Abstract
The addition of hydroxyl radicals to the C8 position of guanine can lead to the formation of 2,6-diamino-4-hydroxy-5-formamido-2′-deoxypyrimidine (Fapy-dG) lesion, whose endogenous levels in cellular DNA rival those of 8-oxo-7,8-dihydroxy-2′-deoxyguanosine. Despite its prevalence, the structure of duplex DNA containing Fapy-dG is unknown. We have prepared an undecameric duplex containing a centrally located β-cFapy-dG residue paired to dC and determined its solution structure by high resolution NMR spectroscopy and restrained molecular dynamic simulations. The damaged duplex adopts a right-handed helical structure all residues in an anti conformation, forming Watson-Crick base pair alignments, and 2-deoxyribose conformations in the C2′-endo/C1′-exo range. The formamido group of Fapy rotates out of the pyrimidine plane and is present on the Z and E configurations that equilibrate with an approximate 2:1 population ratio. The two isomeric duplexes show similar lesion induced deviations from a canonical B-from DNA conformation that are minor and limited to the central three-base-pair segment of the duplex, affecting the stacking interactions with the 5-lesion-neighboring residue. We discuss the implications of our observations for translesion synthesis during DNA replication and the recognition of Fapy-dG by DNA glycosylases.
INTRODUCTION
Cell exposure to ionizing radiation and the process of aerobic respiration generate free radicals that can react with sugars and bases in the DNA, causing oxidative damage. The hydroxyl radical can react with the purine ring generating different base radicals, among them the C8-OH adduct radical. One electron oxidation of the C8-OH guanine radical forms 8-oxo-7,8-dihydroxy 2′-deoxyguanosine (8-oxo-dG), whereas the competing one-electron reduction reaction, accompanied by opening of the imidazole ring, generates the 2,6-diamino-4-hydroxy-5-formamido-2′-deoxypyrimidine (Fapy-dG) lesion.1 Fapy lesions are prevalent under a variety of experimental conditions. Exposure of duplex DNA to ionizing radiation or reactive oxygen species produces both 8-oxo-dG and Fapy-dG adducts; notably, the latter is formed in an oxidizing environment.2-5 Mammalian cells show background amounts of both lesions and their levels rise quickly after exposure to ionizing radiation or treatment with hydrogen peroxide.6-8 Furthermore, individuals with chronic diseases such as Alzheimer or cancer show increased levels of both 8-oxo-dG and Fapy-dG lesions.9-12
Base Excision Repair (BER) eliminates Fapy lesions from genomic DNA. Early studies using DNA substrates containing methyl-Fapy residues or having multiple oxidative lesions showed that bacterial formamidopyrimidine-DNA glycosylase (Fpg, also known as MutM) removes 8-oxo-dG, Fapy-dG and Fapy-dA from double-stranded duplexes.13-15 More recently, substrates containing a site-specific Fapy adduct confirmed the original reports and allowed the evaluation as to how the counter base influence on the excision activity of Fpg.16-17 The specificity constant for removal of Fapy-dG is about 17-fold higher when the adduct is paired with dC, compared to dA. On the other hand, Fpg can excise with similar efficiency Fapy-dA lesions paired to any other base. In eukaryotic cells, several proteins show activity for the removal of Fapy lesions. Ogg1, the functional homolog of Fpg, can cleave Fapy-dG and 8-oxo-dG from duplex DNA, but is fundamentally inactive in the presence of Fapy-dA adducts.18-22 In addition, mammalian Nei-like enzymes (NEIL) can excise Fapy lesions. NEIL1 protein efficiently removes Fapy-dG and Fapy-dA from damaged duplexes but, in contrast to Ogg1 or Fpg, is inactive towards 8-oxo-dG containing substrates.23-25
Recent advance in the preparation of oligodeoxynucleotides containing site-specific Fapy adducts26,27 made possible the characterization of the mutagenic properties of the lesions. In addition to the correct dNTP, the Klenow exo fragment of Pol I misincorporates dA opposite to Fapy-dG and Fapy-dA lesions, events that respectively lead to G → T and A → T transversions in bacteria.28,29 In agreement with these in vitro results, E. coli replication of a plasmid containing Fapy-dG results in the observation of a small number of G → T transversions.30 On the other hand, Fapy-dG causes a high frequency of G → T transversions in COS cells, where it is more mutagenic that the related 8-oxo-dG lesion in the same sequence context.31
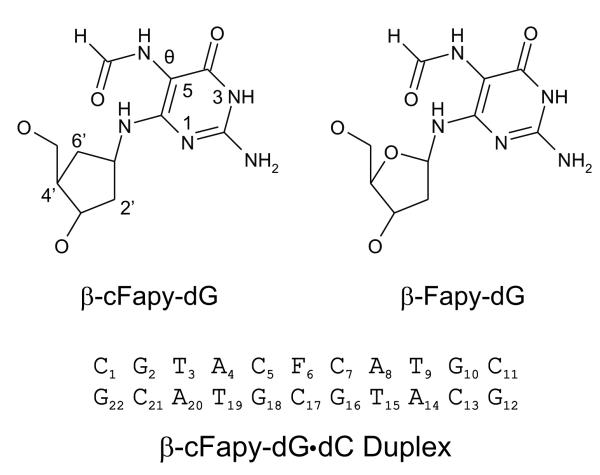
The structural characteristics of DNA duplexes containing Fapy lesion are currently unknown. The lack of efficient synthetic methods has hindered, up to now, the preparation of a sample large enough for NMR structural studies. We have recently reported a new synthesis approach for the incorporation of Fapy-dG in any sequence context, opening the door to future structural studies of lesion containing duplexes.32 However, the high conformational and structural variability of Fapy-dG forecasts the extreme complexity of this ongoing endeavor. In contrast to normal nucleosides, the glycosidic bond of Fapy rearranges upon standing, producing an equilibrium mixture of α and β anomeric forms. Moreover, the formamido group of Fapy can rotate around the C5-N7 bond, and the partial double-bond character of the C8-N7 bond generates geometric (Z and E) isomers. As a result, a Fapy containing duplex is expected to exist in two anomeric forms, each of which can exist as a mixture of Z and E formamido isomers that can adopt different rotameric states.33 Thus, proper assignment of the NMR spectra of such a complex isomeric mixture is a formidable task, even for contemporary NMR techniques. This problem, however, can be partially resolved by using a non epimerizing Fapy analogue. Two different Fapy isosters that block the epimerization reaction are presently available. One of them, called the C-Fapy nucleoside, replaces the N6 amino group of the lesion by a methylene link, thereby converting the labile 1,1-aminoether group of Fapy into a stable ether group.34 An obvious drawback of such modification is that it eliminates a hydrogen bond donor (N6), increases the hydrophobicity of the Fapy nucleotide and changes its electronic properties. The second analog, termed cFapy (carba-Fapy), has the O4′ deoxyribose atom of the lesion replaced by a CH2 group, removing a hydrogen bond acceptor from the sugar ring and minimally altering the electronic characteristics of Fapy.35,36 In the past, our laboratory has successfully used the latter approach for establishing the solution structure of duplexes bearing an α- or β-abasic site observing that carbocyclic sugar analogues were excellent structural mimics of the natural lesion without affecting Ape1 enzyme recognition.37 We report here the solution structure of a damaged duplex, called hereafter the cFapyG duplex, having a centrally located β-cFapy-dG•dC (cFapyG•dC) base pair, as determined by NMR spectroscopy and restrained molecular dynamics simulations. The duplex sequence used in this study and the structures of Fapy-dG and its carbocyclic analogue are shown in Figure 1.
Figure 1.
The top panel shows the chemical structure of β-Fapy-dG and β carba Fapy-dG. The bottom panel depicts the composition and numbering scheme of the β-Fapy-dG•dC duplex.
MATERIALS AND METHODS
Sample Preparation
We prepared the protected cFapy phosphoramidite according to a published procedure.38. Solid phase synthesis of the oligonucleotide containing a cFapy-dG lesion followed standard procedures with minor modifications. To prevent the exchange of the formyl group, we used pivalic anhydride/pyridine without methyl imidazole as the capping reagent. In addition, we added 10% DMF to the standard oxidizing solution to suppress oxidation of the Fapy formyl group. MALDI mass spectrum confirmed the proper composition of the damage-containing oligonucleotide (theoretical mass = 3333.20; obatained mass 3333.70). We purified DNA samples by two rounds of reverse-phase HPLC (C18 column) using as a mobile phase a 0.1 M triethylammonium acetate (TEAA) buffer (pH 6.8) and a linear gradient 0 to 40 % acetonitrile in 40 min. The sample was desalted using a Sephadex G-25 column and converted to the sodium salt by passing it through a Dowex 50W cation exchange column. We obtained a 1:1 stoichiometry by following the NMR intensity of thymine methyl signals during gradual addition of the undamaged strand to the lesion containing strand. Samples for NMR studies consisted of about 1.1 μmoles of duplex DNA dissolved in 0.7 ml of 25 mM phosphate buffer, pH 6.8, containing 50 mM NaCl and 0.5 mM EDTA, in 99.96% D2O or 90% H2O/10% D2O.
NMR Methods
We recorded one- and two-dimensional NMR spectra in spectrometers operating at 500, 600 and 800 MHz field strengths. Proton spectra taken with the duplex dissolved in 99.96% D2O at 25 °C consisted of phase-sensitive39 NOESY (90, 130 and 300 ms mixing time), COSY, DQF-COSY, TOCSY (70, 130 ms isotropic mixing times) and a hetero nuclear [13C 1H] HSQC. We suppressed the intensity of the residual water signal by saturation. We recorded phase sensitive proton NOESY spectra (120 and 220 ms mixing time) in 10% D2O buffer at 4 °C using the ‘excitation sculpting’ sequence to suppress the strong water signal.40 We processed time-domain data using Felix (Accelrys Inc., San Diego, CA) or NMRPipe41 software and visualized the spectra with Felix using Sparky.42 Typical 2D data sets consisted of 2048 by 300 complex points in the t2 and t1 dimensions, respectively. Prior to Fourier transformation, we filtered the time domain data by multiplication with shifted sine bell window functions. We applied no baseline correction, nor other ‘data massaging’ methods, to the frequency domain spectra.
Computational Methods
We used X PLOR 3.143 to run restrained molecular dynamic (rMD) simulations and refine the structure of the cFapy-dG•dC duplex. Simulations run using an all-atom force field derived from CHARMM44 with the dielectric constant set to 445 and the recently published parameters for the Fapy-dG lesion.46 Computation of inter proton distances followed a full relaxation matrix approach. Briefly, we minimized hydrogen position on a canonical B-form duplex by 1,000 steps of energy minimization, using a sole potential energy function that is proportional to the difference between back-calculated and experimental NOE intensities.47 For these calculations, we used experimental NOE intensities derived from D2O NOESY spectra at the three different mixing times. A grid search established that an isotropic correlation time of 2.92 ns fitted best the experimental NOE intensities to the initial B-form duplex and this value was set during computation of the inter-proton distances. Since NOE cross peaks at the center of the duplex differentiated between the E and Z isomers of Fapy, we calculated independent sets of inter proton distances for each of them. rMD simulations for the Z isomer were run restraining 470 inter proton distances by square-well potential energy functions, with boundaries of ±0.6 Å for non overlapping cross peaks and ±0.9 Å for overlapping peaks and peaks observed only on the longest mixing time NOESY. Similarly, refinement of the E isomer proceeded with a total of 422 distance restraints using the same boundary criteria. Following NMR evidence, we also enforced W-C hydrogen bond distances on all undamaged base pairs and backbone dihedral angles within a range encompassing the B and A form conformation on the four terminal base pairs of the duplex. W C hydrogen bonds with bigger distance bounds were also enforced at the damaged cFapyG•dC pair. We built two initial structures having the formyl group with the Z or E configuration and subjected them to a short energy minimization run using the conjugate gradient method. The rMD protocols were identical to those recently implemented for the refinement of duplexes having acrolein derived lesions.48 Briefly, we initiated the simulations at five different temperatures and heated the system to 500 K on 100 ps. Simultaneously, the penalty constant for enforcing inter proton distances increased from 10 to 300 kcal/(mol A2) and remained at this value until the end of the simulation. Simulations run at high temperature for 100 to 120 ps, after which we slowly cooled the system down to 300 K in 100 ps and ran 150 ps of additional rMD at this temperature. We calculated twenty five independent structures for each duplex by starting the simulations at five different temperatures (100, 105, 110, 115, and 120 K) and running the high temperature step for five different time lengths (100, 105, 110, 115, and 120 ps). Atom coordinates of the last 30 ps of each simulation were averaged and energy minimized, generating final refined structures of the cFapy-dG•dC duplex. Based on their pair wise Root Mean Square Deviation (RMSD) in the heavy atom position of the ensemble, we calculated the average model and energy minimized it, generating unique refined structures of the cFapy-dG•dC duplex. We used the HyperChem 8 program (HyperCube, Inc.) in order to compute the energy profile of the C6-C5-N7-C8 torsion angle (θ) of cFapy-dG in the Z and E duplexes. Using the conjugate gradient method, we performed energy minimization of the most representative duplex structure of each ensemble after systematically changing the θ torsion by 10° steps, going from −180° to 180°. The potential energy informed at each θ torsion value corresponded to the central (…C5-F6-C7…)•(…G16-C17-G18…) base pair segment. We visualized and analyzed the final structures with Chimera49 and calculated structural parameters with Curves.50
RESULTS
NMR Spectra
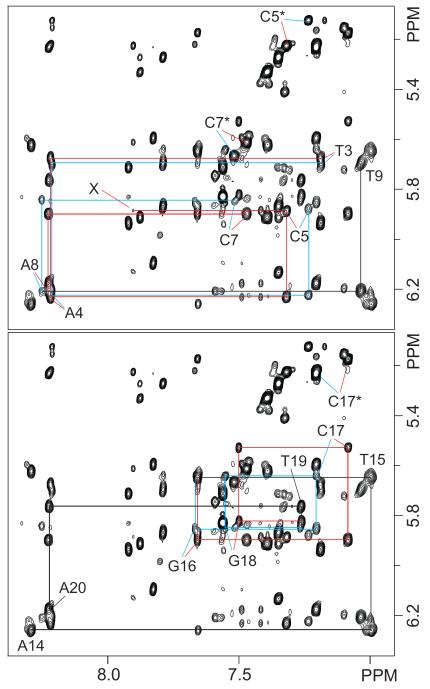
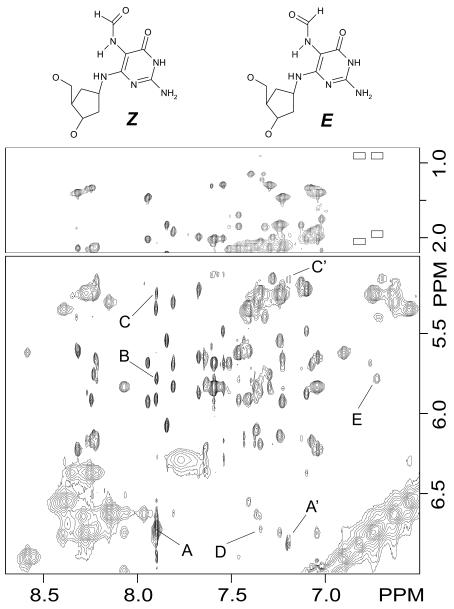
The one dimensional spectrum of the cFapy-dG duplex dissolved in “100%” D2O buffer (Figure 1S, Supporting Information) shows reasonably sharp and well-resolved proton signals, indicating that the NMR structural characterization of the damaged-containing duplex is feasible. Assignment of the non exchangeable protons resulted from the analysis of NOESY, COSY and TOCSY spectra following standard procedures.51,52 Figures 2 and 2S (Supporting Information) show NOE interactions between the base (6.89-8.40 ppm) and sugar-H1′ (5.07-6.33 ppm) protons for the (T3-T9)•(A14-A20) segment (Figure 2) or the whole cFapy-dG•dC duplex (Figure 2S), present on an 800 MHz NOESY spectrum (300 ms mixing time) recorded at 25 °C in 100% D2O buffer. Indicative of a standard right-handed helix, each purine-H8 and pyrimidine-H6 proton on the unmodified strand displays NOE cross-peaks to the H1′ proton of the same and 5′-flanking residues. At the center and on both strands of the duplex, NOE interactions are duplicated (Figure 2, red and blue traces) indicating the presence of, at least, two isomeric forms of the lesion with a 2:1 population ratio, as estimated by comparison of the relative intensity of corresponding HSQC cross peaks. The regular base to H1′ interactions break at the site of the cFapy nucleotide, where the sequential C5(H1′)-F(HCO) NOE is very weak for the more populated (major) lesion isomer (Figure 2, peak X) or missing in the case of the less populated (minor) form. Normal NOE interactions resume with the observation of the intra residue C7(H6)-C7(H1′) cross peak and continue without interruptions until the 3′-end of the modified strand, indicating that the regular distances are only lost at the cFapy residue. In addition, the observation of NOE cross peaks between sugar-H1′ and the adenine-H2 protons (Figure 3, peaks E-P) and between purine H8 and the H5 proton of 3′-flanking cytosine residues (Figure 3, peaks Q-V), confirms that both isomeric forms of the cFapy-dG•dC duplex exist as regular right handed helices in solution.
Figure 2.
Contour plots of an expanded region of an 800 MHz NOESY spectrum (300 ms mixing time) recorded at 25 °C in 100% D2O buffer. The figure shows intra residue and sequential base (purine H8/pyrimidine H6) to sugar H1′ proton interactions for the modified (top) and unmodified (bottom) strands on the central (T3 T9)•(A14 A20) segment of the cFapy-dG•dC duplex. Red and blue colors trace these connectivities on the Z and E isomeric duplexes, respectively. Numbered letters label the intra residue NOE peaks and asterisks indicate cytosine(H5 H6) peaks. X identifies the F6(HCO) C5(H1′) NOE of the Z isomer.
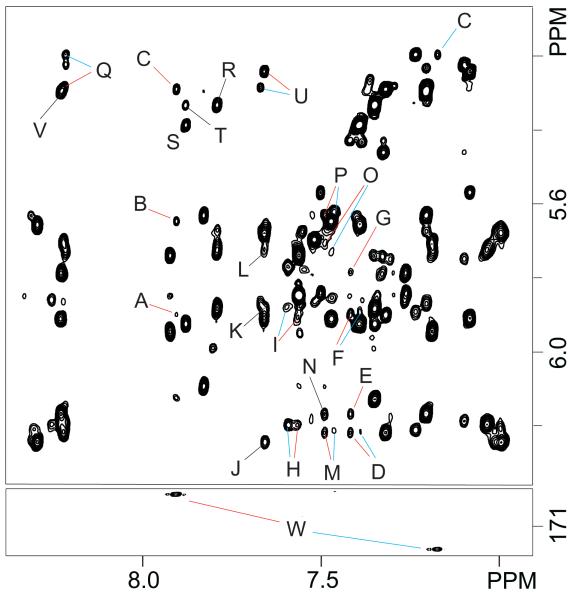
Figure 3.
The top panel depicts additional NOE interactions on the region shown in Figure 2. Red and blue colors differentiate the Z and E isomeric duplexes, respectively. Labeled peaks are assigned as follows: A, F6(HCO) C5(H1′); B, F6(HCO) C7(H5); C, F6(HCO) C5(H5); D, A4(H2) A4(H1′); E, A4(H2) A20(H1′); F, A4(H2) C5(H1′); G, A4(H2) T19(H1′); H, A8(H2) A8(H1′); I, A8(H2) G16(H1′); J, A14(H2) A14(H1′); K, A14(H2) G10(H1′); L, A14(H2) T9(H1′); M, A20(H2) A4(H1′); N, A20(H2) A20(H1′); O, A20(H2) T3(H1′); P, A20(H2) C21(H1′); Q, A4(H8) C5(H5); R, G10(H8) C11(H5); S, G12(H8) C13(H5); T, G12(H8) C11(H5); U, G16(H8) C17(H5); V, A20(H8) C21(H5). Bottom, expanded region of an 800 MHz [1H 13C] HSQC spectrum recorded at 25 °C that was to identify the formyl group of each cFapy-dG isomer by the F6(HCO) F6(CO) interaction (peaks labeled W).
Analysis of an 800 MHz [1H-13C] HSQC spectrum recorded at 25 °C reveals the presence of carbonyl resonances at 169.6 and 172.0 ppm, coupled to proton resonances at 7.92 and 7.21 ppm, respectively, confirming the presence of two isomeric forms for the formyl group of cFapy (Figure 3, bottom panel). The F(COH) proton of the major form displays a very weak NOE cross-peak to C5(H1′) and stronger interactions with C7(H5) and C5(H5) (Figure 3, top panel, peaks A-C, respectively), whereas the minor isomeric form exhibits only one NOE interaction with the C5(H5) proton (Figure 3, peak C). The aromatic proton region (8.7-6.9 ppm) of the spectrum displays an NOE peak between C5(H6) and the formyl proton of the major isomeric form as well as an expected exchange cross peak between the formyl protons of both isomers (Figure 3S, Supporting Information). Other regions of the NOESY spectra do not reveal any additional interaction for the cFapy formyl protons of the damaged duplexes. The chemical shifts of the non exchangeable protons are listed in Table 1S (Supporting Information).
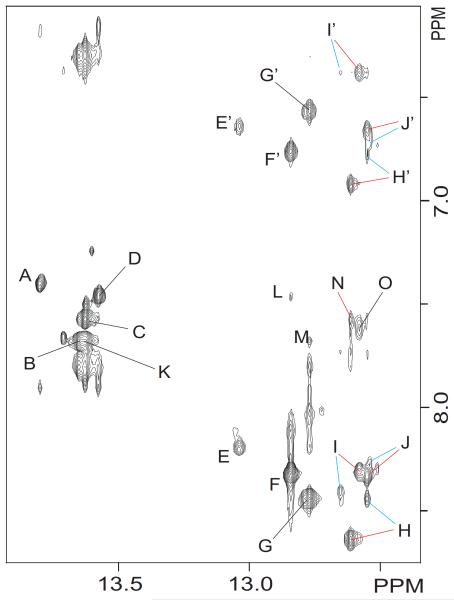
Assignment of the exchangeable protons of the isomeric duplexes follows the analysis of an 800 MHz NOESY (220 ms mixing time) spectrum recorded in 10% D2O buffer at 5 °C. Each thymine imino proton displays a strong cross peak to the H2 proton of its partner adenine in the complementary strand (Figure 4, peaks A-D). Similarly, each guanine imino proton, as well as the cFapy (N3H) protons, exhibit NOE peaks to the hydrogen bonded and non hydrogen bonded amino protons of their counter cytosine nucleotide (Figure 4, peaks E/E’ J/J’). These observations establish A•T and G•C Watson Crick (W C) hydrogen bond alignments for all base pairs of the duplex, including across the cFapy-dG•dC base pair. As previously observed on the NOESY spectra in 100% D2O, the presence of isomeric forms of the lesion causes the duplication of the imino proton signals on the central three base pair segment of the duplex. Further analysis of the same region reveals the presence of NOE interactions between the imino and the H2 proton on flanking adenine residues (Figure 4, peaks K-N), and between the imino protons of adjacent base pairs (data not shown), indicating proper stacking throughout the cFapy-dG•dC duplex. The chemical shift of the exchangeable protons is listed Table 1S (Supporting Information).
Figure 4.
Contour plot of an expanded region of an 800 MHz NOESY (220 ms mixing time) spectrum recorded in 10% D2O buffer at 5 °C, showing interactions between the imino and base/amino proton regions. Red and blue colors differentiate the Z and E isomeric duplexes, respectively. Labeled peaks are assigned as follows: A, T19(N3H) A4(H2); B, T9(N3H) A14(H2); C, T15(N3H) A8(H2); D, T3(N3H) A20(H2); E/E′, G12(N1H) C11(N4H)hb/nhb; F/F’, G2(N1H) C21(N4H)hb/nhb; G/G’, G10(N1H) 13(N4H)hb/nhb; H/H’, G16(N1H) C7(N4H)hb/nhb; I/I’, F(N3H) C17(N4H)hb/nhb; J/J’, G18(N1H) C5(N4H)hb/nhb; K, T15(N3H) A14(H2); L, G2(N1H) A20(H2); M, G10(N1H) A14(H2); N, G16(N1H) A8(H2); O, F(N3H) F(N2H).
Configuration of the formamido group of cFapy-dG
The low temperature NOESY spectrum in 10% D2O buffer also provides key evidence for establishing the configuration of the formamido group of lesion in the isomeric duplexes. The formyl proton of the major isomer displays a very strong NOE cross peak with an exchangeable proton that resonates at 6.73 ppm, whereas the minor form exhibits a corresponding but weaker interaction with a signal at 6.82 ppm (Figure 5, peaks A and A’, respectively). Considering that the glycosidic torsion angle of cFapy-dG is in an anti conformation and the central three-base-pair segment of the duplex adopts regular W C alignments, then F(N7H) and F(N6H) are the only proton candidates to interact with F(COH) and generate those NOE peaks. When cFapy-dG is in anti conformation its F(N6H) proton will be less than 2.8 Å apart from the closest of the F(H6′/H6″) protons, whereas F(N7H) will never be closer than 3.7 Å. Therefore, the absence of NOE interactions between the exchangeable 6.73 and 6.82 ppm signals and the lesion F(H6′/H6″) protons (Figure 5, empty boxes) rules out the possibility that the 6.73 and 6.82 ppm resonances belong to F(N6H). Thus, the strong NOE cross peak of the major isomeric form of cFapy-dG (Figure 5, peak A) belongs to the interaction between its F(N7H) and F(COH) protons that are 2.4 Å apart when the formamido group has aZ configuration. The equivalent NOE peak on the minor form (Figure 5, peak A’) clearly originates from interaction of the same two protons and, in principle, can be ascribed to the E configuration of the lesion formamido group or, alternatively, to a low populated Z form with a different conformation around the θ torsion. Careful examination of the NOE cross peaks from the minor form (Figure 5, peaks A’ and C’) reveals that F(COH) is now a doublet, establishing a large J(N7H) (COH) coupling constant between the N7H and COH protons of the cFapy-dG. Thus, the E configuration of the formamido group, that has a coupling constant of about 11Hz as opposed to less than 2 Hz for the Z form (53), originates the less populated isomer of the cFapy-dG duplex. Further analysis of the same NOESY region indentifies cross peaks between the F(N7H) proton of the Z isomer and the H6 and H1′ protons of C5 (Figure 5, peaks D and E, respectively), establishing a θ torsion value that positions the NH5 proton of cFapy pointing towards the lesion flanking C5 residue. We failed to identify any additional evidence for the configuration of the E-cFapy isomer.
Figure 5.
The figure shows contour plots of the same NOESY experiment shown in Figure 4, depicting interactions between the base/amino and methyl (top), and base/amino and sugar H1′ (bottom) proton regions of the spectrum. Labeled peaks are assigned as follows: A, Z F6(HCO) F6(N5H); A’, E F6(HCO) F6(N5H); B, Z F6(HCO) C5(H1′); C, Z F6(HCO) C5(H5) and C’, E F6(HCO) C5(H5); D, Z F6(N5H) C5(H6); E, Z F6(N5H) C5(H1′).
Structure of cFapy-dG•dC Duplexes
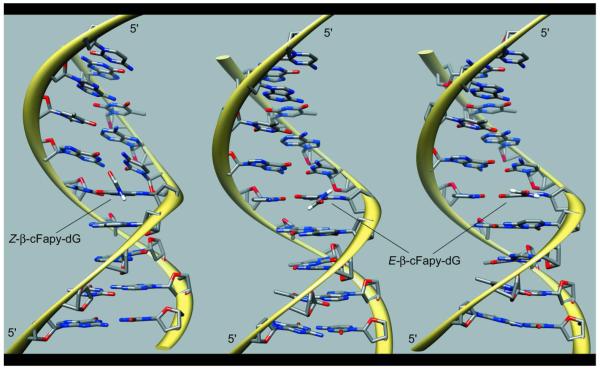
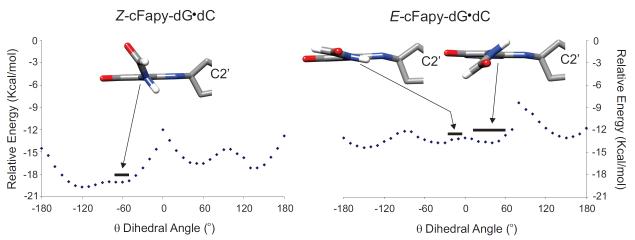
The twenty five refined models of the Z-cFapy-dG•dC duplex comprise an ensemble with pair-wise RMSD smaller than 1.14 Å at the heavy atom positions and no violation of the NOE-derived distances larger than 0.1 Å (Figure 4S, Supporting Information), indicating that the NMR-restrained MD simulations yields a unique structural motif for the Z duplex. As shown in Figure 6, the Z-cFapy-dG•dC duplex is a regular B form helix that is minimally perturbed at its center by the presence of the lesion. All nucleotides are in anti conformation, forming canonical W-C base pairs, and display 2-deoxyribose conformations in the C2′-endo/C1′-exo range. The formyl group and pyrimidine ring of the lesion are not co planar adopting a θ torsion angle (C6-C5-N7-C8) of −59° in the averaged structure. As a result, the formyl group points towards the center of the major groove towards C7, the lesion flanking residue at the 3′-side. Structural adjustments at the lesion site are minor and include a small decrease of the helical-twist at the C5•G18/cFapy-dG•dC step, which is fully compensated in the following step of the duplex, and a small kink on the helical axis mostly originating between the cFapy-dG•dC and C7•G16 base pairs (Figure 6, Table 1). The refined models of the E-cFapy-dG•dC duplex converge to an ensemble with pair wise RMSD smaller than 2.00 Å on the heavy atom positions and no violation of the NOE derived distance boundaries exceeding 0.1 Å (Figure 5S, Supporting Information), establishing somehow a larger structural dispersion among the refined structures than in the case of the Z cFapy-dG•dC duplex. The E-cFapy-dG•dC duplex structure is a regular B form helix with all residues in the anti conformation, adopting canonical W-C base pair alignments and 2 deoxyribose conformations in the C2′-endo/C1′-exo range. Further examination of the refined models at the lesion site reveals that rMD simulations generate two groups of structures, shown in Figure 6, which differ only at the lesion site by the value of the θ torsion angle of cFapy-dG. Nineteen out of the 25 models have θ torsions in the positive 12°-63° range, 41° on the average structure, positioning the formyl group of cFapy-dG towards C5, its 5′-side neighboring residue. The remaining 6 structures display θ torsions in the negative 3°-26° range, −14° on their average, leaving the pyrimidine ring and formamido group of the lesion almost co planar (Figure 6). Additional structural adjustments of the E-cFapy-dG•dC duplexes at the lesion site parallel those observed for the Z cFapy-dG•dC duplex, namely compensatory changes on helical twist at the central three-base-pair segment of the duplex and a slightly larger kink of the helical axis between the cFapy-dG•dC and C7•G16 base pairs (Figure 6, Table 1). We further investigated the conformation of the θ torsion angle of the lesion by computing the energy profile of the C5-N7 rotation within the boundaries of the refined duplex structures. The Z cFapy-dG•dC duplex shows a low energy valley between −50° and −160° that includes the θ angle value measured in the average refined structure (θ = −59°), suggesting that the experimental NMR restraints do not drive the θ angle to an unfavorable conformation (Figure 7). In the case of the E cFapy-dG•dC duplex, the energy profile for rotation of the θ angle is rather shallow displaying four minima separated by small barriers and the θ torsion values obtained by rMD fall within two of the four minima (Figure 7).
Figure 6.
Structure of the β-cFapy-dG•dC duplexes seen with the major groove prominent. The left panel corresponds to the Z isomer (PDB ID: 2lwm, BMRB ID 18638) and the central and right panels to the major (PDB ID: 2lwn, BMRB ID 18639) and minor (PDB ID: 2lwo, BMRB ID 18640) rotameric forms of the E isomer, respectively.
Table 1.
Structural parameters of β-cFapy-dG•dC duplexesa
| Z-Duplex |
E-Duplex (major isomer) |
E-Duplex (minor isomer) |
|
|---|---|---|---|
| BP-twist C5•G18 - X6•C17 (°) | 12.5 (10 -15) | 8.0 (2 – 13) | 7 (3 – 9) |
| BP-twist X6•C17 - C6•G16 (°) | 62.0 (61 - 64) | 61.0 (58 – 63) | 61 (58 – 62) |
| Axis bend (°) | 12.0 (2 – 20) | 22.0 (14 – 32) | 28 (24 – 30) |
| cFapy-dG θ torsion angle (°) | 59.0 (51 – 69) | 41 (12 – 63) | −14 (−3 – −27) |
Values measured on the average refined structures. Between parentheses is the range of values measured on the rMD models.
Figure 7.
Energy profiles for the rotation of the lesion C5 N7 torsion angle (θ) on the refined duplex structures. The thick lines display the range of θ values measured on the refined models and on top is a view of β-cFapy-dG moiety on the averaged structures.
DISCUSION
Identified more than fifty years ago, Fapy lesions still defy their structural characterization in duplex DNA. Central to this challenge was the absence, until recently32, of an efficient synthesis method for the preparation of large amounts of oligodeoxynucleotides containing the Fapy lesion in any sequence context. Furthermore, Fapy equilibration between α and β anomers, the existence of Z and E formamido isomers and the presence of rotameric isomers at room temperature predict that the NMR structure determination of Fapy-containing duplexes will be a quite difficult task. Hence, the characterization of the undecameric duplex containing the isosteric β-cFapy-dG analog (Figure 1) provides the first look into the impact of β-Fapy-dG on DNA structure.
NMR Data
The two dimensional NMR data of the cFapy-dG•dC duplex establish the presence of a regular right-handed helix, stabilized by the formation of W-C base pair alignments throughout the duplex. Duplication of NOE cross peaks for residues on the central four base pair segment readily reveals the presence of two isomeric duplexes that originated from the equilibration between the Z and E forms of the formamido group of the lesion with an approximate 2:1 population ratio. The observation that the Z isomer is the predominant form of the lesion is in agreement with the equilibrium reported for Fapy-dG located at the center of a trimeric single strand oligomer32 and with the previous characterization of Fapy nucleosides33,53 though in the latter cases, the Z:E population ratio (ca 10:1) was significantly higher than the one we observe here for cFapy-dG in duplex DNA. Although we cannot rule out the possibility that differences in the electronic structure of the 2 deoxyribose and cyclopentane rings may account for the observed change, it seems quite unlikely, since minor electronic density variations at the N6 position can hardly propagate to the 5-formylamino group and cause a detectable change in its conformational behavior. A more plausible explanation for the population change of the Z and E isomers likely is the effect of the double stranded DNA environment.
Structure of cFapy-dG•dC Duplexes
Restrained molecular dynamics simulations generated structural ensembles for each isomer of the duplex that were in excellent agreement with the NMR data. The convergence of the Z-cFapy-dG•dC models is slightly better than that of the E-cFapy-dG•dC structures, a fact that most likely reflects the greater number of NMR restraints obtained and enforced during the refinement of the Z duplex by molecular dynamics. Both duplexes are regular right handed helices that belong to the B form DNA structure family, with glycosidic torsions in the anti range, 2 deoxyribose conformations in the C1′exo/C2′ endo range and base pairs stabilized by well-formed W-C alignments. Structural adjustments at the lesion site are only minor and similar for both isomeric duplexes (Table 1), indicating that the Z and E configurations of the amide bond can be readily accommodated within the B form DNA conformation.
As previously reported,46,54 opening of the imidazole ring of the guanine residue reduces the double bond character of the C5 N7 bond and forces the formyl group of Fapy to rotate out of the pyrimidine plane, easing in this way unfavorable electrostatic and steric interactions at the lesion site. The refined models of the Z-cFapy-dG•dC duplex have the formyl group pointing towards C7, displaying θ torsion values in a narrow range, −51° to −69°, that is within the low energy valley computed for rotation of the C5-N7 bond in the duplex structure. In the case of the E-cFapy-dG•dC duplex, the formyl group points towards C5, or is almost co planar with the pyrimidine ring, having θ torsion values that fall in two different low energy valleys of the C5-N7 torsion energy profile (Figures 6 and 7). Interestingly, none of the θ angle values observed on the refined structures of the Z or E-cFapy-dG•dC duplex appears at the lowest energy value. In both duplexes however, the energy differences are very small and most likely within the accuracy of the experimental method. Alternatively, these differences may reflect the formation of water mediated hydrogen bonds stabilizing the slightly higher energy value adopted by the θ torsion angle. Simple computer modeling shows that the formation of water mediated hydrogen bonds is possible on both isomeric duplexes (Figure 6S, Supporting Information).
Stability of cFapy-dG•dC duplexes
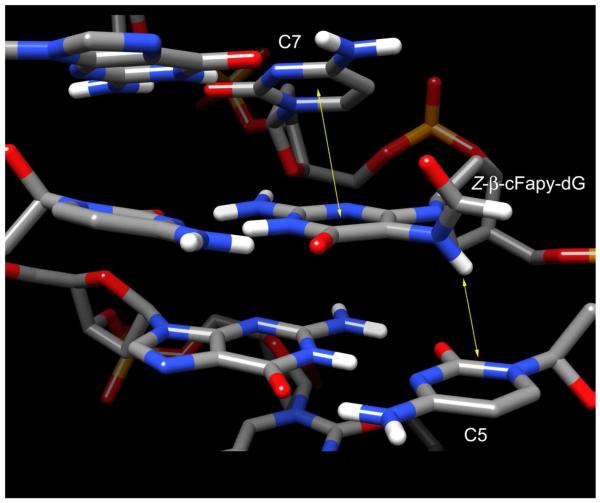
Previous studies have shown that whereas β-cFapy-dG has appreciable preference for pairing with dC, implying the existence of constructive interactions across the cFapy-dG•dC base pair, the lesion causes significant duplex destabilization, which could range from 2.4 to 4 kcal/mol, depending on the sequence context.36 The cFapy-dG•dC duplex used in our study has a lower duplex melting temperature than its undamaged parent control (Figure 7S, Supporting Information), a fact that is in qualitative agreement with the 1.5 kcal/mol destabilization caused by the natural β-Fapy-dG lesion in the same sequence context.32 Inspection of the damaged duplex structure allows the identification of key molecular interactions that explain the preference for pairing to dC as well as the decrease in duplex stability. The main stabilizing contacts include all three Watson-Crick hydrogen bonds across the cFapy-dG•dC pair and proper stacking of the cFapy pyrimidine ring and the 3′ flanking cytosine. Destabilizing interactions comprise the loss of stacking between the damaged base and its 5′ neighboring residue, which results from the conversion of the purine imidazol ring into the non-aromatic and non-planar formylamido group (Figure 8). Further analysis of the three dimensional models reveals no additional distortions that could add to duplex destabilization, suggesting that the energetic and structural impact of cFapy-dG in DNA is localized at the lesion site and flanking base pairs.
Figure 8.
Central part of the Z β-cFapy-dG•dC duplex (PDB ID: 2lwm, BMRB ID 18638) showing stacking interactions between the cFapy-dG and adjacent 2′ deoxycytosine residues.
Biological Implications. Mutagenicity and reading of the lesion by DNA polymerases
Earlier studies have established that DNA polymerases have strong preference for dCTP incorporation opposite the Fapy-dG lesion.29-31,36 This observation is in full accordance with the fact that when β-cFapy-dG is in anti-conformation minimally perturbs the DNA structure, and the isomerism of the 5 formylamino group does not affect W-C hydrogen bonding with the counter base (Figure 8). In addition to dCTP, bacterial and mammalian polymerases can insert dATP opposite the lesion, generating a mutagenic base pair intermediate that, by analogy with 8-oxo-dG, would have Fapy-dG adopting the syn conformation. While our study does not appraise this possibility, the non planarity and lack of aromaticity of the formylamido group suggest that the putative Fapy-dG(syn)•dA(anti) mispair intermediate would be less stable than the one formed by 8-oxo-dG, hence explaining the mutagenic differences between these lesions. The β-cFapy-dG•dC duplex structure may also help to explain the observation that the Klenow fragment of Pol I pauses after dCTP (or dATP) incorporation opposite the lesion.30 As mentioned above, whereas the stacking interaction between β-Fapy-dG and its 3′ neighbour is mostly unaffected by opening of the imidazole ring, in contrast, stacking with the 5′ flanking nucleotide is appreciably impaired (Figure 8). Therefore, we posit that this limited stacking may perturb the optimal location of the ensuing nucleotide affecting the enzyme’s ability to position properly the incoming dNTP for efficient phosphodiester bond formation.
Repair of the Fapy-dG lesion
Duplex DNA bearing a β-cFapy-dG•dC pair shares essential structural features with the 8-oxo-dG•dC containing duplex, including a minimal perturbation of the B form DNA conformation and W C hydrogen bonding at the lesion containing base pair.55,56 This observation serves as a good explanation for the fact that, despite their considerable chemical difference, the same DNA glycosylases can recognize these two lesions. The crystal structure of a β-cFapy-dG•dC duplex in a complex with Fpg showed that only the Z rotameric form of the lesion is present in the active site pocket of the enzyme, establishing a discrimination mechanism against the E isomer.54 Our NMR study, on the other hand, proves that both rotameric isomers of the lesion are present in duplex DNA and can readily interconvert with an equilibration rate on the millisecond time scale at 25 °C (Figure 3S). Therefore, although the existence of rotameric isomers of Fapy-dG may briefly delay enzyme activity, it is largely inconsequential for its ultimate excision by the repair enzyme. Identical considerations can be extended to the rotameric state around the C5 N7 bond that exhibits faster interconversion rates. A distinguishing feature of Fapy lesions is their ability to equilibrate, with the biological properties of the α isomer being quite different from those of the β-Fapy-dG isomer and the oxo dG lesion. Based on the published crystal structure of Fpg in complex with a β-cFapy-dG•dC duplex,54 it is doubtful that the stabilizing contacts observed in the binding pocket of the enzyme could be reproduced with the α isomer of the lesion. Furthermore, even if Fpg does manage to bind the α isomer, the presence of the Fapy-dG ring will most likely hinder the catalytic attack at its C1′ position. We previously demonstrated that both α and β-Fapy isomers may exist in neutral media for a relatively long period of time without undergoing inter conversion.32 Thus, systematic studies of the structure, mutagenic potential and reparability of the lesion α isomer are essential for full comprehension of Fapy-dG toxicity in the cell. Several of these studies are currently underway.
Supplementary Material
Acknowledgments
FUNDING SUPPORT NIH grant ES017368 supported this research.
ABBREVIATIONS
- Fapy
Formamidopyrimidine
- cFapy
carba Fapy
- Fpg
formamidopyrimidine glycosylase
- BER
Base Excision Repair
- NEIL
Nei Like
- NOESY
Nuclear Overhauser Effect Spectroscopy Y
- COSY
COrrelation Spectroscopy Y
- DQF-COSY
Double Quantum Filtered COSY
- TOCSY
TOtal Correlation Spectroscopy Y
- [13C 1H] HSQC
13C 1H Heteronuclear Single Quantum Coherence
- Z
zusammen
- E
entgegen
Footnotes
SUPPORTING INFORMATION Proton chemical shifts (Table 1S) on the β-cFapy-dG•dC duplex measured at 25 °C. One dimensional proton spectrum of the β-cFapy-dG•dC duplex recorded in 100% D2O buffer at 25 °C (Figure 1S), full ‘finger print’ region assignments on a 300 ms mixing time NOESY spectrum, recorded in 100% D2O buffer at 25 °C for the damaged (Figure 2SA) and undamaged (Figure 2SB) strands of the duplex, and expanded contour plot of the aromatic proton region on a 300 ms mixing time NOESY spectrum recorded in 100% D2O buffer at 25 °C (Figure 3S). Three dimensional view of the twenty five Z (Figure 4S) and E (Figure 5S) β-cFapy-dG•dC duplex structures, and examples of water mediated hydrogen bonds in the β-cFapy-dG•dC duplex (Figure 6S). UV260 melting curves for the β-cFapy-dG•dC and dG•dC duplexes (Figure 7S).
REFERENCES
- 1).Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2).Aruoma OI, Halliwell B, Dizdaroglu M. Iron ion dependent modification of bases in DNA by the superoxide radical generating system hypoxanthine/xanthine oxidase. J. Biol. Chem. 1989;264:13024–13028. [PubMed] [Google Scholar]
- 3).Aruoma OL, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases inDNA Induced by hydrogen peroxide and ferric ion chelates. J. Biol. Chem. 1989;264:20509–20512. [PubMed] [Google Scholar]
- 4).Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation generated free radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. 1990. [DOI] [PubMed] [Google Scholar]
- 5).Dizdaroglu M, Rao G, Halliwell B, Gajewski E. Damage to the DNA bases in mammalian chromatin by hydrogen peroxide in the presence of ferric and cupric ions. Arch. Biochem. Biophys. 1991;285:317–324. doi: 10.1016/0003-9861(91)90366-q. [DOI] [PubMed] [Google Scholar]
- 6).Nackerdien Z, Olinski R, Dizdaroglu M. DNA base damage in chromatin of gamma irradiated cultured human cells. Free Radic. Res. Commun. 1992;16:259–273. doi: 10.3109/10715769209049179. [DOI] [PubMed] [Google Scholar]
- 7).Pouget J, Douki T, Richard M, Cadet J. DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC MS and HPLC EC and comet assay. Chem. Res. Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 8).Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 9).Malins DC, Haimanot R. Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Res. 1991;51:5430–5432. [PubMed] [Google Scholar]
- 10).Malins DC, Polissar NL, Gunselman SJ. Progression of human breast cancers to the metastatic state is linked to hydroxyl radical induced DNA damage. Proc. Natl. Acad. Sci. USA. 1996;93:2557–2563. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J. Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 12).Kirkali G, Tunca M, Genc S, Jaruga P, Dizdaroglu M. Oxidative DNA damage in polymorphonuclear leukocytes of patients with familial Mediterranean fever. Free Radic. Biol. Med. 2008;44:386–393. doi: 10.1016/j.freeradbiomed.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 13).Boiteux S, O′Connor TR, Laval J. Formamidopyrimidine DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8 oxoguanine (8 hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 16).Wiederholt CJ, Delaney MO, Greenberg MM. Interaction of DNA containing Fapy.dA or its C nucleoside analogues with base excision repair enzymes: implications for mutagenesis and enzyme inhibition. Biochemistry. 2002;41:15838–15844. doi: 10.1021/bi025903e. [DOI] [PubMed] [Google Scholar]
- 17).Wiederholt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA containing Fapy.dG and its beta C nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 18).Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation or hydrogen peroxide/metal ion generated free radicals. Nucleic Acids Res. 1998;26:1228–1233. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha hOgg1 protein and the polymorphic alpha hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Dherin C, Dizdaroglu M, Doerflinger H, Boiteux S, Radicella JP. Repair of oxidative DNA damage in Drosophila melanogaster: identification and characterization of dOgg1, a second DNA glycosylase activity for 8 hydroxyguanine and formamidopyrimidines. Nucleic Acids Res. 2000;28:4583–4592. doi: 10.1093/nar/28.23.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Morales-Ruiz T, Birincioglu M, Jaruga P, Rodriguez H, Roldan-Arjona T, Dizdaroglu M. Arabidopsis thaliana Ogg1 protein excises 8 hydroxyguanine and 2,6 diamino 4 hydroxy 5 formamidopyrimidine from oxidatively damaged DNA containing multiple lesions. Biochemistry. 2003;42:3089–3095. doi: 10.1021/bi027226u. [DOI] [PubMed] [Google Scholar]
- 22).Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8 oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 2007;282:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 25).Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6 diamino 4 hydroxy 5 formamidopyrimidine and 4,6 diamino 5 formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 26).Haraguchi K, Delaney MO, Wiederholt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and characterization of oligonucleotides containing formamidopyrimidine lesions (Fapy.dA, Fapy.dG) at defined sites. Nucleic Acids Res. Suppl. 2001:129–130. [PubMed] [Google Scholar]
- 27).Haraguchi K, Delaney MO, Wiederholt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and characterization of oligodeoxynucleotides containing formamidopyrimidine lesions and nonhydrolyzable analogues. J. Am. Chem. Soc. 2002;124:3263–3269. doi: 10.1021/ja012135q. [DOI] [PubMed] [Google Scholar]
- 28).Delaney MO, Wiederholt CJ, Greenberg MM. Fapy-dA induces nucleotide misincorporation translesionally by a DNA polymerase. Angew. Chem. Int. Ed. Engl. 2002;41:771–775. doi: 10.1002/1521-3773(20020301)41:5<771::aid-anie771>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 29).Wiederholt CJ, Greenberg MM. Fapy.dG instructs Klenow exo– to misincorporate deoxyadenosine. J. Am. Chem. Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 30).Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, Fapy•dG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 31).Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring opened formamidopyrimidines (Fapy lesions) and 8 oxo purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Lukin M, Minetti CA, Remeta DP, Attaluri S, Johnson F, Breslauer KJ, de los Santos C. Novel post synthetic generation, isomeric resolution, and characterization of Fapy-dG within oligodeoxynucleotides: differential anomeric impacts on dna duplex properties. Nucleic Acids Res. 2011;39:5776–5789. doi: 10.1093/nar/gkr082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Burgdorf LT, Carell T. Synthesis, stability, and conformation of the formamidopyrimidine G DNA lesion. Chem. Eur. J. 2002;8:293–301. doi: 10.1002/1521-3765(20020104)8:1<293::aid-chem293>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 34).Delaney MO, Greenberg MM. Synthesis of oligonucleotides and thermal stability of duplexes containing the β c nucleoside analogue of Fapy•dg. Chem. Res. Toxicol. 2002;15:1460–1465. doi: 10.1021/tx025588x. [DOI] [PubMed] [Google Scholar]
- 35).Büsch F, Pieck JC, Ober M, Gierlich J, Hsu GW, Beese LS, Carell T. Dissecting the differences between the α and β anomers of the oxidative DNA lesion FaPydG. Chem. Eur. J. 2008;14:2125–2132. doi: 10.1002/chem.200701373. [DOI] [PubMed] [Google Scholar]
- 36).Ober M, Müller H, Pieck C, Gierlich J, Carell T. Base pairing and replicative processing of the formamidopyrimidine dG DNA lesion J. Am. Chem. Soc. 2005;127:18143–18149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- 37).de los Santos C, El khateeb M, Rege P, Tian K, Johnson F. Structural impact of C1′(OH) conformation on duplex DNA containing abasic sites. Biochemistry. 2004;43:15349–15357. doi: 10.1021/bi048400c. [DOI] [PubMed] [Google Scholar]
- 38).Ober M, Linne U, Gierlich J, Carell T. The two main DNA lesions 8 oxo 7,8 dihydroguanine and 2,6 diamino 5 formamido 4 hydroxypyrimidine exhibit strongly different pairing properties. Angew. Chem. Int. Ed. 2003;42:4947–4951. doi: 10.1002/anie.200351287. [DOI] [PubMed] [Google Scholar]
- 39).States DJ, Haberkorn RA, Ruben DJ. A two dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Mag. Res. 1982;48:286–292. [Google Scholar]
- 40).Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave forms and pulsed field gradients. J. Mag. Res. 1995;112:275–279. [Google Scholar]
- 41).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42).Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 43).Brünger A. XPLOR Version 3.1 A system for X Ray crystallography and NMR. Yale Univ. Press; New Haven, CT: 1993. [Google Scholar]
- 44).Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy minimization and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 45).Friedman RA, Honig B. The electrostatic contribution to dna base–stacking interactions. Biopolymers. 1992;32:145–159. doi: 10.1002/bip.360320205. [DOI] [PubMed] [Google Scholar]
- 46).Song K, Hornak V, de los Santos C, Grollman AP, Simmerling C. Molecular mechanics parameters for the FapydG DNA lesion. J. Comput. Chem. 2008;29:17–23. doi: 10.1002/jcc.20625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Yip P, Case DA. A new method for refinement of macromolecular structures based on nuclear Overhauser effect spectra. J. Mag. Res. 1989;83:643–648. [Google Scholar]
- 48).Zaliznyak T, Bonala R, Attaluri S, Johnson F, de los Santos C. Structure of duplex DNA containing α OH PdG: the mutagenic adduct produced by acrolein. Nucleic Acids Res. 2009;37:2153–2163. doi: 10.1093/nar/gkp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50).Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:655–667. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 51).van de Ven FJM, Hilbers CW. Nucleic acids and nuclear magnetic resonance. Eur. J. Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 52).de los Santos C. Comprehensive Natural Products Chemistry. Vol. 7. Elsevier Science Ltd.; Oxford: 1999. Probing DNA structure by NMR spectroscopy. Chapter 3. [Google Scholar]
- 53).Raoul S, Bardet M, Cadet J. γ Irradiation of 2′ deoxyadenosine in oxygen free aqueous solutions: identification and conformational features of formamidopyrimidine nucleoside derivatives. Chem. Res. Toxicol. 1995;8:924–933. doi: 10.1021/tx00049a005. [DOI] [PubMed] [Google Scholar]
- 54).Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6 Diamino 4 hydroxy 5 formamidopyrimidine) by formamidopyrimidine DNA glycosylase. J. Biol. Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 55).Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann J, Williams LD. X ray structure of a DNA decamer containing 7,8 dihydro 8 oxoguanine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Crenshaw CM, Wade JE, Arthanari H, Frueh D, Lane BF, Núñez ME. Hidden in plain sight: subtle effects of the 8 oxoguanine lesion on the structure, dynamics, and thermodynamics of a 15 base pair oligodeoxynucleotide duplex. Biochemistry. 2011;50:8463–8477. doi: 10.1021/bi201007t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.