Abstract
The application of established drug compounds to novel therapeutic indications, known as drug repositioning, offers several advantages over traditional drug development, including reduced development costs and shorter paths to approval. Recent approaches to drug repositioning employ high-throughput experimental approaches to assess a compound’s potential therapeutic qualities. Here we present a systematic computational approach to predict novel therapeutic indications based on comprehensive testing of molecular signatures in drug-disease pairs. We integrated gene expression measurements from 100 diseases and gene expression measurements on 164 drug compounds yielding predicted therapeutic potentials for these drugs. We demonstrate the ability to recover many known drug and disease relationships using computationally derived therapeutic potentials, and also predict many new indications for these drugs. We experimentally validated a prediction for the anti-ulcer drug cimetidine as a candidate therapeutic in the treatment of lung adenocarcinoma, and demonstrate both in vitro and in vivo using mouse xenograft models. This novel computational method provides a novel and systematic approach to reposition established drugs to treat a wide range of human diseases.
INTRODUCTION
The identification of novel disease indications for approved drugs, or drug repositioning, offers several advantages over traditional drug development (1). The traditional paradigm of drug discovery is generally regarded as protracted and costly, with studies showing that it takes approximately 15 years and over $1 billion to develop and bring a novel drug to market (2). A substantial portion of drug development costs are incurred during early development and toxicity testing, with more than 90% of drugs failing to move beyond these early stages (3). The repositioning of drugs already approved for human use mitigates the costs and risks associated with early stages of drug development, and offers shorter routes to approval for therapeutic indications. Successful examples of drug repositioning include the indication of sildenafil for erectile dysfunction and pulmonary hypertension, thalidomide for severe erythema nodosum leprosum, and retinoic acid for acute promyelocytic leukemia (4).
The prevailing approach to drug repositioning is based on established tools and techniques developed for screening libraries of lead compounds against biological targets of interest in early stage drug discovery. In the case of drug repositioning, libraries of approved drugs are screened across a broad set of putative biological targets using high-throughput screening (HTS) technologies that incorporate multiplex biological assays (1, 4). Computational approaches to the discovery of novel biological targets for approved drugs have been developed to overcome the high-costs and other logistical limitations associated with experimental HTS approaches, allowing for greatly expanded searches for novel repositioning opportunities (5–10). While these approaches provide comprehensive and systematic means to discover novel biological targets for existing drugs, they do not address the challenge of moving from in silico or in vitro binding of a drug and a novel biological target to realized therapeutic utility in affected patients.
Gene expression microarrays (11, 12) enable the measurement of genome wide expression levels, and they are regularly and broadly applied in clinical studies of human diseases. Comparative gene expression analysis of primary affected, peripheral and secondary organs, tissues and biofluids are used to study the molecular pathophysiology of a disease condition, or to identify expression patterns that serve as prognostic or diagnostic indicators. By examining the sets of genes that are up regulated and down regulated in a disease state as compared to a normal state, it is possible to create a gene expression profile, or signature, of a disease (13–17). When derived from clinical samples, these signatures represent the accurate and consistent gene expression state imparted by immunological, metabolic, and other complex factors comprising the broad physiological manifestation of a disease (18, 19).
Microarrays are also used to discover gene expression patterns that signify the perturbation of biological systems by drug compounds (20–24). A collection of genome-wide transcriptional expression data from cultured human cells treated with a broad range of FDA approved bioactive small molecules (25) has been used to study the molecular basis of drug effects, particular diseases of interest like colon cancer (26) or prostate cancer (27).
In this study, we performed a large-scale integration of expression signatures of human diseases from the public data with the drug effect signatures previously found (25). Using this approach we built a compendium of disease-drug relationship predictions based on matching genome-wide signatures of disease pathophysiology and drug effect. Instead of examining a single drug-disease pair or even looking at reactions of a large set of drugs on a single disease, we focused on discovering connections between drugs and diseases across all the available gene measurements. Here we present evidence that this strategy can confirm already-known therapeutic uses for drugs and uncover new uses for FDA-approved drugs.
RESULTS
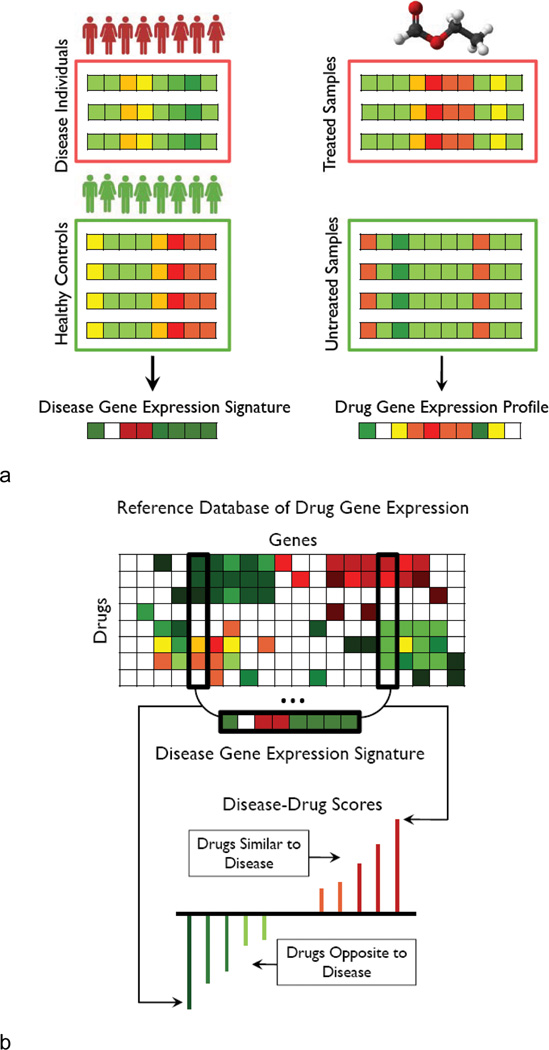
We identified and combined data from publicly available microarray datasets from NCBI Gene Expression Omnibus (GEO) representing 100 diseases with gene expression data from several human cancer cell lines treated with 164 drugs or small molecules to predict previously undescribed therapeutic drug-disease relationships. We then defined a disease signature for each of the 100 diseases as a set of genes that are significantly up and down regulated for that disease compared to normal using Significance Analysis of Microarrays (SAM) (19, 28). Next, we statistically compared each of the disease signatures to each of the reference drug expression signatures from the Connectivity Map (25) to infer possible therapeutic indications based on gene expression. A schematic overview of our methodology is shown in Figure 1. We calculate a similarity score for every pairing of a drug and disease reflecting the similarity of the drug and disease signatures, ranging from +1 indicating a perfect correlation of signatures to −1 indicating exactly opposite signatures. Our hypothesis is that if a disease state is signified by a specific set of genome-wide expression changes, and if exposure to a particular drug causes the reverse set of changes in a model cell line (i.e. negative similarity scores indicating an opposing relationship), then that drug has the potential to have a therapeutic effect on that disease.
Figure 1.
Analytic workflow. A) Two gene expression collections: a set of disease gene expression with corresponding controls and gene expression of tissue treated drugs and small molecules with corresponding controls. Significance Analysis of Microarrays (SAM) is used to obtain a signature of significantly up and down regulated genes for each disease. Rank normalization and the pre-processing procedure previously described is used to create a reference database of drug gene expression. B) A modification to the Connectivity Map method (25) is used to query the disease signature against the drug reference expression set to assign a drug-disease score to each drug-disease pair based on profile similarity. These scores are interpreted resulting in a list of candidate therapeutics for each disease of interest.
In order to evaluate the significance of our predictions, we employed a permutation approach under which random drug signatures were generated and the analysis was repeated 1,000 times for each disease. We computed the false discovery rate for individual drug-disease similarity score values, or q-value by calculating the expected number of false positives, given the actual distribution of similarity scores and the distribution of scores after randomization for each disease signature.
Out of more than 16,000 possible drug-disease pairings, 2,664 were statistically significant (q-value < 0.05), more than half of which suggest a therapeutic (negative or opposite) relationship. Overall, our method provided significant candidate therapeutic drug-disease relationships for 53 out of the 100 diseases at a false discovery rate of 5%. The other 47 diseases could not be significantly associated with any of the 164 drugs we tried and were excluded from further consideration. Each of the 164 drugs was significantly associated with at least one of the 53 remaining diseases.
Many cancers, including gastric cancer, melanoma, and transitional cell carcinoma held the highest number of significant matches to therapies (Table 1). The drug predicted to be efficacious for the largest set of diseases was vorinostat, a histone deacetylase (HDAC) inhibitor with significant therapeutic association with 21 out of the 100 diseases in our dataset. Other HDAC inhibitors, such as trichostatin A and Helminthosporium carbonum (HC) toxin, as well as gefitinib, an EGFR inhibitor used in cancer treatment had over 20 significant predicted therapeutic indications (Table 1).
Table 1.
| Top 10 Drugs with Most Indications |
Top 10 Diseases with Most Indications | ||
|---|---|---|---|
| vorinostat | 21 | Transitional cell cacinoma | 95 |
| Gefitinib | 18 | Melanoma | 79 |
| HC toxin | 18 | Cardiomyopathy | 73 |
| Colforsin | 17 | Adenocarcinoma of lung | 72 |
| 17-dimethylamino-geldanamycin | 16 | Multiple benign melanocytic nevi | 68 |
| trichostatin A | 16 | Squamous cell carcinoma of lung | 67 |
| 3-hydroxy-DL-kynurenine | 15 | Malignant neoplasm of stomach | 66 |
| 5114445 | 15 | Dermatomyositis | 63 |
| dexverapamil | 15 | Malignant mesotheliomaof pleura | 53 |
| prochlorperazine | 15 | Primary cardiomyopathy | 48 |
Global properties of drug-disease connections
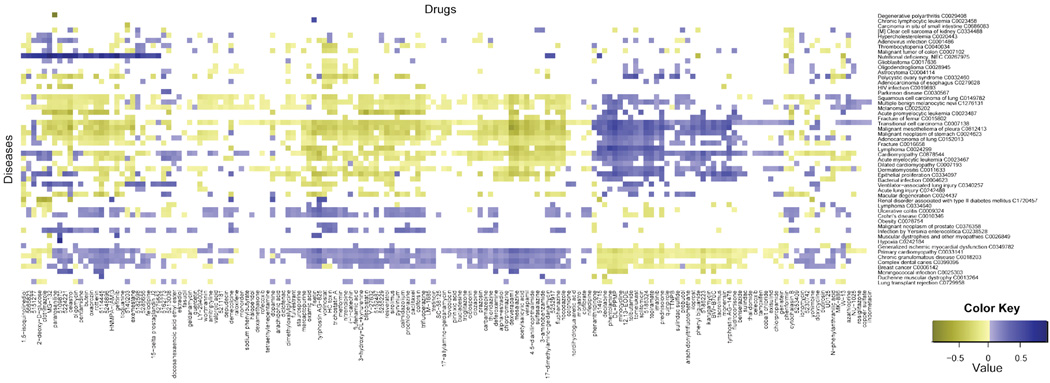
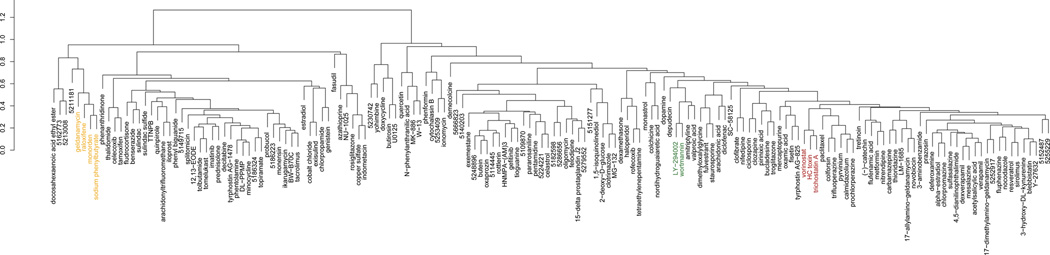
By examining of the relationships between the 164 drugs and 53 diseases based on predicted therapeutic scores, we identified major clusters of predicted therapeutic relationships between drugs and diseases (Fig. 2). We then looked separately at the clustering of drugs based on their similarity scores across diseases (Fig 3a). Drugs with similar mechanisms of action clustered together. For example, the histone deacetylase inhibitors vorinostat, HC toxin, and trichostatin A formed a cluster, as did the phosphatidylinositol-3-kinase (PI3K) inhibitors LY−294002 and wortmannin, and the salicylate anti-inflammatory drugs sulfasalazine, mesalazine, and acetylsalicylic acid. We also found clusters of drugs that are known to affect different aspects of the same biological process. For example, geldanamycin, raloxifene, monorden, and sodium phenylbutyrate are all known to perturb the activity of heat shock protein 90 (HSP90) chaperone complex, either through direct inhibition or perturbation of upstream factors (29) and these agents also clustered together (Table 1).
Figure 2.
Heatmap of drug-disease scores. Most of the heatmap is white, indicating the majority of drug and gene expression profiles are not significantly concordant. Yellow indicates a negative (therapeutic) drug-disease score meaning the expression profiles of the two are opposing, and that the drug might be a potential treatment option of the disease. Blue indicates a positive drug-disease score meaning the expression profiles of the two are similar, and therefore the drug may be antagonistic towards the disease.
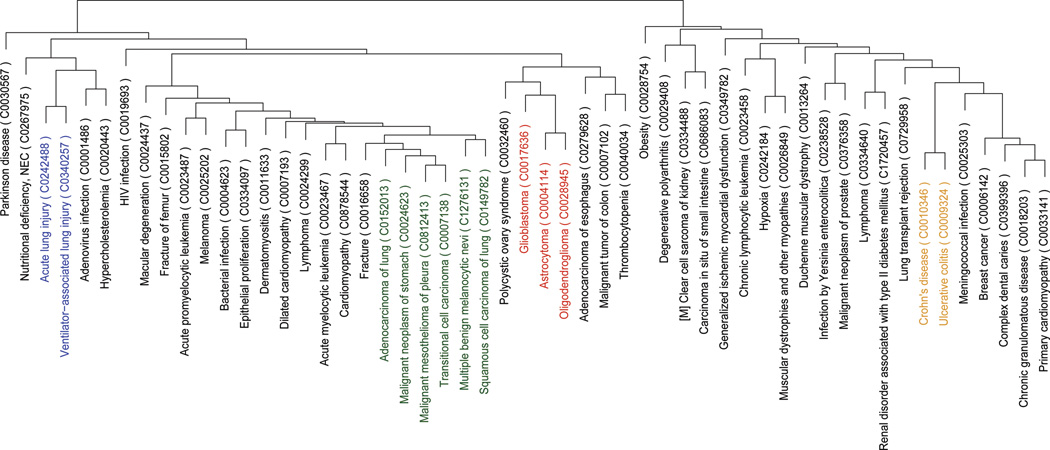
Figure 3.
Hierarchical clustering of drugs and diseases by predicted therapeutic scores. A) Drugs are clustered based on their prediction scores. Several groups are highlighted in color representing clusters of drugs with known shared mechanisms of action. B) Diseases are clustered based on their prediction scores. Groups highlighted with color are known to share characteristic pathophysiology.
Diseases were similarly clustered based on their computed therapeutic scores across the panel of drugs (Fig. 3b). We find a large cluster of cancers, which includes lung, stomach and other cancers pointing to potential commonality of predicted therapeutic response between these conditions (Fig. 3b; green). The inflammatory bowel diseases (IBD) ulcerative colitis (UC) and Crohn’s disease (CD) appeared together as part of a larger cluster of diseases for which corticosteroids and other immunosuppressive drugs are broadly indicated (Fig. 3b; yellow). IBD was clustered near infection by Yersinia enterocolitica, which can present clinical symptoms similar to those of IBD (30).
The reported clusters were determined to be significant by bootstrap analysis (31).
Prediction of known and novel drugs for individual diseases
In addition to examining overarching relationships between groups of drugs and diseases, we also present individual therapeutic predictions based on the hypothesis that if a drug has a gene expression signature that is opposite of a disease signature, that drug could potentially be used as a treatment for that disease. One of the strongest therapeutic predictions for Crohn’s disease and UC is the corticosteroid prednisolone, a well-known treatment for these conditions (32), with a score of −0.216. Interestingly, we found that topiramate, an anti-convulsant drug currently used to treat epilepsy, has a stronger therapeutic score for Crohn’s than the established therapeutic prednisolone, with a score of −0.220, shown by the comparison of the Crohn’s disease signature to the gene expression profile of topiramate. Genes that are up-regulated in disease are down-regulated by the drug and vice-versa, reflecting the significant score. Topiramate is also one of the strongest predicted therapies for ulcerative colitis based on our analysis, and the efficacy of topiramate in ameliorating an IBD phenotype was established in an independent study using a TNBS rat model of colitis (Dudley J and Sirota M et al. co-appearing in this issue).
Prediction and validation of cimetidine as a novel indication for lung adenocarcinoma
To perform an initial experimental evaluation of our approach, we chose to evaluate one of the therapeutic predictions for lung adenocarcinoma (LA), as lung cancer contributes the greatest burden of cancer mortality and incidence in Europe and the US (33). Although our methodology predicted multiple novel therapeutic relationships for LA, we chose to test cimetidine as it is an off-patent and inexpensive drug available over-the-counter in the United States, and has a favorable side-effect profile (34). Our prediction score of −0.088 for cimetidine was moderate, but still more significant than the score of −0.075 for gefitinib, a well-known therapy for LA.
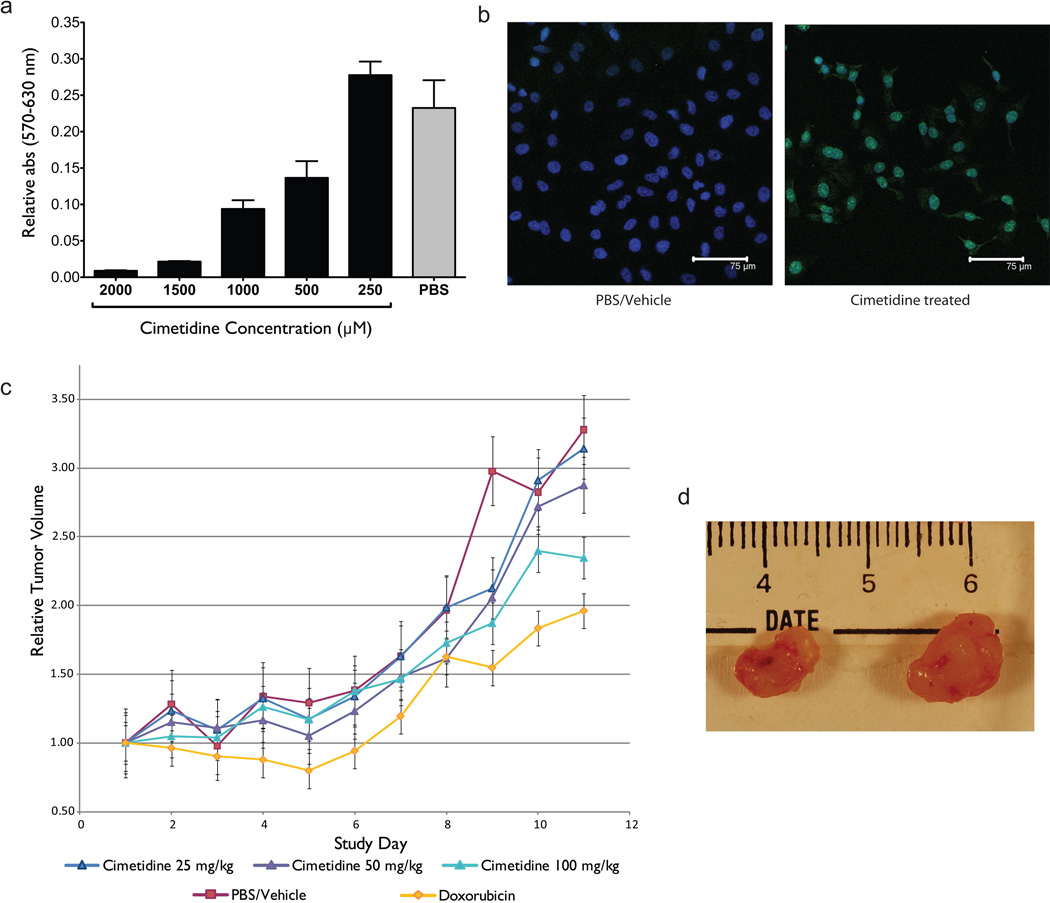
To evaluate the efficacy of cimetidine against LA in vitro, we performed an MTT colorimetric assay to assess growth and proliferation of LA cells (A549) after exposure to cimetidine, and assessed apoptosis through detection of DNA fragmentation by TUNEL assay. LA cells exposed to increasing concentrations of cimetidine exhibited a dose dependent reduction in growth and proliferation compared to cells treated with PBS vehicle (Fig. 4a). LA cells also exhibited extensive apoptosis one day after direct exposure to 2,000 µM of cimetidine (Fig. 4b right; green=TUNEL-positive) whereas apoptosis was not detectable by TUNEL fluorescent microscopy in LA cells from the same line after exposure to PBS/vehicle solution.
Figure 4.
Experimental validation of cimetidine for lung adenocarcinoma. A) MTT calorimetric assay showing dose-dependent inhibition of lung adenocarcinoma cell growth after exposure to cimetidine in vitro. B) Evaluation of apoptosis by TUNEL assay. Lung adenocarcinoma cells treated with 2000 µM cimetidine exhibit a significant increase in TUNEL-positive (green) nuclei compared to vehicle (PBS) treated control. C) Results from a tumor xenograft experiment testing the efficacy of H2-agonist cimetidine in inhibiting the growth of A549 lung adenocarcinoma cell line tumors in SCID mice. Three treatment groups (25/50/100 mg/kg/injection) and one control group (PBS) was used. Another group was treated with doxorubicin as a positive control. Concordant with our prediction, the results demonstrate that cimetidine shows efficacy in inhibiting the growth of NSCLC xenograft implant tumors in a dose-dependent manner. D) Representative images of tumors treated with high dose of cimetidine (left) and control (right).
To further evaluate the efficacy of cimetidine against LA in vivo, we tested three doses of cimetidine in mouse models explanted with a human A549 LA cell line, and followed the growth of these tumors for 12 days. Tumors in mice treated with vehicle (Fig. 4c, red line) grew to 3.25× original volume, while tumors in mice treated with our positive-control dose of the chemotherapeutic doxorubicin grew to 2× original volume (Fig. 4c, orange line). Cimetidine caused a statistically significant reduction in growth as compared to control (100 mg/kg/day group vs. PBS control on day 12; t-test P<0.001), with the highest dose of cimetidine approaching the efficacy of the positive control therapy doxorubicin. Cimetidine exhibited its anti-neoplastic properties in a dose-dependent manner, with tumors in mice treated with 50 mg/kg/day (Fig. 4c, purple line) growing to 2.8× original size, and those treated with 100 mg/kg/day (Fig. 4c, light blue line) growing only 2.3× (Tukey’s HSD P-value < 0.05). Figure 4d shows representative examples of tumors treated with a 100 mg/kg/day of cimetidine (left) in comparison to control (right).
To test the specificity of our predictions, we performed a similar mouse explant experiment using the ACHN renal cell carcinoma cell line (Fig. S1). We chose ACHN as a control for this experiment because it did not have any significant association with cimetidine according to our analysis (score 0, P-value=1.0). Concordant with our computational prediction, we found that cimetidine did not exhibit efficacy against renal cell carcinoma tumors in a mouse xenograft model (Fig. S1).
DISCUSSION
In this study, we sought to discover novel therapeutic relationships between drug compounds and diseases through the large-scale integration of public gene expression data for drugs and disease. Our hypothesis was that a comprehensive and systematic comparison of drug and disease gene expression signatures could be used to build a global space of drug-disease relationships, which could be explored to identify novel therapeutic relationships between drugs and diseases conditions. The global clusterings of drugs and diseases by predicted therapeutic scores revealed known thereapeutic clusters of drugs and diseases, and also provided pathophysiological context to support interpretation of novel predicted therapeutic relationships.
In clusters of drugs based on their gene signature similarity across diseases, we observe shared mechanisms of action (e.g. HDAC inhibition) or shared physiological processes (e.g. immunomodulation). In some cases the axis of commonality is not apparent within drug clusters, and such cases may serve to illuminate our understanding of a drug’s mechanism of action where the mechanism is presently unknown, or even suggest new biology. For example, we found a cluster containing the salicylate drugs sulfasalazine, mesalazine (a metabolite of sulfasalazine), and acetylsalicylic acid grouped with the calcium channel blocker verapamil and its R-enantiomer, dexverapamil. The two distinct drug types represented in this cluster are not known to share a mechanism of activity and are not generally considered to be interchangeable or complementary therapeutic options for a common disease indication. Although the underlying mechanism is unclear, both mesalazine and sulfasalazine are used as maintenance drugs in the treatment of the inflammatory bowel disorders Crohn’s disease and ulcerative colitis. A possible therapeutic role for verapamil in inflammatory bowel disease (IBD) is established by several studies, where it has been shown to reduce inflammation in ulcerative colitis through leukotriene inhibition (35) and promote mucosal healing in experimental colitis (36). Further study into the genes and potential additional drug targets driving this clustering could reveal new information about IBD biology and new therapeutic directions.
In the clustering of diseases based on their profile across drug signatures, we found consistency within the clustering based on established therapeutic conventions and likely shared pathophysiology. The major cluster containing Crohn’s disease and lung transplant rejection also harbors a number of diseases for which corticosteroids and other immunosuppressants are regularly prescribed. The large cluster of cancers share many of the same anti-neoplastic and immunomodulatory therapeutics. Less evident groupings, such as that between polycystic ovary syndrome (PCOS) and glioblastoma, may present opportunities for drug repositioning within a cluster. However in some cases, such groupings may have alternative explanations that are not necessarily indicative of an opportunity for drug repositioning. For example, the peculiar clustering of cardiomyopathies with cancers could be a reflection of the fact that many chemotherapeutics are known to induce cardiomyopathies as a side effect (37).
In addition to examining broader relationships between groups of drugs and diseases, we also discovered individual therapeutic predictions for 53 out of 100 diseases in our dataset. It is not surprising that a large proportion of the diseases we studied do not have any significant therapeutic predictions, since the set of drugs we examined was limited, and the baseline assumption is that most drugs will not promiscuously exhibit therapeutic potential for many other diseases.
Many of our therapeutic predictions recapitulate established therapeutic knowledge. For instance, we predict that HDAC inhibitors (trichostatin A, valproic acid, vorinostat, and HC toxin) have therapeutic effects in treatment of different types of brain tumors (astrocytoma, glioblastoma, oligodendroglioma) as well as other cancers (esophagus, lung, colon). HDAC inhibitors are known to be anticancer agents that act by inhibiting cancer cell proliferation and inducing apoptosis (38–40). Trichostatin A was recently shown to restore the loss of NECL1, a tumor suppressor in glioma (41). Valproic acid is a drug used to treat epilepsy and bipolar disorder but has been more recently reported to be effective in the treatment of various cancers including glioma (42), myeloma (43) and melanoma (44) often in combination with other therapies. Vorinostat is currently used for the treatment of several lymphomas, but has recently undergone successful phase II trials for treatment of recurrent glioblastoma multiforme (GBM) showing that vorinostat is well tolerated in patients with recurrent GBM and has modest single-agent activity (45). Recent studies have also shown that HC toxin effectively suppresses the malignant phenotype of neuroblastoma cells (46, 47). Our prediction of celastrol, an antioxidant and anti-inflammatory drug, for the treatment of melanoma is also supported by previous research, which shows that the compound resembles the well-characterized ATF2 peptide and may therefore offer new approaches for treatment (48).
After screening a larger set of compounds, we also predicted and validated cimetidine as a possible therapeutic agent for lung adenocarcinoma. Cimetidine is a histamine H2 receptor (H2R) antagonist that acts as an inhibitor of acid production in the stomach. It is used and prescribed most often as an over-the-counter drug that is used in treatment of heartburn and peptic ulcers. Cimetidine has previously demonstrated efficacy in other adenocarcinoma models (49–52). A molecular mechanism or other molecular basis for the observed efficacy of cimetidine against lung adenocarcinoma has not been previously reported or otherwise described. Initial reports of cimetidine’s anti-tumor activity proposed that cimetidine inhibits tumor growth via immunomodulation (53–56). However, as demonstrated by the in vitro and in vivo experimental results of this study, cimetidine inhibits tumor growth and cellular proliferation in immune-deficient hosts and by direct exposure to LA cells. More recent studies have demonstrated that cimetidine is able to inhibit vascular development in tumors, suggesting that cimetidine might work to inhibit tumor-associated angiogenesis (51, 57). Additional studies into the antineoplastic properties of cimetidine have revealed putative roles in the inhibition of tumor cell adhesion and induction of apoptosis (58, 59). It is also possible that the efficacy of cimetidine for lung adenocarcinoma is mediated by some yet unknown interaction between cimetidine and a target outside of its canonical H2R target.
Despite computational and experimental validation of our approach, there are several caveats to consider. One potential drawback to the approach described here is the need to have gene expression profile measurements on the candidate drugs being evaluated. In addition, it is not clear how drug performance in a breast cancer cell line, used extensively to measure transcriptional response of drugs in ConnectivityMap, is relevant to all types of diseases. However, the number of publicly available data reporting the effects of drugs on gene expression across disease tissues continues to grow (60). Furthermore, disease-related microarray data could be combined with other types of knowledge on drugs (61, 62) for computational prediction of drug side effects.
Therapeutic efficacy is always more complex than just a simple matching of expression profiles. For example compounds have to reach the desired or appropriate tissue to have an effect. However, the tissue-agnostic methodology used in this analysis might be suitable to find both direct and distant effects of drugs. Although our finding for cimetidine will need further preclinical testing and demonstration in larger randomized prospective clinical trials, our results validate the concept of computational analysis of public gene expression databases as a potentially useful approach to drug discovery that may uncover additional uses for approved drugs.
MATERIALS AND Methods
We combined data from publicly available microarray datasets representing 100 diseases and gene expression data from human cell lines treated with 164 drugs or small molecules in order to predict therapeutic drug-disease interactions.
Drug and Disease Gene Expression Data
Disease expression data was obtained from the NCBI Gene Expression Omnibus (GEO) (63, 64), using methods previously described (65–67). We used 176 gene expression microarray data sets, each with samples reflecting disease tissue and control non-disease tissue (Supplementary Table 1). These data sets covered 100 diseases, totaling 3113 microarrays (Supplementary Table 1). A data set with measurements for lung adenocarcinoma and adjacent normal tissues measured from human patients (GSE2514) was among the 176 data sets used for the study.
Rank normalization was applied to the disease expression data in order to carry out robust cross-platform analysis (68, 69). Since experiments were run on different platforms, we standardize gene identifiers from chip specific probe identifiers to NCBI GeneID identifiers using AILUN (70) in order to be able to reason across multiple experiments. In cases where multiple microarray probes mapped to the same NCBI GeneID, we averaged across individual probe expression values. On each data set, we performed Significance Analysis of Microarrays (SAM) (28) comparing control samples to disease samples to generate a list of up-regulated and down-regulated genes for each disease state. We used a false discovery rate threshold of 0.05 for q-values. The list of significantly up and down regulated genes for each disease-control comparison constitutes a disease signature (Fig. 1).
Drug-exposure gene expression microarray data measured on several types of cancer cell lines in the context of 164 distinct small molecules was obtained from Lamb et al. (25). Most of the experiments were carried out on the breast cancer epithelial cell line MCF7, but a subset of the compounds were also profiled in the prostate cancer epithelial cell line PC3 and the nonepithelial lines HL60 (leukemia) and SKMEL5 (melanoma). The original experiments were carried out on two platforms: GEO Platform (GPL) 96 Affymetrix GeneChip Human Genome U133 Array Set HG-U133A and GPL3921 Affymetrix GeneChip HT-HG_U133A Arrays. In order to be able to reason between drug and disease expression data, we standardized gene identifiers from microarray specific probe identifiers to NCBI GeneID identifiers, averaging across individual probe expression values (Fig. 1). We followed the pre-processing and normalization steps described in the Supplemental materials in Lamb et al. (25).
Computational Pipeline
We used the drug-exposure expression data as a reference database and queried this database with each individual disease signature by applying a non-parametric, rank-based pattern-matching strategy based on the methodology originally introduced by Lamb et al. (25) in order to generate a ranked list of potential treatments for each of the diseases of interest (Fig. 1). Given a disease signature, we evaluated its similarity to each of the reference expression drug profiles in the drug expression set by computing an enrichment score for the up-regulated and the down-regulated disease genes. If the up-regulated disease genes appear near the top (up-regulated) of the rank-ordered drug gene expression list and the down-regulated disease genes fall near the bottom (down-regulated) of the rank-ordered drug gene expression list, we can conclude that the drug and the disease expression profiles are similar and thus the drug might cause a change in tissue expression similar to having the disease. More interestingly, if the up-regulated disease genes fall near the bottom of the rank-ordered drug gene expression list and the down-regulated disease genes are near the top of the rank-ordered drug gene expression, then the given drug and disease have complimentary expression profiles and the drug might be a possible treatment option for the disease of interest.
For each disease we compute an enrichment score (es) separately for the set of up- or down- regulated genes in the signature: esup and esdown. We construct a reference drug expression set by taking the difference between the gene expression values of the samples treated with a compound of interest and untreated samples. Furthermore we carry out rank normalization on the resulting gene expression differences. Let r be the total number of genes in the reference drug expression set and let s be the number of up- or down- regulated of each gene in the disease signature. We first construct a vector V of the position (1… r) for each of the genes in the disease signature based on the values from the reference drug expression set. Those are sorted in ascending order such that V(j) is the position of disease gene j, where j = 1, 2, … s. Then, for each set of up- and down- regulated disease genes, we compute aup, adown, bup and bdown defined as:
If aup/down > bup/down, we set esup/down to aup/down, and if bup/down > aup/down, we set esup/down to -bup/down. The drug-disease score dds is set to zero where esupand esdown have the same algebraic sign. Otherwise, we set dds = esup - esdown. For more detail, see Supplemental materials in Lamb et al. (25).
In the original method proposed by Lamb et al. the final drug-disease score was scaled from +1 to −1. In using this scaling, every disease signature was able to match some drug profile at a maximal level. Because the scores for each disease were assigned a unique scaling factor, the strength or confidence of one drug-disease association could not be compared with another drug-disease association. We thus chose not to carry out this final scaling step, instead using the resulting raw values as drug-disease scores. In order to present the results in a more readable fashion, we choose to average the scores by drug dose and cell line although we do realize that this might dampen some of the signal. Finally, we ranked the list of drug and small molecule instances for each disease from most correlated to most anti-correlated, according to the computed similarity drug-disease score.
As a control, for every disease signature in our dataset, we chose a random signature of the same size and recomputed the drug-disease scores for each drug. We repeated this experiment 100 times. The standard deviation of the original scores is statistically significantly greater than the scores generated from random signatures (Levene’s Test p-value < 2.2 × 10−16) meaning that there were more significant therapeutic indications in our resulting dataset then expected by chance.
In order to obtain a measure of significance on the drug-disease scores, we start by computing a p-value for each drug-disease score by comparing the distribution of actual scores in comparison to those obtained on randomized data for each disease signature. We further used the qvalue (71) statistical package to compute tail area-based false discovery rate (FDR) for each drug-disease score in our result given the previously computed p-values. Based on this analysis, we are able to establish the cutoff for significance for drug-disease scores for each disease signature yielding a false discovery rate of 5%.
To identify disease and drug classes, hierarchical cluster analysis is applied to the data using the computed Pearson correlation coefficients as a distance metric between disease or drug pairs. Initially, each disease or drug is assigned to its own cluster. The algorithm proceeds iteratively, at each stage joining the two most similar clusters, until there is just a single cluster left. We use the Pvclust R package (31) to compute a bootstrap analysis of the clusters. The bootstrap probability of a cluster corresponds to the frequency with which the cluster appears in bootstrap samples of the data. Approximately Unbiased (AU) probability values are computed using bootstrap samples of various sizes and indicate how strongly the cluster is supported by data (AU>95%).
Evaluation of Cimetidine as a Chemotherapeutic Agent for Non-small Cell Lung Carcinoma (NSCLC)
To evaluate our prediction we performed experimental validation using tumor xenograft models of non-small cell lung carcinoma. Our experimental design was based on two human cancer cell lines that were implanted into SCID mice. 30 mice were implanted with the A549 human adenocarcinoma cell line (5 million/µl PBS injection into upper flanks) and divided into 5 groups comprising 6 mice each. Three groups received varying doses of cimetidine ranging from 25–100 mg/kg daily via IP injection. One group was injected with 2 mg/kg doxorubicin IP bi-weekly as a positive control, and the final group received only PBS/vehicle as a control. The experiment was carried out for 12 days after the tumor implants reached a volume of 100 mm3. Tumor volumes were measured by caliper using the following equation (mm3=0.52× (width (cm))2 × height (cm)).
To evaluate the specificity of our approach, we performed a parallel experiment using the ACHN renal carcinoma cell line (5 million/µl PBS injection into upper flanks), as our system predicted that cimetidine would not be efficacious against this cancer type. The same experimental design and protocol used to evaluate mice implanted with the A549 cell line was used for the animals implanted with ACHN. Differences in tumor volume between groups were evaluated on the final day of dosing using Tukey’s Honest Significant Difference (HSD) test, and also Student’s t-test between the highest does of cimetidine vs. the PBS control. Mouse xenograft experiments were performed by the Transgenic Mouse Research Center core facility at the Stanford Comprehensive Cancer Center. Cell lines were verified to be free of murine pathogens by RADIL (MO, USA). All chemicals and reagents were purchased from Sigma-Aldrich Corporation (MO, USA).
Colorimetric MTT assay for cell survival and proliferation
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay reagents from Millipore (Temecula, CA) were used for cell survival and proliferation. The assay was carried out according to the manufacturer’s instructions. A549 cells were seeded at 5,000 cells per well in 96 well plate 15–18 hours before the start of the treatment. Cimetidine solution (250mM) was prepared by adding 0.63g of cimetidine in 8ml of PBS then adding 10N of HCl (200µl) and 10N of NaOH (150µl). Then the pH was adjusted to the physiological level and total volume was adjusted to 10ml. Cells were treated 3 times with cimetidine (250µM, 500µM, 1,000µM, 1,500µM and 2,000µM) using 250mM stock solution) at 11am, 6pm and at 11am the next day followed by MTT assay at 4pm. After the treatments, 0.01ml of MTT (Millipore CT01-5, 50mg/ml in PBS) solution was added to each well and the cells were incubated for 4 hours at 37°C for the cleavage of MTT to occur. Then, color development solution (isopropanol with 0.04N HCl, 0.1ml each) was added and mixed thoroughly. Within an hour, absorbance was measured at 570nm and at a reference wavelength at 630nm. For the calculation, absorbances measured at 570nm were subtracted with those measured at 630nm.
TUNEL assay
TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) assay was carried out according to the manufacturer’s instructions (Roche, Cat No. 11684809910). A549 cells were seeded at 20,000 cells per well in a Nunc 4 well Lab Tek chamber slides in DMEM with 10% FBS. Cells were seeded 15–18 hours before the start of the treatment. Cells were treated 2 times with cimetidine (500µM–3000µM using 250mM stock solution) at 11am and at 6pm and were stained the next day. Briefly, medium was removed, cells were washed in cold PBS and the cells were air-dried. Then, freshly made 4% paraformaldehyde solution was added and cells were fixed for 15 minutes in room temperature. Cells were rinsed with PBS and were incubated with permeabilization solution (0.1% triton X-100) for 2 min on ice. Next, Terminal deoxynucleotidyl transferase and label solution were mixed and 50µl each of the mixture was added to each well. Cells were incubated for 1 hour in a humidified chamber at 37°C. For negative controls, no label solution was added. Slides were covered with cover slips to ensure proper staining. Cells were then washed with cold PBS 3 times and dried. Finally, Hoechst solution for nuclei staining (Sigma, St. Louis, MO) was diluted 1:10,000 in PBS and was added to the cells for 1–2 minutes. Cells were rinsed with PBS 3 times and 2 times in water. Cells were applied with aqueous mounting medium and cover slipped. Immunofluorescence was measured using a Leica confocal microscope at the Cell Sciences Imaging Center at Stanford Cancer center.
Supplementary Material
Results from a tumor xenograft experiment testing the efficacy of H2-agonist cimetidine in inhibiting the growth of ACHN renal carcinoma cell line tumors in SCID mice. Three treatment groups (25/50/100 mg/kg/injection) and two groups of controls were used. One control group was administered the PBS vehicle, and the other control group was left untreated. The results demonstrate that cimetidine lacks efficacy in inhibiting the growth of renal carcinoma xenograft implant tumors which is in agreement with the prediction rendered by our in silico repositioning method.
Table 2.
Significant Drug Clusters Described in Text. Drugs were clustered in an unsupervised manner by their predicted therapeutic score across 100 diseases (See Fig.3a)
| Drug Group | Drugs |
|---|---|
| PI3K Inhibitors | LY−294002 and wortmannin |
| HSP90 | geldanamycin, raloxifene, monorden, and sodium phenylbutyrate |
| HDAC Inhibitors | vorinostat, HC toxin, and trichostatin A |
| Salicylate anti-inflammatory | sulfasalazine, mesalazine, and acetylsalicylic acid |
Table 3.
Significant Disease Clusters Described in Text. Diseases were clustered in an unsupervised manner according to their predicted therapeutic response across 164 drug compounds (See Fig. 3b).
| Canonical | Non-canonical |
|---|---|
| Cancers | Crohn’s Disease and Lung Transplant |
| Ulcerative Colitis and Crohn’s Disease | Polycystic Ovary and Glioblastoma |
| Cardiomyopathy and Cancer |
Summary.
We developed a systematic computational method to predict novel therapeutic indications through integration of public gene expression signatures of drugs and diseases, and demonstrate the ability recover many known drug and disease relationships, as well as predict novel therapeutic relationships -- one of which is experimentally validated in a preclinical model for lung adenocarcinoma.
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences (R01 GM079719), the National Cancer Institute (R01 CA138256), Howard Hughes Medical Institute, Pharmaceutical Research and Manufacturers of America Foundation, Lucile Packard Foundation for Children's Health, the Hewlett Packard Foundation, US National Library of Medicine (T15 LM007033). We thank Alex Skrenchuk and Boris Oskotsky from Stanford University for computer support, and Eugene Davydov, Chuong Do, Samuel Gross and Marc Schaub from Stanford University for constructive discussion. We thank Qi Zheng of the Transgenic Mouse Research Center at the Stanford Comprehensive Cancer Center for help in conducting the mouse xenograft experiments.
LIST OF ABBREVIATIONS
- AILUN
Array Information Library Universal Navigator
- FDA
Food and Drug Administration
- FDR
False Discovery Rate
- GEO
Gene Expression Omnibus
- HDAC
Histone Deacetylases
- NCBI
National Center for Biotechnology Information
- SAM
Significance Analysis of Microarrays
- MDS
Macroscopic Damage Score
- TNBS
trinitrobenzenesulfonic acid
- UC
Ulcerative Colitis
- CD
Crohn’s Disease
- IBD
Inflammatory Bowel Disease
- H&E
Hematoxylin and Eosin
- GPL
GEO Platform
- PCOS
Polycystic Ovary Syndrome
- GBM
Glioblastoma Multiforme
- ER
Estrogen Receptor
- HC
Helminthosporium Carbonum
- PI3K
Phosphatidylinositol-3-Kinase
- HSP90
Heat Shock Protein 90
- NSCLC
Non-small Cell Lung Carcinoma
- H2R
Histamine H2 Receptor
REFERENCES
- 1.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004 Aug;3:673. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 2.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003 Mar;22:151. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 3.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007 Aug 9;448:645. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 4.Aronson JK. Old drugs--new uses. Br J Clin Pharmacol. 2007 Nov;64:563. doi: 10.1111/j.1365-2125.2007.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lum PY, Derry JM, Schadt EE. Integrative genomics and drug development. Pharmacogenomics. 2009 Feb;10:203. doi: 10.2217/14622416.10.2.203. [DOI] [PubMed] [Google Scholar]
- 6.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009 Apr;8:286. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 7.Qu XA, Gudivada RC, Jegga AG, Neumann EK, Aronow BJ. Inferring novel disease indications for known drugs by semantically linking drug action and disease mechanism relationships. BMC Bioinformatics. 2009;10(Suppl 5):S4. doi: 10.1186/1471-2105-10-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009 Nov 12;462:175. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie L, Li J, Xie L, Bourne PE, Nussinov R. Drug Discovery Using Chemical Systems Biology: Identification of the Protein-Ligand Binding Network To Explain the Side Effects of CETP Inhibitors. PLoS Comput Biol. 2009 May 15;5 doi: 10.1371/journal.pcbi.1000387. e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinnings SL, et al. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput Biol. 2009 Jul;5 doi: 10.1371/journal.pcbi.1000423. e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 12.Lipshutz RJ, et al. Using oligonucleotide probe arrays to access genetic diversity. BioTechniques. 1995 Sep;19:442. [PubMed] [Google Scholar]
- 13.Yap YL, Zhang XW, Smith D, Soong R, Hill J. Molecular gene expression signature patterns for gastric cancer diagnosis. Comput Biol Chem. 2007 Aug;31:275. doi: 10.1016/j.compbiolchem.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Milano A, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3:e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang WZ, et al. Comparative analysis of gene expression profiles between the normal human cartilage and the one with endemic osteoarthritis. Osteoarthritis Cartilage. 2008 Jun 24; doi: 10.1016/j.joca.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Walsh CS, et al. ERCC5 is a novel biomarker of ovarian cancer prognosis. J Clin Oncol. 2008 Jun 20;26:2952. doi: 10.1200/JCO.2007.13.5806. [DOI] [PubMed] [Google Scholar]
- 17.Antonacopoulou AG, et al. POLR2F, ATP6V0A1 and PRNP expression in colorectal cancer: new molecules with prognostic significance? Anticancer Res. 2008 Mar-Apr;28:1221. [PubMed] [Google Scholar]
- 18.Dobrin R, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley JT, Tibshirani R, Deshpande T, Butte AJ. Disease signatures are robust across tissues and experiments. Mol Syst Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Rio M, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 2007 Mar 1;25:773. doi: 10.1200/JCO.2006.07.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Wood WH, 3rd, Becker KG, Weeraratna AT, Morin PJ. Gene expression response to cisplatin treatment in drug-sensitive and drug-resistant ovarian cancer cells. Oncogene. 2007 May 3;26:2860. doi: 10.1038/sj.onc.1210086. [DOI] [PubMed] [Google Scholar]
- 22.Fichtner I, et al. Anticancer drug response and expression of molecular markers in early-passage xenotransplanted colon carcinomas. Eur J Cancer. 2004 Jan;40:298. doi: 10.1016/j.ejca.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Holleman A, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004 Aug 5;351:533. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 24.Robert J, Vekris A, Pourquier P, Bonnet J. Predicting drug response based on gene expression. Crit Rev Oncol Hematol. 2004 Sep;51:205. doi: 10.1016/j.critrevonc.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313:1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 26.Garman KS, et al. A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc Natl Acad Sci U S A. 2008 Dec 9;105:19432. doi: 10.1073/pnas.0806674105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Setlur SR, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008 Jun 4;100:815. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 Apr 24;98:5116. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano H, Jayachandran M, Yoshikawa A, Miller VM. Differential effects of chronic treatment with estrogen receptor ligands on regulation of nitric oxide synthase in porcine aortic endothelial cells. J Cardiovasc Pharmacol. 2006 Apr;47:621. doi: 10.1097/01.fjc.0000211749.24196.98. [DOI] [PubMed] [Google Scholar]
- 30.Ryan KJ, Ray CG, Sherris JC. ebrary Inc. New York: McGraw-Hill; 2004. p. xiii.p. 979. [Google Scholar]
- 31.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006 Jun 15;22:1540. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 32.Irving PM, Gearry RB, Sparrow MP, Gibson PR. Review article: appropriate use of corticosteroids in Crohn's disease. Aliment Pharmacol Ther. 2007 Aug 1;26:313. doi: 10.1111/j.1365-2036.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- 33.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011 Mar-Apr;61:69. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 34.Sabesin SM. Safety issues relating to long-term treatment with histamine H2-receptor antagonists. Aliment Pharmacol Ther. 1993;7(Suppl 2):35. doi: 10.1111/j.1365-2036.1993.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 35.Gertner DJ, Rampton DS, Stevens TR, Lennard-Jones JE. Verapamil inhibits in-vitro leucotriene B4 release by rectal mucosa in active ulcerative colitis. Aliment Pharmacol Ther. 1992 Apr;6:163. doi: 10.1111/j.1365-2036.1992.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 36.Fedorak RN, Empey LR, Walker K. Verapamil alters eicosanoid synthesis and accelerates healing during experimental colitis in rats. Gastroenterology. 1992 Apr;102:1229. [PubMed] [Google Scholar]
- 37.Cheng H, Force T. Molecular mechanisms of cardiovascular toxicity of targeted cancer therapeutics. Circ Res. Jan 8;106:21. doi: 10.1161/CIRCRESAHA.109.206920. [DOI] [PubMed] [Google Scholar]
- 38.Drummond DC, et al. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 39.Schneider-Stock R, Ocker M. Epigenetic therapy in cancer: molecular background and clinical development of histone deacetylase and DNA methyltransferase inhibitors. IDrugs. 2007 Aug;10:557. [PubMed] [Google Scholar]
- 40.Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- 41.Gao J, et al. Loss of NECL1, a novel tumor suppressor, can be restored in glioma by HDAC inhibitor-Trichostatin A through Sp1 binding site. Glia. 2009 Jul;57:989. doi: 10.1002/glia.20823. [DOI] [PubMed] [Google Scholar]
- 42.Morita K, Gotohda T, Arimochi H, Lee MS, Her S. Histone deacetylase inhibitors promote neurosteroid-mediated cell differentiation and enhance serotonin-stimulated brain-derived neurotrophic factor gene expression in rat C6 glioma cells. J Neurosci Res. 2009 Aug 15;87:2608. doi: 10.1002/jnr.22072. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz C, et al. Valproic acid induces non-apoptotic cell death mechanisms in multiple myeloma cell lines. Int J Oncol. 2007 Mar;30:573. [PubMed] [Google Scholar]
- 44.Valentini A, Gravina P, Federici G, Bernardini S. Valproic acid induces apoptosis, p16INK4A upregulation and sensitization to chemotherapy in human melanoma cells. Cancer Biol Ther. 2007 Feb;6:185. doi: 10.4161/cbt.6.2.3578. [DOI] [PubMed] [Google Scholar]
- 45.Galanis E, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009 Apr 20;27:2052. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deubzer HE, et al. Anti-neuroblastoma activity of Helminthosporium carbonum (HC)-toxin is superior to that of other differentiating compounds in vitro. Cancer Lett. 2008 Jun 8;264:21. doi: 10.1016/j.canlet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Deubzer HE, et al. Histone deacetylase inhibitor Helminthosporium carbonum (HC)-toxin suppresses the malignant phenotype of neuroblastoma cells. Int J Cancer. 2008 Apr 15;122:1891. doi: 10.1002/ijc.23295. [DOI] [PubMed] [Google Scholar]
- 48.Abbas S, et al. Preclinical studies of celastrol and acetyl isogambogic acid in melanoma. Clin Cancer Res. 2007 Nov 15;13:6769. doi: 10.1158/1078-0432.CCR-07-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eaton D, Hawkins RE. Cimetidine in colorectal cancer |[ndash]| are the effects immunological or adhesion-mediated? British Journal of Cancer. 2002 Jan 21;86:159. doi: 10.1038/sj.bjc.6600097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuta K, et al. Anti-tumor effects of cimetidine on hepatocellular carcinomas in diethylnitrosamine-treated rats. Oncol Rep. 2008 Feb 1;19:361. [PubMed] [Google Scholar]
- 51.Natori T, Sata M, Nagai R, Makuuchi M. Cimetidine inhibits angiogenesis and suppresses tumor growth. Biomedicine & Pharmacotherapy. 2005 Jan 1; doi: 10.1016/j.biopha.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 52.Sürücü O, et al. Tumour growth inhibition of human pancreatic cancer xenografts in SCID mice by cimetidine. Inflamm Res. 2004 Mar 1;53(Suppl 1):S39. doi: 10.1007/s00011-003-0318-1. [DOI] [PubMed] [Google Scholar]
- 53.Osband ME, et al. Successful tumour immunotherapy with cimetidine in mice. Lancet. 1981 Mar 21;1:636. doi: 10.1016/s0140-6736(81)91554-3. [DOI] [PubMed] [Google Scholar]
- 54.Gifford RR, Ferguson RM, Voss BV. Cimetidine reduction of tumour formation in mice. Lancet. 1981 Mar 21;1:638. doi: 10.1016/s0140-6736(81)91555-5. [DOI] [PubMed] [Google Scholar]
- 55.Gifford RR, Voss BV, Ferguson RM. Cimetidine protection against lethal tumor challenge in mice. Surgery. 1981 Aug;90:344. [PubMed] [Google Scholar]
- 56.Kumar A. Cimetidine: an immunomodulator. DICP. 1990 Mar;24:289. doi: 10.1177/106002809002400316. [DOI] [PubMed] [Google Scholar]
- 57.Jiang CG, Liu FR, Xu HM, Wu T, Gao J. [Effects of cimetidine on the biological behaviors of human gastric cancer cells] Zhonghua Yi Xue Za Zhi. 2006 Jul 11;86:1813. [PubMed] [Google Scholar]
- 58.Kobayashi K, Matsumoto S, Morishima T, Kawabe T, Okamoto T. Cimetidine inhibits cancer cell adhesion to endothelial cells and prevents metastasis by blocking E-selectin expression. Cancer Res. 2000 Jul 15;60:3978. [PubMed] [Google Scholar]
- 59.Fukuda M, Kusama K, Sakashita H. Cimetidine inhibits salivary gland tumor cell adhesion to neural cells and induces apoptosis by blocking NCAM expression. BMC Cancer. 2008;8:376. doi: 10.1186/1471-2407-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Y, et al. paper presented at the AMIA Annual Symposium; 2007; Chicago, IL. [Google Scholar]
- 61.Chiang AP, Butte AJ. Data-driven methods to discover molecular determinants of serious adverse drug events. Clin Pharmacol Ther. 2009 Mar;85:259. doi: 10.1038/clpt.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008 Jul 11;321:263. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 63.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 Jan 1;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butte AJ, Chen R. Finding disease-related genomic experiments within an international repository: first steps in translational bioinformatics; AMIA … Annual Symposium proceedings / AMIA Symposium; 2006. p. 106. [PMC free article] [PubMed] [Google Scholar]
- 66.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006 Jan;24:55. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dudley J, Butte AJ. Pac Symp Biocomput; 2008. pp. 580–591. [PMC free article] [PubMed] [Google Scholar]
- 68.Warnat P, Eils R, Brors B. Cross-platform analysis of cancer microarray data improves gene expression based classification of phenotypes. BMC Bioinformatics. 2005;6:265. doi: 10.1186/1471-2105-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abba MC, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen R, Li L, Butte AJ. AILUN: reannotating gene expression data automatically. Nat Methods. 2007 Nov;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society. 2002;Series B:479. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results from a tumor xenograft experiment testing the efficacy of H2-agonist cimetidine in inhibiting the growth of ACHN renal carcinoma cell line tumors in SCID mice. Three treatment groups (25/50/100 mg/kg/injection) and two groups of controls were used. One control group was administered the PBS vehicle, and the other control group was left untreated. The results demonstrate that cimetidine lacks efficacy in inhibiting the growth of renal carcinoma xenograft implant tumors which is in agreement with the prediction rendered by our in silico repositioning method.