwang H.G., et al. (2008) QTL mapping of agronomic traits using an RIL population derived from a cross between temperate japonica cultivars in rice (Oryza sativa L.). Breed. Sci. 58: 271–279 [Google Scholar]
- Kwon S.W., Cho Y.C., Lee J.H., Suh J.P., Kim J.J., Kim M.K., Choi I.S., Hwang H.G., Koh H.J., Kim Y.G. (2011) Identification of quantitative trait loci associated with rice eating quality traits using a population of recombinant inbred lines derived from a cross between two temperate japonica cultivars. Mol. Cells 31: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Green P., Abrahamson J., Barlow A., Daly M.J., Lincoln S.E., Newburg L. (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Mackill D.J. (1995) Classifying japonica rice cultivars with RAPD markers. Crop Sci. 35: 889–894 [Google Scholar]
- Mao B., Cai W.J., Zhang Z.H., Hu Z.L., Li P., Zhu L.H., Zhu Y.G. (2003) Characterization of QTLs for harvest index and source-sink characters in a DH population of rice (Oryza sativa L.). Yi Chuan Xue Bao 30: 1118–1126 [PubMed] [Google Scholar]
- Matsubara K., Kono I, Hori K, Nonoue Y, Ono N, Shomura A, Mizubayashi T, Yamamoto S, Yamanouchi U, Shirasawa K, et al. (2008) Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between Japonica rice cultivars. Theor. Appl. Genet. 117: 935–945 [DOI] [PubMed] [Google Scholar]
- McCouch S.R., Teytelman L, Xu Y, Lobos K.B., Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 257–279 [DOI] [PubMed] [Google Scholar]
- Mei H.W., Luo L.J., Ying C.S., Wang Y.P., Yu X.Q., Guo L.B., Paterson A.H., Li Z.K. (2003) Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two testcross populations. Theor. Appl. Genet. 107: 89–101 [DOI] [PubMed] [Google Scholar]
- Monden Y., Naito K, Okumoto Y, Saito H, Oki N, Tsukiyama T, Ideta O, Nakazaki T, Wessler S.R., Tanisaka T. (2009) High potential of a transposon mPing as a marker system in japonica × japonica cross in rice. DNA Res. 16: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H., Ebana K, Shibaya T, Yonemaru J, Yano M. (2010) Core single-nucleotide polymorphisms—a tool for genetic analysis of the Japanese rice population. Breed. Sci. 60: 648–655 [Google Scholar]
- Sasahara H., Fukuta Y, Fukuyama T. (1999) Mapping of QTLs for vascular bundle system and spike morphology in rice, Oryza sativa L. Breed. Sci. 49: 75–81 [Google Scholar]
- Shibaya T., Nonoue Y, Ono N, Yamanouchi U, Hori K, Yano M. (2011) Genetic interaction is involved in inhibition of heading by Heading date 2 in rice under long day conditions. Theor. Appl. Genet. 123: 1133–1143 [DOI] [PubMed] [Google Scholar]
- Tabata M., Hirabayashi H, Takeuchi Y, Ando I, Iida Y, Ohsawa R. (2007) Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.). Breed. Sci. 57: 47–52 [Google Scholar]
- Takeuchi Y., Hori K, Suzuki K, Nonoue Y, Takemoto-Kuno Y, Maeda H, Sato H, Hirabayashi H, Ohta H, Ishii T, et al. (2008) Major QTLs for eating quality of an elite Japanese rice cultivar, Koshihikari, on the short arm of chromosome 3. Breed. Sci. 58: 437–445 [Google Scholar]
- Wada T., Ogata T, Tsubone M, Uchimura Y, Matsue Y. (2008) Mapping of QTLs for eating quality and physicochemical properties of the japonica rice ‘Koshihikari’. Breed. Sci. 58: 427–435 [Google Scholar]
- Yamamoto R., Horisue N, Ikeda R. (1996) Rice Breeding Manual. Yokendo, Tokyo, Japan [Google Scholar]
- Yamamoto T., Nagasaki H., Yonemaru J., Ebana K., Nakajima M., Shibaya T., Yano M. (2010) Fine definition of the pedigree haplo-types of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics 11: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo M., Hirao M, Imai T. (2005) Annual change in leading rice varieties between 1956 and 2000 in Japan. Bull. Natl. Inst. Crop Sci. 7: 19–125 [Google Scholar]
- Yonemaru J., Yamamoto T., Fukuoka S., Uga Y., Hori K., Yano M. (2010) Q-TARO: QTL Annotation Rice Online Database. Rice 3: 194–203 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.