Abstract
Background
In marsupials, growth and development of the young occur postnatally, regulated by milk that changes in composition throughout the long lactation. To initiate lactation in mammals, there is an absolute requirement for insulin (INS), a gene known to be imprinted in the placenta. We therefore examined whether INS is imprinted in the mammary gland of the marsupial tammar wallaby (Macropus eugenii) and compared its expression with that of insulin-like growth factor 2 (IGF2).
Results
INS was expressed in the mammary gland and significantly increased, while IGF2 decreased, during established milk production. Insulin and IGF2 were both detected in the mammary gland macrophage cells during early lactation and in the alveolar cells later in lactation. Surprisingly, INS, which was thought only to be imprinted in the therian yolk sac, was imprinted and paternally expressed in the liver of the developing young, monoallelically expressed in the tammar mammary gland and biallelic in the stomach and intestine. The INS transcription start site used in the liver and mammary gland was differentially methylated.
Conclusions
This is the first study to identify tissue-specific INS imprinting outside the yolk sac. These data suggest that there may be an advantage of selective monoallelic expression in the mammary gland and that this may influence the growth of the postnatal young. These results are not consistent with the parental conflict hypothesis, but instead provide support for the maternal–infant co-adaptation hypothesis. Thus, imprinting in the mammary gland maybe as critical for postnatal growth and development in mammals as genomic imprinting in the placenta is prenatally.
Keywords: Genomic imprinting, Mammary gland, Lactation, Marsupial, Insulin, Co-adaptation
Background
Genomic imprinting is an epigenetic modification to the DNA that regulates the expression of selected genes from only one parental allele. In vertebrates, imprinting is restricted to the therian (marsupial and eutherian) mammals but as yet no imprinted genes have been identified in monotremes [1,2]. Thus mammalian genomic imprinting is thought to have evolved after the therian–monotreme divergence. In mice and humans, most imprinted genes are expressed in the placenta, some of which are exclusively imprinted in this organ [3-7]. Although the significance of imprinted gene expression is still debated, many imprinted genes regulate growth and nutrient provisioning to the developing fetus [8-11]. Potentially, therefore, any organ that regulates growth via nutrient exchange with the developing young may have imprinted gene expression. Imprinted gene expression occurs in the hypothalamus to regulate maternal behaviour, metabolism and milk letdown [12-14]. Paternally expressed gene 3 (Peg3) and MAGE-like gene 2 are examples of genes that are imprinted in the neonatal hypothalamus. Neonatal Peg3 knockout mice lose their ability to find the teat and feed, while MAGE-like gene-2-deficient mice markedly reduce their activity, metabolism and food intake [13,15,16]. In the adult, heterozygous Peg3 knockout mothers have impaired milk letdown and fail to allow sucking by the pups while heterozygous males have naive sexual behaviour [12-14,17-19]. Paternally expressed gene 1 (Peg1) deficient females have abnormal maternal behaviour and impaired placentophagia, sometimes leaving their pups untouched after parturition [20]. A large number of autosomal genes with sex-specific imprinting in the cortex and hypothalamus have been identified recently, but as yet there are no data on their possible functions [21,22].
Mammary glands, lactation, maternal and neonatal behaviour involved in postnatal feeding are all essential mammalian characteristics that regulate and enhance the growth and survival of young. The mouse X chromosome is nonrandomly maternally inactivated in mammary epithelial cells [23]. In the same study, the X-linked gene Rnf12, which encodes the ubiquitin ligase Rnf12/RLIM, is identified to be a critical survival factor for milk-producing alveolar cells [23]. Limited breast cancer studies have similarly demonstrated monoallelic expression in the mammary gland [24-26]. For example, there is monoallelic expression of insulin-like growth factor 2 (IGF2) in normal breast tissue and in all but two cases of breast cancer [27]. These data indicate that certain growth and survival factors are selectively monoallelically expressed in the eutherian mammary gland.
INS encodes insulin, a polypeptide hormone that regulates carbohydrate metabolism, cell growth and survival, protein synthesis, vascularisation and vasodilation [28-31]. Alternative INS transcripts, with either an extended 5′UTR or a retained intron, and chimaeric transcripts with exons from the upstream tyrosine hydroxylase(TH) gene have been identified in the developing chick. These alternative transcripts all result in decreases in the efficiency of protein translation, which may control cell survival in the early developing embryo [32-35].
Insulin is essential for the induction of milk protein synthesis in mammals [36-41]. Elevated insulin levels in the goat mammary gland increase milk production and the milk protein content [42]. Similarly in dairy cows, the milk protein yield increases by approximately 15% after insulin administration [43,44]. INS is located upstream of the growth-promoting, paternally expressed gene IGF2, and functions in the mammary gland in conjunction with IGF2 to induce growth and alveologenesis [38]. IGF2 was the first imprinted gene identified in both eutherians and marsupials [45,46]. In eutherians, IGF2 is imprinted in most fetal and adult tissues –but in the marsupial, although the maternal IGF2 allele is completely silenced in the fetal and pouch young liver, silencing is incomplete in the placenta and IGF2 is biallelically expressed in the adult liver [47,48]. In the eutherian mammary gland, IGF2 acts with cyclin D1 to mediate the prolactin-induced proliferation of mammary epithelial cells during alveolar formation [38,49].
Insulin and IGF2 are both found in the milk of humans, dairy cows and rats [50-52]. They are present at the highest concentrations in colostrum, but are still present at low concentrations in milk produced later in lactation [52]. Milk IGFs may support nutrient transfer to the young by increasing mucosal cell turnover and enhancing villus growth of the newborn’s gastrointestinal tract [51,52].
INS imprinting has been analysed previously, but only in the pancreas and the yolk sac of mice and humans. Paternal expression, and therefore imprinting, is detected only in the yolk sac, but there is biallelic expression in the pancreas and fetal head and body and so it is not imprinted in these tissues [53-56]. Similarly, there is paternal INS expression in the tammar wallaby (Macropus eugenii) yolk sac placenta [57]. Imprinting of the mouse Ins2 (the homologue of human INS) gene and the Igf2 gene is disrupted by maternal inheritance of a targeted deletion of the H19 gene and its flanking sequence, so imprinting of INS is regulated by the same imprinting control region as IGF2 and H19[58].
The identification of Peg3, a hypothalamic gene that regulates maternal suckling behaviour in mice, prompted the development of the maternal–infant co-adaptation hypothesis [14,19,59] as an alternative to the parental conflict hypothesis to explain the evolution and maintenance of genomic imprinting in mammals [60,61]. The conflict hypothesis predicted that imprinting in the fetus and placenta evolved as a consequence of competition between the male and female genomes to optimise their respective reproductive success [60]. In contrast, the maternal–infant co-adaptation hypothesis predicts that imprinting evolved to enhance the genetic integration of the intimate maternal–offspring interactions [19,59]. For example, genes in the offspring that regulate the offspring’s requirements and behaviour (for example, nutritional demand and sucking) and genes in the mother that regulate her response (for example, nutritional supply and suckling) may acquire tissue-specific imprinted expression (for example, placenta, mammary gland and brain) to allow a greater potential for rapid fixation of beneficial traits in the population [12,13,19,59,62]. This hypothesis provides a clear explanation for the presence of imprinting before and after birth and could apply to genes expressed in the mammary gland that regulate both maternal milk production and supply. INS, which is imprinted in the placenta, is also important for the development and function of the mammary gland, and so there may be imprinted expression in this unique mammalian organ to regulate nutrient production and transport to the neonate after birth as the placenta does before birth.
To examine this hypothesis we investigated the allelic expression of INS in the marsupial mammary gland. Marsupials give birth at a much earlier developmental stage than most eutherians and have only a short-lived chorio-vitelline placenta. The vast majority of growth and development of the young occurs postnatally through a long, complex and physiologically sophisticated period of lactation [63,64]. In the tammar the length of lactation is 9 months, during which the young increase from a birth weight of ~450 mg to ~2.5 kg before they are fully weaned. Three broad phases can be recognised, during which the size of the mammary gland and the composition of the milk change. In the tammar, as in other marsupials, milk composition changes in concert with the developmental stage of the young to directly regulate its growth. Developmental acceleration or retardation can be experimentally induced when young are reared on either late-stage or early-stage milk, respectively [65-67]. Thus the mammary gland is an obvious target for imprinting under the maternal–infant co-adaptation hypothesis. Furthermore, both mammary gland size (correlated to the volume of milk produced) and the composition of the milk could potentially be regulated by both INS and IGF2. We hypothesised that if imprinting enhances the production and transport of nutrients and growth factors, as the co-adaptation hypothesis predicts, the mammary gland, particularly in marsupials, would be a primary site of imprinted gene expression. We examined this hypothesis by analysing the allelic expression of INS and compared it with that of IGF2 in the tammar wallaby throughout lactation.
Results
Characterisation of INS and IGF2 mRNA expression in the tammar mammary gland
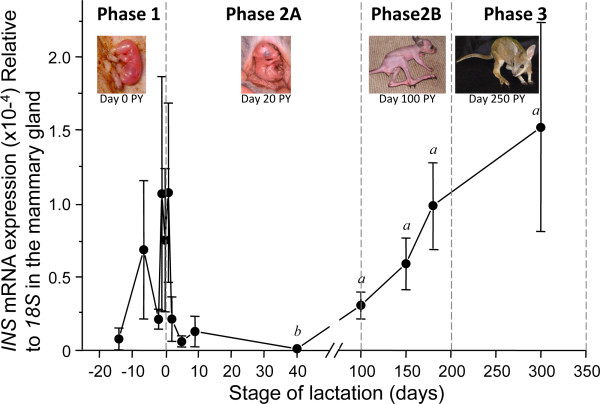
INS and IGF2 were expressed throughout the three broad phases of lactation: phase 1 (during pregnancy), phase 2A (day 0 to day 100 of lactation), phase 2B (day 100 to day 200 of lactation) and phase 3 (day 200 to day 350 of lactation). There was no statistically significant variation in INS expression throughout the early stages of lactation, before day 5, when the gland is developing and producing colostrum. However, there was a significant increase in INS expression during established milk production between day 9 and day 300 of lactation (P < 0.05) (Figure 1).
Figure 1.
INSmRNA expression relative to18SmRNA expression in the mammary gland.INS mRNA expression (mean ± standard error of the mean × 10-4) compared with18S expression in the mammary gland across the four phases of lactation. After birth, INS remained low until after 100 days of lactation.a significantly higher than b (P < 0.05). INS, insulin gene.
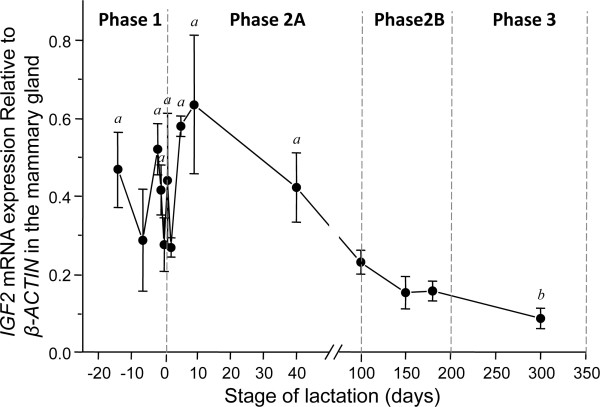
IGF2 expression was high during early lactation and decreased from phase 2 onwards (P < 0.05) (Figure 2). There was a significant negative correlation (P < 0.05) between INS and IGF2 after day 5 of lactation.
Figure 2.
IGF2mRNA expression relative toβ-ACTINmRNA expression in the mammary gland.IGF2 mRNA expression (mean ± standard error of the mean) compared with β-ACTIN expression in the mammary gland across the four phases of lactation. IGF2 was highest in the perinatal period, but steadily decreased throughout the remainder of lactation. a significantly higher than b(P < 0.05). IGF, insulin-like growth factor.
Localisation of insulin and IGF2 protein in the tammar mammary gland
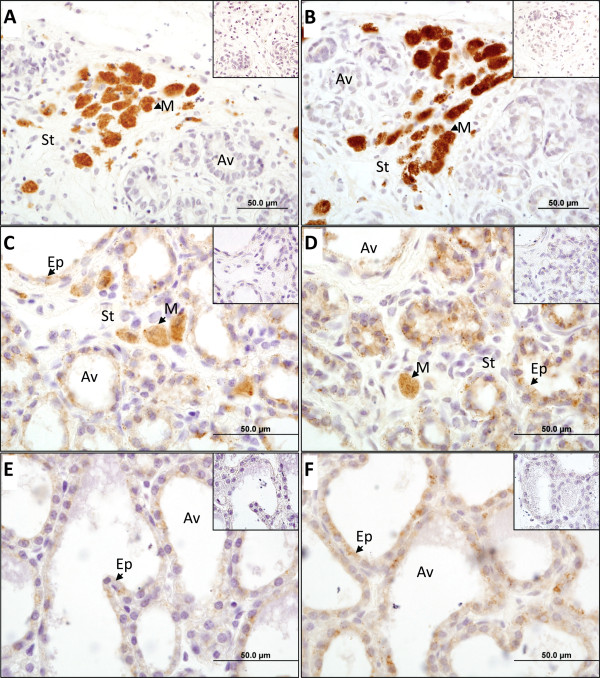
Both insulin and IGF2 were strongly stained and localised in the cytoplasm of macrophage cells in the stromal cells during lactogenesis phase 1 (Figure 3A,B). At later stages, protein was detected predominantly in the cytoplasm of the alveolar epithelial cells in the lactating mammary gland, with some macrophages containing both insulin (Figure 3C,E) and IGF2 (Figure 3D,F). There was no cytoplasmic or nuclear staining in the IgG antibody controls (Figure 3, insets) or in the no-antibody (diluent-only) negative controls. The cytoplasmic location of insulin in the tammar placenta was similar to that previously reported [57]. In the pancreas, the insulin antibody was detected only in the cytoplasm of the cells in the islets of Langerhans (see Additional file 1).
Figure 3.
Immunolocalisation of insulin and IGF2 protein in the tammar mammary gland. Insulin (A), (C) and (E) and insulin-like growth factor 2 (IGF2)(B), (D) and (F) protein (brown staining) localisation in mammary glands at three different stages of lactation. (A), (B) One day before birth (day −1), macrophage cells (M) in the stroma (S) were intensely stained and there was no staining in the alveolar cells (Av). (C), (D) At day 95 of lactation, insulin and IGF2 was detected in the cytoplasm of the alveoli epithelial cells (Ep), with infrequent macrophage cell staining. (E), (F) At day 200 of lactation, staining was concentrated in the epithelial cells of the alveoli. There was no staining in any of the IgG antibody control (top right insert). There was no staining in any of the no-antibody controls (data not shown). Scale bars shown at the bottom right.
Monoallelic expression in the tammar mammary gland
The INS gene
Two previously identified INS SNPs located 16 base pairs apart [57] were used to identify the imprint status of INS. Of 20 adult individuals, seven animals were polymorphic (two animals were polymorphic at both SNP sites, one animal was polymorphic at site 1 and four animals were polymorphic at site 2). cDNA amplified from the mRNA of all seven polymorphic individuals showed monoallelic expression (Figure 4A). A population of 32 animals were genotyped for the two SNPs and it was concluded that three different INS alleles (G-C, G-T and A-T) exist in the tammar population (see Additional file 2: Tables S1 and S2). Of these three alleles, allele 1 (G-C) and allele 2 (G-T) were monoallelically expressed in the mammary gland (see Additional file 2: Table S3).
Figure 4.
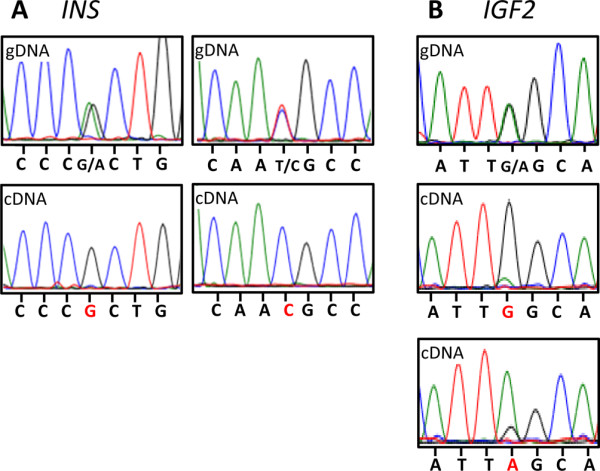
Sequence chromatographs for imprint analysis in the mammary gland. Direct sequence analysis for insulin (INS) and insulin-like growth factor 2 (IGF2) imprinting in the mammary gland. (A) INS chromatogram traces (viewed in Finch TV version 5.1) for genomic DNA (gDNA) and cDNA. Two polymorphic sites were identified in gDNA, distinguishing the two alleles. In the mammary gland of seven individuals tested, two animals had monoallelic expression at both SNPs (site 1:G/A and site 2: C/T) and five animals had monoallelic expression in a single SNP (one individual at site 1and four individuals at site 2). (B) IGF2 chromatogram traces for gDNA (3′ to 5′) and cDNA (5′ to 3′). A single polymorphic site was identified in gDNA to distinguish the two alleles. Both alleles of IGF2 were monoallelically expressed in four different individuals (three animals expressed the G allele and one animal the A allele).
To determine whether INS monoallelic expression was a global phenomenon in the tammar or was selectively maintained in the mammary gland, the allelic expression of INS was analysed in various adult and pouch young tissues. cDNA was amplified from four different tissues from five pouch young, all heterozygous at the identified SNPs and between the age of 10 and 100 days old. Biallelic expression was detected in the stomach and intestine, biased expression was observed in the adult adrenal gland and monoallelic expression was observed in nine out of 10 livers (three out of three adults and six out of seven pouch young) examined (Table 1). All three alleles were monoallelically expressed in the pouch young liver: two pouch young expressed allele 1, two pouch young expressed allele 2 and two pouch young expressed allele 3 (A-T). Two pouch young with known maternal genotype showed only paternal INS expression of allele 3 in the liver in two separate RNA extractions (Figure 5A). A third pouch young showed paternal expression of allele 2 in the liver (Figure 5B).
Table 1.
Allelic expression ofINSin different adult and pouch young tammar tissues
| Sex | Age (days) | Brain | Stomach | Intestine | Liver | Adrenal gland |
|---|---|---|---|---|---|---|
| F |
15 to 16 |
> |
Absent |
Biallelic |
Monoallelic (allele 1) |
nd |
| M |
30 |
nd |
nd |
nd |
Monoallelic (Pat, allele 2) |
nd |
| M |
36 |
> |
Biallelic |
Biallelic |
Monoallelic (allele 1) |
nd |
| M |
45 to 46 |
Low |
Biallelic |
Biallelic |
Biallelic |
nd |
| M |
52 |
Low |
Biallelic |
Biallelic |
Monoallelic (allele 2) |
nd |
| M |
81 |
Low |
Biallelic |
Biallelic |
Monoallelic (Pat, allele 3) |
nd |
| F |
182 |
nd |
nd |
nd |
Monoallelic (Pat, allele 3) |
nd |
| F |
Adult |
nd |
nd |
nd |
Monoallelic (allele 2) |
> |
| F |
Adult |
nd |
nd |
nd |
Monoallelic (allele 1) |
> |
| F | Adult | nd | nd | nd | Monoallelic (allele 1) | > |
Pouch young and adult tissues analysed for INS imprinting via direct sequencing. Both female (F) and male (M) samples had monoallelic, biallelic or biased (>) expression. Some tissues had very low RT-PCR amplification (low) or no amplification (absent) so imprinting could not be determined. Three pouch young with a known maternal genotype showed paternal (Pat) monoallelic expression in the liver. nd, not determined.
Figure 5.
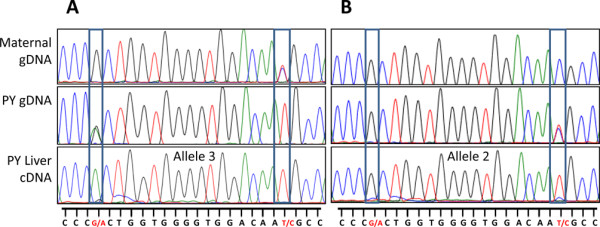
INSsequence chromatographs for imprint analysis in the pouch young liver. Direct sequence analysis for INS in the pouch young liver. Chromatogram traces of genomic DNA (gDNA) from the mother and pouch young and of cDNA from the pouch young liver. (A) The pouch young inherited allele 2 (G-T) from its mother, and the clear monoallelic expression of allele 3 (A-T) in the liver was inherited from the father. (B) The pouch young inherited allele 1 (G-C) from its mother, and the clear monoallelic expression of allele 2 (G-T) in the liver was inherited from the father. RNA was extracted twice from the same liver sample and direct sequencing produced the same results in both samples. These results indicate that INS expression in the liver is a result of parent-of-origin specific genomic imprinting and not random monoallelic expression. INS, insulin gene.
The IGF2 gene
Of 20 individuals analysed, seven animals were polymorphic at one of the previously identified IGF2 SNP sites in the tammar [68]. Monoallelic expression was observed in the mammary gland from two different IGF2 alleles (Figure 4B).
The INS transcription start site was not differentially methylated in the mammary gland
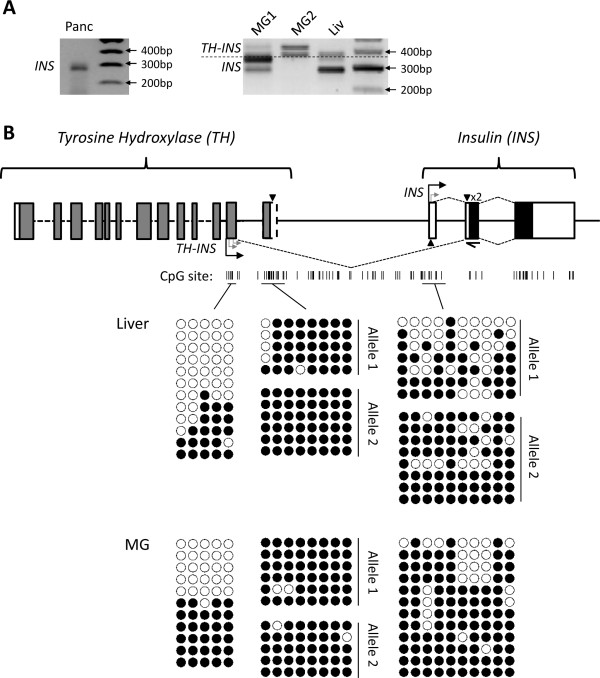
5′-Rapid amplification of cDNA ends (5′-RACE) was used to identify the transcription start sites (TSSs) of INS. A single transcript was identified in the adult pancreas, while multiple transcripts were identified in the mammary gland and liver (Figure 6A). TSSs were located in the INS first exon and in the second to last predicted exon of the tammar TH gene (Figure 6B), separated by approximately 3.6 kb. Only INS transcripts were identified in the pancreas whereas both INS and TH-INS transcripts were detected in the mammary gland and liver (Figure 6A). Repeated imprint analysis, using transcript-specific primers, showed conserved paternal expression of TH-INS in two pouch young livers, and conserved monoallelic expression of INS in two mammary glands. The genomic region around the TH-INS and INS TSSs were relatively CpG-rich, and a CpG island (CGI) was located downstream of the TH-INS TSS, at the location of the predicted last TH exon (Figure 6B). The methylation status of these three regions was analysed by bisulphite sequencing, using primers designed by MethPrimer [69], to determine whether INS allelic expression was regulated by differential methylation. Methylation was assessed in both liver and mammary tissue (Figure 6). A SNP was located in the CGI and near the INS TSS. These were used to distinguish between alleles in heterozygous samples. The TH-INS TSS appears to have a differentially methylated region (DMR)-like methylation pattern, but no usable SNP was identified within this region to determine allele specificity of the methylation. No allele-specific methylation was observed at the INS TSS and the CGI was completely methylated (Figure 6).
Figure 6.
Structure and methylation of tammarINS. (A) 5′-Rapid amplification of cDNA ends (5′-RACE) was performed on RNA derived from one pancreas (Panc), two mammary glands (MG) and one liver (Liv). Five INS transcripts were amplified using a primer designed in the first INS coding exon (half-arrow). Three transcripts were chimeras and contained an exon derived from the neighbouring tyrosine hydroxylase (TH) gene and two were transcribed from the INS noncoding exon. The mammary gland 1 (MG1; lactation phase 1) and liver expressed both types of transcripts, the pancreas expressed only the INS-derived transcripts, and the mammary gland 2 (MG2; lactation phase 3) expressed only the TH-INS transcripts. (B) Schematic of predicted tammar TH and INS genes (not to scale). Predicted coding exons (grey), verified coding exons (black) and noncoding exons (white) are represented by boxes. Transcription start sites identified by 5′-RACE are indicated with turned arrows. CpGs are indicated by short vertical black lines. SNPs are indicated by black triangles. Bisulphite sequenced regions (black horizontal lines) are shown with individual bisulphite sequences underneath: open and closed circles are unmethylated and methylated CpGs, respectively. Each row represents the methylation pattern on a separate DNA fragment from the same sample. Both methylated and unmethylated alleles were present in the liver and mammary gland tissues at the TH-INS TSS. Only methylated alleles were present at the CpG Island and the INS TSS had a variable methylation pattern.INS, insulin gene.
Discussion
The tissue-specific monoallelic expression of INS in the marsupial mammary gland suggests that there may be a role for genomic imprinting during lactation. Marsupials have a long and physiologically sophisticated lactation that provides fine control of the growth and development of the marsupial young by the mother. Thus imprinting in the mammary gland could regulate postnatal growth in mammals, just as imprinting in mammalian placenta regulates prenatal growth and development.
Both INS and IGF2 were expressed in mammary glands throughout lactation, but the expression patterns differed. The increase in INS mRNA between phase 2A and phase 3 may reflect a role for endogenous insulin in the hyperplasia that characterises late lactation. This increase also coincides with a marked increase in volume and a change in milk composition. Insulin is required for the initiation of lactation in mammals and induces casein gene expression [36,41,43]. More specifically, insulin plays a crucial role in the transcription of at least 18 milk genes including Stat5a and Elf5, two major milk protein gene transcription factors and key components of prolactin signalling [40]. The increase of INS expression is therefore consistent with it regulating the increase in volume and the change to mature milk in the mammary gland. Relative IGF2 mRNA expression was highest in phase 1, consistent with paracrine signalling of IGF2 regulating the initial proliferation of mammary gland epithelial cells [49,70].
During phase 1 of lactation, insulin and IGF2 protein were localised in the stroma, specifically in the macrophages. Macrophages are critical for normal mammary gland branching and alveolar morphogenesis [71,72]. Insulin and IGF2 may therefore regulate epithelial cell proliferation and morphogenesis as the mammary gland prepares for lactation [38]. Later in lactation, both insulin and IGF2 were detected in the cytoplasm of the epithelial cells of the alveoli, so it is likely that both proteins are secreted into the tammar milk, as in the cow and rat [51,52].
Is INS imprinted in the marsupial mammary gland?
In the tammar, there was clear monoallelic INS expression in the mammary gland of seven individuals from phase 1 through to phase 2B of lactation. As reported previously in the tammar yolk sac placenta [57], INS was paternally expressed and therefore imprinted in the pouch young liver. INS was biallelically expressed in both the stomach and intestine and was biased in the adrenal gland, demonstrating that INS monoallelic expression is not widespread but is specifically maintained in the marsupial liver, mammary gland and placenta. As most specimens were collected from the wild, the parental genotypes were unknown. Because paternal expression of INS was established in the liver (Figure 5) and previously in the yolk sac [57] and we detected monoalleleic expression of three different INS alleles in the mammary gland and liver and a differentially methylated TSS, our data suggest that INS monoallelic expression is most probably due to genomic imprinting and not due to random monoallelic expression or allelic difference [73]. However, this cannot be definitively proven without a reciprocal cross. Based on limited analyses in mice and humans, the INS gene was presumed to be exclusively imprinted in the yolk sac [55,74]. Interestingly, there is an alternatively spliced INS-IGF2 transcript that is imprinted and paternally expressed in the human fetal eye and limb [75]. INS monoallelic expression may therefore exist in tissues other than the yolk sac in eutherians, as it does in marsupials.
The marsupial mammary gland is the primary site for nutrient exchange between mother and infant. Our results suggest that INS may directly initiate and maintain alveologenesis and play a part in controlling the change in tammar milk composition by up regulating major milk protein gene transcription and translation factors [40,41]. As INS is critical for mammalian lactation, imprinting may have been maintained in this tissue to control mammary gland development and the transcription of vital milk proteins. Such a role is unlikely to be restricted to marsupials since eutherian mammals also depend on lactation to support their developing young after birth.
Methylation of the transcription start site may regulate the expression of INS
The methylation pattern seen at the TH-INS TSS strongly implies that this is a differentially methylated region, although this remains to be confirmed. Tammar INS expression could be regulated by the conserved differentially methylated imprinting control region located between IGF2 and H19[47], which regulates INS imprinting in both humans and rodents [58,76]. The putative DMR located at the THINS TSS may regulate imprinting in a similar manner to the DMRs located at each of the human IGF2 promoters [77]. The high level of methylation at the CGI may be in place to prevent the inclusion of the last TH exon into the TH-INS mRNA. The variable methylation pattern observed at the INS TSS suggests that methylation at this TSS could regulate tissue-specific expression in the tammar as it does in eutherians [78]. INS imprinting could be transcript specific, as it is in the mouse imprinted gene Dopa decarboxylase [79] and the human imprinted GRB10[80] and IGF2[77,81] genes. However, preliminary analysis indicates that both INS and TH-INS transcripts are monoallelically expressed. INS may be the fourth marsupial imprinted gene to be associated with a DMR [47,82,83] and, if so, provides further evidence of a common origin of imprinting mechanisms in therian mammals.
Conclusions
Both INS and IGF2 are imprinted and paternally expressed in the marsupial placenta and in the liver (this study) [57,68]. Both genes are also monoallelically expressed in the mammary gland, suggesting that both may be imprinted in this tissue. The differential methylation of the TH-INS TSS in both the liver and mammary gland strongly supports the suggestion that INS is also imprinted in the mammary gland. This is the first indication that genomic imprinting may occur in a marsupial mammary gland and is consistent with the predictions of the maternal–infant co-adaptation hypothesis that genomic imprinting is involved in regulating the growth and development of young postnatally. INS was biallelically expressed in the developing digestive tract. Retention of monoallelic expression of INS therefore appears to be under selection in the mammary gland as well as in the placenta and liver. The mammary gland is a unique mammalian organ that regulates postnatal nutritional transfer by a positive feedback loop with the mother’s brain in response to the sucking stimulus. This interaction is similar to that observed between the placenta, fetus and the maternal hypothalamus. Genomic imprinting in the mammary gland may therefore be as critical for regulating postnatal growth as it is for regulating prenatal growth in the placenta.
Methods
All experiments and wild animal collection were approved by the University of Melbourne Animal Experimentation Ethics Committee, and the animal handling and husbandry procedures were in accordance with the National Health and Medical Research Council of Australia 2004 guidelines. Animals were collected under approval from the South Australian Department of Environment and Natural Resources.
Animals
Tammar wallabies of Kangaroo Island, South Australia origin were held at our colony in Melbourne. Pregnancy was initiated in females carrying an embryo in diapause by the removal of their pouch young [63]. Adult females carrying fetuses in the final third of gestation (day 19 to day 26 of the 26.5 day pregnancy) or pouch young (day 0 to day 350 postpartum) were killed either by shooting in the wild, stunning and cervical dislocation or by an anaesthetic overdose (sodium pentobarbitone, 60 mg/ml to effect). The lactating mammary gland and adult and pouch young tissues were collected and snap frozen in liquid nitrogen. As the animals were collected from the wild, the parental genotypes for most samples were unknown. Tammar wallabies are seasonal breeders, and males do not reach sexual maturity until approximately 2 years of age and females are unreliable breeders until they attain at least 2 years of age [63]. In addition, all females carry a diapausing blastocyst conceived a year before the birth of the new young, so it is extremely difficult to determine paternity in the wild population. Executing a targeted mating between homozygous individuals and producing an informative heterozygous adult lactating female was therefore beyond the scope of the present study.
RNA and DNA extraction and RT-PCR
Individual genotypes were identified using PCR and direct sequencing of genomic DNA extracted from approximately 20 mg snap-frozen tissue using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Total RNA was extracted from mammary glands using the RNeasy Lipid Tissue Mini Kit (Qiagen,Hilden, Germany) or from other tissues using Tri-Reagent (Ambion, Austin, Texas, USA) as described by the manufacturer with a final elution of RNA in 60 to 80 μl RNAsecure H2O (a dilution of 1/24 μl RNAsecure to water;Ambion). Total RNA was DNase treated (DNA-free™; Ambion) to remove contaminating DNA, run on a 1% agarose gel to assess the quality, quantified with a nano-spectrometer (NanoDrop ND-1000 Spectrophotometer; NanoDrop Technologies Inc., Wilmington, DE, USA) and cDNA was synthesised using the SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen, Carisbad, CA, USA). Typically, 2 μg or a maximum of 8 μl total RNA was used in each cDNA synthesis reaction, with 1 μl Oligo (dT)12-18 (50 μM). cDNA integrity was immediately assessed with GAPDH RT-PCR. All primers were designed using Primer3 (v. 0.4.0) [84] and were synthesised by Sigma-Aldrich (Castle Hill, NSW, Australia) (see Additional file 3).
Immunohistochemistry
Insulin and IGF2 protein distribution was assessed in 10 mammary glands: four glands from phase 1 of lactation (day 25 of pregnancy, 1 day before birth), and six glands from phase 2 (three each from days 100 and 200 of lactation,post partum). IGF2 immunohistochemistry was performed as previously described [85] (Sc-7435; Santa Cruz, Santa Cruz, CaliforniaUSA). Insulin B N-20(sc-7838; Santa Cruz) antibody, raised against an epitope with 88% identity (15/17) to the tammar insulin predicted protein, was used to assess insulin distribution. Pancreas and placental tissues were used as controls to assess specificity of the antibody (see Additional file 1). The immuno histochemistry protocol was optimised in the mammary gland for both antibodies. Small pieces of mammary gland (placenta and pancreas for positive controls) were fixed in 4% paraformaldehydebefore paraffin embedding. Sections (6 to 7 μm) were mounted onto polylysine-coated slides (polysine;Menzel-Glaser; Lomb Scientific, Thermo Fisher, Waltham, MA, USA) before dewaxing, rehydrating and washing in 0.1% Triton-X-100. A 10-minute incubation in 0.05% Pronase (type XXIV; Sigma-Aldrich) preceded blocking in 10% normal rabbit serum/Tris-buffered saline/0.1% BSA. Primary antibodies (Insulin B N-20,sc-7838 at 0.4 μg/ml;and IGF-II F-20, sc-7435 at 0.4 μg/ml) were incubated overnight at 4°C. Negative controls were incubated with either goat IgG (sc-2028;Santa Cruz), at the same concentration as the target antibody, or with no antibody (diluent only). Goat anti-rabbit biotinylated secondary antibody (DAKOGlostrup, Denmark) was used before incubation with streptavidin/horseradish peroxidase-conjugated (DAKO) and was colour developed with liquid 3, 3'-diaminobenzidine (DAB)(DAKO) for 2 to 5 minutes. Sections were counterstained with a 1:10 dilution of Lillie-Mayer Haemotoxylin (AHLMAustralian Biostain Pty. Ltd, Tralagon, VIC, Australia).
Quantitative RT-PCR analysis
Quantitative real-time PCR was used to quantify INS and IGF2 expression in approximately 61 to 65 different mammary gland samples:20 to 22 from phase 1 of lactation (during pregnancy), and 25 to 27 from phase 2A (day 0 to day 100 of lactation), 11 to 12 from phase 2B (day 100 to 200 of lactation) and 4 to 5 from phase 3 (day 200 to day 350 of lactation). There were at least three samples (usually four or five) at each time point. For each sample, 800 ng total RNA was reverse transcribed using the SuperScript III First Strand Synthesis System for RT-PCR kit (Invitrogen). All primers crossed intron–exon boundaries (see Additional file 3). The INS probe, labelled at the 5′ end with 6-carboxy fluorescein, was designed using Biosearch Technologies Real Time Design software (Novato, CA, USA). 18 S and β-ACTIN were used as reference genes. Reactions were performed in triplicate in 20 μl volumes consisting of 1 × Brilliant® II QRT-PCR Master Mix (Stratagene, Agilent Technologies, Santa Clara, CA, USA), 0.3 μM forward and reverse INS primers, 0.2 μM MTR probe, and 1 μl cDNA template. The 18 S primers and probe were used at approximately 0.25 μM. IGF2 PCRs were performed as above but used FastStart Universal SYBR Green Master (Rox) (Roche Products Pty Limited Dee Why, NSW, Australia), and 0.4 μM forward and reverse IGF2 or 0.3 μM forward and reverse β-ACTIN primers were added without a MTR probe. Primer, probe, and cDNA concentrations were optimised in preliminary experiments.
Real-time PCR was carried out in an Stratagene Mx3000PTM Sequence Detector (Integrated Sciences, Chatswood, NSW, Australia) using the following conditions: 95°C for 10 minutes, followed by 50 (INS) or 45 (IGF2) cycles at 95°C for 30 seconds, 63°C (INS) or 61°C (IGF2) for 1 minute and 72°C for 30 seconds. A pancreas (INS) or liver (IGF2) sample triplicate and a negative template (water) triplicate were included on each plate as a calibrator and negative control, respectively. The data were analysed in Microsoft Excel and R [86]. The amplification efficiency was calculated from the standard curve and the cycle threshold values were corrected [87].
Allelic expression analysis
Direct sequencing of cDNA (as described above) was used to confirm allelic expression of animals heterozygous at one or both polymorphic sites. Approximately 0.5 to 1 μl template was used with 0.2 μM each primer for INS or IGF2 with either GoTaq Green Master Mix (Promega) or TaKaRa Ex Taq HS (Takara Bio Inc. Otsu, Shiga, Japan). RT-PCR cycles consisted of 94 or 96°C for 1 minute, followed by 30 to 35 cycles of 30 seconds at 94 or 96°C, 1 minute at 63°C, and 30 seconds at 72°C, and a final extension at 72°C for 5 minutes. PCR products from cDNA and genomic DNA were resolved by gel electrophoreses and the bands were extracted (QIAquick Gel Extraction Kit; Qiagen). The purified product was then sequenced using the Seq primers, designed for optimal sequence results (see Additional file 3). Sequences were assessed using FinchTV (v.1.3.1) DNA sequence chromatogram trace viewer software. The relative peak height for each allele indicates biallelic (equal peak heights) or imprinted (unequal peak heights) expression.
5′-Rapid amplification of cDNA ends
To acquire the full-length transcript for tammar INS we performed 5′-RACE, using both the SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) and 5′ Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen). GSP1 primer was used to synthesise first-strand cDNA from 5 μg total RNA. GSP2 was used in conjunction with the provided Abridged Anchor Primer to amplify the 5′ end of the transcript. PCR products were cloned using a pGEM®-T Easy vector and JM109competent cells (Promega). Plasmids were purified using Wizard® Plus SV Minipreps DNA Purification System (Promega) and sequenced.
Methylation analysis
Using MethPrimer [69], 10 CpG sites were identified upstream of exon1. Then 1 μg DNA was treated with a sodium bisulphite solution at 50°C for 4 hours before ethanol precipitating and eluting in 50 μl tris-EDTA buffer. As a template we used 20 ng of bisulphite treated DNA with 0.2 μM each bisulphite primer in a TaKaRa Ex Taq HS 25 μl reaction. A 3-minute extension time in the PCR thermal cycling was used to reduce PCR bias by permitting the polymerase to read through CG-rich regions. As no SNP was available to test for experimental biases at the TH-INS TSS, a solution containing 5 mg/mlBSA and 5% glycerol was added to the PCR reactions as a denaturant to reduce PCR bias during amplification [88]. PCR products were cloned as described above and sequences were analysed using the Quma quantification tool for methylation analysis [89]. A G/A SNP site was located in the CGI downstream of TH-INS TSS and a G/T SNP was located in the INS TSS region (Figure 6).
Abbreviations
BSA: Bovine serum albumin; CGI: CpG island; DMR: Differentially methylated region; GSP: Gene-specific primer; IGF: Insulin-like growth factor; INS: Insulin gene; PCR: Polymerase chain reaction; Peg3: Paternally expressed gene 3; 5′-RACE: 5′-rapid amplification of cDNA ends; RT: Reverse transcriptase; SNP: Single nucleotide polymorphism; TH: Tyrosine hydroxylase; TSS: Transcription start site; UTR: Untranslated region.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MBR conceived the idea. MBR, JMS, AJP and GS designed the experiments. JMS performed the experiments. JMS, SS, MBR, GS and AJPanalysed the data. JMS, MBR, SS, GS and AJP wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Insulin immunolocalisation positive controls. The immunolocalisation of insulin in the pancreas and placenta, providing positive controls to verify authentic protein recognition.
Genotyping andINSimprinting in the tammar wallaby. Additional tables and figures that display the number and proportion of INS alleles within a population and extra sequence chromatographs of INS in the mammary gland.
Primers used in this study. A table listing all of the primers used in this study.
Contributor Information
Jessica M Stringer, Email: stringer@student.unimelb.edu.au.
Shunsuke Suzuki, Email: ssuzuki@shinshu-u.ac.jp.
Andrew J Pask, Email: andrew.pask@uconn.edu.
Geoff Shaw, Email: g.shaw@unimelb.edu.au.
Marilyn B Renfree, Email: m.renfree@unimelb.edu.au.
Acknowledgements
This study was supported by the Australian Research Council (ARC) Centre of Excellence in Kangaroo Genomics; an ARC Federation Fellowship to MBR and a National Health and Medical Research Council R.D. Wright Fellowship to AJP. The authors thank Kerry Martin, Alison Bradfield and Scott Brownlees for assistance with the animals, Helen Clark and Bonnie Dopheide for technical assistance, and members of the Tammar Research Group for their help in collecting tissue. They also thank Dr David V Kaufman (Department of Pathology, University of Melbourne, Australia) for his advice on mammary gland histology and Professor Anne Ferguson-Smith for her helpful comments.
References
- Killian JK, Byrd JC, Jirtle JV, Munday BL, Stoskopf MK, MacDonald RG, Jirtle RL. M6P/IGF2R imprinting evolution in mammals. Mol Cell. 2000;5:707–716. doi: 10.1016/S1097-2765(00)80249-X. [DOI] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Stewart N, Munday BL, Andersen NA, Nicol S, Jirtle RL. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J Exp Zool. 2001;291:205–212. doi: 10.1002/jez.1070. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenet Genome Res. 2006;113:90–98. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta – a review. Placenta. 2005;26:S10–S20. doi: 10.1016/j.placenta.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189:109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Watkins M, Burton GJ, Ferguson-Smith AC. Roles for genomic imprinting and the zygotic genome in placental development. Proc Natl Acad Sci USA. 2001;98:4522–4527. doi: 10.1073/pnas.081540898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Ager EI, Shaw G, Pask AJ. Genomic imprinting in marsupial placentation. Reproduction. 2008;136:523–531. doi: 10.1530/REP-08-0264. [DOI] [PubMed] [Google Scholar]
- Moore G, Oakey R. The role of imprinted genes in humans. Genome Biol. 2011;12:1–2. doi: 10.1186/gb-2011-12-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LL, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–334. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Curley JP, Barton S, Surani A, Keverne EB. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc Biol Sci. 2004;271:1303–1309. doi: 10.1098/rspb.2004.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney WT, Curley JP, Champagne FA, Keverne EB. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc Natl Acad Sci U S A. 2007;104:6084–6089. doi: 10.1073/pnas.0609471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader–Willi syndrome. Hum Mol Genet. 2007;16:2713–2719. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, Muglia LJ, Van Gelder RN, Herzog ED, Stewart CL. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-X. [DOI] [PubMed] [Google Scholar]
- Curley JP, Pinnock SB, Dickson SL, Thresher R, Miyoshi N, Surani MA, Keverne EB. Increased body fat in mice with a targeted mutation of the paternally expressed imprinted gene Peg3. FASEB J. 2005;04:3216fje. doi: 10.1096/fj.04-3216fje. [DOI] [PubMed] [Google Scholar]
- Keverne EB, Curley JP. Epigenetics, brain evolution and behaviour. Front Neuroendocrinol. 2008;29:398–412. doi: 10.1016/j.yfrne.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao B, Ma H, Shokhirev MN, Drung A, Yang Q, Shin J, Lu S, Byron M, Kalantry S, Mercurio AM, Lawrence JB, Hoffmann A, Bach I. Paternal RLIM/Rnf12 is a survival factor for milk-producing alveolar cells. Cell. 2012;149:630–641. doi: 10.1016/j.cell.2012.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yballe CM, Vu TH, Hoffman AR. Imprinting and expression of insulin-like growth factor-II and H19 in normal breast tissue and breast tumor. J Clin Endocrinol Metab. 1996;81:1607–1612. doi: 10.1210/jc.81.4.1607. [DOI] [PubMed] [Google Scholar]
- Miller N, McCann AH, O'Connell D, Pedersen IS, Spiers V, Gorey T, Dervan PA. TheMASProto-oncogene is imprinted in human breast tissue. Genomics. 1997;46:509–512. doi: 10.1006/geno.1997.5063. [DOI] [PubMed] [Google Scholar]
- Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, O'Shea D, Costello P, McGoldrick A, Keating G, Tobin B, Gorey T, McCann A. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–5451. [PubMed] [Google Scholar]
- Yun K, Soejima H, Merrie AEH, McCall JL, Reeve AE. Analysis ofIGF2gene imprinting in breast and colorectal cancer by allele specific-PCR. J Pathol. 1999;187:518–522. doi: 10.1002/(SICI)1096-9896(199904)187:5<518::AID-PATH276>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Fehlmann M, Le Cam A, Freychet P. Insulin and glucagon stimulation of amino acid transport in isolated rat hepatocytes. Synthesis of a high affinity component of transport. J Biol Chem. 1979;254:10431–10437. [PubMed] [Google Scholar]
- Chabowski A, Coort SLM, Calles-Escandon J, Tandon NN, Glatz JFC, Luiken JJFP, Bonen A. Insulin stimulates fatty acid transport by regulating expression of FAT/CD36 but not FABPpm. Am J Physiol Endocrinol Metab. 2004;287:E781–E789. doi: 10.1152/ajpendo.00573.2003. [DOI] [PubMed] [Google Scholar]
- Hernández-Sánchez C, Mansilla A, de la Rosa E, de Pablo F. Proinsulin in development: new roles for an ancient prohormone. Diabetologia. 2006;49:1142–1150. doi: 10.1007/s00125-006-0232-5. [DOI] [PubMed] [Google Scholar]
- Rowzee AM, Ludwig DL, Wood TL. Insulin-like growth factor type 1 receptor and insulin receptor isoform expression and signaling in mammary epithelial cells. Endocrinology. 2009;150:3611–3619. doi: 10.1210/en.2008-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanchez C, Mansilla A, de la Rosa EJ, Pollerberg GE, Martinez-Salas E, de Pablo F. Upstream AUGs in embryonic proinsulin mRNA control its low translation level. EMBO J. 2003;22:5582–5592. doi: 10.1093/emboj/cdg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanchez C, Rubio E, Serna J, de la Rosa EJ, de Pablo F. Unprocessed proinsulin promotes cell survival during neurulation in the chick embryo. Diabetes. 2002;51:770–777. doi: 10.2337/diabetes.51.3.770. [DOI] [PubMed] [Google Scholar]
- Mansilla A, Lopez-Sanchez C, de la Rosa EJ, Garcia-Martinez V, Martinez-Salas E, de Pablo F, Hernandez-Sanchez C. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep. 2005;6:1182–1187. doi: 10.1038/sj.embor.7400539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa EJ, de Pablo F. Proinsulin: from hormonal precursor to neuroprotective factor. Front Mol Neurosci. 2011;4:20. doi: 10.3389/fnmol.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolander FF, Nicholas KR, Van Wyk JJ, Topper YJ. Insulin is essential for accumulation of casein mRNA in mouse mammary epithelial cells. Proc Natl Acad Sci USA. 1981;78:5682–5684. doi: 10.1073/pnas.78.9.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KR, Collet C, Joseph R, Sankaran L. Hormone-responsive survival of mammary gland explants from the pregnant tammar wallaby (Macropus eugenii) in the absence of exogenous hormones and growth factors. Comp Biochem Physiol Part A: Physiology. 1991;100:163–167. doi: 10.1016/0300-9629(91)90201-M. [DOI] [PubMed] [Google Scholar]
- Berlato C, Doppler W. Selective response to insulin versus insulin-like growth factor-I and -II and up-regulation of insulin receptor splice variant B in the differentiated mouse mammary epithelium. Endocrinology. 2009;150:2924–2933. doi: 10.1210/en.2008-0668. [DOI] [PubMed] [Google Scholar]
- Burgos SA, Dai M, Cant JP. Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J Dairy Sci. 2010;93:153–161. doi: 10.3168/jds.2009-2444. [DOI] [PubMed] [Google Scholar]
- Menzies K, Lee H, Lefèvre C, Ormandy C, Macmillan K, Nicholas K. Insulin, a key regulator of hormone responsive milk protein synthesis during lactogenesis in murine mammary explants. Funct Integr Genomics. 2010;10:87–95. doi: 10.1007/s10142-009-0140-0. [DOI] [PubMed] [Google Scholar]
- Menzies KK, Le Fevre C, Macmillan KL, Nicholas KR. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct Integr Genomics. 2009;9:197–217. doi: 10.1007/s10142-008-0103-x. [DOI] [PubMed] [Google Scholar]
- Bequette BJ, Kyle CE, Crompton LA, Buchan V, Hanigan MD. Insulin regulates milk production and mammary gland and hind-leg amino acid fluxes and blood flow in lactating goats. J Dairy Sci. 2001;84:241–255. doi: 10.3168/jds.S0022-0302(01)74474-8. [DOI] [PubMed] [Google Scholar]
- McGuire MA, Griinari JM, Dwyer DA, Bauman DE. Role of insulin in the regulation of mammary synthesis of fat and protein. J Dairy Sci. 1995;78:816–824. doi: 10.3168/jds.S0022-0302(95)76693-0. [DOI] [PubMed] [Google Scholar]
- Griinari JM, McGuire MA, Dwyer DA, Bauman DE, Palmquist DL. Role of insulin in the regulation of milk fat synthesis in dairy cows. J Dairy Sci. 1997;80:1076–1084. doi: 10.3168/jds.S0022-0302(97)76032-6. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-X. [DOI] [PubMed] [Google Scholar]
- O'Neill MJ, Ingram RS, Vrana PB, Tilghman SM. Allelic expression of IGF2 in marsupials and birds. Dev Genes Evol. 2000;210:18–20. doi: 10.1007/PL00008182. [DOI] [PubMed] [Google Scholar]
- Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, McCarrey JR, Renfree MB, Reik W, Dunham I. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- Stringer JM, Suzuki S, Pask AJ, Shaw G, Renfree MB. Promoter-specific expression and imprint status of marsupial IGF2. PLoS ONE. 2012. in press. [DOI] [PMC free article] [PubMed]
- Brisken C, Ayyannan A, Nguyen C, Heineman A, Reinhardt F, Jan T, Dey SK, Dotto GP, Weinberg RA. IGF-2 is a mediator of prolactin-induced morphogenesis in the breast. Dev Cell. 2002;3:877–887. doi: 10.1016/S1534-5807(02)00365-9. [DOI] [PubMed] [Google Scholar]
- Malven PV, Head HH, Collier RJ, Buonomo FC. Periparturient changes in secretion and mammary uptake of insulin and in concentrations of insulin and insulin-like growth factors in milk of dairy cows. J Dairy Sci. 1987;70:2254–2265. doi: 10.3168/jds.S0022-0302(87)80285-0. [DOI] [PubMed] [Google Scholar]
- Prosser C. Insulin-like growth factors in milk and mammary gland. J Mammary Gland Biol Neoplasia. 1996;1:297–306. doi: 10.1007/BF02018082. [DOI] [PubMed] [Google Scholar]
- Blum JW, Baumrucker CR. In: Bioactive Components of Milk. Volume 606. Advances in Experimental Medicine and Biology. Bösze Z, editor. Springer, New York; 2008. Insulin-like growth factors (IGFs), IGF binding proteins, and other endocrine factors in milk: role in the newborn; pp. 397–422. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandels M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Giddings SJ, King CD, Harman KW, Flood JF, Carnaghi LR. Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet. 1994;6:310–313. doi: 10.1038/ng0394-310. [DOI] [PubMed] [Google Scholar]
- Deltour L, Montagutelli X, Guenet J-L, Jami J, Páldi A. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol. 1995;168:686–688. doi: 10.1006/dbio.1995.1114. [DOI] [PubMed] [Google Scholar]
- Moore T. Genetic conflict, genomic imprinting and establishment of the epigenotype in relation to growth. Reproduction. 2001;122:185–193. doi: 10.1530/rep.0.1220185. [DOI] [PubMed] [Google Scholar]
- Ager EI, Suzuki S, Pask AJ, Shaw G, Ishino F, Renfree MB. Insulin is imprinted in the placenta of the marsupial, Macropus eugenii. Dev Biol. 2007;309:317–328. doi: 10.1016/j.ydbio.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram R, Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R. A maternal–offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006;4:e380. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Haig D. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 2000;31:9–32. doi: 10.1146/annurev.ecolsys.31.1.9. [DOI] [Google Scholar]
- Keverne EB. Olfactory learning. Curr Opin Neurobiol. 1995;5:482–488. doi: 10.1016/0959-4388(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Tyndale-Biscoe H. Renfree MB: Monographs on Marsupial Biology: Reproductive Physiology of Marsupials. Cambridge University Press, Cambridge; 1987. [Google Scholar]
- Renfree MB. Review: Marsupials: placental mammals with a difference. Placenta. 2010;31:S21–S26. doi: 10.1016/j.placenta.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Green B, Merchant JC, Tyndale-Biscoe H, Janssens PA. In: The developing marsupial. Models for biomedical research. Tyndale-Biscoe CH, Janssens PA, editor. Springer, Berlin; 1988. The composition of marsupial milk; pp. 41–54. [Google Scholar]
- Trott JF, Simpson KJ, Moyle RL, Hearn CM, Shaw G. Maternal regulation of milk composition, milk production, and pouch young development during lactation in the tammar wallaby (Macropus eugenii) Biol Reprod. 2003;68:929–936. doi: 10.1095/biolreprod.102.005934. [DOI] [PubMed] [Google Scholar]
- Menzies BR, Shaw G, Fletcher TP, Renfree MB. Perturbed growth and development in marsupial young after reciprocal cross-fostering between species. Reprod Fertil Dev. 2007;19:976–983. doi: 10.1071/RD07142. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Renfree MB, Pask AJ, Shaw G, Kobayashi S, Kohda T, Kaneko-Ishino T, Ishino F. Genomic imprinting of IGF2, p57 (KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech Dev. 2005;122:213–222. doi: 10.1016/j.mod.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Grimm SL, Seagroves TN, Kabotyanski EB, Hovey RC, Vonderhaar BK, Lydon JP, Miyoshi K, Hennighausen L, Ormandy CJ, Lee AV, Stull MA, Wood TL, Rosen JM. Disruption of steroid and prolactin receptor patterning in the mammary gland correlates with a block in lobuloalveolar development. Mol Endocrinol. 2002;16:2675–2691. doi: 10.1210/me.2002-0239. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- Lin EY, Gouon-Evans V, Nguyen AV, Pollard JW. The macrophage growth factor, CSF-1, in mammary gland development and tumor progression. J Mammary Gland Biol Neoplasia. 2002;7:147–162. doi: 10.1023/A:1020399802795. [DOI] [PubMed] [Google Scholar]
- Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore GE, Abu-Amero SN, Bell G, Wakeling EL, Kingsnorth A, Stanier P, Jauniaux E, Bennett ST. Evidence that insulin is imprinted in the human yolk sac. Diabetes. 2001;50:199–203. doi: 10.2337/diabetes.50.1.199. [DOI] [PubMed] [Google Scholar]
- Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P, Constancia M, Moore GE. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, Kandeel F, Mullen Y, Pfeifer GP, Ferreri K. Insulin gene expression is regulated by DNA methylation. PLoS One. 2009;4:e6953. doi: 10.1371/journal.pone.0006953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menheniott TR, Woodfine K, Schulz R, Wood AJ, Monk D, Giraud AS, Baldwin HS, Moore GE, Oakey RJ. Genomic imprinting of Dopa decarboxylase in heart and reciprocal allelic expression with neighboring Grb10. Mol Cell Biol. 2008;28:386–396. doi: 10.1128/MCB.00862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, Eggermann T, Ropers H, Kalscheuer VM. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum Mol Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- Ekstrom TJ, Cui H, Li X, Ohlsson R. Promoter-specific IGF2 imprinting status and its plasticity during human liver development. Development. 1995;121:309–316. doi: 10.1242/dev.121.2.309. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, Ishino F, Renfree MB, Kaneko-Ishino T. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton B, Carone B, Obergfell C, Ferreri G, Gondolphi C, VandeBerg J, Imumorin I, O'Neill R, O'Neill M. Genomic imprinting of IGF2 in marsupials is methylation dependent. BMC Genomics. 2008;9:205–216. doi: 10.1186/1471-2164-9-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ, Krawetz S, Misener S. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Human Press, Totowa; 2000. Primer3; pp. 365–386. [Google Scholar]
- Ager E, Pask A, Shaw G, Renfree MB. Expression and protein localisation of IGF2 in the marsupial placenta. BMC Dev Biol. 2008;8:17–29. doi: 10.1186/1471-213X-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Development_Core_Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna; 2011. R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss KO, Roos KP, Nonay RL, Dovichi NJ. Combating PCR Bias in Bisulfite-Based Cytosine Methylation Analysis. Betaine-Modified Cytosine Deamination PCR. Anal. Chem. 1998;70:3818–3823. [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucl Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Insulin immunolocalisation positive controls. The immunolocalisation of insulin in the pancreas and placenta, providing positive controls to verify authentic protein recognition.
Genotyping andINSimprinting in the tammar wallaby. Additional tables and figures that display the number and proportion of INS alleles within a population and extra sequence chromatographs of INS in the mammary gland.
Primers used in this study. A table listing all of the primers used in this study.