Abstract
Despite significant treatment advances over the past decade, multiple myeloma (MM) remains largely incurable. In this study we found that MM cells were remarkably sensitive to the death-inducing effects of a new class of sangivamycin-like molecules (SLMs). A panel of structurally related SLMs selectively induced apoptosis in MM cells but not other tumor or non-malignant cell lines at sub-micromolar concentrations. SLM6 was the most active compound in vivo, where it was well-tolerated and significantly inhibited growth and induced apoptosis of MM tumors. We determined that the anti-MM activity of SLM6 was mediated by direct inhibition of cyclin-dependent kinase 9 (CDK9), which resulted in transcriptional repression of oncogenes that are known to drive MM progression (c-Maf, cyclin D1, and c-Myc). Furthermore, SLM6 demonstrated superior in vivo anti-MM activity over the CDK inhibitor flavopiridol, which is currently in clinical trials for MM. These findings demonstrate that SLM6 is a novel CDK9 inhibitor with promising preclinical activity as an anti-MM agent.
Keywords: Multiple myeloma, CDK9, P-TEFb, sangivamycin
Introduction
Multiple myeloma (MM) is the second most common hematological cancer and accounts for approximately 10,000 deaths per year in the United States (1). The development of immunomodulatory drugs (IMiDs; thalidomide and lenalidomide) and the proteasome inhibitor bortezomib has revolutionized the clinical management of MM. Despite these advances, the disease remains largely incurable. It is therefore critical to develop new treatments, particularly ones with clinical activity in the refractory setting, in order to extend treatment options, prolong survival, and improve the quality of life for MM patients.
MM is characterized by the accumulation of plasma cells within the bone marrow and may spread to extraosseus sites in its late stages. Plasma cells are terminally differentiated B cells that are specialized for production and secretion of immunoglobulins (Ig). An early transformation event in the genesis of MM involves an erroneous class switch recombination involving the Ig heavy chain (IgH) gene (14q32). These illegitimate gene translocations juxtapose proto-oncogenes with potent enhancer sequences within the IgH locus leading to dysregulation of their expression (2). The most commonly dysregulated oncogenes in MM include FGFR3/MMSET, MAF, and CCND1 (3). Additional genetic events may be required to drive MM to a pathological stage, as these translocations are also present in monoclonal gammopathy of undetermined significance (MGUS), an asymptomatic precursor stage to MM (4). Whole genome sequencing analysis has revealed that MM is characterized by a diverse array of genetic abnormalities (5). Among these anomalies are somatic mutations in KRAS, NRAS, and TP53, as well as members of the NF-kB signaling pathway. In addition, rearrangement or translocation of the MYC gene is detected in 50% of MM patients and nearly all established MM cell lines (6).
The genetic heterogeneity of MM and the multitude of oncogenes and signaling pathways that drive MM development and progression pose a challenge to the development of molecular targeted therapies. Inhibitors of cyclin dependent kinase-9 (CDK9) may simultaneously target multiple oncogenic pathways by disrupting gene transcription - a potentially advantageous therapeutic strategy in a heterogeneous disease like MM. CDK9 is a subunit of the Positive-Transcription Elongation Factor b (P-TEFb) complex, which regulates mammalian gene transcription by phosphorylating the carboxy-terminus of RNA polymerase II at Ser2, a modification that initiates the elongation phase of transcription (7). Inhibitors of CDK9 and P-TEFb have shown preclinical activity in MM. For example the BET bromodomain inhibitor JQ1 demonstrated preclinical anti-MM activity through a mechanism that involves displacing BRD4 and blocking the recruitment of P-TEFb to c-Myc target genes (8). Also, broad-spectrum CDK inhibitors with activity against CDK9 have shown activity against MM and are currently in clinical development (9-11).
Sangivamycin is a nucleoside analog that was isolated from Streptomyces rimosus (12), and subsequently found to possess potent anti-tumor and anti-retroviral activity (13,14). A phase I trial of sangivamycin in the 1960s demonstrated the safety of this compound in humans, however no follow-up clinical studies were conducted (15). The anti-cancer activity of sangivamycin has been attributed to pleiotropic effects including inhibition of protein kinase C (PKC; 16). We recently identified a class of small molecules with structural homology to sangivamycin (Sangivamycin-Like Molecules; SLMs) in a high throughput cell-based drug screen for compounds that overcome hypoxia-induced resistance to apoptosis in preclinical models of colon cancer (17). The mechanistic effects of SLMs closely resembled those of dual GSK-3β/CDK1 inhibitors, although the precise molecular targets of SLMs have not been conclusively elucidated. Furthermore, the activity of SLMs in tumor types other than colorectal cancer has not been thoroughly examined. Here we demonstrate a selective sensitivity of MM cells to SLMs, and in vivo screening of a panel of SLM-related structures identified SLM6 as an active and well tolerated lead compound for further development. A candidate approach led us to identify CDK9 as the critical molecular target responsible for mediating the potent anti-MM activity of SLM6. This work demonstrates the mechanism, molecular target, and potential of SLM6 as a novel agent for the treatment of MM, a disease that recurs nearly 100% of the time and requires additional therapies to improve patient survival and therapeutic options.
Materials and Methods
Cell lines, reagents, and antibodies
Cancer cell lines were purchased from ATTC and maintained in the growth media recommended by the supplier at 37°C and 5% CO2. Human fetal osteoblasts (hFOB) were kindly provided by Dr. Alessandro Fatatis (Drexel University College of Medicine) and were grown in DMEM/F12 media supplemented with 10% FBS and Geneticin (400 μg/ml). Cell lines were routinely verified using the following tests: morphology was evaluated by microscopic examination, growth curve analysis was performed, and the plasma cell immunophenotype of MM cells was confirmed by flow cytometric analysis of cell surface CD138 expression. SLM3 (NSC 188491), SLM3 HCl (NSC 749838), SLM5 (NSC 107512), SLM6 (NSC 107517), SLM7 (NSC 131663), and sangivamycin (NSC 65346) were provided by the NIH Developmental Therapeutics Program (DTP). Gemcitabine, Cladribine, and 5′fluorouracil (5-FU) were provided by the Penn State Milton S. Hershey Medical Center infusional pharmacy. Flavopiridol and bortezomib were purchased from Selleck Chemicals. Purvalanol A was from Sigma. Antibodies to PARP, caspase-8, caspase-9, phopho-CDK9, CDK9, cyclin D1, Mcl-1, and c-Myc were purchased from Cell Signaling Technology. Antibodies to L-Myc, c-Maf, and RNA polymerase-2 were from Santa Cruz Biotechnology. Antibodies to phospho-RNA polymerase II (Ser2) and RNA polymerase II (Ser5) were from Bethyl Laboratories Inc. Antibodies to Ran were from BD Bioscience.
Cell viability and apoptosis assays
Cell viability was measured in 96-well cell culture plates using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) per the manufacturer’s instructions. Linear regression analysis was performed to calculate inhibitory concentration 50 (IC50) values. For analysis of apoptosis, cells were collected, fixed and stained with an active caspase-3 specific antibody (BD Pharmingen) or an antibody that specifically recognizes single stranded DNA fragments (Enzo Life Sciences) and analyzed by FACS analysis.
In vivo studies
Hairless SCID mice were housed and maintained in accordance with the Institutional Animal Care and Use Committee and state and federal guidelines for the humane treatment and care of laboratory animals. NCI-H929 cells were injected subcutaneously into the rear flank of mice at a density of 5 × 106 cells per injection in PBS/Matrigel (v:v) and 200 μl total volume. When tumors were between 300 and 400 mm3 in volume, tumor-bearing mice were randomized, and treatments were initiated. All drugs were delivered by intraperitoneal injection in PBS containing less than 0.1% DMSO. Tumor volumes were monitored over time by caliper measurements. For short-term in vivo experiments, animals were given one dose of SLM6, then tumors were harvested after 48 hours, fixed and embedded in paraffin, sectioned and stained with hematoxylin and eosin or immunostained with antibodies specific for cleaved caspase-3 and cleaved caspase-8 (both from Cell Signaling Technology). For RPMI-8226/NIH-3T3 tumors, 5 × 106 RPMI-8226 cells stably expressing GFP were mixed with 1 × 106 NIH-3T3 mouse fibroblasts in a PBS/Matrigel solution (v:v) and 200 μl total volume, and then injected subcutaneously. Treatments were initiated after 2 weeks when tumors were palpable and had grown to a volume of 100-200 mm3. For non-invasive fluorescence imaging studies, mice were anesthetized and imaged using the CRi Maestro system and related quantitative multispectral imaging software.
RT-PCR
RNA was isolated at the indicated time point using RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol. Reverse transcription was performed with 1μg of RNA using SuperScript II (Invitrogen) according to the manufacturer’s protocol. PCR was performed using FastStart Taq DNA Polymerase (Roche) according to the manufacturer’s protocol. Primers were: cyclin D1 (sense, 5′-CCTGCTTTGGCGGGCAGACA-3′; antisense 5′-ACGCCGTGGTGGCACGTAAG-3′) and c-myc (sense, 5′-CGACCCGGACGACGAGACCT-3′; antisense, 5′-GTTCGGGCTGCCGCTGTCTT-3′. PCR products were electrophoresed on a 2% agarose gel and visualized using ethidium bromide.
Isolation of CD138-positive MM patient plasma cells
Aspirates from bone marrow needle biopsies were collected and de-identified for research use with approval from the University of Pennsylvania Institutional Review Board. White blood cells were isolated from whole bone marrow by Ficoll-Paque gradient separation. CD138-positive plasma cells were isolated using a magnetic bead-conjugated anti-CD138 antibody (Miltenyi Biotec) and MininMACS Separator (Miltenyi Biotec). The purity of CD138-positive populations was verified by FACS analysis. CD138-positive plasma cells were cultured in RPMI 1640 media supplemented with 10% FBS and gentamycin, and grown at 37°C under 5% CO2.
In Vitro Kinase Assays
In vitro kinase assays with CDK1/cyclin B, CDK2/cyclin A, CDK4/cyclin D1, CDK7/cyclin H, CDK9/cyclin K, and CDK9/cyclin T1 recombinant protein complexes were performed using the “HotSpot” assay platform as described previously (18).
Results
Multiple myeloma cell lines are most sensitive to the cytotoxic effects of SLMs
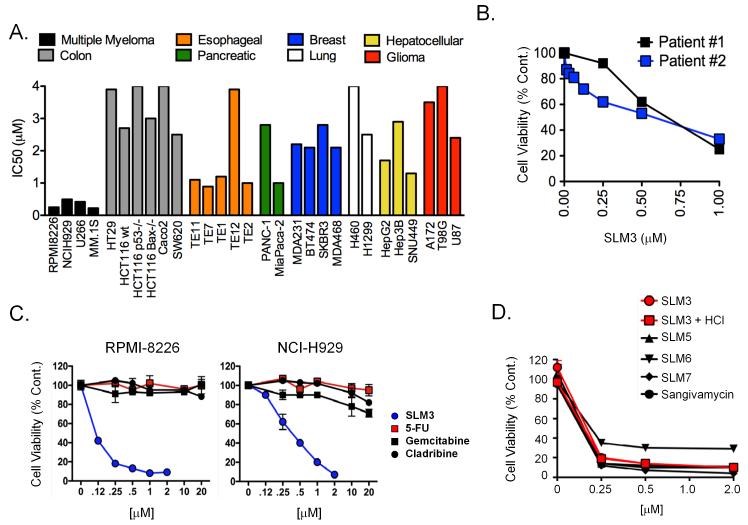
To test the activity of SLMs across multiple tumor types, we determined inhibitory concentration 50 values (IC50s) for SLM3 in cell viability assays using human cell lines derived from multiple tumor types, including colon (HCT116, HCT116 p53−/−, HCT116 Bax−/−, SW620, HT29, Caco-2), esophageal (TE1, TE2, TE7, TE11, TE12), pancreatic (MiaPaca-2, Panc-1), lung (H460, H1299), breast (MDA-MB-231, MDA-MB-468, BT474, SKBR3), hepatocellular (SNU449, Hep3B, HepG2), glioma (A172, U87, T98G), and multiple myeloma (RPMI-8226, NCI-H929, U266B1, MM.1S). We chose a 24-hour treatment time to focus the assay read-out on the cytotoxic effects of SLM3 and minimize cytostatic effects. The potency of SLM3 varied considerably between tumor types of different origin (Fig. 1A; IC50 range: 0.2 - >4 μM). Breast, colon, lung, hepatocellular, glioma, and one of two pancreatic cancer cell lines were relatively resistant to SLM3, with IC50s >2 μM (Fig. 1A and S1). Esophageal cells were relatively more sensitive to SLM3 with IC50s of μ1 μM. Most notably, SLM3 exhibited striking cytotoxic effects in MM cells, where IC50s ranged from 200-400 nM (Fig. 1A). We also confirmed this activity in primary CD138-positive plasma cells from MM patient bone marrow biopsies (Fig. 1B). SLM3 showed superior activity compared to other nucleoside analogs (5′-fluorouracil, gemcitabine, and cladribine; Fig. 1C), demonstrating a specific sensitivity of MM cells to SLM3 as opposed to a general sensitivity to cytotoxic agents of similar structure. We tested the anti-MM activity of other SLM molecules that were identified in the NCI Developmental Therapeutics Program (DTP) library. We found that SLM3 (NSC 188491), the HCl salt of SLM3 (NSC 742838, a.k.a. SMA-838), SLM5 (NSC 107512), SLM6 (NSC 107517), SLM7 (NSC 131663), and sangivamycin (NSC 65346) significantly reduced MM cell viability at concentrations less than 250nM (Fig. 1D).
Figure 1. MM cells are more sensitive to SLM3 than cells of other tumor types.
(A) Bioluminescence-based cell viability assays were performed with multiple doses of SLM3 over a period of 24 hours. The graph shows inhibitory concentration 50 (IC50) values for the indicated cell lines which were determined by linear regression analysis. Cell lines are grouped by color based on their tissue of origin.
(B) CD138-positive plasma cells were isolated from the bone marrow aspirates of MM patients and treated with SLM3 at the indicated concentrations. Cell viability data from 2 different patients is shown after 24 hours of treatment. Each data point represents mean ± S.E (n=3).
(C) RPMI-8226 and NCI-H929 cells were treated with the indicated concentrations of SLM3, gemcitabine, cladribine, and 5′-fluorouracil (5-FU) for 24 hours. Cell viability dose response curves are shown. Data points represent mean cell viability ± S.E (n=3).
(D) RPMI-8226 cells were treated with increasing concentrations of the indicated SLMs and sangivamycin for 24 hours. Cell viability curves are shown. Each data point represents mean cell viability ± S.E.
SLMs selectively induces apoptosis of MM cells but not other tumor or normal cell types
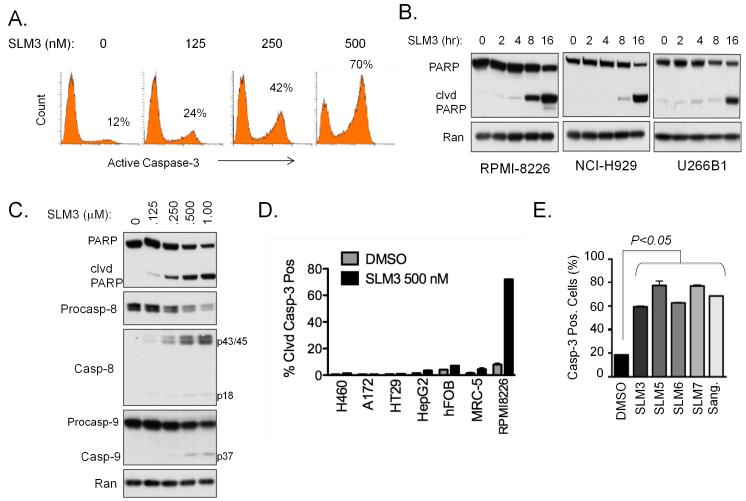
The fact that SLMs rapidly and robustly reduced MM cell viability suggested they actively induced cell death, possibly by apoptosis. We tested this hypothesis by investigating the expression of apoptotic markers in response to SLM3 treatment. We found that SLM3 induced a dose-dependent increase in the apoptotic markers cleaved caspase-3 (Fig. 2A) and cleaved poly (ADP-ribose) polymerase (PARP; Fig. 2B). SLM3 activated caspase-8 and caspase-9 in a dose-dependent manner, further demonstrating the activation of apoptotic effectors (Fig. 2C). In comparison to MM cells, SLM3 induced negligible levels of apoptosis in cell lines derived from other tumor types (H460, A172, HT29, HepG2) and non-transformed cells [human fetal osteoblasts (hFOB) and normal human lung fibroblasts (MRC-5); Fig. 2D]. Furthermore, other SLMs and sangivamycin induced significant levels of MM cell apoptosis (60-80%; Fig. 2E). Thus, SLMs as an entire class of small molecules with structural homology to sangivamycin have robust single agent anti-MM activity with the ability to selectively induce apoptosis in MM cells.
Figure 2. SLM3 selectively induces apoptosis in MM cells.
(A) RPMI-8226 cells were treated with increasing concentrations of SLM3 for 16 hours. Cells were analyzed by flow cytometry for cleaved (active) caspase-3 immunostaining. Representative flow cytometric histograms are shown. The inset percentages indicate the proportion of cleaved caspase-3 positive cells.
(B) RPMI-8226, NCI-H929, and U266B1 cells were treated with 500 nM SLM3 for the indicated time points. Western blots for the caspase-3 substrate PARP are shown. Processing of PARP and the appearance of the cleaved PARP fragment indicates caspase-3 enzymatic activity. Ran blots are included as a loading control.
(C) RPMI-8226 cells were treated with the indicated concentrations of SLM3 for 16 hours. Western blots are shown.
(D) Tumor cells (H460, A172, HT29, HepG2, and RPMI-8226) from multiple tissues of origin and non-malignant cells (hFOB and MRC-5) were treated with 500 nM SLM3 for 16 hours. The percentage of cells that stained positively for active caspase-3 were quantified by flow cytometry. Data represent mean ± S.E (n=3).
(E) RPMI-8226 cells were treated with the indicated SLMs and sangivamycin at concentrations of 500 nM for 16 hours. Cleaved caspase-3 positive cells were quantified using flow cytometry. Data represent mean ± S.E. A student’s t-test was used for statistical analysis.
SLM6 shows the most anti-MM efficacy in vivo
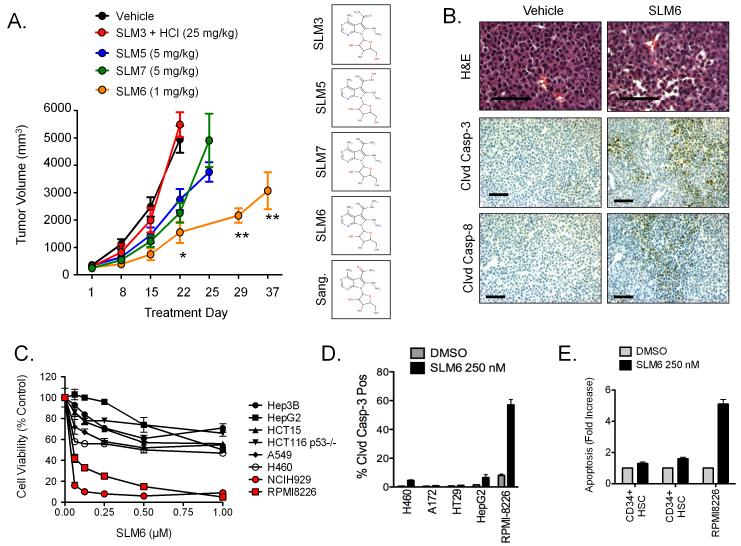
In order to identify the SLM(s) with the most efficacy in vivo we tested the anti-tumor activity of several SLM structures in an MM subcutaneous plasmacytoma model. For dose selection, we used a dose range based on available toxicity data provided by NCI DTP, and performed pilot studies in a small cohort of mice to qualitatively assess toxicity. Based on body condition scoring, we observed no overt toxicity after a single dose of 25 mg/kg SLM3 HCl, 5 mg/kg for SLM5 and SLM7, and 1 mg/kg for SLM6. These doses were then given by weekly intraperitoneal injections in tumor-bearing mice. SLM5 and SLM7 delayed tumor growth by 7 days relative to vehicle-treated controls, and although these responses reached statistical significance they were short-lived (Fig. 3A). By comparison, SLM6 showed the most significant anti-tumor activity, which was sustained for 5 weeks after only a single dose on day 1. The HCl salt of SLM3 had no effect on the growth of MM tumors. We then tested for apoptotic markers in tumors from mice that received short-term SLM6 treatment (48 hours). We detected regions of intense cleaved caspase-3 and cleaved caspase-8 immunostaining in tumors from SLM6-treated mice, whereas negligible staining was found in control tumors (Fig. 3B). Like SLM3, SLM6 showed selective killing and induction of apoptosis in MM cells in vitro compared to tumor cells from other tissues of origin (Fig. 3C and 3D) and CD34+ hematopoietic stem cells from normal human donors (Fig. 3E). SLM6 showed no signs of systemic toxicity, which we determined by serum biochemistry in immunocompetent mice (C57BL/6; Fig. S2A). Furthermore, SLM6 showed no effects on normal hematopoiesis in vivo other than modest thrombocytopenia, which was determined by complete blood count with differential in C57BL/6 mice (Fig. S2B). Due to the superior in vivo profile of SLM6, we chose to focus our subsequent mechanistic studies on this molecule.
Figure 3. Other SLMs induce death and apoptosis of MM cells with SLM6 demonstrating superior efficacy in vivo.
(A) NCI-H929 subcutaneous xenografted tumors were established in hairless SCID mice. Mice were treated (i.p.) with the indicated doses of SLMs and tumor volume was measured over time by caliper measurement. All SLMs were given once weekly, except for SLM6 which was given only once on day 1. Data points represent mean tumor volume ± S.E. (n=5 mice). Statistical significance was determined using a student’s t-test (*P<0.05 for SLM5, SLM6, and SLM7 compared to vehicle on day 22. **P<0.05 for SLM6 compared to vehicle on day 22). The chemical structures for each of the SLMs are shown at the right.
(B) NCI-H929 xenograft-bearing mice were treated with SLM6 (1 mg/kg; i.p.). After 48 hours, tumors were collected and analyzed by immunohistochemistry for cleaved caspase-3 and cleaved caspase-8. Scale bars equal 100 μm.
(C) Various tumor cell types were treated with increasing concentrations of SLM6 for 24 hours. Cell viability dose response curves are shown. Each data point represents the mean ± S.E (n=3).
(D) The indicated tumor cell lines were treated with SLM6 (250 nM) for 16 hours. Cleaved caspase-3 positive cells were quantified by flow cytometry. Data represent mean ± S.E (n=3).
(E) CD34+ hematopoietic stem cells (HSCs) were isolated from the peripheral blood of healthy donors and treated with 250 nM SLM6 for 24 hours. Apoptosis was measured by FACS analysis of cleaved caspase-3 positive cells. The results obtained from 2 different donors are shown in comparison to RPMI-8226 MM cells. Data represent mean ± S.E. (n=3).
Inhibition of CDK9 is critical to the anti-MM activity of SLM6
We next took a candidate approach to identify molecular targets of SLM6 that mediate its anti-MM activity. Previous work on the molecular effects of SLM3 and sangivamycin, by our group and others, suggested that the anti-tumor activity of these compounds may be mediated by inhibition of multiple molecular targets. Sangivamycin is known to inhibit PKC (16), and SLM3 (also known as ARC; 19) was shown to have modest activity against PKC and CDK9 in vitro (20). In addition, we previously showed that SLM3 sensitizes hypoxic cancer cells to apoptosis-inducing agents via a mechanism involving GSK-3β and CDK1 (17). We first took a pharmacological approach to test the role of GSK-3β and CDK1 in SLM6-mediated MM cytotoxicity. We measured the effects on MM cell viability of selective GSK-3β (lithium chloride, LiCl) and CDK1 (purvalanol A) inhibitors alone and in combination. By comparison to SLM6, neither LiCl nor purvalanol A reduced MM cell viability by themselves or in combination at concentrations that effectively inhibited their designated molecular targets (Fig. S3). Similar results were obtained with GSK-3β inhibitor sc-24020 (data not shown). Because the combined inhibition of GSK-3β and CDK1 was not toxic to MM cells, we deduced that these molecular targets alone could not account for the anti-MM activity of SLM6.
To test the activity of SLM6 against PKC, we stimulated MM cells with phorbol-12-myristate-13-acetate (phorbol ester), an analog of diacylglycerol that directly binds to and activates PKC. We then measured the effects of SLM6 on PKC activity by analyzing the autophosphorylation of pan PKC isoforms (a site homologous to Ser660 on PKC beta II) and phosphorylation of extracellular signal-regulated kinase 1/2 (ERK), which is activated downstream of PKC. SLM6 had no effect on phorbol ester-induced PKC or ERK phosphorylation, whereas sangivamycin significantly reduced phosphorylation of both kinases (Fig. S4). Thus, SLM6 does not affect PKC signaling at concentrations that induce MM cell death, leading us to conclude that PKC was not a critical mediator of SLM6-induced MM cell death.
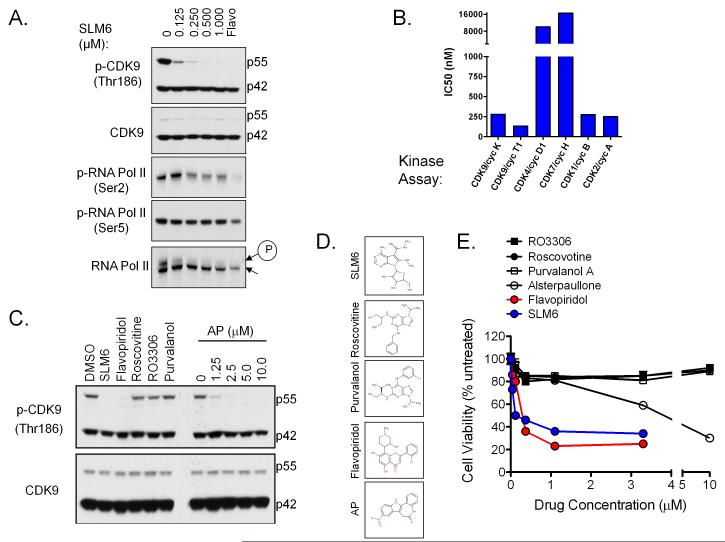
We next tested the activity of SLM6 against CDK9 signaling. We found that SLM6 inhibited phosphorylation of CDK9 at Thr186, an autophosphorylation site critical to the kinase activity of P-TEFb (21, 22). SLM6 also inhibited phosphorylation of RNA polymerase II at the CDK9-specific Ser2 site at concentrations as low as 250 nM (Fig. 4A). Interestingly, SLM6 as well as flavopiridol, an inhibitor of CDK9 and other CDKs, more effectively blocked autophosphorylation of the 55-kDa isoform of CDK9 compared to the 44-kDa form. Unlike flavopiridol, SLM6 had no effect on the phosphorylation of RNA polymerase II at Ser5, a CDK7 specific site (Fig. 4A). In vitro kinase assays confirmed that SLM6 potently and directly inhibited of CDK9/cyclin K and CDK9/cyclin T1 kinase activity with IC50s of 280 and 133 nM, respectively (Fig. 4B and S5). SLM6 also inhibited CDK1/cyclin B and CDK2/cyclin A with IC50s less than 300 nM, but demonstrated little activity toward CDK4/cyclin D1 and CDK7/cyclin H complexes (IC50 >10μM; Fig. 4B and S5). By comparison, flavopiridol inhibited all CDKs tested, with high potency against CDK1, CDK2, CDK4, and CDK9 (i.e. IC50 <50 nM) and less potency toward CDK7 (IC50 of approximately 4 μM; Fig. S5). We treated MM cells with various CDK inhibitors (i.e. alsterpaullone, roscovitine, purvalanol A, RO3306, flavopiridol, and SLM6) and found that only the inhibitors with activity toward CDK9 (i.e. SLM6, flavopiridol, and micromolar concentrations of alsterpaullone) were able to reduce the viability of MM cells (Fig. 4C and 4D). These findings show that inhibition of CDK9 is paramount to the selective anti-MM activity of SLM6.
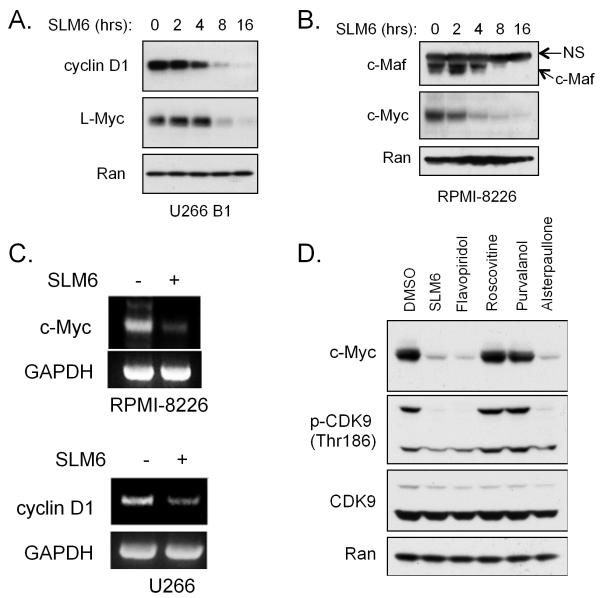
Figure 4. SLM6 inhibits CDK9 and represses oncogenic gene translocation products.
(A) NCI-H929 cells were treated with the indicated concentrations of SLM6 for 2 hours. Western blots are shown. Flavopiridol (250 nM) was included as a positive control for inhibition of CDK9 and CDK7. Arrows at the right of RNA pol. II mark a higher molecular band corresponding to the hyperphosphorylated form of the enzyme (encircled P) and a lower molecular weight band that corresponds to the hypophosphorylated form.
(B) In vitro kinase assays were performed using the indicated CDK/cyclin complexes. SLM6 IC50s are shown.
(C) RPMI-8226 cells were treated with SLM6 (250 nM), flavopiridol (250 nM), roscovitine (5 μM), RO3306 (10 μM), purvalanol A (10 μM), or a dose range of alsterpaullone for 6 hours. Western blots are shown.
(D) RPMI-8226 cells were treated with increasing concentrations of the indicated CDK inhibitors for 24 hours. Cell viability data are shown.
SLM6 represses oncogenic gene translocation products and demonstrates combinatorial killing with bortezomib
Given the role of CDK9 in the regulation of transcription, we next investigated the impact of SLM6 treatment on the expression of oncogenes that are known to drive the progression of MM. With regard to the MM cell lines used in this study, U266B1 cells overexpress cyclin D1 due to an insertion of the IgH gene sequence upstream of the CCND1 transcriptional start site (23), and RPMI-8226 express high levels of c-Maf due to a gene rearrangement (24). In addition, c-Myc is highly expressed in NCI-H929 and RPMI-8226 cells, and L-Myc is highly expressed in U266B1 cells due to rearrangements of these respective genes6. We found that treatment with SLM6 repressed the expression of cyclin D1 in U266B1 cells and c-Maf expression in RPMI-8226 cells in a time-dependent manner (Fig. 5A and 5B). SLM6 also repressed c-Myc protein levels in RPMI-8226 and NCI-H929 cells, and suppressed L-Myc expression in U266B1 cells (Fig. 5A, data not shown, and 5B). RT-PCR analysis showed that SLM6 decreased c-Myc and cyclin D1 mRNA transcripts in RPMI-8226 and U266B1 cells, respectively (Fig. 5C), suggesting that SLM6 represses these oncogenes at the transcriptional level. Only CDK inhibitors with activity toward CDK9 were capable of down-regulating c-Myc in RPMI-8226 cells (Fig. 5D), suggesting that CDK9 inhibition is critical to the repression of MM oncogenes by SLM6.
Figure 5. SLM6 represses multiple myeloma oncogenic gene translocation products.
(A) U266 B1 cells were treated with 250 nM SLM6 for the indicated time points. Western blots are shown. Ran is included as a loading control.
(B) RPMI-8226 cells were treated with 250 nM SLM6 for the indicated time points. Western blots are shown. The arrow to the right of the c-Maf blot marks a non-specific band (NS) with molecular weight of 55 kDa.
(C) RPMI-8226 and U266B1 cells were treated with SLM6 (250 nM) for 6 hours. RT-PCR analysis of c-Myc and Cyclin D1 is shown. GAPDH was included as a loading control.
(D) RPMI-8226 cells were treated with the indicated CDK inhibitors for 8 hours. Western blots are shown. The following drug concentrations were used: SLM6 at 250 nM, flavopiridol at 250 nM, roscovitine at 5 μM, purvalanol A at 10 μM, and alsterpaullone at 5 μM.
SLM6 has more potent anti-MM activity in vivo than flavopiridol and is highly active in combination with bortezomib
The multi-CDK inhibitor flavopiridol potently induces apoptosis of MM cells in vitro and is being evaluated in human trials, although objective clinical responses have thus far been limited (25, 26). We next performed in vivo studies comparing the anti-MM activity of SLM6 to flavopiridol. We developed a model whereby GFP-expressing RPMI-8226 MM cells were subcutaneously co-injected with NIH3T3 mouse fibroblasts. This co-injection strategy increased tumor take rate to >90% (n=100) compared to 0% for RPMI-8226 cells alone (n=20) or 40% for NCI-H929 cells alone (n=70). As GFP was stably expressed in RPMI-8226 cells, MM tumor burden could be visualized and distinguished from unlabeled fibroblasts using non-invasive fluorescence imaging techniques (Fig. 6A). Immunohistochemical analysis revealed that tumors were composed of >95% CD138+ MM plasma cells (Fig. 6A). We treated mice with weekly doses of SLM6 (0.5 mg/kg), flavopiridol (5.0 mg/kg) or vehicle (PBS) and measured tumor growth over time by caliper measurements and non-invasive fluorescence imaging. With results similar to what was observed in the NCI-H929 xenograft model, SLM6 significantly reduced the size of MM tumors relative to vehicle-treated controls, with anti-MM effects being evident within 7 days of the first treatment (Fig. 6B and C). Flavopiridol, by comparison, at a dose 10 times higher than SLM6 showed no anti-MM activity in this model. The repeated dosing of SLM6 had no effect on mouse body weight or body condition scoring (data not shown), suggesting that multiple doses of SLM6 at 0.5 mg/kg were efficacious as well as non-toxic to mice.
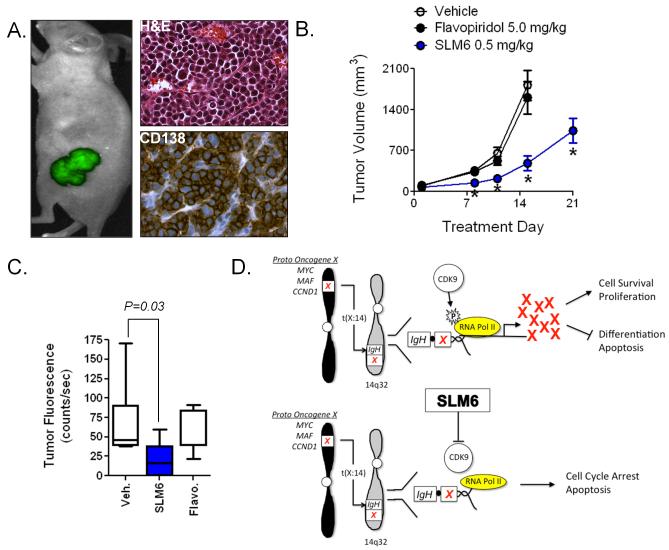
Figure 6. SLM6 is a more potent anti-MM agent than flavopiridol in vivo.
(A) RPMI-8226-GFP cells were co-injected subcutaneously with unlabeled NIH-3T3 mouse fibroblasts into hairless SCID mice. MM tumor burden was non-invasively imaged in live animals using the CRi Maestro multispectral imaging system. Immunohistochemical analysis of CD138 expression was performed on paraffin-embedded sections. Representative images demonstrate that RPMI-8226-GFP/NIH-3T3 co-injections give rise to tumors that are composed of >95% CD138+ plasma cells.
(B) Mice bearing tumors described in (A) were treated with weekly i.p. doses of SLM6 (0.5 mg/kg), flavopiridol (5.0 mg/kg), or vehicle. Tumor volume was measured over time using calipers. Data represent mean ± S.E. *P<0.001 compared to vehicle-treated controls by student’s t-test (N=10).
(C) Tumor burden from mice in (B) was measured by non-invasive fluorescence imaging on day 15 of the trial. Data points represent the mean GFP fluorescence per tumor (N=10). A student’s t-test was used for statistical analysis.
(D) Model of SLM6 activity: Chromosomal abnormalities involving the IgH gene (14q32) are common in MM and often lead to the overexpression of several oncogenes. In the schematic, oncogene x represents one of several possible oncogenes that are induced in MM due to juxtaposition with the IgH gene, which is actively expressed in post-germinal B cells and plasma cells. Complete transcription of oncogene x requires the elongation phase of transcription, which is governed by CDK9 phosphorylation of the c-terminal tail of RNA polymerase II. The resulting oncoprotein X drives MM transformation and progression by inhibiting plasma cell death and terminal differentiation while promoting proliferation. SLM6 induces MM cell apoptosis by inhibiting CDK9 function, which blocks transcription and leads to rapid down-regulation of oncogenes that have short protein half-lives and require perpetual synthesis to maintain MM survival and proliferation.
The proteasome inhibitor bortezomib is FDA-approved for front-line MM treatment. Despite high initial response rates, bortezomib eventually loses its efficacy. Therefore, we next determined if SLM6 could enhance the activity of bortezomib in MM cells. We treated NCI-H929 as wells as CD138+ MM patient bone marrow cells with increasing concentrations of SLM6 in the presence of bortezomib. In both cases the combination of SLM6 and bortezomib more effectively reduced MM cell viability than either single agent alone (Fig. S6). MM cell lines and primary patient plasma cells were sensitive to both SLM6 and bortezomib as single agents, and the combined effects of the drugs was additive rather than synergistic. Nevertheless, the combination of SLM6 and bortezomib was highly toxic to MM cells, suggesting that this may be an effective therapeutic combination.
Discussion
The primary objective of this study was to continue the development of new class of SLMs by testing their activity across tumor cell lines from different tissues of origin. Our cell screening approach revealed that MM cell lines and primary patient plasma cells were remarkably more sensitive (3 to 10 times) to single agent SLM3 than other tumor types, and we therefore focused our study on characterizing and investigating the molecular mechanism of activity of SLMs in MM. MM cells were relatively insensitive to other nucleoside analogs (5′-fluorouracil gemcitabine, and cladribine) as well as drugs with specific molecular targets (LiCl, purvalanol A, RO3306, and roscovitine), demonstrating a unique sensitivity to SLMs rather than a general hypersensitivity to therapeutic agents. Our initial screening approach investigated the effects of SLM3, which was the focal molecule from our previous work (17). We expanded our drug candidates to include other structurally related SLMs and found that like SLM3, other SLMs effectively reduced viability and induced apoptosis of MM cells. In vitro drug activity does not necessarily translate to in vivo efficacy. To prioritize individual SLM structures based on their drug-like properties, we tested each compound in an in vivo MM plasmacytoma model. SLM6 demonstrated the most efficacy in vivo, producing robust and sustained anti-tumor responses with limited toxicity after a single dose of 1 mg/kg. Further in vivo testing revealed that repeated dosing of SLM6 at a dose of 0.5 mg/kg was also tolerable and active against MM tumors. Future studies should expand the dosing regimens of SLM6 to test more doses, schedules, and routes of delivery.
We took a candidate approach to determining the anti-MM mechanism of action of SLM6. We found that SLM6 potently and directly inhibited CDK9 and repressed the expression of oncogenes that are overexpressed in MM cell lines as a result of gene translocations involving the IgH gene locus (c-Maf, cyclin D1, c-Myc, and L-Myc). It is likely that the simultaneous down-regulation of multiple genes contributes to the anti-MM activity of SLM6. However, given the known roles of c-Maf, cyclin D1, and c-Myc in the maintenance and progression of MM, their transcriptional repression likely plays a pivotal role in the anti-MM activity of SLM6. We found that SLM6 also inhibits CDK1 and CDK2, although pharmacologic studies using other CDK inhibitors revealed that only those with activity against CDK9 were capable of inducing MM cell death. This demonstrates that the activity of SLM6 against CDK9 is critical to its apoptosis-inducing effects in MM cells.
A biological explanation for the high sensitivity of MM cells to SLM6 may be related to the instability of the oncoproteins that are over-expressed in MM. We found the protein half-lives of c-Maf, cyclin D1, c-Myc, to range between 15 minutes and 1 hour (Fig. S7), whereas the half-lives of non-oncogenic “housekeeping” proteins, such as glycogen synthase, the nuclear GTPase Ran, ribosomal S6 protein (S6), and the retinoblastoma tumor suppressor gene (Rb), all exceeded 24 hours (Fig. S7). This suggests that MM-associated oncoproteins are in a constant state of turnover and require perpetual replenishment by de novo synthesis, and therefore their expression and activity may be affected more rapidly and significantly by an agent that disrupts transcription. We propose a model for the anti-MM activity of SLM6 (Fig. 6D), where unstable oncoproteins are expressed at high rates due to recombination errors involving the IgH gene. These oncogenes induce transformation and drive cell proliferation while preventing the differentiation and apoptosis of MM plasma cells. Inhibition of CDK9 function by SLM6 interrupts gene transcription, quickly leading to degradation of MM oncoproteins due to their instability and short protein half-lives. As MM cells are highly dependent on these oncogenic signals, their loss of expression results in rapid apoptosis.
Our study focused on a new class of experimental small molecules, SLMs, and the observation that these compounds were highly cytotoxic to MM cells at low nanomolar concentrations. We determined that SLM6 was the most promising molecule of this class given its favorable activity in vivo. Our mechanistic studies found CDK9 was the critical molecular target of SLM6 that was responsible for mediating its anti-MM activity. In support of the role for targeting CDK9 in the treatment of MM, other CDK9 inhibitors have demonstrated similar activity against MM cells. These compounds include flavopiridol, SNS-032, AT7519, and the BET bromodomain inhibitor JQ1, which appears to indirectly inhibit CDK9 by disrupting the recruitment of the P-TEFb to c-Myc target genes (8-11). We show that through CDK9 inhibition, SLM6 represses multiple oncogenic gene translocation products in MM cells, a favorable therapeutic strategy in a disease that is driven by a diverse array of genetic abnormalities involving numerous oncogenes. In conclusion, SLM6 is a novel CDK9 inhibitor and a promising agent for the treatment of MM that warrants further development - efforts that are currently underway in our group.
Supplementary Material
Acknowledgements
We thank the Penn State Hershey College of Medicine Microscopy and Histology Core Facility for technical assistance with our histological analyses.
Grant Support
Funding: W.S. El-Deiry is an American Cancer Society Research Professor. The work was funded in part by support from NIH grants CA141395, CA123258, CA105008, and CA135273, as well as from Penn State Hershey Cancer Institute laboratory start-up funds to W.S. El-Deiry.
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34. [PubMed] [Google Scholar]
- 3.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–22. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 4.Bergsagel PL, Kuehl WM. Molecular pathogenesis and consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000;97:228–33. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 8.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin Cancer Res. 2002;8:3527–38. [PubMed] [Google Scholar]
- 10.Conroy A, Stockett DE, Walker D, Arkin MR, Hoch U, Fox JA, et al. SNS-032 is a potent and selective CDK 2, 7 and 9 inhibitor that drives target modulation in patient samples. Cancer Chemother Pharmacol. 2009;64:723–32. doi: 10.1007/s00280-008-0921-5. [DOI] [PubMed] [Google Scholar]
- 11.Santo L, Vallet S, Hideshima T, Cirstea D, Ikeda H, Pozzi S, et al. AT7519, A novel small molecule multi-cyclin-dependent kinase inhibitor, induces apoptosis in multiple myeloma. Oncogene. 2010;29:2325–36. doi: 10.1038/onc.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uematsu T, Suhadolnik RJ. In vivo and enzymatic conversion of toyocamycin to sangivamycin by Streptomyces rimosus. Arch Biochem Biophys. 1974;162:614–19. doi: 10.1016/0003-9861(74)90223-9. [DOI] [PubMed] [Google Scholar]
- 13.Rao KV, Renn DW. BA-90912: an antitumor substance. Antimicrob Agents Chemother. 1963;161:77–9. [PubMed] [Google Scholar]
- 14.Bergstrom DE, Brattesani AJ, Ogawa MK, Reddy PA, Schweickert MJ, Balzarini J, De Clercq E. Antiviral activity of C-5 substituted tubercidin analogues. J Med Chem. 1984;27:285–92. doi: 10.1021/jm00369a010. [DOI] [PubMed] [Google Scholar]
- 15.Cavins JA, Hall TC, Olson KB, Khung CL, Horton J, Colsky J, et al. Initial toxicity study of sangivamycin (NSC-65346) Cancer Chemother Rep. 1967;51:197–200. [PubMed] [Google Scholar]
- 16.Loomis CR, Bell RM. Sangivamycin, a nucleoside analogue, is a potent inhibitor of protein kinase C. J Biol Chem. 1988;263:1682–92. [PubMed] [Google Scholar]
- 17.Mayes PA, Dolloff NG, Daniel CJ, Liu JJ, Hart LS, Kuribayashi K, et al. Overcoming hypoxia-induced apoptotic resistance through combinatorial inhibition of GSK-3β and CDK1. Cancer Res. 2011;71:5265–75. doi: 10.1158/0008-5472.CAN-11-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–45. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishnan SK, Gartel AL. A novel transcriptional inhibitor induces apoptosis in tumor cells and exhibits antiangiogenic activity. Cancer Res. 2006;66:3264–70. doi: 10.1158/0008-5472.CAN-05-3940. [DOI] [PubMed] [Google Scholar]
- 20.Stockwin LH, Yu SX, Stotler H, Hollingshead MG, Newton DL. ARC (NSC 188491) has identical activity to Sangivamycin (NSC 65346) including inhibition of both P-TEFb and PKC. BMC Cancer. 2009;9:63. doi: 10.1186/1471-2407-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylatied at threonine 186. J Biol Chem. 2005;280:28819–26. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 22.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO. 2008;27:1907–18. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabrea A, Bergsagel PL, Chesi M, Shou Y, Kuehl WM. Insertion of excised IgH switch sequences causes overexpression of cyclin D1 in a myeloma tumor cell. Mol Cell. 1999;3:119–23. doi: 10.1016/s1097-2765(00)80180-x. [DOI] [PubMed] [Google Scholar]
- 24.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–63. [PubMed] [Google Scholar]
- 25.Dispenzieri A, Gertz MA, Lacy MQ, Geyer SM, Fitch TR, Fenton RG, et al. Flavopiridol in patients with relapsed or refractory multiple myeloma: a phase 2 trial with clinical and pharmacodynamic end-points. Haematologica. 2006;91:390–3. [PubMed] [Google Scholar]
- 26.Holkova B, Perkins EB, Ramakrishnan V, Tombes MB, Shrader E, Talreja N, et al. Phase I trial of bortezomib (PS-341; NSC 681239) and alvocidib (flavopiridol; NSC 649890) in patients with recurrent or refractory B-cell neoplasms. Clin Cancer Res. 2011;17:3388–97. doi: 10.1158/1078-0432.CCR-10-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.