Abstract
Since the food-borne pathogen Listeria monocytogenes is common in dairy farm environments, it is likely that phages infecting this bacterium (“listeriaphages”) are abundant on dairy farms. To better understand the ecology and diversity of listeriaphages on dairy farms and to develop a diverse phage collection for further studies, silage samples collected on two dairy farms were screened for L. monocytogenes and listeriaphages. While only 4.5% of silage samples tested positive for L. monocytogenes, 47.8% of samples were positive for listeriaphages, containing up to >1.5 × 104 PFU/g. Host range characterization of the 114 phage isolates obtained, with a reference set of 13 L. monocytogenes strains representing the nine major serotypes and four lineages, revealed considerable host range diversity; phage isolates were classified into nine lysis groups. While one serotype 3c strain was not lysed by any phage isolates, serotype 4 strains were highly susceptible to phages and were lysed by 63.2 to 88.6% of phages tested. Overall, 12.3% of phage isolates showed a narrow host range (lysing 1 to 5 strains), while 28.9% of phages represented broad host range (lysing ≥11 strains). Genome sizes of the phage isolates were estimated to range from approximately 26 to 140 kb. The extensive host range and genomic diversity of phages observed here suggest an important role of phages in the ecology of L. monocytogenes on dairy farms. In addition, the phage collection developed here has the potential to facilitate further development of phage-based biocontrol strategies (e.g., in silage) and other phage-based tools.
INTRODUCTION
Listeria monocytogenes is a Gram-positive pathogenic bacterium that can cause a severe food-borne disease, listeriosis, in humans and farm ruminants. L. monocytogenes has been isolated from a variety of environmental sources, e.g., water, soil, silage, vegetation, and food processing plants (3, 17, 18, 23, 29, 42). A number of studies have reported a high prevalence of L. monocytogenes in dairy farm environments (5, 19, 21, 33, 55). In addition, a previous study has found a considerably higher prevalence of L. monocytogenes in dairy farm environments than in urban and natural environments (45). Ruminants, including cattle, sheep, and goats, are not only often fecal shedders of L. monocytogenes but are also hosts in which L. monocytogenes can cause a severe disease (41). Silage (i.e., fermented plant material that is commonly used as feed for ruminants), if spoiled or improperly fermented, has often been found to contain L. monocytogenes (1, 20), including at high numbers (>107 CFU/g silage) (61). Spoiled silage has also been reported to be the most important source of L. monocytogenes responsible for listeriosis cases and outbreaks in ruminants (5, 20). The high prevalence of L. monocytogenes on dairy farms and particularly in silage not only suggests that these environments may represent a major reservoir for L. monocytogenes (34) but also suggests that silage may be a superior source for listeriaphage isolation.
Bacteriophages infecting L. monocytogenes and other Listeria spp. have been isolated from diverse sources (e.g., sewage, silage, water, and food processing plant environments) and from lysogenic L. monocytogenes strains (30, 35, 40). Listeriaphages isolated from different sources have also previously been evaluated for host range diversity. For example, Loessner and Busse (40) observed 16 different lysis patterns, which could be classified into four lysis groups, among 16 listeriaphages isolated from sewage or lysogenic strains. While most L. monocytogenes serotype 1/2a and 4b strains were lysed by at least one of these phages, the majority of serotype 3a, 3b, and 3c strains were resistant to all phages. In another study, Hodgson (30) found that 6/59 phages represented a broad host range, exhibiting the ability to lyse all 4 strains of serotype 1/2 and all 11 strains of serotype 4b tested. Similarly, Kim et al. (35) reported that 9/12 listeriaphages isolated from two turkey processing plants were characterized as broad-host-range phages, exhibiting the ability to lyse the majority of L. monocytogenes serotype 1/2a strains (16/26) and 4b strains (38/39). A number of listeriaphages from these and other studies have been well characterized, including by genome sequencing (10, 36, 64), and have been developed for biocontrol and other applications, such as phage A511 (27, 28) and P100 (10, 28, 51).
Recent studies suggest potential uses of listeriaphage as a biocontrol agent for L. monocytogenes in a variety of ready-to-eat (RTE) foods (10, 28, 31, 37, 38, 51). Some studies have also suggested the suitability of phage applications in controlling food-borne pathogens at the preharvest level and reducing shedding in animals (8, 9, 52). Only one study, by Kim et al. (35), has evaluated phage diversity in food processing plant environments; a better understanding of ecology and diversity of listeriaphage, including in primary food production environments, is thus still needed. Further establishment of diverse phage collections will also facilitate the development, improvement, and evaluation of listeriaphage-based biocontrol strategies. In this study, we used dairy farms as a model system in a longitudinal study to do the following: (i) gain a better understanding of the ecology and diversity of listeriaphages in farm environments, particularly in silage, and (ii) further develop listeriaphage collections.
MATERIALS AND METHODS
Sample collection.
A total of 134 silage samples were collected from silage bunkers of two dairy farms in New York State between October 2007 and July 2009. The two dairy farms were selected based on owners' willingness to allow frequent sample collection. No information on the previous prevalence of Listeria spp. or listeriaphages was available for these farms. For farm 1, two preliminary sampling visits were completed in October 2007 and January 2008, with 19 samples collected (Table 1). Phage recovery results for the preliminary visits are not reported here because these collected samples were used to optimize phage isolation procedures. At each sampling visit, 7 to 10 silage samples were collected from silage bunkers and placed in a sterile Whirl-Pak bag (Nasco, Modesto, CA). Only silage samples with a pH of >5.5 were collected, since a pH at this level indicates improperly fermented silage, increasing the likelihood of Listeria spp. and listeriaphage isolation. Silage samples used for isolation here showed pH values of 6 to 6.5.
Table 1.
Recovery of Listeria spp., L. monocytogenes, and listeriaphages from silage samples collected
| Farm and visit no. (sample collection date [mo/yr]) | No. of silage samples tested | No. of samples positive for: |
No. of samples that yielded plaques (ng) |
||
|---|---|---|---|---|---|
| Listeria spp.a | L. monocytogenes (nf) | Direct isolation | Enrichment method | ||
| Farm 1, preliminary samplingb | |||||
| 3 (10/2007) | 9 | 2 | 1 (2) | NA | NA |
| 4 (01/2008) | 10 | 2 | 3 (5) | NA | NA |
| Total | 19 | 4 | 4 (7) | NA | NA |
| Farm 1 | |||||
| 5 (04/2008) | 10 | 7 | 0 | 4 (7) | 4 (6) |
| 6 (08/2008) | 10 | 0 | 0 | 4 (7) | 3 (7) |
| 7 (09/2008) | 10 | 0 | 0 | 0 | 5 (0)c |
| 8 (10/2008) | 7 | 5 | 0 | 1 (2) | 1 (0)c |
| 9 (11/2008) | 8 | 3 | 0 | 3 (3) | 4 (6) |
| 10 (12/2008) | 9 | 2 | 0 | 1 (2) | 2 (3) |
| 11 (01/2009) | 8 | 1 | 0 | 2 (2) | 4 (4) |
| Total | 62 | 18 | 0 | 15 (23)d | 23 (26)d |
| Farm 2 | |||||
| 1 (02/2009) | 10 | 2 | 0 | 3 (6) | 5 (7) |
| 2 (03/2009) | 9 | 3 | 0 | 4 (6) | 4 (7) |
| 3 (04/2009) | 10 | 0 | 0 | 3 (3) | 1 (1) |
| 4 (05/2009) | 9 | 3 | 0 | 4 (7) | 5 (7) |
| 5 (06/2009) | 7 | 6 | 2 (2) | 2 (4) | 3 (5) |
| 6 (07/2009) | 8 | 0 | 0 | 2 (5) | 4 (7) |
| Total | 53 | 14 | 2 (2) | 18 (31)e | 22 (34)e |
Listeria spp. refers to Listeria spp. other than L. monocytogenes.
Preliminary sampling visits 1 to 4 were used to collect samples for optimizing phage isolation procedures; results for phage isolation from these preliminary efforts are not reported. Visits 3 and 4 also included silage samples that were tested for Listeria spp. and L. monocytogenes; results reported here as L. monocytogenes isolates were obtained only during visits 3 and 4 to farm 1.
Positive samples yielded no phages that could be propagated.
For farm 1, 12 samples were positive after enrichment only, while 4 samples were positive only by direct isolation and 11 samples were positive by both methods.
For farm 2, 10 samples were positive after enrichment only, while 6 samples were positive only by direct isolation and 12 samples were positive by both methods.
n, no. of isolates.
n, no. of phage isolates.
Isolation of L. monocytogenes.
Each silage sample (10 g) was transferred to a sterile Whirl-Pak bag and mixed with 90 ml of Listeria enrichment broth (LEB) (Difco, Becton, Dickinson, Sparks, MD). After 24 h and 48 h of incubation at 30°C, 50 μl of the enrichment was streaked onto Oxford plating medium (Difco, Becton, Dickinson, Sparks, MD), followed by incubation at 30°C for 48 h. For each sample, up to four Listeria-like colonies were substreaked onto L. monocytogenes plating medium (LMPM) (R-F Laboratories, Downers Grove, IL). Plates were incubated at 37°C for 48 h. On LMPM, L. monocytogenes and Listeria ivanovii appear as blue colonies, indicating phospholipase activity, while other Listeria spp. appear as white colonies (48). Blue colonies on LMPM plates were further characterized as detailed below to classify them as species and subtypes. Samples with white colonies representing Listeria-like characteristics were classified as positive for Listeria spp. other than L. monocytogenes.
Subtype characterization of putative L. monocytogenes isolates.
Isolated blue colonies from LMPM were substreaked on brain heart infusion (BHI) (Difco, Becton, Dickinson, Sparks, MD) agar plates for characterization by sigB allelic typing (14) and automated EcoRI ribotyping using the RiboPrinter system (Dupont Qualicon, Wilmington, DE). The RiboPrinter software classifies ribotype patterns into DuPont identifications (IDs) (e.g., DUP-1043), and a given DuPont ID can contain more than one distinct ribotype pattern (i.e., patterns that differ by a single weak band within a given DuPont ID). Different patterns within a given DuPont ID were designated with an additional letter (e.g., DUP-1043A and DUP-1043B).
L. monocytogenes isolates were also characterized using the standard CDC L. monocytogenes pulsed-field gel electrophoresis (PFGE) protocol (24, 25) with two restriction enzymes (ApaI and AscI). PFGE was performed using the Bio-Rad Chef Mapper electrophoresis unit. Images of PFGE patterns were acquired using the Bio-Rad Gel Doc software program, version 1.1, and analyzed using the BioNumerics software program, version 4.2 (Applied Maths, Sint-Martens-Latem, Belgium).
Bacterial strains and cultures for listeriaphage isolation.
Four L. monocytogenes strains, representing serotypes 1/2a, 1/2b, 4a, and 4b, were consistently used as hosts for listeriaphage isolation and enrichment (Table 2). These serotypes include the most common L. monocytogenes serotypes and have been used for listeriaphage isolation in other studies (30, 35, 40). While inclusion of L. monocytogenes isolates found on either farm would potentially improve detection of phages on a specific farm, this approach would have affected our ability to compare isolation frequencies or levels of phages between farms without bias.
Table 2.
L. monocytogenes strains used for listeriaphage isolation and phage host range determination
| L. monocytogenes strain (previous ID)a | Lineage | Source | Serotype | Ribotype | Reference(s) |
|---|---|---|---|---|---|
| FSL J1-175* | I | Water | 1/2b | DUP-1042A | 2 |
| FSL J1-169 | I | Human | 3b | DUP-1052A | 22, 26 |
| FSL J1-049 | I | Human | 3c | DUP-1042C | 22, 63 |
| FSL R2-574 (F2365)* | I | Food | 4b | DUP-1038B | 44 |
| FSL F6-367 (MACK)* | II | Lab strain | 1/2a | DUP-1030A | 30 |
| FSL C1-115 | II | Human | 3a | DUP-1039C | 22, 26 |
| FSL J1-094 | II | Human | 1/2c | 116-1501-S-4 | 4, 22 |
| FSL F2-695 | IIIA | Human | 4a | DUP-1061A | 49 |
| FSL F2-501 | IIIA | Human | 4b | DUP-18606 | 49 |
| FSL J2-071 | IIIA | Animal | 4c | DUP-1061A | 47, 49 |
| FSL W1-110 | IIIC | Unknown | 4b | DUP-1055 | 13, 22 |
| FSL J1-208* | IV | Animal | 4a | DUP-10142 | 49 |
| FSL J1-158 | IV | Animal | 4b | DUP-10142 | 13, 22 |
L. monocytogenes strains used as host strains for listeriaphage isolation are indicated with “*”; strains FSL J2-071, FSL J1-208, and FSL J1-158 were isolated from ruminants with clinical listeriosis symptoms.
An overnight broth culture of each host strain was prepared by inoculating an isolated colony from a BHI agar plate into 5 ml of LB MOPS (LB medium buffered with 50 mM morpholinepropanesulfonic acid [MOPS], pH 7.6). Cultures were incubated for 18 h at 30°C, with shaking at 220 rpm, to reach an optical density at 600 nm (OD600) of 0.5 to 0.6 (approximately 1 × 109 CFU/ml).
Isolation of listeriaphages.
Listeriaphage isolation was performed with the same samples used for L. monocytogenes isolation, using two methods: (i) direct phage isolation and (ii) phage isolation after enrichment. Phage isolation after enrichment was used to isolate phages that may be present at low levels, while direct isolation facilitated isolation of phages with distinct plaque morphologies and allowed phage quantification.
For direct phage isolation, silage samples (10 g) were mixed with 90 ml of phosphate-buffered saline (PBS), pH 7.4, in a sterile Whirl-Pak bag with a filtered screen (Nasco), followed by a manual homogenization. Each sample was then filtered through a 0.45-μm bottle-top filter, followed by filtration of a 1-ml aliquot through a 0.2-μm syringe filter. While we appreciate that recovery of some large phages may be jeopardized when using a 0.2-μm filter, this pore size has been used by others to isolate listeriaphages (35, 40). Filtrates from the 0.2-μm filter were used for phage isolation using the double-layer plate method (40), with minor modifications. Briefly, an overlay was prepared by mixing 300 μl of a 1:10 dilution of an overnight culture of a host strain (approximately 3 × 107 CFU/ml) with 100 μl of the sample filtrate and 4 ml of the soft agar, 0.7% LB MOPS/Glu/salts agarose (LB medium buffered with 50 mM MOPS, pH 7.6, and 10 mM [each] MgCl2 and CaCl2) (30). This overlay mixture was poured onto a freshly prepared bottom agar plate (1.5% LB MOPS/Glu/salts agarose). For each filtrate, this double-layer isolation was performed separately with each of the four host strains. Overlay plates were incubated at 30°C for 24 h, followed by phage purification as detailed below.
For phage isolation after enrichment, 10 g of silage was mixed with 90 ml of LB MOPS in a sterile Whirl-Pak bag with a filtered screen, followed by addition of 250 μl of separate overnight cultures for the four host strains, representing approximately 2.5 × 108 CFU of each host strain. The sample enrichment was incubated at 30°C for 24 h. An aliquot (100 μl) of each sample enrichment was used for sequential filtration and phage isolation as detailed above.
Phage purification and preparation of high-titer phage lysate stock.
One representative of each plaque morphology present on a given plate was used for phage purification. An isolated plaque was picked with a sterile Pasteur pipette and suspended in 100 μl of PBS. Four 10-fold serial dilutions of the plaque-PBS suspension were used to prepare overlay plates as described above. After incubation for 24 h at 30°C, the overlay plate yielding the lowest number of isolated plaques was used for two more phage purification passages. An isolated plaque from the third passage was used to prepare three overlay plates. After 24 h of incubation at 30°C, 5 ml of PBS was used to harvest the overlay, followed by addition of chloroform to a final concentration of 2% (vol/vol), centrifugation at 4,200 × g for 15 min, and filtration of the supernatant using a 0.2-μm syringe filter. While we appreciate that some phages may be sensitive to chloroform, phage titers sufficient for our experiments were obtained with this approach. Titers for each phage were determined by a spot test, performed at room temperature, using the respective host strain used for phage growth and eight 10-fold serial dilutions of the phage lysate (10 μl each). Phage titers were also used to determine the routine test dilution (RTD), which was defined as the highest dilution that just fails to give confluent lysis. Phage lysate stocks were stored at 4°C.
Listeriaphage host range determination.
Spot tests of the 114 phages isolated here were performed, as two independent replicates, with 13 L. monocytogenes reference strains (Table 2). These strains were chosen to represent the nine most common serotypes, as well as all four currently recognized L. monocytogenes lineages. Lawns for each reference strain were prepared as described above, and spot tests were performed with 10 μl of phage lysates adjusted to a 100× RTD, representing approximately 1 × 105 to 5 × 106 PFU/ml (see Table S1 in supplemental material). The absence of bacterium- inhibitory effects caused by high-titer phage suspensions was confirmed in the serial dilution spot tests detailed above. After 24 h of incubation at room temperature, each spot on the lawn was evaluated for lysis (+) or no lysis (−). Lysis was defined as the occurrence of multiple single plaques or turbid or confluent lysis at a spot.
Phage lysis profiles on the 13 host strains were used to identify clusters of phages with similar host ranges. For this analysis, a phage was considered to be lysing a host if plaquing was observed in at least one replicate. Hierarchical clustering was performed using Ward's method and binary distance in the R software program (version 2.14.0; R Development Core Team, Vienna, Austria [http://www.R-project.org]). Clusters with a reference approximately unbiased (AU) value of >45% were assigned a cluster designation (e.g., cluster A).
Listeriaphage genome size determination.
At least 25% of phage isolates obtained from each visit to a given farm (and at least one phage isolate from each visit) were selected for genome size determination. To the extent possible, phage isolates were selected from different silage samples and to represent multiple isolation hosts. DNA extraction was performed using phage lysates prepared as described above, except that SM (NaCl-MgSO4) buffer, pH 7.4, was used for phage harvest. Phages were precipitated using polyethylene glycol 8000 in the presence of 1 M NaCl, followed by resuspension in SM buffer. DNase I (Promega BioScience, San Luis Obispo, CA) (5 μg/ml, final concentration) and RNase A (Sigma) (30 μg/ml, final concentration) were added to digest nucleic acids from lysed bacterial cells. After addition of EDTA to a final concentration of 20 mM, phage DNA was purified using digestion with proteinase K (0.2 mg/ml) and SDS (0.5%), followed by extraction with phenol-chloroform and ethanol precipitation. Genome sizes were then estimated using PFGE as previously described (32, 56). Briefly, the gel was run for 22 h in 1× TBE buffer (Tris-borate-EDTA) (pH 8.0) at a 0.5-s to 5-s switch time, 6 V/cm, and an angle of 120°. Size standards of 8 to 48 kb and a λ PFGE marker (both from Bio-Rad, Hercules, CA) were used to facilitate estimation of genome sizes, which was performed using the software program BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium).
Statistical analysis.
To estimate odds ratios for phage susceptibility of serotype 4 and non-serotype 4 strains and of strains of different lineages, logistic regression was performed using a generalized linear model. The final model was then used to predict prevalences of phage susceptibility for strains with different characteristics, including 95% confidence intervals. All statistical analyses were performed using the R software program (version 2.14.0; R Development Core Team, Vienna, Austria [http://www.R-project.org]).
RESULTS
Despite infrequent isolation of L. monocytogenes, listeriaphages are commonly isolated from silage samples collected on dairy farms.
Among the 134 silage samples collected on two dairy farms (81 and 53 samples from farms 1 and 2, respectively), 4 samples from farm 1 and 2 samples from farm 2 were positive for L. monocytogenes. For farm 1, seven L. monocytogenes isolates obtained from four different samples collected during the two preliminary visits (October 2007 and January 2008; Table 1) were further characterized. These seven isolates represented four PFGE types, three sigB allelic types, and three ribotypes (see Fig. S1 in the supplemental material) and were classified into lineages I (three isolates) and II (four isolates). The two isolates from farm 2 represented the same sigB allelic types, the same ribotype, and the same PFGE type. In addition, Listeria spp. other than L. monocytogenes were isolated from a number of silage samples (Table 1).
Excluding the 19 samples collected during the two preliminary sampling visits to farm 1, a total of 115 silage samples (62 and 53 samples from farms 1 and 2, respectively) were screened for listeriaphages. Of these, 55 samples were positive for phages and 114 listeriaphage isolates were recovered, using four L. monocytogenes hosts and two phage isolation methods (i.e., direct isolation and isolation after enrichment). For farm 1, 27/62 samples were positive for phages, yielding 49 phage isolates (Table 1); 12 samples were positive after enrichment only, while 4 samples were positive only by direct isolation and 11 samples were positive by both methods. For this farm, 23 and 26 of the 49 phage isolates were obtained from direct isolation and isolation after enrichment, respectively. For farm 2, 28/53 samples were positive for phages, yielding 65 phage isolates (Table 1); 10 samples were positive after enrichment only, while 6 samples were positive only by direct isolation and 12 samples were positive by both methods. For this farm, 31 and 34 of the 65 phage isolates were obtained from direct isolation and isolation after enrichment, respectively.
The direct phage isolation method also allowed for enumeration of listeriaphages present in a given sample, with a detection limit of 1.0 × 102 PFU/g (Table 3). Phage levels in 15 samples from farm 1 that were positive by direct isolation ranged from 1.0 × 102 to 1.5 × 104 PFU/g, with two samples showing phage levels that were “too numerous to count” (TNTC) on at least one host strain (Table 3). For farm 2, phage levels in 18 samples that were positive by direct isolation ranged from 1.0 × 102 to 1.2 × 104 PFU/g, with 8 samples showing phage levels that were TNTC for at least one host strain. Due to variations in plaque sizes, TNTC could represent between 100 and 200 plaques per plate, and therefore TNTC is estimated to represent >2.0 × 104 PFU/g in our study.
Table 3.
Enumeration of listeriaphages for samples positive by direct phage isolation
| Farm and sample collection date (mo/yr) | Sample | Enumerationa (PFU/g) of listeriaphages on host strain (serotype) |
|||
|---|---|---|---|---|---|
| J1-175 (1/2b) | F2365 (4b) | MACK (1/2a) | J1-208 (4a) | ||
| Farm 1 | |||||
| 04/2008 | H-S5-S31D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 4.0 × 102 |
| H-S5-S32D | <1.0 × 102 | <1.0 × 102 | 2.0 × 102 | 5.3 × 103 | |
| H-S5-S39D | <1.0 × 102 | 4.3 × 103 | <1.0 × 102 | 1.2 × 103 | |
| H-S5-S40D | <1.0 × 102 | >2.0 × 104 | <1.0 × 102 | >2.0 × 104 | |
| 08/2008 | H-S6-S44D | <1.0 × 102 | 2.0 × 102 | <1.0 × 102 | 2.0 × 102 |
| H-S6-S46D | <1.0 × 102 | 5.0 × 103 | 1.0 × 103 | >2.0 × 104 | |
| H-S6-S47D | <1.0 × 102 | 3.0 × 102 | 1.0 × 102 | 1.0 × 103 | |
| H-S6-S50D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 4.0 × 102 | |
| 09/2008 | None | None | None | None | None |
| 10/2008 | H-S8-S64Db | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | (i) 7.0 × 102 |
| (ii) 2.6 × 103 | |||||
| 11/2008 | H-S9-S68D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 2.5 × 103 |
| H-S9-S72D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.1 × 103 | |
| H-S9-S73D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.2 × 103 | |
| 12/2008 | H-S10-S80D | <1.0 × 102 | 1.0 × 102 | <1.0 × 102 | 2.0 × 102 |
| 01/2009 | H-S11-S85D | <1.0 × 102 | 1.0 × 102 | <1.0 × 102 | <1.0 × 102 |
| H-S11-S90D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.5 × 104 | |
| Farm 2 | |||||
| 02/2009 | A-S1-S1D | <1.0 × 102 | 6.0 × 102 | 6.0 × 102 | 8.0 × 102 |
| A-S1-S8D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.2 × 103 | |
| A-S1-S10D | <1.0 × 102 | <1.0 × 102 | >2.0 × 104 | 1.0 × 104 | |
| 03/2009 | A-S2-S15D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | >2.0 × 104 |
| A-S2-S16D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | >2.0 × 104 | |
| A-S2-S17D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.2 × 104 | |
| A-S2-S18D | <1.0 × 102 | 1.0 × 102 | 1.0 × 102 | >2.0 × 104 | |
| 04/2009 | A-S3-S22D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.0 × 102 |
| A-S3-S23D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | >2.0 × 104 | |
| A-S3-S24D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | >2.0 × 104 | |
| 05/2009 | A-S4-S30D | <1.0 × 102 | <1.0 × 102 | <1.0 × 102 | 1.0 × 102 |
| A-S4-S31D | <1.0 × 102 | 4 × 102 | 1.2 × 103 | 3.2 × 103 | |
| A-S4-S34D | <1.0 × 102 | <1.0 × 102 | 2.0 × 102 | <1.0 × 102 | |
| A-S4-S36D | <1.0 × 102 | 1.0 × 102 | <1.0 × 102 | 1.0 × 102 | |
| 06/2009 | A-S5-S42D | <1.0 × 102 | 3.0 × 103 | 3.8 × 103 | >2.0 × 104 |
| A-S5-S43D | <1.0 × 102 | <1.0 × 102 | 1.0 × 102 | <1.0 × 102 | |
| 07/2009 | A-S6-S47D | <1.0 × 102 | 3.0 × 102 | 1.0 × 102 | <1.0 × 102 |
| A-S6-S48D | <1.0 × 102 | 8.0 × 102 | 3.3 × 103 | >2.0 × 104 | |
Samples that did not yield plaques on a given host were reported as <1.0 × 102 PFU/g of silage, the detection limit of the method used. Due to variations in plaque sizes, the presence of 100 to 200 plaques typically represented the cutoff for countable plaque numbers; samples that yielded plaques too numerous to be counted were thus reported as >2.0 × 104 PFU/g of silage.
This sample showed two plaque morphologies; number of PFU/g was reported for each type of plaque morphology, indicated as “(i)” and “(ii).”
Listeriaphages isolated here represent a wide diversity of host range characteristics.
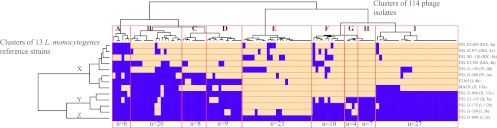
Host range determination of all 114 phage isolates, with 13 diverse L. monocytogenes strains (Table 2), classified these phage isolates into 56 different lysis profiles. Cluster analysis classified these lysis profiles into nine distinct lysis groups (Fig. 1; see also Table S2 in the supplemental material). Each lysis group included between 4 (lysis group G) and 27 (lysis group I) phage isolates. While most lysis groups were comprised of similar numbers of phage isolates from each farm, three groups (E, G, and H) included phage isolates predominantly from farm 2, and group I included phage isolates predominantly from farm 1 (see Table S2). Among the nine lysis groups, two groups (E and F, representing about 28.9% of phages tested) demonstrated a broad host range, exhibiting the ability to lyse 11 or 12 of the 13 L. monocytogenes strains. Only 5/49 phage isolates from farm 1 but 28/65 phage isolates from farm 2 fell into these two broad-host-range groups (see Table S2). Two lysis groups (A and C), representing narrow-host-range phages with the ability to lyse 1 to 5 strains, included 12.3% of the 114 phages characterized. The majority of the 114 phages (58.7%) showed the ability to lyse 6 to 10 strains tested and were classified into five lysis groups (B, D, and G to I).
Fig 1.
Heat map and hierarchical clustering of lysis profiles from the host range determination of 114 listeriaphages on 13 L. monocytogenes reference strains (Table 2). Beige represents lysis and blue represents no lysis on a given strain. Clusters of phage isolates are shown on the horizontal axis; clusters are designated A to I based on similarities of the lysis profiles with approximately unbiased (AU) values of >45%. Numbers of phage isolates grouped into each cluster are shown below the heat map. Host strains are shown on the vertical axis; clusters are designated X to Z based on similarities in susceptibility to phages.
Most listeriaphages lyse all serotype 4 strains, as well as the serotype 1/2a strain Mack.
Among the 13 reference strains, 7 strains, representing serotypes 4a (n = 2), 4b (n = 4), and 4c (n = 1), were lysed by 63.2 to 88.6% of the 114 phages (Table 4). Among the “non-serotype 4” strains, only the serotype 1/2a strain Mack was also lysed by a large proportion of phage isolates (74.6%), while the other, serotype 1/2b, 1/2c, 3a, and 3b strains were lysed by 22.8 to 40.4% of phage isolates. The serotype 3c strain FSL J1-049 was not lysed by any phage isolates (Table 4). Hierarchical clustering of these reference strains based on similarities in phage susceptibility was consistent with these findings. The seven serotype 4 strains and the serotype 1/2a strain Mack were classified into the same major cluster (X), while the serotype 3c strain FSL J1-049, which was highly resistant to all phages, was classified into its own cluster (Z) (Fig. 1). The other, serotype 1/2b, 1/2c, 3a, and 3b strains were grouped into cluster Y. The overall prevalence of phage susceptibility was 51.9% (95% confidence interval [CI], 35.5 to 67.8) among the non-serotype 4 strains and 88.9% (95% CI, 80.3 to 94.0) among the serotype 4 strains (Table 4), indicating a significant difference in phage susceptibility among these two groups (P < 0.001).
Table 4.
Susceptibilities of L. monocytogenes reference strains to listeriaphages
| L. monocytogenes strain | Serotype (lineage) | No. (%) of phage lysis groupsa lysing specific strain | No. (%) of phages lysing specific strain | % prevalence of phage susceptibilityb (95% CI) |
|---|---|---|---|---|
| Non-serotype 4 strains | 51.9 (35.5–67.8) | |||
| MACK | 1/2a (II) | 8 (89) | 85 (74.6) | |
| FSL J1-175 | 1/2b (I) | 4 (44) | 29 (25.4) | |
| FSL J1-094 | 1/2c (II) | 6 (67) | 46 (40.4) | |
| FSL C1-115 | 3a (II) | 6 (67) | 35 (30.7) | |
| FSL J1-169 | 3b (I) | 4 (44) | 26 (22.8) | |
| FSL J1-049 | 3c (I) | 0 | 0 | |
| Serotype 4 strains | 88.9 (80.3–94.0) | |||
| FSL F2-695 | 4a (IIIA) | 8 (89) | 101 (88.6) | |
| FSL J1-208 | 4a (IV) | 9 (100) | 101 (88.6) | |
| F2365 | 4b (I) | 8 (89) | 72 (63.2) | |
| FSL F2-501 | 4b (IIIA) | 8 (89) | 93 (81.6) | |
| FSL J1-158 | 4b (IV) | 8 (89) | 84 (73.7) | |
| FSL W1-110 | 4b (IIIC) | 7 (78) | 88 (77.2) | |
| FSL J2-071 | 4c (IIIA) | 8 (89) | 99 (86.8) |
See Table S2 in the supplemental material for details on the 9 lysis groups. A phage lysis group was classified as lysing a reference strain if any phages in a given lysis group showed lysis on a given host strain.
Prevalence of phage susceptibility (P < 0.001) among reference strains that were classified into non-serotype 4 strains and serotype 4 strains.
Strains of lineages III and IV were lysed by a large proportion of phage isolates (77.2 to 88.6%) (Table 4). This is consistent with the fact that all strains from these two lineages represent serotype 4. Lineage I strains showed considerable diversity regarding phage susceptibility, which ranged from 0 to 63.2%. Overall, the prevalence of phage susceptibility was higher among strains in lineages III (98.5%; 95% CI, 75.2 to 89.2) and IV (83.3%; 95% CI, 75.2 to 89.2) than among those in lineages I (68.1%; 95% CI, 47.4 to 83.4) and II (25.4%; 95% CI, 12.3 to 45.3). Consistent with the high phage susceptibility of serotype 4 as well as lineage III and IV strains, the majority of phages were isolated on the lineage IV serotype 4a host strain FSL J1-208 (60/114 phage isolates) and the lineage I serotype 4b host strain F2365 (25/114 phage isolates) (see Table S1 and Fig. S2 the in supplemental material).
Listeriaphages differ markedly in genome size, indicating genetic diversity of phages on dairy farms.
Phage genome sizes were determined for at least one phage isolate per visit to a given farm. Among 72 phage isolates tested (30 and 42 from farms 1 and 2, respectively), 10 (4 from farm 1 and 6 from farm 2) did not yield a clear band (or bands) after PFGE analysis, even though OD260 measurements suggested the presence of appropriate amounts of nucleic acid to yield a detectable band. These 10 phage isolates represented four different lysis groups. While further analysis on a 0.7% agarose gel showed a nucleic smear, suggesting a single-stranded RNA or DNA genome, additional experiments will be needed to characterize the genomes of these phage isolates.
The other 62 phage isolates showed a genome size range from 26 to 140 kb (Table 5). One phage from farm 2, classified into lysis group F, initially showed three bands, of approximately 41, 83, and 115 kb; PFGE analysis of this phage DNA after heating at 75°C for 15 min showed a single band at 40 kb, indicating the presence of cohesive ends that facilitated genome multimerization. For 35 phage isolates, PFGE analysis revealed two slightly blurred bands of similar sizes. The size difference of these two bands was approximately 3 to 6 kb. Twenty-three phage isolates from farm 1 showed the “two-band” pattern, with sizes of 58 to 64 kb for the small band and 63 to 68 kb for the large band; for farm 2, 12 phage isolates showed the “two-band” patterns, with sizes of 57 to 63 kb for the small band and 61 to 68 kb for the large band. These phages represented seven lysis groups (A to F and I). Although all phage lysates were prepared after purification for three passages, selected phage isolates with these two-band patterns were repurified but still maintained the same patterns. PFGE after heat treatment at 72°C for 15 min (performed for selected phages) also yielded the same patterns, suggesting that secondary structures (or the presence of cohesive ends) may not be responsible for the observed two-band patterns. While both bands typically showed different DNA concentrations, there was no consistent pattern such that either the larger or smaller band was always at a higher concentration (see Fig. S3 in the supplemental material). Full genome sequencing of four phage isolates from different lysis groups with these banding patterns (unpublished data) allowed assembly into a single genome of a size nearly the same as that of the larger band, suggesting the presence of a single phage. Phage genome size estimation of two selected phages by PFGE following an alternative protocol described in reference 39, which did not require phage DNA extraction prior to PFGE analysis, confirmed the double-band patterns in these two phages that previously showed these banding patterns.
Table 5.
Genome size diversity of selected listeriaphagesa
| Phage lysis group | Genome sizeb (kb) of representative phage isolates from each farm [visit no.c] |
|
|---|---|---|
| Farm 1 | Farm 2 | |
| A | 61/65 [6] | 57/61 [4] |
| 31 [9] | 58/63 [5] | |
| B | 62/65 [5] | 66; 60/65 [1] |
| 61/66 [6] | 65 [2] | |
| 58/63 [8] | 58/63 [3] | |
| 60/66 [10] | 57/62 [4] | |
| 61/65 [11] | 60/65 [5] | |
| 60/63 [6] | ||
| C | 64/68 [6] | 63 [1] |
| 62/67 [10] | 61/65; 63/68 [2] | |
| D | 62/67 [6] | 62; 63; 60/65 [1] |
| 61/66; 63/68 [11] | 61/65 [2] | |
| 68 [6] | ||
| E | ND | 97; 119 [1] |
| 140 [5] | ||
| 59/63; 70; 117; 127; 131; 132; 134; 135; 136 [6] | ||
| F | 61/65 [5] | 121 [1] |
| 59/63; 61/67 [9] | 64 [2] | |
| 41/83/115 [4] | ||
| G | None | 123 [4] |
| H | 123 [9] | 32 [2] |
| 26 [4] | ||
| I | 59/63 [5] | 32 [2] |
| 33; 60/64; 61/65 [6] | ||
| 58/64 [8] | ||
| 62/66 [9] | ||
| 61/65 [10] | ||
At least 25% of phage isolates obtained from each visit to a given farm were selected for genome size estimation; for each visit, at least one isolate was characterized. To the extent possible, phage isolates were selected to represent multiple isolation hosts.
Phage genome sizes were estimated by PFGE analysis and size estimation using the BioNumerics software program. “None” indicates that no phage isolate was classified into this lysis group; “ND” indicates that genome size determination was not performed with phage isolates of this lysis group. For some phage isolates, two bands of similar sizes were observed by PFGE analysis, and the estimated sizes for both bands are indicated (e.g., 60/65 kb).
See Table 1 for details on farm sampling visits.
Overall, all nine lysis groups included phages with various genome sizes (Table 5). Genome size diversity was also observed among phages from the same farm that grouped into a given lysis profile. For example, phages in lysis group F from farm 2 revealed three distinct genome sizes (Table 5). These findings suggest that phages exhibiting similar host ranges, even among phages from the same farm, still show considerable genetic diversity. Genome sizes of phages from farm 1 ranged from approximately 31 kb (one phage of lysis group A) to 123 kb (one phage of lysis group H). Among phages from farm 2, the smallest phage genome size was approximately 26 kb (one phage of lysis group H), while 12/42 phage isolates, classified into three lysis groups (E to G), showed large genome sizes, with the range of 97 to 140 kb.
Combined analysis of phage genome size and lysis patterns of phages from a given farm showed that phages with the same genome size and lysis pattern were isolated over multiple sampling visits. For example, for farm 1, phages representing genomes of the “two-band” patterns (approximately 60 kb and 65 kb), classified into lysis group I, were isolated from samples collected during five visits to farm 1 (Table 5). For farm 2, phages that grouped into lysis group B and showed these two-band patterns were also isolated over multiple visits. While data on genome sizes and host range patterns indicate reisolation of the same or similar phages from a given farm over time, further analysis of these phages (e.g., restriction fragment length polymorphism [RFLP] analysis) is needed to assess their similarities.
DISCUSSION
In this study, we used dairy farms as a model system to develop a better understanding of the ecology and diversity of listeriaphages, with a focus on silage, which is well established to support growth of L. monocytogenes to high levels and to be a source associated with animal listeriosis. Our data specifically demonstrate the following: (i) listeriaphages are abundant in silage available on dairy farms, (ii) L. monocytogenes lineage III and IV and serotype 4 strains are highly susceptible to phages, and (iii) except for a largely conserved ability to lyse serotype 4 strains, listeriaphages show considerable host range and genome size diversity. The diverse phage collection described here also represents a promising resource for further development of listeriaphages as a biocontrol agent (e.g., to control L. monocytogenes in silage) and other phage-based applications and for further genomic studies of listeriaphages.
Listeriaphages are abundant in farm environments.
While phages are in general well known to be the most abundant entities in the environment (6, 7, 50), the relative abundance of species-specific phages (e.g., listeriaphages) in different environments is less well studied. In our study, listeriaphages were isolated from the majority of silage samples, with some samples representing phage levels of >1.5 × 104 PFU/g of silage. A possible explanation for detection of phages, in some samples, by direct isolation but not by enrichment would be either degradation of phages during enrichment (e.g., due to proteases or nucleases present in the enrichment or produced by bacteria other than the host strains) or entry into a lysogenic cycle during enrichment. Interestingly, a high prevalence of phages infecting L. monocytogenes was observed despite the fact that the majority of silage samples were not positive for L. monocytogenes, possibly suggesting that L. monocytogenes host populations were regulated by lysis through the phages present. On the other hand, since Listeria spp. other than L. monocytogenes were isolated from a number of silage samples, other Listeria spp. may be hosts that facilitated replication of these phages. In addition, it is possible that members of other, closely related Gram-positive bacterial genera could serve as natural hosts of listeriaphages, as supported by the finding that some Staphylococcus aureus phages had been shown to facilitate horizontal transfer of DNA into Listeria (11). Further phage host range characterization with other potential hosts, particularly Listeria spp. isolates, would be needed to better understand whether hosts other than L. monocytogenes could facilitate propagation of phages isolated here.
While silage samples have previously been used to isolate listeriaphages for further characterization (30) and while it is well known that poorly fermented silage is commonly contaminated with high levels of L. monocytogenes (17, 18, 21, 29), prevalences and levels of listeriaphages in silage have not previously been reported. In one previous study that reported listeriaphage prevalence among samples collected from two turkey processing plants, 12 listeriaphage isolates were obtained from 8 out of 113 samples tested (35). The high prevalence of listeriaphages observed in silage samples here not only suggests that improperly fermented silages and possibly dairy farm environments in general are good substrates for listeriaphage isolation but also suggests that phage-mediated horizontal gene transfer in L. monocytogenes may be particularly frequent in these environments. This hypothesis is consistent with the previous finding that lineage III and IV L. monocytogenes strains, which are highly susceptible to phages (see below) and are most common among ruminants, also show a comparatively high level of horizontal gene transfer (43, 46, 49).
L. monocytogenes lineage III and IV strains (serotypes 4a, 4b, and 4c) are highly susceptible to phages and represent superior hosts for phage isolation.
Host range determination of the 114 phage isolates showed that L. monocytogenes lineage III and IV strains (these strains represent serotype 4a, 4b, or 4c), as well as the only lineage I serotype 4b strain included in our host strain set, were lysed by the majority of our phage isolates. These observations are consistent with a number of previous studies (30, 35, 40, 58), including a study by Loessner and Busse (40), who reported that most serotype 4 strains (96%) were sensitive to at least 1 of the 16 phages tested. Kim et al. (35) also found that serotype 4b strains were typically sensitive to most phages isolated from the turkey processing plants. Somewhat contradictory to our findings, Shen et al. (54) reported that 5/8 L. monocytogenes isolates, classified into serogroup 4b based on PFGE typing, showed resistance to a listeriaphage cocktail consisting of 6 phages. Our study also showed that the serotype 1/2a strain Mack (classified into lineage II) was lysed by most phages. This finding is consistent with the study by Kim et al. (35) that found the majority of serotype 1/2a strains (16/26) to be sensitive to most phages tested. The findings that the one serotype 3c strain evaluated was resistant to all phages tested here and that the serotype 3a and 3b host strains were resistant to a considerable number of phages are consistent with a previous report by Loessner and Busse (40) that serotype 3a, 3b, and 3c strains were typically untypeable by phage typing due to their resistance to all 16 phages tested. Kim et al. (35) also found that all three isolates from turkey processing plants representing serotypes 3c or 1/2c were not lysed by phage A511 and two broad-host-range listeriaphages obtained from the same environment. Moreover, Shen et al. (54) found that 11/51 L. monocytogenes isolates classified, based on PFGE typing, into serogroup 3b or 1/2b were resistant to a listeriaphage cocktail. While specific mechanisms of phage resistance for serotype 3 and 1/2c strains remain unknown, cell wall teichoic acids (TA) and glucosamine in particular have been shown to be receptors for listeriaphages, and the absence or alteration of this TA substituent can convey phage resistance (e.g., see the work of Wendlinger et al. [60]).
Overall, our data not only provide further evidence that, on a population basis, L. monocytogenes serotypes differ in phage resistance but also suggest that selection of an L. monocytogenes strain(s) as a host(s) for phage isolation can considerably affect phage isolation frequency. Lineage III and IV and serotype 4b strains, as well as the serotype 1/2a strain Mack, are likely to facilitate better phage recovery and thus are highly recommended as hosts for phage isolation. In addition, the use of serotypes that are typically resistant to phages as hosts for phage isolation will facilitate isolation of phages that may be able to lyse these strains, which would be important for biocontrol and other applications.
Except for a largely conserved ability to lyse serotype 4 strains, listeriaphages show considerable host range and genome size diversity.
Host range determination of the 114 phage isolates showed that these phage isolates could be classified into nine lysis groups. Lysis groups E and F, which included broad-host-range phages with the ability to lyse 11 or 12 strains, accounted for 28.9% of the 114 phages. By comparison, Loessner and Busse (40) found that only 3/16 phages characterized in their study were classified into the broad-host-range phage group, whereas most phages in their collection represented a narrow host range (lysis of 9 to 21 of 57 strains). Interestingly, all broad-host-range phages in their study (40) were isolated from environmental samples (i.e., sewage). Similarly, all six broad-host-range phages described by Hodgson (30) were isolated from sewage and silage samples. However, a study by Kim et al. (35) reported that the majority of phages (i.e., 9/12) from the turkey processing plants were classified in the broad-host-range group, with the ability to lyse all 27 L. monocytogenes strains and 4/5 Listeria spp. tested. Differences in sources of phages and protocols, including host strains used for enrichment and phage isolation, may contribute to the differences in host ranges observed among the phages from these studies.
While a considerable number of listeriaphages (>400 phages) have previously been isolated and characterized, genome sizes of <20 listeriaphages have been determined using PFGE analysis or genome sequencing (10, 15, 30, 36). The majority of previously reported listeriaphages showed genome sizes with a range of 35.6 kb (phage P40; accession no. EU855793) to 48.2 kb (phage B054; accession no. DQ003640). No previous listeriaphage genome between 50 and 130 kb has been reported, except in the most recent study of phage P70, which showed the genome size of approximately 67.1 kb (53). Two Myoviridae-family listeriaphages showed large genome sizes of 131.4 kb (phage P100; accession no. DQ004855) and 137.6 kb (phage A511; accession no. DQ003638). By comparison, the 72 phages whose genome sizes were determined here showed genome sizes ranging from approximately 26 to 140 kb, including several phages with genome sizes between 55 and 70 kb. A number of phages isolated in the current study thus show genome sizes that have rarely been found among listeriaphages. Interestingly, a number of phages characterized here showed two bands of similar sizes in the genome size determination experiments. While we cannot completely exclude that these two bands represent an experimental artifact (e.g., the presence of single- and double-stranded DNA in the DNA prep), we have excluded the presence of cohesive ends and have found that sequence generated from several of these phages assembled into a single genome, excluding a contaminating phage as an explanation. We thus propose that these double bands may be due to a packaging mechanism that yields two chromosome variants. For example, the phages with these patterns may represent two capsid size variants of “headful packaging” phages, which could lead to packaging of two chromosome lengths. Packaging of different chromosome sizes can occur in genomes that are terminally redundant and circularly permuted, as observed in phages P1, P22, and T4 (12, 57, 59, 62). While this hypothesis is consistent with the data for T4, which has been shown to form a petite variant that could be more or less common than the full-size capsid, e.g., depending on time after infection (16), further characterization of these phages will be necessary.
The phage collection developed here will provide opportunities for further studies of the genomics and biology of listeriaphages, in addition to providing a potential initiation of further development of phage-based biocontrol strategies (e.g., control of L. monocytogenes in silage) and other applications. However, additional comprehensive characterization of these phages is necessary for identification of specific phages appropriate for these applications. For example, full-genome sequencing is particularly needed to confirm that phages to be used as a biocontrol agent do not carry antibiotic resistance or putative virulence genes.
Supplementary Material
ACKNOWLEDGMENTS
The project was supported by USDA-CSREES Hatch Funds (NYC-143445) and the Royal Thai Government Fellowship (to K.V.).
We thank Richard Calendar of UC Berkeley for providing a strain of L. monocytogenes and for his helpful suggestions on phage work. We also thank Matthew Stasiewicz and Pajau Vangay for their help with statistical analysis.
Footnotes
Published ahead of print 5 October 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Arimi SM, Ryser ET, Pritchard TJ, Donnelly CW. 1997. Diversity of Listeria ribotypes recovered from dairy cattle, silage, and dairy processing environments. J. Food Prot. 60:811–816 [DOI] [PubMed] [Google Scholar]
- 2. Bergholz TM, den Bakker HC, Fortes ED, Boor KJ, Wiedmann M. 2010. Salt stress phenotypes in Listeria monocytogenes vary by genetic lineage and temperature. Foodborne Pathog. Dis. 7:1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beuchat LR. 1996. Listeria monocytogenes: incidence on vegetables. Food Control 7:223–228 [Google Scholar]
- 4. Bille J, Rocourt J. 1996. WHO international multicenter Listeria monocytogenes subtyping, study-rationale and set-up of the study. Int. J. Food Microbiol. 32:251–262 [DOI] [PubMed] [Google Scholar]
- 5. Borucki MK, et al. 2005. Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm. Appl. Environ. Microbiol. 71:5893–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278–284 [DOI] [PubMed] [Google Scholar]
- 7. Brüssow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16 [DOI] [PubMed] [Google Scholar]
- 8. Callaway TR, et al. 2003. Preslaughter intervention strategies to reduce food-borne pathogens in food animals. J. Anim. Sci. 81:E17–E23 [Google Scholar]
- 9. Callaway TR, et al. 2004. Recent pre-harvest supplementation strategies to reduce carriage and shedding of zoonotic enteric bacterial pathogens in food animals. Anim. Health Res. Rev. 5:35–47 [DOI] [PubMed] [Google Scholar]
- 10. Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301–312 [DOI] [PubMed] [Google Scholar]
- 11. Chen J, Novick RP. 2009. Phage-mediated intergeneric transfer of toxin genes. Science 323:139–141 [DOI] [PubMed] [Google Scholar]
- 12. Coren JS, Pierce JC, Sternberg N. 1995. Headful packaging revisited: the packaging of more than one DNA molecule into a bacteriophage P1 head. J. Mol. Biol. 249:176–184 [DOI] [PubMed] [Google Scholar]
- 13. De Jesus AJ, Whiting RC. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611–1617 [DOI] [PubMed] [Google Scholar]
- 14. den Bakker HC, Fortes ED, Wiedmann M. 2010. Multilocus sequence typing of outbreak-associated Listeria monocytogenes isolates to identify epidemic clones. Foodborne Pathog. Dis. 7:257–265 [DOI] [PubMed] [Google Scholar]
- 15. Dorscht J, et al. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eiserling FA, Geiduschek EP, Epstein RH, Metter EJ. 1970. Capsid size and deoxyribonucleic acid length: the petite variant of bacteriophage T4. J. Virol. 6:865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fenlon DR. 1985. Wild birds and silage as reservoirs of Listeria in the agricultural environment. J. Appl. Bacteriol. 59:537–543 [DOI] [PubMed] [Google Scholar]
- 18. Fenlon DR. 1999. Listeria monocytogenes in the natural environment, p 21–38 Listeria, listeriosis, and food safety, 2nd ed Marcel Dekker, Inc., New York, NY [Google Scholar]
- 19. Fenlon DR, Stewart T, Donachie W. 1995. The incidence, numbers and types of Listeria monocytogenes isolated from farm bulk tank milks. Lett. Appl. Microbiol. 20:57–60 [DOI] [PubMed] [Google Scholar]
- 20. Fenlon DR, Wilson J, Donachie W. 1996. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Microbiol. 81:641–650 [DOI] [PubMed] [Google Scholar]
- 21. Fox E, et al. 2009. Listeria monocytogenes in the Irish dairy farm environment. J. Food Prot. 72:1450–1456 [DOI] [PubMed] [Google Scholar]
- 22. Fugett E, Fortes E, Nnoka C, Wiedmann M. 2006. International Life Sciences Institute North America Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929–2938 [DOI] [PubMed] [Google Scholar]
- 23. Gianfranceschi M, Gattuso A, Tartaro S, Aureli P. 2003. Incidence of Listeria monocytogenes in food and environmental samples in Italy between 1990 and 1999: serotype distribution in food, environmental and clinical samples. Eur. J. Epidemiol. 18:1001–1006 [DOI] [PubMed] [Google Scholar]
- 24. Graves LM, Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55–62 [DOI] [PubMed] [Google Scholar]
- 25. Graves LM, Swaminathan B. 2006. PulseNet's step-by-step laboratory protocol for molecular subtyping of Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis methods. Biotechnology 21:57–72 [DOI] [PubMed] [Google Scholar]
- 26. Gray MJ, et al. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guenther S, Loessner MJ. 2011. Bacteriophage biocontrol of Listeria monocytogenes on soft ripened white mold and red-smear cheeses. Bacteriophage 1:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guenther S, Huwyler D, Richard S, Loessner MJ. 2009. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho AJ, Ivanek R, Grohn YT, Nightingale KK, Wiedmann M. 2007. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev. Vet. Med. 80:287–305 [DOI] [PubMed] [Google Scholar]
- 30. Hodgson DA. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312–323 [DOI] [PubMed] [Google Scholar]
- 31. Holck A, Berg J. 2009. Inhibition of Listeria monocytogenes in cooked ham by virulent bacteriophages and protective cultures. Appl. Environ. Microbiol. 75:6944–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holmfeldt K, Middelboe M, Nybroe O, Riemann L. 2007. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microbiol. 73:6730–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Husu JR. 1990. Epidemiological studies on the occurrence of Listeria monocytogenes in the feces of dairy cattle. Zentralbl. Veterinarmed. B 37:276–282 [DOI] [PubMed] [Google Scholar]
- 34. Ivanek R, Gröhn YT, Wiedmann M. 2006. Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog. Dis. 3:319–336 [DOI] [PubMed] [Google Scholar]
- 35. Kim JW, Siletzky RM, Kathariou S. 2008. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl. Environ. Microbiol. 74:6623–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klumpp J, et al. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leverentz B, Conway WS, Janisiewicz W, Camp MJ. 2004. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. J. Food Prot. 67:1682–1686 [DOI] [PubMed] [Google Scholar]
- 38. Leverentz B, et al. 2003. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 69:4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lingohr E, Frost S, Johnson RP. 2009. Determination of bacteriophage genome size by pulsed-field gel electrophoresis. Methods Mol. Biol. 2:19–25 [DOI] [PubMed] [Google Scholar]
- 40. Loessner MJ, Busse M. 1990. Bacteriophage typing of Listeria species. Appl. Environ. Microbiol. 56:1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loken T, Aspoy E, Gronstol H. 1982. Listeria monocytogenes excretion and humoral immunity in goats in a herd with outbreaks of listeriosis and in a healthy herd. Acta Vet. Scand. 23:392–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lyautey E, et al. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73:5401–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meinersmann RJ, Phillips RW, Wiedmann M, Berrang ME. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson KE, et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nightingale KK, et al. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orsi RH, Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301:79–96 [DOI] [PubMed] [Google Scholar]
- 47. Pohl MA, Wiedmann M, Nightingale KK. 2006. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am. J. Vet. Res. 67:616–626 [DOI] [PubMed] [Google Scholar]
- 48. Restaino L, Frampton EW, Irbe RM, Schabert G, Spitz H. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food Prot. 62:244–251 [DOI] [PubMed] [Google Scholar]
- 49. Roberts A, et al. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685–693 [DOI] [PubMed] [Google Scholar]
- 50. Rohwer F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 51. Rossi LPR, et al. 2010. Occurrence of Listeria spp. in Brazilian fresh sausage and control of Listeria monocytogenes using bacteriophage P100. Food Control 22:954–958 [Google Scholar]
- 52. Sargeant JM, Amezcua MR, Rajic A, Waddell L. 2007. Pre-harvest interventions to reduce the shedding of E. coli O157 in the faeces of weaned domestic ruminants: a systematic review. Zoonoses Public Health 54:260–277 [DOI] [PubMed] [Google Scholar]
- 53. Schmulki MM, Erne D, Loessner MJ, Klumpp J. 19 September 2012. Bacteriophage P70: unique morphology and unrelatedness to other Listeria bacteriophages. J. Virol. doi:10.1128/JVI.02350-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shen Y, et al. 2006. Isolation and characterization of Listeria monocytogenes isolates from ready-to-eat foods in Florida. Appl. Environ. Microbiol. 72:5073–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skovgaard N, Morgen CA. 1988. Detection of Listeria spp. in faeces from animals, in feeds, and in raw foods of animal origin. Int. J. Food Microbiol. 6:229–242 [DOI] [PubMed] [Google Scholar]
- 56. Stenholm AR, Dalsgaard I, Middelboe M. 2008. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 74:4070–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Streisinger G, Emrich J, Stahl MM. 1967. Chromosome structure in phage T4, iii. Terminal redundancy and length determination. Proc. Natl. Acad. Sci. U. S. A. 57:292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sword CP, Pockett MJ. 1961. The isolation and characterization of bacteriophages from Listeria monocytogenes. J. Gen. Microbiol. 25:241–248 [DOI] [PubMed] [Google Scholar]
- 59. Tye BK, Huberman JA, Botstein D. 1974. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 85:501–527 [DOI] [PubMed] [Google Scholar]
- 60. Wendlinger G, Loessner MJ, Scherer S. 1996. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 142:985–992 [DOI] [PubMed] [Google Scholar]
- 61. Wiedmann M, et al. 1999. Molecular investigation of a listeriosis outbreak in goats caused by an unusual strain of Listeria monocytogenes. J. Am. Vet. Med. Assoc. 215:369–371 [PubMed] [Google Scholar]
- 62. Wu H, Sampson L, Parr R, Casjens S. 2002. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45:1631–1646 [DOI] [PubMed] [Google Scholar]
- 63. Zhang W, Jayarao BM, Knabel SJ. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.