Abstract
Liquid chromatography (LC) separation combined with electrochemical coulometric array detection (EC), is a sensitive, reproducible, and robust technique that can detect hundreds of redox-active metabolites down to the level of femtograms on column, making it ideal for metabolomics profiling. EC detection cannot, however, structurally characterize unknown metabolites that comprise these profiles. Several aspects of LC-EC methods prevent a direct transfer to other structurally-informative analytical methods, such as LC-MS and NMR. These include system limits of detection, buffer requirements, and detection mechanisms. To address these limitations, we developed a workflow based on the concentration of plasma, metabolite extraction, and offline LC-UV fractionation. Pooled human plasma was used to provide sufficient material necessary for multiple sample concentrations and platform analyses. Offline parallel LC-EC and LC-MS methods were established that correlated standard metabolites between the LC-EC profiling method and the mass spectrometer. Peak retention times (RT) from the LC-MS and LC-EC system were linearly related (r2=0.99); thus LC-MS RTs could be directly predicted from the LC-EC signals. Subsequent offline microcoil-NMR analysis of these collected fractions was used to confirm LC-MS characterizations by providing complementary, structural data. This work provides a validated workflow that is transferrable across multiple platforms and provides the unambiguous structural identifications necessary to move primary mathematically-driven LC-EC biomarker discovery into biological and clinical utility.
Keywords: Metabolite characterization, NMR, mass spectrometry, metabolomics, LC-UV fractionation, electrochemical coulometric array detection
INTRODUCTION
A metabolomics profiling study often has two major analytical goals 1) to create metabolite profiles that distinguish between two biological systems (e.g., disease state1–3, medication state3, 4, age, environmental stresses5–7 or diet3, 8–12) and; 2) to structurally characterize the specific metabolites that comprise these profiles4, 13–16. It is this second goal -- the identification and structural characterization of the unknown metabolites of interest that define the biological and clinical importance of the study and that are generally present in limited (ng-μg) quantities -- that is often a major hurdle and bottleneck facing metabolomics research.
The ability of a platform to characterize metabolites of interest in a profiling study is directly related to the analytical methodology being used. Liquid chromatography coupled with electrochemical coulometric array detection (LC-EC), for example, is a sensitive, reproducible, and robust technique17–20 with the ability to detect hundreds of metabolites down to femtograms of material and across a dynamic range of 9 orders of magnitude, making it ideal for profiling21–26. The detection mechanism used in LC-EC is quite specific, focusing on compounds containing easily oxidizable moieties. This specificity allows for a unique subset of the metabolome to be profiled that other commonly used profiling techniques, such as NMR and LC-MS, do not comprehensively detect, such as purines27–29, tryptophan metabolites24, 30, DNA oxidative damage products31–33, and neurotransmitters20, 34, 35. Additionally, these methods have provided insights into drug metabolism36, 37 and efficacy38, mechanisms of addiction39, diet10, 19, 40–43, disease pathogenesis1, 17, 31, 44 and progression45, 46.
EC detection uses an array of porous graphite electrodes to detect the current output as analytes are oxidized, and allows for 100% analyte conversion, a property that underlies the technique’s sensitivity. Under its optimal conditions, LC-EC detects metabolites by first separating by hydrophobicity in the LC dimension using buffers containing a high concentration of salts and pentane sulfonic acid (PSA)22, 23, 25, 26. The salts are necessary to maintain proper conductivity in the EC electrodes and PSA is an ion-pairing reagent used to aid both the retention of the hydrophilic metabolites and enhance the analytical reproducibility of the method26, 47. A given metabolite is detected based on that compound’s oxidation potential 20, 22 as the LC eluent flows through an array of up to 16 EC cells connected in tandem and held at increasing voltages. This detector orientation provides a second separation dimension, in which co-eluting species are distinguished based on their distinct oxidation potentials. Although sensitive, reproducible, and robust, this method does not provide any structural information regarding the compounds that it detects. The use of analytical standards and spiking experiments to compare retention time and oxidation potential profile, provides provisional identification of peaks of interest but requires some prior knowledge of the metabolites, making the characterization of unknowns impossible.
This report demonstrates a workflow that enables the concentration of LC-EC-detected, easily-oxidized species and that enables the transfer of the same metabolite peak(s) across different instrument systems and buffers. Supplementary analysis of metabolites identified in LC-EC profiling studies with structurally-rich analytical tools, e.g., LC-MS and NMR, can provide the complementary information necessary to fully characterize unknown metabolites of interest. There are, however, several analytical challenges that make the LC-EC method difficult to transfer to these techniques, including: system limits of detection, buffer requirements, and detection mechanisms. We circumvent these problems by utilizing an offline LC-UV fractionation method, after plasma concentration and metabolite extraction. Fractionation and concentration are necessary to address the sensitivity disparities between the analytical detectors and by doing so fractions can be more readily analyzed across multiple platforms with varying limits of detection. This fractionation method was developed to maintain the integrity of the LC-EC profiling method without the ion-pairing reagents and salt buffers that interfere with both LC-MS and NMR analyses. Herein, we describe a workflow that provides analytical characterization of small molecule metabolites, identified by LC-EC profiling and multivariate statistics, using a combination of NMR- and MS-based analytical techniques, including LC-MS with accurate mass and high energy collisional dissociation (HCD) fragmentation and microcoil-NMR spectroscopy. This method is applied here to the characterization of human plasma metabolites, but would be generally applicable for other targets of electrochemical/coulometric electrode array analysis, e.g., mitochondrial samples25, cerebrospinal fluid,31, 48 DNA damage products32, 33 and natural product extracts49, 50.
MATERIALS AND METHODS
Chemicals
LC-MS grade acetonitrile (ACN), methanol (MeOH), and isopropyl alcohol (IPA) as well as ammonium acetate and acetic acid were obtained from Fisher Scientific (Pittsburgh, PA). Triethylamine (TEA) and perfluoropentanoic acid (PFA) were purchased from SigmaAldrich (St. Louis, MO), as were all chemicals used to produce our biochemical standard (termed DSV), a mixture of 54 endogenous serum metabolites found at varying concentrations (see supplement for complete list). All deuterated solvents were purchased from Cambridge Isotopes (Andover, MA), and 1-pentanesulfonate, sodium salt (PSA) was purchased from Regis Technologies (Morton Grove, IL). A human plasma pool (POOL) comprised of both males and females was purchased from Interstate Blood Bank, INC (Memphis, TN).
Metabolite Extraction for LC-EC Profiling and LC-UV Fractionation
The proteins from either 125 μL (standard profiling preparation) or 2.5 mL (concentrated method) of POOL were precipitated with 500 μL and 10 mL, respectively, of cold (−20° C) acetonitrile/0.4% glacial acetic acid, while on ice. The samples were then centrifuged for 15 minutes at 4°C, at 12,000 rpm; the supernatant was transferred into microcentrifuge tubes and dried under vacuum in a cold centrivap. The dried extract in each tube was reconstituted in 100 μl of LC-EC mobile phase A immediately prior to injection26 for profiling, while the 2.5 mL prep was resuspended in 200 μL of the LC-UV mobile phase A.
LC-EC Profiling Method26
Separation was performed by gradient elution on two reversed-phase C18 META 250 × 4.6, 5μm columns (Thermo Scientific) connected in tandem with PEEK tubing. The column temperature was held at 32 °C. The mobile phase A consisted of 100% water modified with 60 mM PSA, 0.1% methanol, and 1 mg/L citric acid adjusted to pH 3.1 with acetic acid. Mobile phase B consisted of 80:10:10 MeOH: ACN: IPA modified with 40 mM lithium acetate and 2.0% acetic acid in 10 mg/L citric acid. The gradient was increased between 0–95%B over 112 minutes (see supplement for details). 50 μL of sample was injected per run and detection was performed on an ESA (ThermoFisher Scientific, Chelmsford, MA) LC-EC system with 16-channel coulometric array detector operated with potentials incremented in 60 mV steps (0–900 mV). All LC-EC system functions were controlled by CoulArray® software (CoulArray® for Windows® software version 3.10)
HPLC Fractionation for Metabolite Identification
Separation of the concentrated metabolite extract was performed on an Agilent 1200 series HPLC system consisting of a binary pump, an auto-sampler, a degasser, a variable wave detector and a fraction collector. Mobile phase A consisted of 100% water, modified with 100 mM TEA and 25 mM ammonium acetate with acetic acid to pH 3; mobile phase B was a mixture of 80:10:10 MeOH:ACN:IPA modified with 25 mM ammonium acetate and acetic acid. Separation was performed by the same LC method as used for the LC-EC profiling. Time-dependent fractions were collected from 3 to 104 minutes for a total of 96 fractions, in a 96-well plate (Waters, Milford, MA), yielding approximately 1 mL/fraction. The fractions were named according to the well they were collected in, A–H numbered 1–12. Thus the name of any given fraction can be directly determined from its collection position. The variable wave detector was monitored at 280 nm. After collection, fractions were transferred to 1.5 ml microcentrifuge tubes and 20 μl of DMSO was added prior to drying under vacuum.
Fraction Analysis by LC-MS and LC-EC
For the LC-MS and LC-EC analysis, the POOL fractions were reconstituted in either 100 or 50 μL of LC-MS mobile phase A (25 mM ammonium acetate in deionized water using a Millipore water filtration system (Billerica, MA), adjusted to pH 3.1 using acetic acid) and separation was performed using two Shiseido C-18 columns (4.6 × 150 mm) connected in tandem and maintained at room temperature. The gradient was increased between 0–95%B over 100 minutes (see supplement for details). The LC-MS injected 80 μL while the LC-EC injected 50 μL.
LC-MS Instrumentation
LC-MS was performed on a HPLC system consisting of an autosampler and an Accela quaternary HPLC pump (ThermoFisher, San Jose, CA). The HPLC system was connected to an Exactive bench-top Orbitrap mass spectrometer (ThermoFisher, San Jose, CA) equipped with a heated electrospray ionization (HESI) probe. The spray voltage was set to 4 kV. The heated capillary and HESI probe were held at 300° and 400° C, respectively. The sheath gas flow was set to 60 units and that of the auxiliary gas at 20 units. The instrument was tuned and calibrated as specified by the vendor, in both positive and negative ion mode, and operated in high resolution mode, corresponding to 50K resolution and 2Hz scan speed. Spectra were acquired using four scan events in sequence, which alternated between full scan between m/z 50 and 1000 and HCD at 60 eV in both the positive and negative ion mode. The instrument was controlled using Xcalibur software version 2.1.
For identification, the exact mass of the analytes of interest was searched against the METLIN51 database and HMDB 52 with a mass tolerance of either 0.005 daltons or 5 ppm. Both sites allow the user to search masses in both the positive and negative ion mode, and multiple adduct possibilities are calculated based on the exact mass.
NMR Analysis
NMR spectra were acquired on a Bruker Avance II 700 MHz spectrometer (Bremen, Germany) operating at 699.97 MHz (1H frequency). The spectrometer was equipped with a triple resonance inverse gradient capillary microcoil NMR probe (MRM/Protasis, Savoy, IL). The probe has a 5 μl flow cell.
POOL fractions were dissolved in 10 μL of DMSO-d6 and 8μl was loaded into the capillary probe using One-Minute-NMR (Version 2.18.43) automation (Protasis, Savoy, IL) controlling a CTC-PAL autosampler (LEAP Technologies, Carrboro, NC) with dimethyl sulfoxide-d6 (DMSO-d6) as the push solvent. 40 μl of DMSO-d6 was used to deliver the sample to the probe’s active volume, using a 40 μl/minute flow rate. The spectrometer was set to automatically start 1 minute after the sample was delivered to the probe.
Spectra were acquired with a 45 degree tip angle, 1.45 s acquisition time and a 1 s recycle delay. The receiver gain was set to 256 and the spectral width was set to 8 KHz, depending on the estimated analyte concentration. The FIDs were processed by zero filling to 64K, baseline corrected, and Fourier transformed with a 1 Hz exponential line broadening. All chemical shifts were referenced to deuterated sodium 2,2, dimethyl-silapentane-1-sulfonate (DSS-d6) as 0 ppm. Topspin version 2.1 was used to process the data and to control the spectrometer. All samples were recovered after NMR analysis.
RESULTS AND DISCUSSION
The goal of this work is to establish a workflow, using structurally-rich analytical detectors such as MS and NMR, to characterize metabolites that were chromatographically detected and identified by LC-EC profiling. Previous studies have focused on the online-simultaneous detection of unknowns using a parallel configuration of LC-EC/MS36, 37 and although this method has its advantages, the common disadvantage encountered is the compromise of detector optimal working conditions, which inherently limits the specificity and sensitivity of the method. Subsequently, all detectors used herein (LC-EC, LC-MS and NMR) were run independently of each other to ensure that each could be used at its optimal working conditions. This configuration requires the correlation of metabolite peaks between several LC methods and detectors and it is this correlation which defines the workflow’s utility in metabolomics research.
Sample Concentration and Fractionation
Depending on the oxidation potential of a metabolite, the conductivity of the LC mobile phase, and the type of electrode being used, the LC-EC profiling method can require as little as femtograms of material to achieve a signal-to-noise ratio greater than three. The sample requirements for both MS and NMR, are at least 10 to 10,000 times this amount, respectively, require sample concentration to be performed prior to analysis. Commercially-obtained human plasma pools (POOL) are expected to contain most or all of the endogenous metabolites commonly encountered in a human plasma profiling study. Such a pooled plasma sample was therefore used to provide sufficient material necessary for multiple sample concentration steps and platform development. These are the same samples used during profiling studies as QC checks for any analytical variations encountered.
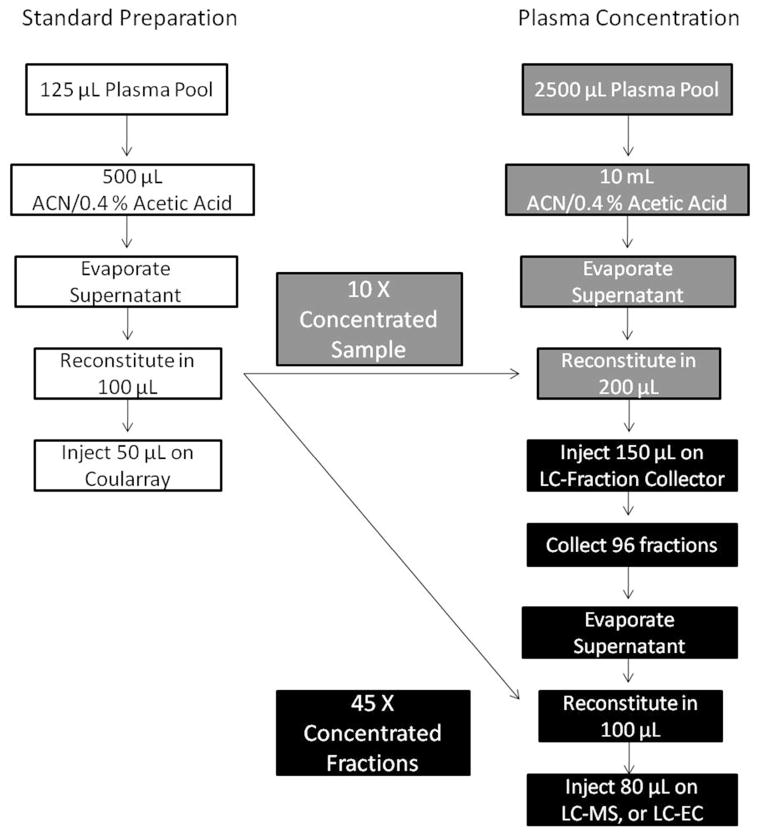
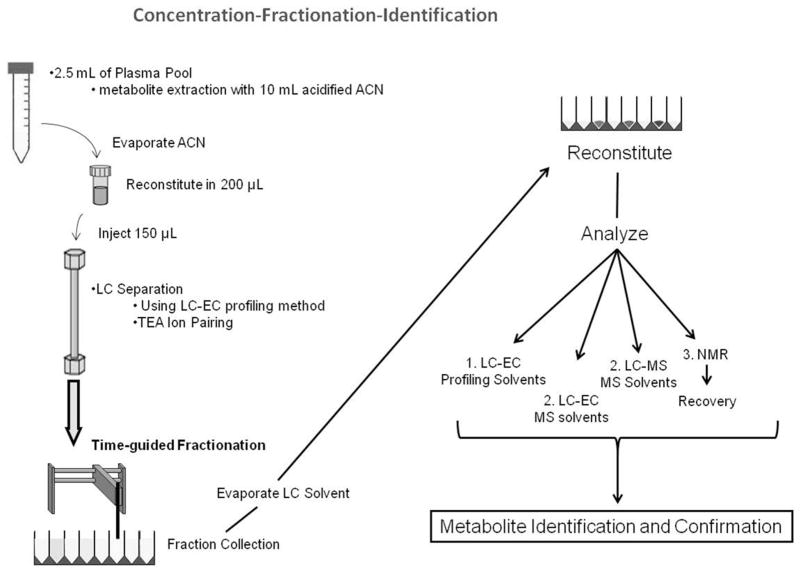
Standard LC-EC profiling extracts the metabolites from 125 μL of plasma into a final re-suspended volume of 100 μL. Therefore, this amount was the baseline level to which the initial concentration was compared. To avoid overloading our analytical column, and to be able to dissolve the extracts in 200 μL or less for LC injection purposes, we started with an initial volume of 2.5 mL of POOL dissolved in 200 μL of solvent, a 10 X concentration from the standard procedure (Figure 1). The standard plasma metabolite extraction method is listed on the left (entirely in white); on the right is the concentration method where the grey squares indicate a theoretical 10X concentration compared to the standard prep as described above, and the black squares show a theoretical 45X concentration. The concentrated pool extracts were reconstituted in 200 μL with 150 μL being injected on the LC-UV system. This injection amount is 3 times greater than the standard 50μL that is injected on the LC-EC profiling system and at this point, the fractions are 30X more concentrated than the standard analysis. Additionally, after fraction collection the LC eluent was evaporated and samples reconstituted in 100 μL, or two-thirds the extract starting volume, yielding a theoretical 45X concentration.
Figure 1.
Sample Concentration Scheme. The standard plasma metabolite extraction method is listed on the left entirely in white. The grey method on the right achieves a theoretical 10X concentration of sample; the black squares below it show a theoretical 45X concentration based on sample fractionation and reconstitution.
The human plasma metabolome is complex with compounds covering many chemistries and at different concentrations, and therefore requires reducing this complexity prior to determining metabolite structures. Sample fractionation reduces this inherent sample complexity, preferably achieving one compound per fraction, which also reduces the associated potential for ion suppression in MS and signal overlap in NMR. Ideally, to directly relate collected fractions back to the original LC-EC profile, either post EC fractionation or LC-EC guided fractionation should be used. LC-EC, however, is a destructive analytical technique that alters the analyte structure during detection. When using post-detection fractionation, this oxidative change in metabolite structure would complicate correlation between the primary LC-EC profiles and any secondary analytical technique. Additionally, because the LC-EC profiling buffers contain large quantities of non-volatile salts and PSA, a post-fraction-collection clean-up would be required to remove these interfering species. In pilot studies, post fraction-collection clean-up using solid phase extraction (SPE) led to sample losses (data not shown). Taken together, these problems make direct LC-EC fractionation an impractical option.
Because post-analysis sample clean-up seemed unlikely to be useful for sample concentration, we pursued an alternative fractionation method that maintained the integrity of the LC-EC profiles without requiring a post-collection clean-up. Metabolites extracted from plasma using ACN represent primarily hydrophilic compounds that are poorly retained on standard C18 reversed phase (RP) chromatography columns, such as those used during our LC-EC profiling experiments. To effectively separate and retain these hydrophilic species, minimize co-elution, and achieve separations that closely resemble the original LC-EC profiles, an ion-pairing reagent is necessary for fraction collection. Although beneficial to the chromatographic separation during fraction collection, the ion-pairing reagent can potentially cause MS ionization suppression; detrimentally affecting the subsequent LC-MS analysis of the fractions.
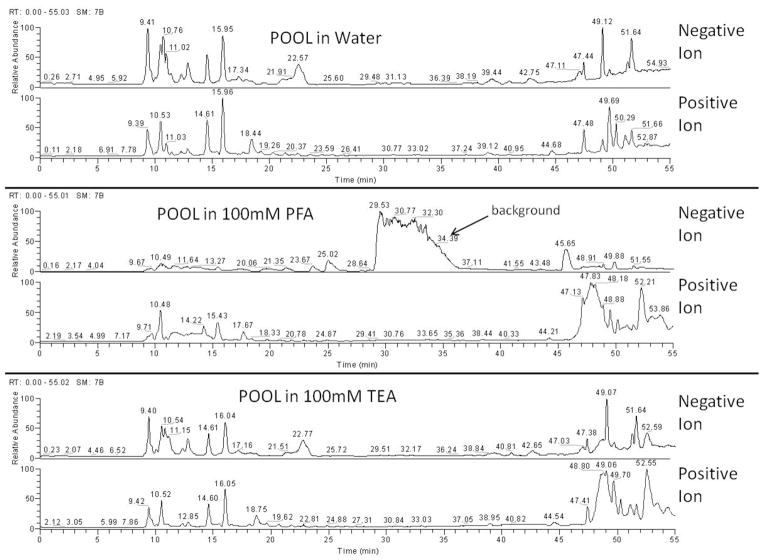
To circumvent this ion-suppression problem, two commonly used, volatile, and LC-MS friendly additives were tested as alternatives to the non-volatile PSA used in the LC-EC profiling experiments: (1) perfluoropentanoic acid (PFA), a negatively charged reagent, commonly used to enhance the retention of cationic metabolites in LC-MS analysis53 and (2) triethylamine (TEA), a volatile cationic compound that ion-pairs with negatively-charged analytes, commonly used to improve both retention and separation in LC-MS54. Only two ion-pairing reagents, out of many that can yield similar results, were tested because we were focused on evaluating the difference between positively and negatively charged additives, and how each would affect our dual-polarity LC-MS method. The two that were chosen were expected to have the best volatility and therefore best suited for LC-MS analysis. Metabolite extracts from 125 μL of POOL were reconstituted in either (i) water containing 20 mM ammonium acetate, pH 3 (ii) 100 mM PFA with 20 mM ammonium acetate, pH 3 or (iii) 100 mM TEA with 20 mM ammonium acetate, pH 3. These samples were examined via LC-MS, without any mobile phase additives, using positive and negative ion switching to provide greater metabolome coverage with a single injection. This experiment mimics the conditions used during LC-UV fraction collection, and therefore the sample solvent composition during LC-MS analysis of those fractions, establishing what interferences, if any, the TEA will provide. With the water injection serving as a control, the chromatograms in Figure 2 show that PFA generated an intense background signal between 30–36 minutes in negative ion mode, suppressing most other signals. In contrast, TEA produced very little interference in either positive or negative TIC and maintained the same chromatographic profile as the water control. From these profiles, it is clear that the TEA ion-pairing reagent provides a cleaner LC-MS analysis, similar to the water control, without the significant background interferences observed in the PFA experiment. These results led us to choose TEA as the LC fractionation ion-pairing reagent for reproducing the LC-EC chromatograms under MS-friendly conditions.
Figure 2.
Comparison of Ion-Pairing Reagents for LC-MS Analysis. The top two chromatograms show the LC-MS separation and detection of plasma extractions dissolved in water and detected via negative ion (upper trace) and positive ion (lower trace) full scan profiling. The center pair shows the LC-MS analysis of extracts dissolved in 100 mM PFA, and the bottom pair is for the extracts dissolved in 100mM TEA.
For sample fractionation, with the intention of achieving a similar metabolite separation as attained in the LC-EC profiling experiments, the same LC columns and LC method was used substituting 100 mM TEA with 20 mM ammonium acetate pH 3 for the PSA and lithium acetate buffer. Time-guided fractionation was performed every minute and LC-UV retention time (RT) was used to correlate the fractions back to the original LC-EC profiles. Each fraction was checked by re-injection onto the LC-EC, using the original LC-EC profiling method and buffers, to confirm general metabolite hydrophobicity and retention time in comparison with a non-concentrated/non-fractionated POOL sample. This check also confirmed our prediction of which fractions contained the LC-EC peaks of interest.
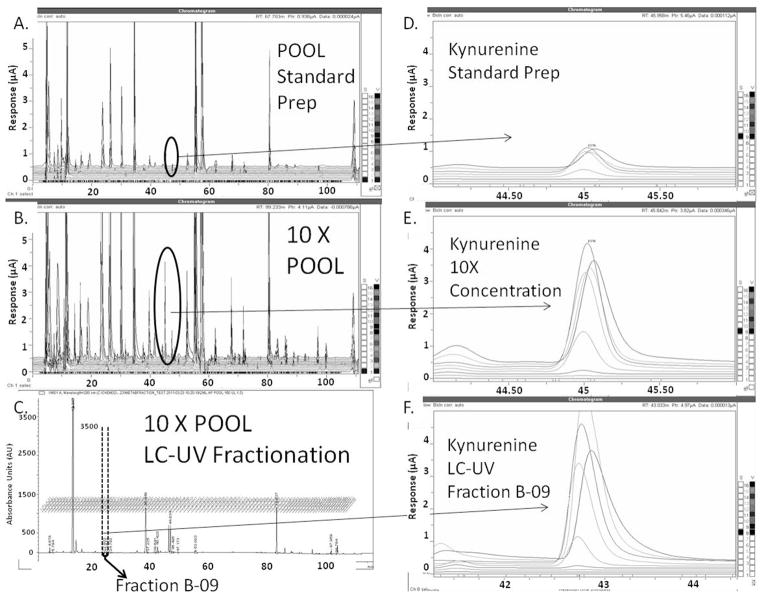
The separation, fractionation, and sample concentration steps were tested by comparing a 125 μL POOL extract run via LC-EC profiling to: (i) a 2.5 mL POOL extract and (ii) the LC-UV fraction (B09) that corresponded to a RT of 45 minutes in the LC-EC profiling method. Prior to this analysis, the DSV standard was injected on the fraction collection platform, and these fractions were analyzed by offline LC-MS and LC-EC profiling (data not shown). Because the DSV standard contained the tryptophan pathway metabolite kynurenine (KYN), this previous experiment suggested fraction B09 should contain a concentrated amount of KYN. From the chromatograms in Figure 3, it is clear that, although the LC-UV RTs are slightly shifted, the LC-EC separation integrity is generally maintained when using TEA as an ion-pairing reagent. Figure 3, panels A–C, show the relationship between the different sample preparation methods and the sample concentration. The circled portions of the LC-EC chromatograms are the regions where KYN is found; these data indicate its signal increase. Figure 3, panels D–F, are expansions which highlight the concentration change in KYN by expanding the detail of its specific RT and oxidative channel response. It should be noted that, in these LC-EC profiles, the KYN peak produces several signals across multiple channels. This oxidation profile is reflective of the multiple coulometric electrodes in sequence being held at increasing potentials and is unique to the metabolite being analyzed, its concentration in the sample, and the conditions under which the LC-EC is being run. In this comparison, the signal from the most intense channel is being used. The differences in signal from KYN extracted from 125 μL of plasma and the 2.5 mL of plasma (Figure 3 panels C and D) indicate a 4X increase in signal-to-noise (from 1 μA–4μA) was achieved from our initial preparation. The collected fraction produced an even larger signal of 6FA indicating additional concentration. Because 10X and 40X increases were expected, these data indicate that there is ~40% sample loss occurring within the fractionation scheme for this specific metabolite.
Figure 3.
POOL Concentration and Fractionation Test with KYN. Panels A and B show the LC-EC chromatogram for a standard and concentrated POOL sample, respectively, with the circled areas representing the metabolite KYN. Panel C shows the LC-UV chromatogram of a concentrated POOL fractionation experiment, with the portion of the chromatogram representing fraction B–09 highlighted. Panels D–F are expansions of the circled areas in Panels A–C showing the systematic increase in the KYN signal based on sample concentration and fractionation.
Offline Correlation or LC-MS with LC-EC
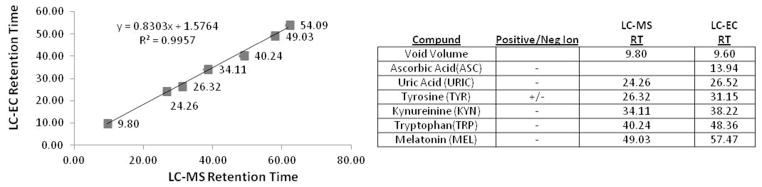
LC-MS was used to determine unknown metabolite molecular weight information and run offline from the LC-EC to facilitate optimal detector conditions as noted above. Unlike the LC fractionation method, ion-pairing reagents are not being added to the LC-MS analysis buffer in order to minimize any potential background interference; therefore changes to the metabolite separation may occur including RT shifts and elution order. To compensate for these changes, and to correlate LC-MS data to the LC-EC peaks of interest, an offline-parallel LC-EC and LC-MS method that compromises between MS and EC detector needs was developed. In past studies, where the LC-MS and LC-EC were run in parallel from a single injection,36, 37 the more MS-friendly salt ammonium acetate was utilized to maintain sufficient EC conductivity. We therefore adopted the same solvent additive. Additionally, due to variations in instrument dead-volumes, shifts in metabolite RT between the LC-MS and LC-EC methods are expected. Due to the gradient elution, however, these shifts are not expected to be a constant time difference, but rather to follow the gradient in a linear relationship. Using six metabolites from the DSV standard that covered a large portion of the LC chromatogram and that showed strong, distinct LC-MS and LC-EC signals and the void volume from both instruments, a linear relationship between peak RT from the LC-MS and LC-EC system with an R2 value of 0.99 was determined (Figure 4). These equations were then used to relate peaks observed in the LC-EC chromatograms to LC-MS RTs where these metabolites should be found.
Figure 4.
Offline LC-EC and LC-MS Correlation and Regression. The RTs of six endogenous metabolite standards and the void volumes from both the LC-EC and LC-MS instruments were used to create a linear regression that correlates the RTs between each method. The RTs showed a linear relationship (R2=0.99), and this regression equation was then used to predict where metabolites would be expected between the methods.
Endogenous Metabolite Identification with LC-MS and NMR Confirmation
A comprehensive schematic of the entire concentration, fractionation, and characterization workflow is shown in Figure 5. Initially, the metabolites from a 2.5 mL POOL sample are extracted and reconstituted in 200 μL of solvent, showing a 10X concentration from the standard 125μL preparation. This concentrated POOL is then fraction collected using the same LC-EC method used in all profiling studies, but with TEA and ammonium acetate as the buffer additives. After fractionation, the solvent from each well is evaporated and reconstituted based on the analytical method to be used. Fractions to be analyzed via each method, LC-EC (profiling method and solvents), offline-parallel LC-MS/LC-EC (MS-friendly method and solvents), and NMR, were separately prepared, yielding 4 fractionations in total.
Figure 5.
Concentration-Fractionation-Identification Schematic. Beginning with 2.5mL of POOL, each sample is extracted and fractionated via LC-UV. This is repeated for each subsequent analytical method, LC-EC, LC-MS, and NMR.
As labeled in Figure 5, the LC-EC profiling method is run first, after fractionation, to confirm the location of the metabolites of interest. This fraction is directly compared to the LC-EC chromatogram of a non-concentrated/non-fractionated POOL sample to assure (comparing RT and oxidation profile) that the peaks observed in the fraction represent previously identified LC-EC peaks of interest. Next, the LC-EC and LC-MS are run using MS-friendly solvents to correlate any MS signal to LC-EC detected peaks. The analysis at this stage is bidirectional. If a major peak of interest is found in the LC-EC chromatogram, its RT is then used to calculate where in the MS this peak may be found. If a major peak is found in the MS, the LC-EC RT is calculated to determine if there is a comparable LC-EC signal that indicates a redox-active metabolite. This aspect of the analysis is quite important because the workflow is being used to characterize LC-EC identified species, primarily, and the diversity of the metabolome provides thousands of compounds that are not necessarily redox-active, but may produce appreciable LC-MS signals. It is important to be able to distinguish between those metabolites that are redox-active and potentially LC-EC profiling metabolites of interest and those that distinctly are not.
After LC-EC and LC-MS analysis, NMR can be used to generate secondary structural information. For example, NMR can distinguish positional isomers on a substituted benzene ring that would not be possible from MS results alone. For NMR studies, a solenoidal microcoil probe was used. This type of probe has high mass sensitivity when compared to conventional 5 mm NMR probes 55. With careful sample loading, this method provides a limit of detection of 2 nanomoles for 1H NMR spectra when acquired overnight. After NMR, all samples were recovered from the probe and preserved for future analyses. Finally, all analytical data was combined to structurally characterize the LC-EC peaks found in each fraction.
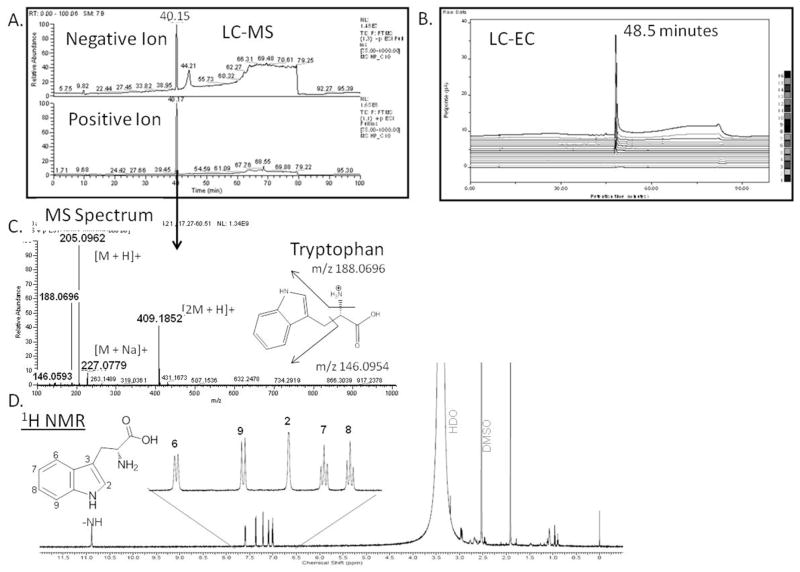
The application of this workflow for the identification of an endogenous metabolite is shown in Figure 6. Fraction C-10 from a concentrated POOL sample, which includes at least one major peak observed eluting at ~ 40 minutes in the LC-UV chromatogram (Figure 3C), was chosen for analysis. This fraction was analyzed by the LC-EC via the profiling method and produced a peak at a RT of around 60 minutes that showed an increased signal relative to the standard preparation POOL (data not shown). This is another example of the type of analysis shown with KYN in Figure 3, highlighting our ability to concentrate and fractionate plasma and to relate specific fractions back to the initial LC-EC profile. Here, we extend the workflow. Specifically, after additional concentration and fractionations, fraction C-10 was analyzed using the offline-parallel LC-MS and LC-EC using MS solvents. Figure 6 compares the chromatogram from this analysis, specifically, the large peak at 48 minutes in the LC-EC was predicted to correspond with the LC-MS peak at 40 min by the regression equation determined above. The LC-MS data in positive ion mode indicated, by both exact mass (the m/z 205.0952 corresponding to the [M+H] + species) and HCD fragmentation (the m/z 188.0656 fragment representing the loss of –NH3 and the m/z 146.0593 fragment corresponding to beta cleavage adjacent to the indole group) that this peak was most likely the metabolite tryptophan (TRP). The 1H NMR spectrum of fraction C10 (Figure 6, bottom panel) was used to support the assignment of the metabolite as TRP. The aromatic region (Figure 6, inset) of the 1H NMR spectrum exhibited five protons, two triplets at 7.01 ppm and 7.09 ppm (J=8.32 Hz), two ortho-coupled doublets at 7.36 ppm and 7.58 ppm (J=8Hz) and a singlet at 7.21 ppm. A highly de-shielded proton was also observed at 10.89 ppm which was the characteristic –NH proton in the indole moiety. Unambiguous structural confirmation was established by comparison of all the analytical results, including the LC-EC profiling method, with those of an authentic standard.
Figure 6.
Fraction C-10 Metabolite Characterization. Chromatograms of the LC-MS (A) and LC-EC (B) analyses in MS-friendly buffer are shown. Peaks at 40.15 minutes in LC-MS and 48.5 minutes in the LC-EC correlated via the regression equation to represent the same compound. The MS spectrum (C) provides evidence that the compound is TRP; the NMR (D) confirms this characterization.
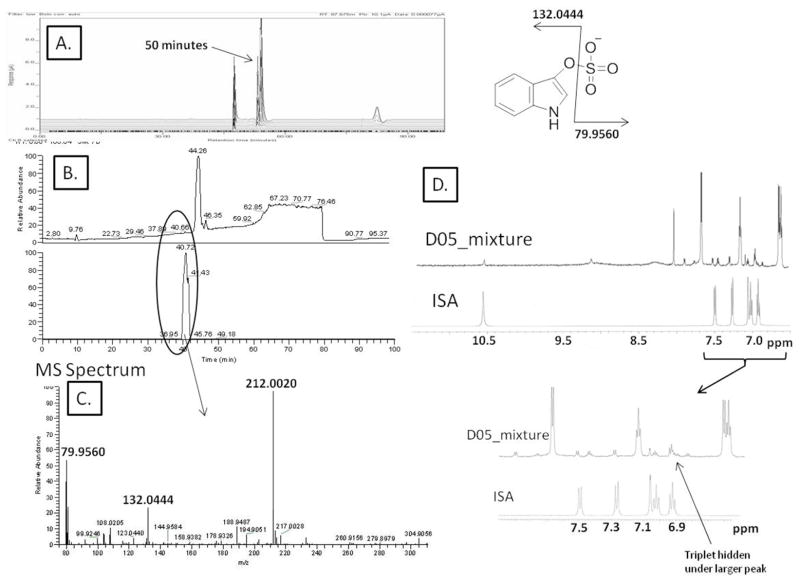
The significance of this workflow is most apparent when LC-EC profiling has revealed a metabolite that is novel, low in concentration, challenging to purify and not easily detected by either LC-MS or NMR alone. The analysis of fraction D05 provides such an example (Figure 7, panels A and B). These figures show the offline/parallel LC-EC and LC-MS chromatograms that correlate a peak at 50.77 minutes in the LC-EC to very low intensity negative ion LC-MS peak at 40.72 minutes. The mass spectrum under this peak, Figure 7C, revealed a peak with m/z 212.0020 with two fragment ions at m/z 132.0444 and m/z 79.9560, suggesting the metabolite indoxyl sulfate (ISA). Due to the multiple species present in fraction D05, direct comparison of the 1H NMR spectra from the fraction to the ISA standard is complicated (Figure 7 panel D expanded section). This section of the NMR trace suggests the presence of an indole moiety in the mixture, based on two ortho-coupled doublets in the aromatic region and a highly de-shielded proton at 10.78 ppm that is attributed to the –NH proton on the pyrole moiety. Due to spectral overlap, only 5 of the 6 indole protons were observed and although, the remaining resonances matched peak-to-peak with those of the authentic standard, the mixture masks the triplet at 6.9 ppm seen in the standard. Spiking studies with the ISA standard and fraction D05 using the LC-EC profiling method (see supplementary Figure 1) further confirmed the characterization of the metabolite.
Figure 7.
Fraction D05 Metabolite Characterization. Chromatograms of the LC-EC (A) and LC-MS (B) analyses in MS-friendly buffer are shown. Peaks at 50.42 minutes in LC-EC and 40.72 minutes in the LC-MS correlated via the regression equation to represent the same compound. The MS spectrum (C) suggests ISA metabolite from the [M-H]-ion at m/z 212.0020 and fragments at m/z 132.0444 and 79.9560. The NMR spectrum (D) supports this characterization by direct comparison with the ISA standard.
In this example, LC-MS provided both exact mass and structural composition of the unknown metabolite, with NMR providing complementary information. The complexity of the metabolome and the multiple species present in the fraction complicate the analysis, and direct comparison of the NMR to the ISA standard suggests the species is present; however, key resonances are masked by higher-concentration metabolites in the mixture. Because NMR is a non-destructive analytical technique, samples are recovered post-analysis and can be used for subsequent testing, such as additional fractionation or analytical detection steps. Moving forward, we are seeking to integrate this workflow with a complementary GC-MS system that can provide additional details regarding the functionality of the metabolites of interest using electron impact ionization when, LC-MS sensitivity fails to provide adequate information (Gathungu, in preparation).
The metabolite identification scheme that we have laid out relies on a correlation between three chromatographic methods for true unambiguous unknown characterization. To assure the chromatography is robust and that the two LC-EC chromatograms (with profiling solvents or with LC-MS solvents) can be consistently relied upon to cross-map peaks of interest, we utilize our DSV standard and the non-concentrated/non-fractionated POOL sample. The correlation between the offline parallel LC-EC and LC-MS is established using the DSV standard. This enables the DSV standard to be used to ensure the peaks used to correlate the two instruments still provide the same correlation equation. This method tests whether either set of columns are going bad, or if there is a problem with either LC system. Additionally, LC-EC profiling and offline parallel LC-EC/LC-MS will contain retention time disparities. By analyzing a non-concentrated/non-fractionated POOL sample using both the LC-EC profiling method and the LC-EC method with LC-MS solvents, we can directly map the fractions between the two systems and assure the peaks we are characterizing are the LC-EC peaks of interest.
Ongoing work is focused on improving this workflow to streamline the metabolite characterization process and the integration with GC-MS as noted above (Gathungu et al., in preparation). Another issue is the identification of very low level trace analytes, requiring the development of more efficient sample concentration approaches. The inherent diversity of the plasma metabolome generates multiple obstacles toward achieving efficient sample concentration, fractionation and detection. These obstacles include, but are not limited to, initial metabolite concentration in the sample and metabolite chemistry that determines the overall stability and solubility of the compound as well as its optimal detection mechanism (i.e. MS vs. NMR vs. UV vs. EC). In theory, the method we have presented here should achieve a 45X concentration of each LC fraction. In practice, however, metabolites of different chemistries, hydrophobicity, and/or solubility, reveal different concentration efficiencies. Future work will focus on those species that remain both below the current system limit of detection, or with chemistries that limit current sample concentration procedures (Gathungu et al., work in progress).
CONCLUSION
Metabolite identification and structural confirmation continues to be a major problem and bottleneck plaguing metabolomics research. In this work, a workflow was designed to structurally characterize metabolites previously identified in an run LC-EC profiling study, using the complementary strengths of MS and NMR. The ability to structurally confirm these species of interest provides the information necessary to move past biomarker discovery and to define the biological and clinical importance of species of interest. This identification workflow not only benefits the unambiguous characterization of LC-EC identified metabolites of interest, but can also supplement any profiling study which provides analytically strong quantitative data without the necessary qualitative characterization. In the future, this workflow will be applied to the characterization of LC-EC identified metabolite markers of caloric restriction (CR) previously determined in both rat and human studies.
Supplementary Material
Acknowledgments
The studies reported were funded by U01-ES16048 (BSK, PI), a part of the NIH Genes and Environment Initiative (GEI), R01-HL109239 (BSK, PI), R01-AG28996 (BSK, PI), Brigham and Women’s Hospital Department of Neurosurgery (BSK), Hydrocephalus Research Initiative (BSK) and R01CA69390 (PV, PI). The authors also thank ThermoFisher for the loan of an Exactive Benchtop orbitrap for demonstration testing and financial support for scientific meeting attendance.
References
- 1.Rozen S, Cudkowicz ME, Bogdanov M, Matson WR, Kristal BS, Beecher C, Harrison S, Vouros P, Flarakos J, Vigneau-Callahan K, Matson TD, Newhall KM, Beal MF, Brown RH, Jr, Kaddurah-Daouk R. Metabolomics. 2005;1:101–108. doi: 10.1007/s11306-005-4810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kind T, Tolstikov V, Fiehn O, Weiss RH. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 5.Viant MR. Mol Biosyst. 2008;4:980–986. doi: 10.1039/b805354e. [DOI] [PubMed] [Google Scholar]
- 6.Hines A, Oladiran GS, Bignell JP, Stentiford GD, Viant MR. Environ Sci Technol. 2007;41:3375–3381. doi: 10.1021/es062745w. [DOI] [PubMed] [Google Scholar]
- 7.Poynton HC, Taylor NS, Hicks J, Colson K, Chan S, Clark C, Scanlan L, Loguinov AV, Vulpe C, Viant MR. Environ Sci Technol. 2011;45:3710–3717. doi: 10.1021/es1037222. [DOI] [PubMed] [Google Scholar]
- 8.Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ. Am J Clin Nutr. 2006;84:531–539. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- 9.Vinayavekhin N, Homan EA, Saghatelian A. ACS Chemical Biology. 2009;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 10.Paolucci U, Vigneau-Callahan KE, Shi H, Matson WR, Kristal BS. Omics. 2004;8:221–238. doi: 10.1089/omi.2004.8.221. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd AJ, Beckmann M, Fave G, Mathers JC, Draper J. Br J Nutr. 2011;106:812–824. doi: 10.1017/S0007114511001164. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd AJ, Fave G, Beckmann M, Lin W, Tailliart K, Xie L, Mathers JC, Draper J. Am J Clin Nutr. 2011;94:981–991. doi: 10.3945/ajcn.111.017921. [DOI] [PubMed] [Google Scholar]
- 13.Kind T, Fiehn O. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abate S, Ahn YG, Kind T, Cataldi TR, Fiehn O. Rapid Commun Mass Spectrom. 2010;24:1172–1180. doi: 10.1002/rcm.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindon JC, Holmes E, Nicholson JK. Analytical Chemistry. 2003;75:384 A–391 A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- 16.Brown M, Wedge DC, Goodacre R, Kell DB, Baker PN, Kenny LC, Mamas MA, Neyses L, Dunn WB. Bioinformatics. 2011;27:1108–1112. doi: 10.1093/bioinformatics/btr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaddurah-Daouk R, Boyle SH, Matson W, Sharma S, Matson S, Zhu H, Bogdanov MB, Churchill E, Krishnan RR, Rush AJ, Pickering E, Delnomdedieu M. Transl Psychiatry. 2011:1. doi: 10.1038/tp.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redman LM, Ravussin E. Wiley-VCH Verlag GmbH & Co. KGaA; 2010. pp. 423–439. [Google Scholar]
- 19.Vigneau-Callahan KE, Shestopalov AI, Milbury PE, Matson WR, Kristal BS. J Nutr. 2001;131:924S–932S. doi: 10.1093/jn/131.3.924S. [DOI] [PubMed] [Google Scholar]
- 20.Matson WR, Langlais P, Volicer L, Gamache PH, Bird E, Mark KA. Clinical Chemistry. 1984;30:1477–1488. [PubMed] [Google Scholar]
- 21.Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. Methods Mol Biol. 2007;371:393–409. doi: 10.1007/978-1-59745-361-5_25. [DOI] [PubMed] [Google Scholar]
- 22.Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. Methods Mol Biol. 2007;358:159–174. doi: 10.1007/978-1-59745-244-1_10. [DOI] [PubMed] [Google Scholar]
- 23.Shurubor YI, Matson WR, Willett WC, Hankinson SE, Kristal BS. BMC Clin Pathol. 2007;7:9. doi: 10.1186/1472-6890-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condray R, Dougherty GG, Jr, Keshavan MS, Reddy RD, Haas GL, Montrose DM, Matson WR, McEvoy J, Kaddurah-Daouk R, Yao JK. Int J Neuropsychopharmacol. 2011;14:756–767. doi: 10.1017/S1461145710001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristal BS, Vigneau-Callahan KE, Matson WR. Anal Biochem. 1998;263:18–25. doi: 10.1006/abio.1998.2831. [DOI] [PubMed] [Google Scholar]
- 26.Kristal BS, Vigneau-Callahan K, Matson WR. Methods Mol Biol. 2002;186:185–194. doi: 10.1385/1-59259-173-6:185. [DOI] [PubMed] [Google Scholar]
- 27.Kristal BS, Vigneau-Callahan KE, Moskowitz AJ, Matson WR. Arch Biochem Biophys. 1999;370:22–33. doi: 10.1006/abbi.1999.1387. [DOI] [PubMed] [Google Scholar]
- 28.Yao JK, Dougherty GG, Jr, Reddy RD, Keshavan MS, Montrose DM, Matson WR, McEvoy J, Kaddurah-Daouk R. PLoS One. 2010;5:e9508. doi: 10.1371/journal.pone.0009508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansen KK, Wang L, Aasly JO, White LR, Matson WR, Henchcliffe C, Beal MF, Bogdanov M. PLoS One. 2009;4:e7551. doi: 10.1371/journal.pone.0007551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaddurah-Daouk R, Rozen S, Matson W, Han X, Hulette CM, Burke JR, Doraiswamy PM, Welsh-Bohmer KA. Alzheimers Dement. 2011;7:309–317. doi: 10.1016/j.jalz.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogdanov M, Brown RH, Matson W, Smart R, Hayden D, O’Donnell H, Flint Beal M, Cudkowicz M. Free Radic Biol Med. 2000;29:652–658. doi: 10.1016/s0891-5849(00)00349-x. [DOI] [PubMed] [Google Scholar]
- 32.Bogdanov MB, Beal MF, McCabe DR, Griffin RM, Matson WR. Free Radic Biol Med. 1999;27:647–666. doi: 10.1016/s0891-5849(99)00113-6. [DOI] [PubMed] [Google Scholar]
- 33.Paul S, Bogdanov MB, Matson WR, Metakis L, Jacobs J, Beal MF. Free Radic Res. 2003;37:499–502. doi: 10.1080/1071576031000076268. [DOI] [PubMed] [Google Scholar]
- 34.Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, Flint Beal M. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 35.Stack EC, Ferro JL, Kim J, Del Signore SJ, Goodrich S, Matson S, Hunt BB, Cormier K, Smith K, Matson WR, Ryu H, Ferrante RJ. Biochim Biophys Acta. 2008;1782:151–162. doi: 10.1016/j.bbadis.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Ebbel EN, Leymarie N, Schiavo S, Sharma S, Gevorkian S, Hersch S, Matson WR, Costello CE. Anal Biochem. 2010;399:152–161. doi: 10.1016/j.ab.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiavo S, Ebbel E, Sharma S, Matson W, Kristal BS, Hersch S, Vouros P. Anal Chem. 2008;80:5912–5923. doi: 10.1021/ac800507y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith KM, Matson S, Matson WR, Cormier K, Del Signore SJ, Hagerty SW, Stack EC, Ryu H, Ferrante RJ. Biochim Biophys Acta. 2006;1762:616–626. doi: 10.1016/j.bbadis.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Patkar AA, Rozen S, Mannelli P, Matson W, Pae CU, Krishnan KR, Kaddurah-Daouk R. Psychopharmacology (Berl) 2009;206:479–489. doi: 10.1007/s00213-009-1625-1. [DOI] [PubMed] [Google Scholar]
- 40.Paolucci U, Vigneau-Callahan KE, Shi H, Matson WR, Kristal BS. Omics. 2004;8:209–220. doi: 10.1089/omi.2004.8.209. [DOI] [PubMed] [Google Scholar]
- 41.Shi H, Paolucci U, Vigneau-Callahan KE, Milbury PE, Matson WR, Kristal BS. Omics. 2004;8:197–208. doi: 10.1089/omi.2004.8.197. [DOI] [PubMed] [Google Scholar]
- 42.Shi H, Vigneau-Callahan KE, Shestopalov AI, Milbury PE, Matson WR, Kristal BS. J Nutr. 2002;132:1031–1038. doi: 10.1093/jn/132.5.1031. [DOI] [PubMed] [Google Scholar]
- 43.Shi H, Vigneau-Callahan KE, Shestopalov AI, Milbury PE, Matson WR, Kristal BS. J Nutr. 2002;132:1039–1046. doi: 10.1093/jn/132.5.1039. [DOI] [PubMed] [Google Scholar]
- 44.Kaddurah-Daouk R, Yuan P, Boyle SH, Matson W, Wang Z, Zeng ZB, Zhu H, Dougherty GG, Yao JK, Chen G, Guitart X, Carlson PJ, Neumeister A, Zarate C, Krishnan RR, Manji HK, Drevets W. Sci Rep. 2012;2:667. doi: 10.1038/srep00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long JD, Matson WR, Juhl AR, Leavitt BR, Paulsen JS. Neurobiol Dis. 2012;46:625–634. doi: 10.1016/j.nbd.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ascherio A, LeWitt PA, Xu K, Eberly S, Watts A, Matson WR, Marras C, Kieburtz K, Rudolph A, Bogdanov MB, Schwid SR, Tennis M, Tanner CM, Beal MF, Lang AE, Oakes D, Fahn S, Shoulson I, Schwarzschild MA. Arch Neurol. 2009;66:1460–1468. doi: 10.1001/archneurol.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matson WR, Langlais P, Volicer L, Gamache PH, Bird E, Mark KA. Clin Chem. 1984;30:1477–1488. [PubMed] [Google Scholar]
- 48.Jeitner TM, Bogdanov MB, Matson WR, Daikhin Y, Yudkoff M, Folk JE, Steinman L, Browne SE, Beal MF, Blass JP, Cooper AJ. J Neurochem. 2001;79:1109–1112. doi: 10.1046/j.1471-4159.2001.00673.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu L, Li S, Chen Z. J Pharm Biomed Anal. 2012;61:252–255. doi: 10.1016/j.jpba.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 50.Shao X, Lv L, Parks T, Wu H, Ho CT, Sang S. J Agric Food Chem. 58:12608–12614. doi: 10.1021/jf1029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 52.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang FQ, Li DQ, Feng K, Hu DJ, Li SP. J Chromatogr A. 1217:5501–5510. doi: 10.1016/j.chroma.2010.06.062. [DOI] [PubMed] [Google Scholar]
- 54.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson DL, Lacey ME, Sweedler JV. Analytical Chemistry. 1998;70:645–650. doi: 10.1021/ac970972y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.