Abstract
The importance of anaerobic oxidation of methane (AOM) as a methane sink in freshwater systems is largely unexplored, particularly in peat ecosystems. Nitrite-dependent anaerobic methane oxidation (n-damo) was recently discovered and reported to be catalyzed by the bacterium “Candidatus Methylomirabilis oxyfera,” which is affiliated with the NC10 phylum. So far, several “Ca. Methylomirabilis oxyfera” enrichment cultures have been obtained using a limited number of freshwater sediments or wastewater treatment sludge as the inoculum. In this study, using stable isotope measurements and porewater profiles, we investigated the potential of n-damo in a minerotrophic peatland in the south of the Netherlands that is infiltrated by nitrate-rich ground water. Methane and nitrate profiles suggested that all methane produced was oxidized before reaching the oxic layer, and NC10 bacteria could be active in the transition zone where countergradients of methane and nitrate occur. Quantitative PCR showed high NC10 bacterial cell numbers at this methane-nitrate transition zone. This soil section was used to enrich the prevalent NC10 bacteria in a continuous culture supplied with methane and nitrite at an in situ pH of 6.2. An enrichment of nitrite-reducing methanotrophic NC10 bacteria was successfully obtained. Phylogenetic analysis of retrieved 16S rRNA and pmoA genes showed that the enriched bacteria were very similar to the ones found in situ and constituted a new branch of NC10 bacteria with an identity of less than 96 and 90% to the 16S rRNA and pmoA genes of “Ca. Methylomirabilis oxyfera,” respectively. The results of this study expand our knowledge of the diversity and distribution of NC10 bacteria in the environment and highlight their potential contribution to nitrogen and methane cycles.
INTRODUCTION
Wetlands are the largest single source of methane, with estimated emissions of 103 Tg per year, which account for about 20 to 40% of the global annual atmospheric methane flux (1, 8, 19). It is estimated that about 50% of the methane produced in wetlands is consumed before it reaches the atmosphere; this significant microbial methane sink is usually considered to consist exclusively of aerobic methanotrophic bacteria, which degrade methane using oxygen as the electron acceptor (2, 5, 19, 39). In ecosystems where oxygen is depleted but sufficient alternative electron acceptors, e.g., sulfate or nitrate, are present, methane can also be converted anaerobically (25, 38). Anaerobic oxidation of methane (AOM) coupled to sulfate reduction is performed by a consortium of anaerobic methanotrophic archaea (ANME) and sulfate-reducing bacteria (SRB) (25, 47). Its significance is well established for marine ecosystems, where it may consume more than 90% of the produced methane (39). In freshwater wetlands, and especially peatlands, electron acceptors are more scarce, with concentrations typically in the low μM range (37). For this reason, redox processes are mostly limited by electron acceptor supply and are very dynamic and highly susceptible to alterations, e.g., by influx of polluted groundwater and atmospheric deposition of nitrogen and sulfur species (18, 46). The influence of nitrogen pollution on methane oxidation is complex, and not all feedback loops are well understood (2, 3, 16, 29). In principal, the role of the alternative electron acceptors nitrate and sulfate for diverting carbon fluxes away from methane production is better established, given that sulfate and nitrate reduction are thermodynamically more favorable than methanogenesis (17, 30, 31, 51). However, these alternative electron acceptors can also enable methane oxidation (47, 54), but this topic has received very little attention with respect to methane cycling in peatlands (43).
In the meantime, for other freshwater ecosystems, more and more evidence about the occurrence of AOM coupled to sulfate (11, 40), Fe(III) (42), and nitrate reduction (9, 38, 44, 50) has become available. Whereas nothing is known about the microorganisms mediating ferric iron reduction with methane, for sulfate reduction a consortium of methanotrophic Archaea and SRB very similar to that in marine ecosystems is hypothesized to be responsible (11, 40). Nitrate- or nitrite-dependent AOM (n-damo), when linked to organisms, so far has always been found to be performed by one bacterial species affiliated with the NC10 phylum (9, 13). Genome sequencing, expression studies, and physiological experiments indicated that this bacterium, named “Candidatus Methylomirabilis oxyfera,” is an intra-aerobic methanotroph that produces its own oxygen from the dismutation of nitric oxide into dinitrogen gas and oxygen. The produced oxygen is then used for canonical aerobic methane oxidation starting with the methane monooxygenase enzyme complex (12). Although 16S rRNA sequences similar to that of “Ca. Methylomirabilis oxyfera” were found in various environments (14), so far n-damo enrichment cultures have only been obtained from two types of ecosystems: eutrophic freshwater sediments and wastewater treatment sludge. The dominant bacteria in all described cultures were closely related (≥97% identity of the 16S rRNA gene sequence) to “Ca. Methylomirabilis oxyfera” (13, 14, 20, 33). Currently it is unclear, however, if “Ca. Methylomirabilis oxyfera”-related species are the only nitrite-dependent methane-oxidizing microorganisms, if methane oxidation is a general feature of NC10 phylum bacteria or is limited to (close relatives of) “Ca. Methylomirabilis oxyfera,” and how important these bacteria are for methane cycling in various ecosystems.
In this paper, we studied a minerotrophic peatland infiltrated by nitrate-containing groundwater. At the sampling site, no methane emission was detectable. Porewater profiling revealed a nitrate-methane transition zone below the oxic layer that could provide an ecological niche for n-damo microorganisms. NC10 bacterial abundance in soil cores was analyzed using quantitative PCR, and the section with the highest cell numbers of “Ca. Methylomirabilis oxyfera,” coinciding with the methane-nitrate transition zone, was used as the inoculum for the enrichment of n-damo bacteria. By mimicking field conditions as much as possible by using nitrite-amended peatland water in continuous cultivation, a new cluster of “Ca. Methylomirabilis oxyfera”-like bacteria was enriched.
MATERIALS AND METHODS
Site description.
The Brunssummerheide peatland (50°55′39.63″N, 5°59′50.73″E) is a small (15 ha) spring fen located in an oligotrophic sandy valley fed by locally upwelling, weakly buffered, nitrate-polluted groundwater. The peat layer is relatively thin (maximum of 2.5 m), and vegetation is dominated by Sphagnum species, Narthecium ossifragum, and Molinia caerulea. At the sampling site, nitrate-enriched groundwater overflows the peatland surface and infiltrates into the peat layer.
Porewater profile determination and soil sampling.
Nitrate and methane profiles were determined by measuring the concentrations in porewater samples collected using 5-cm ceramic cups (Eijkelkamp Agrisearch Equipment, Giesbeek, the Netherlands) connected to Teflon tubes. Porewater samples were obtained at least in duplicate from a depth of 20 to 220 cm at 5- or 10-cm intervals in December 2009 and June 2010. Porewater for methane analyses was collected in vacuumed anaerobic glass bottles (40 ml) prefilled with 5 g sodium chloride and sealed with butyl rubber stoppers. For chemical analyses, porewater was collected in 60-ml syringes. Samples were transported to the laboratory within 2 h in a cooling box and stored at 4°C for a maximum of 14 days before analysis. Methane in the bottle headspace was measured after pressure equilibration with argon using gas chromatography as described previously (14). Nitrate was analyzed colorimetrically on a Traacs 800+ autoanalyzer as described previously (48). Redox potential measurements were performed by gently pushing platinum electrodes into predrilled holes and allowing them to equilibrate. Stable readings were obtained after 30 min (15). Soil samples were obtained from 50- to 130-cm depth with a Russian peat corer, sliced into 5- to 20-cm intervals in the field, immediately put into self-sealing plastic bags, stored in air-tight bins with an oxygen scavenger (Anaerogen; Oxoid), and then transported to the laboratory and stored anaerobically at 4°C until further analysis.
Incubation.
Initially, 200 ml soil slurry of the depth layers of 80 to 100 cm, 100 to 120 cm, and 120 to 135 cm (sampled in July 2009) were incubated in separate bottles (500 ml). Surface water from the peatland was collected and used for medium preparation after removal of particles by filtering through a hemofilter (Hemoflow HF80S; Fresenius Medical Care). The medium contained 2 mM KHCO3, 0.2 mM Na15NO2 (99.6% 15N; Isotec), and 0.5 mM NaNO3. The bottles were made anaerobic by 6 cycles of vacuuming and gassing with Ar-CO2 (75:25), followed by 5 min of flushing with Ar-CO2. 13CH4 (10 ml) then was injected into the headspace (final concentration, ca. 20%). The pH in the bottles was around 6.0, and the bottles were incubated at 25°C with gentle shaking at 100 rpm. The production of 13CO2 was measured by gas chromatography-mass spectrometry (GC-MS) in the headspace (see below).
After 3 months of incubation, the bottle with the strongest 13CO2 production was used as the inoculum for continuous culturing in a 3-liter glass bioreactor (working volume, 1.5 liters; Applikon, Schiedam, the Netherlands) that was operated in sequencing batch mode to prevent biomass loss. One cycle constituted 23 h of a continuous supply of medium and 0.5 h of settling, and finally 0.5 h of discharging with a level-controlled pump. To keep the culture anaerobic, the reactor was continuously flushed with 20 ml min−1 Ar-CO2 (95:5) and 5 ml min−1 methane. The temperature was controlled at 25°C and the pH at 6.0 to 6.2. Dissolved oxygen, temperature, and pH in the reactor were monitored by respective electrodes. Medium was prepared as described above, except unlabeled nitrite was used. The nitrite concentration in the reactor was estimated daily with Merckoquant test strips (0 to 80 mg liters−1; Merck, Germany), and the concentration in the medium was slowly increased from 0.2 to 2.5 mM depending on the activity of the continuous culture. Nitrite concentrations in the reactor were kept below 20 mg liter−1 (0.44 mM). The medium loading to the reactor was between 200 and 500 ml per day.
Activity analysis.
Methane-oxidizing activity in bottles was measured by determining the amount of 13CO2 produced from 13CH4 oxidation with GC-MS (Agilent 5975C inert MSD) as previously described (14). Activity in the reactor was tested in batch experiments with the whole culture. First, medium supply was stopped and unlabeled nitrite was allowed to be depleted. The reactor was flushed with Ar-CO2 (95:5) for 1 h while stirring and was checked for residual methane in the headspace. When undetectable, 0.2 mM 15NO2− and 50 ml 13CH4 were added. Gas samples (20 μl) were taken every hour for analysis of 13CO2, 15,15N2, and 15,14N2. At the same time, 1 ml culture liquid was taken and centrifuged; the supernatant was kept at 4°C for nitrite analysis. Nitrite concentrations were determined with colorimetric methods as described elsewhere (23). The influence of pH on activity was determined in batch incubations of 10 ml biomass in 40-ml serum bottles and buffered with MES [2-(N-morpholino)ethanesulfonate; 20 mM] to pH values between 5.9 and 6.7 and with MOPS [3-(N-morpholino)propanesulfonate; 20 mM] to pH values between 6.75 and 7.4 (measured at the end of incubation).
DNA isolation.
Total DNA from soil samples was isolated with the PowerSoil DNA isolation kit (MO BIO Laboratories Inc.) according to the manufacturer's manual. Approximately 0.3 g homogenized soil was used for DNA isolation, and two independent isolations were carried out for each depth interval. DNA was eluted three times with prewarmed Milli-Q water from the column to ensure that all of the DNA had been collected. DNA in the third elution was undetectable by agarose gel electrophoresis (<0.2 ng μl−1). DNA obtained from the same depth interval was pooled for qPCR analysis to minimize the influences from soil inhomogeneities. DNA from enrichment cultures was isolated with a method based on bead beating and SDS lysis as described previously (14). DNA quality was checked on an agarose gel, and concentrations were measured in triplicate with NanoDrop (ND-1000; Isogen Life Science, the Netherlands).
qPCR.
In order to quantify n-damo bacteria and all bacteria at different depths of the soil cores, quantitative PCR (qPCR) targeting the 16S rRNA gene was performed. To account for imperfect primer matching and known variability of results (14), two different primer pairs were used for each group. For NC10 phylum bacteria, primer pairs p1F and p1R and p2F and p2R and, for all bacteria, primer pairs 1100F and 1492R and 533F and 805R (Table 1) were applied. All qPCR assays were performed according to the MIQE guidelines (minimum information for publication of quantitative real-time PCR experiments) (4). qPCR experiments were carried out with the Bio-Rad IQTM 5 cycler real-time detection system using IQTM SYBR green Supermix (Bio-Rad) in a 25-μl reaction volume as previously described (14), except using 65°C for n-damo-specific primer pairs and 58°C for universal primer pairs as the annealing temperatures, which had been determined as most suitable for the present samples by gradient PCR. The qPCRs were carried out in 96-well plastic plates (Bio-Rad) sealed with Opti-Seal optical disposable adhesive (BIOplastics, the Netherlands). Fluorescence signals were obtained at 72°C at the end of the elongation step of each cycle. PCR products obtained with n-damo-specific and universal bacteria primer pairs were cloned and sequenced using the vector pGEM-T Easy (Promega). The sequences retrieved were of the correct length (201 bp for p1F and p1R, 292 bp for p2F and p2R, 291 bp for 515F and 805R, and 410 bp for 1100F and 1492R), and the obtained n-damo sequences were similar (>97.2% identity) to the sequence of “Ca. Methylomirabilis oxyfera” (accession no. FP565575). Standard curves for n-damo bacteria and general bacteria were constructed with plasmids containing corresponding inserts, taking into account the molecular mass of the plasmid, including the insert, and the plasmid concentration. Plasmid copy numbers used as the standard were between 3.07 × 101 and 3.07 × 108 μl−1 for NC10 bacteria and 8.69 × 101 and 8.69 × 108 μl−1 for all bacteria. Two soil cores with partial overlap were analyzed. Both cores were sliced in sections between 5 and 10 cm in the field (see the section on soil sampling and DNA isolation). In Fig. 1, each depth interval is represented by its average depth. DNA isolated from soil of a depth of 85 to 90 cm was used to test dilution effects; 10 and 100 times dilutions had a maximum difference from nondiluted ones of 8.7%. For NC10 bacteria, nondiluted DNA was used as templates, but for primers targeting all bacteria, 100-times-diluted DNA was used. PCR efficiencies calculated based on standards were between 90.6 and 99.2%. Both standards and samples were run in triplicates. The copy numbers in samples were calculated based on comparison to the threshold cycle values of the standard curve, taking into account the dilution and the amount of total DNA obtained per gram of soil.
Table 1.
Primer pairs used for qPCR analysis and clone library construction
| Designationa and purpose | Sequence (5′–3′) | Reference | Annealing temp (°C) | Target |
|---|---|---|---|---|
| qPCR analysis | ||||
| p1F | GGGCTTGACATCCCACGAACCTG | 14 | 65 | NC10 bacterial 16S rRNA |
| p1R | CGCCTTCCTCCAGCTTGACGC | 14 | ||
| p2F | GGGGAACTGCCAGCGTCAAG | 14 | 65 | NC10 bacterial 16S rRNA |
| p2R | CTCAGCGACTTCGAGTACAG | 14 | ||
| 533F | GTGCCAGCMGCCGCGGTAA | 49 | 58 | All bacterial 16S rRNA |
| 805R | GACTACCAGGGTATCTAATC | 28 | ||
| 1100F | YAACGAGCGCAACCC | 10 | 58 | All bacterial 16S rRNA |
| 1492R | GGTTACCTTGTTACGACTT | 53 | ||
| Clone library construction | ||||
| 8F | AGAGTTTGATYMTGGCTCAG | 21 | 55–65 | NC10 bacterial 16S rRNA |
| 193F | GACCAAAGGGGGCGAGCG | 14 | ||
| 1043R | TCTCCACGCTCCCTTGCG | 14 | ||
| A189bF | GGNGACTGGGACTTYTGG | 34 | 55–65 | NC10 bacterial pmoA |
| cmo182F | TCACGTTGACGCCGATCC | 34 | ||
| cmo682R | AAAYCCGGCRAAGAACGA | 34 |
F, forward; R, reverse.
Fig 1.
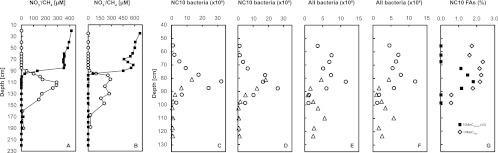
Depth profiles of the Brunssummerheide peatland. (A and B) Nitrate (filled square) and methane (open circle) concentrations in porewater sampled in December 2009 (A) and in June 2010 (B). (C to F) Bacterial cell numbers (cells g−1 wet soil) as assessed by qPCR on DNA extracted from two overlapping soil cores from 51 to 102 cm (open circles) and from 77 to 127 cm (open triangles). NC10 bacterial abundance was determined with primer pairs p1F and p1R (C) and p2F and p2R (D). Total bacterial abundance was determined with primer pair 535F and 805R (E) and 1100F and 1492R (F). (G) Relative abundance of the fatty acids 10-methyl-hexadecanoic acid (10MeC16:0; open diamonds) and 10-methyl-hexadec-7-enoic acid (10MeC16:1Δ7; multiplied by 3; closed squares), which are diagnostic of NC10 bacteria (data are from Kool et al. [24a]).
Phylogenetic analysis.
PCR was performed with DNA isolated from the soil layer used as an inoculum (80- to 100-cm depth), the enrichment culture after 3 months of incubation in bottles, and the continuous culture after 1 and 17 months of enrichment in the reactor. 16S rRNA sequences of n-damo bacteria were obtained with universal bacteria primer 8F or n-damo-specific primer 193F in combination with n-damo-specific primer 1043R (Table 1). PCR products of the correct size were ligated into the pGEM-T Easy cloning vector (Promega) and amplified in Escherichia coli DH5α. Plasmids were isolated from 10 to 15 randomly selected white colonies per library using the GeneJet miniprep kit (Fermentas, Lithuania) and were sequenced at the DNA Diagnostics Center of Nijmegen University Medical Center. The sequences were aligned to reference sequences with the MUSCLE algorithm. Phylogenetic trees were constructed with MEGA5 using the neighbor-joining method, and the robustness of tree topology was tested by bootstrap analysis (1,000 replicates).
Functional gene (particulate methane monooxygenase subunit A, pmoA) clone libraries were also constructed with the same DNA samples. The particulate methane monooxygenase catalyzes the first step of methane oxidation and is well conserved in methane-oxidizing bacteria, therefore pmoA is widely accepted as a marker gene for assessing diversity of aerobic and “Ca. Methylomirabilis oxyfera”-like anaerobic methanotrophs in the environment (34, 36). Two different forward primers targeting either most methanotrophs (A189b) or only close relatives of “Ca. Methylomirabilis oxyfera” (cmo182) were combined with a specific reverse primer (cmo682) (Table 1). A pmoA phylogenetic tree based on nucleotide sequences was constructed as described above.
FISH.
On a monthly basis, 1.5 ml biomass was harvested from the reactor and forced through a 0.5-mm needle to break big cell aggregates. The sample then was centrifuged and the pellet was washed twice with 1 ml 1× phosphate-buffered saline (PBS) and fixed with paraformaldehyde on ice for 3 h. Fluorescence in situ hybridization (FISH) was performed as previously described (13) using a 40% formamide concentration. The following oligonucleotide probes were used: S-*-DBACT-0193-a-A-18 and S-*-DBACT-1027-a-A-18, which are specific for n-damo bacteria (38), and a mixture of EUB I, II, III, and V for most Bacteria (7). Images were acquired with a Zeiss Axioplan 2 epifluorescence microscope equipped with a charge-coupled-device camera together with the Axiovision software package (Zeiss, Germany).
Nucleotide sequence accession numbers.
Representative 16S rRNA and pmoA gene sequences were deposited at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) under the accession numbers JX262153 to JX262155 (pmoA) and JX262156 to JX262161 (16S rRNA).
RESULTS
Porewater profiles.
Porewater depth profiles of the Brunssummerheide sampling location were determined on five occasions between July 2009 and May 2011, with a pattern that was very similar overall. Representative winter (December 2009) and summer (June 2010) profiles are shown in Fig. 1. Nitrate concentration decreased with depth and became undetectable below 100 cm. No methane was detected in the upper 80 cm, but methane gradually increased below the depth of 80 cm and reached the maximum concentration at around 120 cm (Fig. 1A and B). Redox data indicated that the soil was completely anoxic below 50 cm depth, and living roots of vascular plants were not found below 60 cm depth. The maximum concentration of nitrate (0.6 mM) in June 2010 (Fig. 1B) was about 0.2 mM higher than in December 2009 (Fig. 1A), possibly due to relatively stronger evaporation of surface water and higher groundwater influx in summer. The maximum concentration of methane remained similar in both seasons, as did the overall pattern: an opposing gradient at around 80- to 100-cm depth.
Quantifying abundance of NC10 bacteria in different soil depths.
Total bacterial and NC10 phylum abundances at different soil depths were determined in two overlapping cores by qPCR using primers targeting the 16S rRNA genes. The highest cell numbers (1.3 × 107 to 3.2 × 107 g−1 wet soil) of NC10 bacteria were found at 80- to 85-cm depth (Fig. 1C and D), coinciding with the concomitant decrease of methane and nitrate (Fig. 1A and B) and a peak in abundance of NC10 phylum-characteristic fatty acids (Fig. 1G) (24a). In contrast, total bacteria cell numbers, ranging from 0.9 × 108 to 11.8 × 108 cells g−1 wet soil, did not show a depth-related pattern (Fig. 1E and F).
Enrichment and activity.
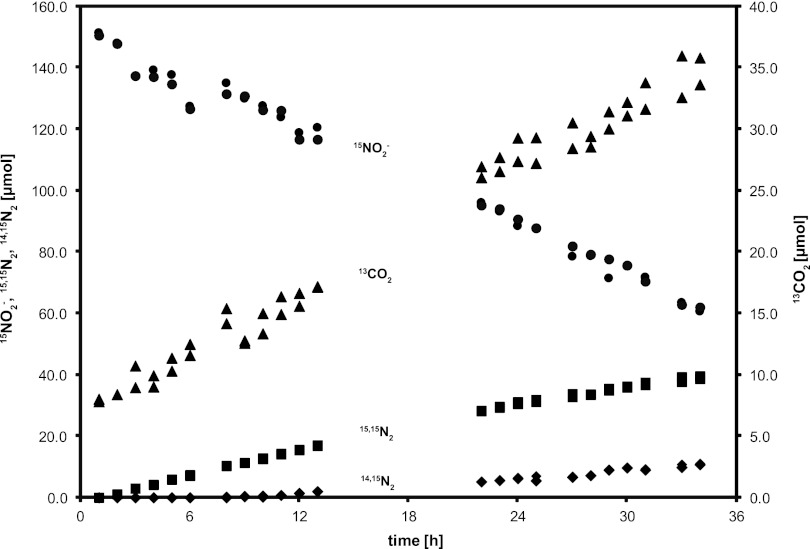
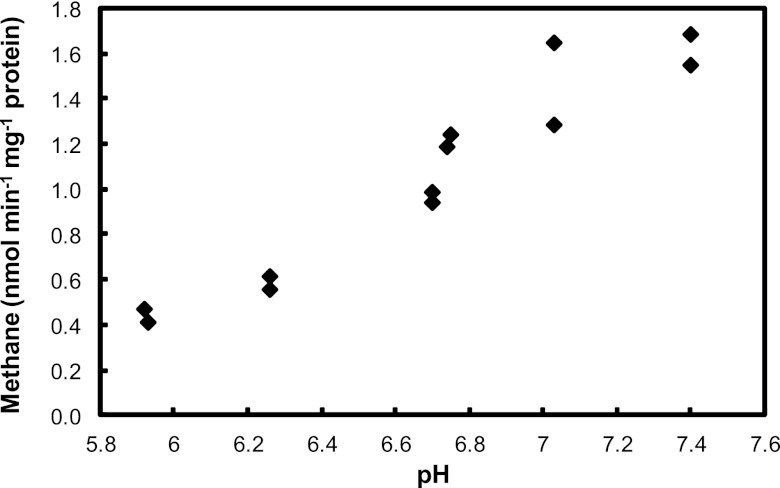
Nitrite-dependent methane-oxidizing activity was initially determined by measuring the fraction of 13CO2 in total CO2 after supplying 13CH4 and nitrite to three soil sections (80 to 100, 100 to 120, and 120 to 135 cm). Despite the addition (and permanent presence) of nitrate (0.5 mM), all soil cores produced some methane in the first 2 weeks of incubation, but no methane oxidation could be detected (detection limit of approximately 0.5 nmol day−1 g−1 soil). After about 3 months of incubation, the 80- to 100-cm section showed methane oxidation activity (9.0 nmol day−1 g−1 soil, assessed as CO2 production), and an increase in this rate indicated microbial growth. This incubation (80 to 100 cm) was used as the inoculum to start a sequencing batch reactor for the enrichment of the responsible microorganism. Batch tests and experience with previous NC10 bacterial enrichment cultures had indicated that nitrite was preferred over nitrate; consequently the medium, prepared with in situ water, was amended not only with nitrate but also with nitrite. Over the first 9 months of enrichment, activity remained low with a nitrite reduction rate of about 50 μmol day−1 liter−1 and then started to increase to about 1.0 mmol (NO2−) day−1 liter−1 in month 15. To test the coupling of nitrite reduction to methane oxidation, both activities were tested in batch experiments after 10 months with 15N- and 13C-labeled substrates during the enrichment period (Fig. 2). Nitrite-N was completely recovered as nitrogen gas, and methane was fully oxidized to CO2. The ratio of 13CO2 to 15,15N2 production was 3:4.3, similar to the theoretical stoichiometry of 3:4 (38). An activity test at different pH values demonstrated that the culture preferred circumneutral conditions but was active down to the lowest tested value of 5.9 (Fig. 3).
Fig 2.
Activity test of the enrichment culture at month 10 with 15NO2− and 13CH4. Nitrite (filled circle) was consumed and 15,15N2 (filled square), 14,15N2 (filled diamonds), and 13CO2 (filled triangle) were produced. The 13CO2 production rate was 20.2 μmol day−1, and the rate of 15,15N2 production was 29.0 μmol day−1.
Fig 3.
Methane-oxidizing activity of the n-damo enrichment culture incubated at different pH values.
FISH analysis of the enriched bacteria.
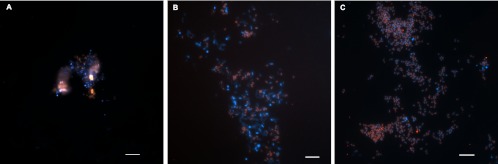
FISH was performed on biomass of the enrichment culture fixed every month, but no clear hybridization with NC10-specific probes was observed until after 8 months of medium supply. Even though small numbers of NC10 bacteria must have been present, they remained undetectable at first due to strong autofluorescent background and hybridization inhibition, presumably caused by peat material. Starting at month 9, NC10 cells could be detected (Fig. 4A). With the progression of incubation, both total cell numbers visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining and the percentage belonging to the NC10 phylum gradually increased (Fig. 4B and C) and coincided with an increased activity of the culture. At month 14 about 50% and at month 19 more than 80% of the population did hybridize with the NC10-specific probes (Fig. 4).
Fig 4.
Fluorescence in situ hybridization of the enrichment culture at different times of incubation. (A) Month 9; (B) month 14; (C) month 19. NC10 bacteria appear pink due to cohybridization of NC10 bacterium-specific probes 193-Cy3 and 1027-Cy3 (red) and a mixture of probes EUBI to III and IV-Cy5 (light blue) for most eubacteria and DAPI (dark blue). (Scale bars: 5 μm).
16S rRNA and pmoA gene phylogenetic analysis.
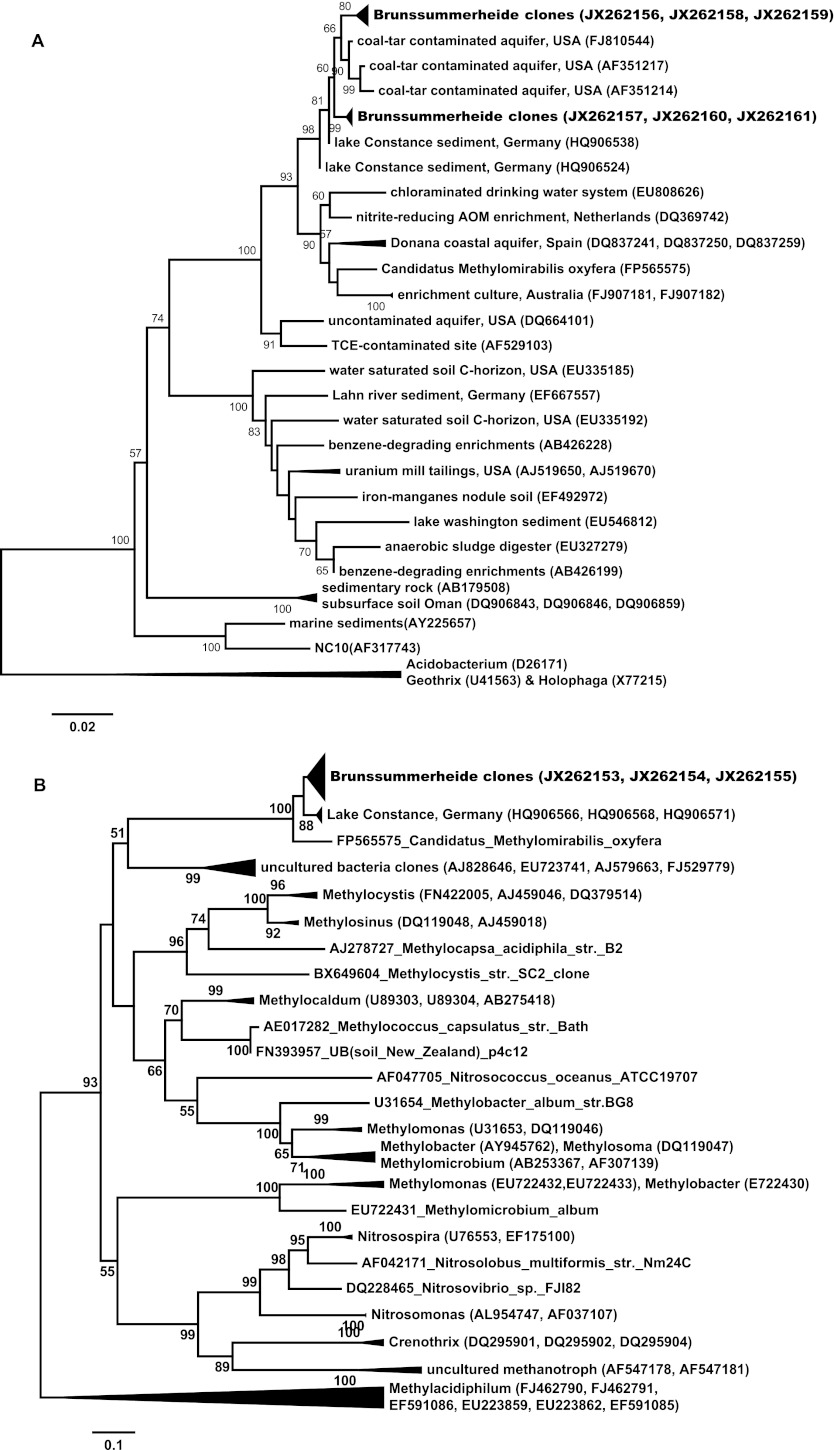
“Ca. Methylomirabilis oxyfera”-related 16S rRNA and pmoA genes were successfully obtained from both inoculum soil and the enrichment after 1 or 17 months of incubation. Long (>1,000 bp) 16S rRNA sequences obtained with primers 8F (universal) and 1043R (NC10 specific) were used for phylogenetic analysis. Results showed that the 16S rRNA sequences belong to group A of NC10 bacteria (14), forming a cluster (differences between 0.1 and 2.7%) with sequences retrieved from a coal-tar-contaminated aquifer (AF351214, AF351217, and FJ810544) and Lake Constance sediment (HQ906524 and HQ906538) (9). These sequences share only 94.9 to 95.5% identity with “Ca. Methylomirabilis oxyfera” (Fig. 5A).
Fig 5.
Phylogenetic trees of the 16S rRNA (A) and the pmoA genes (including amoA and pxmA sequences) (B). The trees were calculated in MEGA5 using the neighbor-joining method. Bootstrap support values (1,000 replicates) greater than 50% are indicated at the nodes. The sequences obtained in this study from inoculum soil and enrichment after 1 or 17 months of incubation are shown in boldface.
The phylogenetic analysis of the pmoA gene showed similar results. pmoA sequences from both soil and enrichment culture again cluster together with pmoA sequences retrieved from Lake Constance sediment (HQ906571, HQ906568, and HQ906566) (9). These pmoA sequences had an identity to those of “Ca. Methylomirabilis oxyfera” of 86.2 to 90.9% on the nucleotide level, but identity of 95.8 to 97.9% on the amino acid level indicated functional conservation (Fig. 5B). No significant difference could be observed between the inoculum and the 17-month-old enrichment culture, indicating that no population shift within the NC10 phylum had occurred. Both the 16S rRNA and pmoA gene phylogenetic results suggested that a new cluster of NC bacteria had been enriched.
DISCUSSION
The Brunssummerheide peatland is a spring fen in an oligotrophic sand valley fed by nitrate-polluted groundwater, therefore it contains nitrate concentrations in the upper peat layer which are uncommonly high for pristine peatlands (52). Also in contrast to many other peatlands (6, 24, 26, 27), methane was not detected in the upper 70 to 80 cm of the depth profile at 5 sampling occasions in different seasons from 2009 to 2011, even though methane was produced in the deep anoxic zone (below 100 cm) (Fig. 1A and B). As roots of vascular plants do not reach that deep in the Brunssummerheide (maximum of 60 cm), this suggested the existence of an anoxic methane sink in the peat independent from oxygen and aerenchymal transport by roots, for which oxidized nitrogen species could serve as the electron acceptors. The countergradient of methane and nitrate at the depth of 80 cm may provide an ideal niche for, and may be at least partly created by, the recently characterized n-damo bacteria. Targeting their 16S rRNA gene in DNA extracts from different depths confirmed that the highest n-damo cell numbers (up to 3.2 × 107 cells g−1 soil) and percentages (3 to 8% of the total bacterial community) were observed at a depth of 80 to 90 cm (Fig. 1C and D), coinciding with the methane-nitrate transition zone (Fig. 1A and B). At this depth, a peak in the abundance of fatty acids diagnostic for NC10 phylum bacteria also was detected (Fig. 1G) (24a). The n-damo cell number and lipid profiles also agreed with the finding that among soil samples from a depth of 80 to 100, 100 to 120, and 120 to 135 cm, only the 80- to 100-cm sample showed anaerobic methane-oxidizing activity upon incubation. Despite the relatively high numbers of n-damo bacteria detected at a depth 80 to 90 cm, it took several months to obtain an enrichment culture with measurable activity. Also, detection by fluorescence in situ hybridization using NC10 phylum-specific probes, hampered by a strongly autofluorescent background from the organic-rich inoculum, was possible only after 9 months of continuous cultivation with a constant supply of nitrite and methane. This may be due to the dilution of the naturally NC10 phylum-enriched soil layer with less active deeper layers (90 to 100 cm) in the inoculum and a very low growth rate under the prevailing conditions, especially the pH (6.0 to 6.2). The pH optimum test showed that the NC10 phylum bacteria enriched in the continuous culture were only acidotolerant to a certain extent and were not acidophilic. They were active down to a pH below 6, but their physiological optimum was clearly higher, above 7 (Fig. 3). This is a prime example for the discrepancy between physiological and ecological optima. In contrast to previous “Ca. Methylomirabilis oxyfera” enrichment cultures from neutral, eutrophic sediments (14), which had a similar optimum (around 7.5) but were not active at a pH below 7 (as assessed under similar conditions by O. Rasigraf [unpublished data]), a different ecotype was dominant in the more acidic and low-nutrient environment. According to the species delineation of 97% identity of the 16S rRNA gene for bacteria in general and 93% of the pmoA gene diagnostic for methanotrophic bacteria (35), the NC10 phylum bacterium dominating the Brunssummerheide enrichment culture even constitutes a new species within the genus “Ca. Methylomirabilis.”
As in other NC10 enrichment cultures (14, 20, 33), the enrichment period was characterized by a long phase without measureable activity, followed by a period of slow but exponential increase in nitrite consumption rate. In the present case, nitrite-reducing activity remained low for the first 9 months and then started to increase to about 1.0 mmol (NO2−) day−1 liter−1 in month 15. After this increase it was not possible to stimulate the growth of the culture further, and a sort of stationary phase similar to that of other enrichments of NC10 bacteria was reached (14, 20, 22). The doubling time of the Brunssummerheide “Ca. Methylomirabilis” strain was estimated to be about 2 months, which is 4- to 8-fold lower than the values reported before (14). It is difficult to predict whether this reflects the growth rate under field conditions. On one hand, some factors, like a higher temperature (25°C, the optimum temperature of methanotrophs in most peat soils [19], in contrast to 10 to 15°C in situ) and constant substrate supply, may be beneficial, but other factors, like stirring, use of surface water instead of porewater, or a decrease in microbial partner communities, may also be disadvantageous for growth in the laboratory.
However, once established, the methanotrophic community does not need to grow fast to constitute a relevant methane sink in the environment. According to previous estimations, “Ca. Methylomirabilis” cells in an enrichment culture have an activity of 0.1 to 0.4 fmol CH4 cell−1 day−1 (14), indicating that the Brunssummerheide soil of 80- to 85-cm depth with about 1.3 × 107 to 3.2 × 107 cells g−1 soil converts between 1.3 and 12.8 nmol CH4 day−1 g−1 soil. This range is at the lower end of methane oxidation rates reported for aerobic methanotrophs (41) in wetlands but apparently is high enough to balance the methane diffusing upwards from deeper, methanogenic soil layers.
Nitrite is clearly the preferred electron acceptor of previously reported “Ca. Methylomirabilis oxyfera” enrichments (13, 14, 20, 38). When nitrite was depleted in the present “Ca. Methylomirabilis” enrichment culture, methane-oxidizing activity in the presence of nitrate (1 mM) ceased; upon addition of fresh nitrite, methane consumption started again (data not shown), demonstrating that the methane-oxidizing activity of Brunssummerheide enrichment is also nitrite dependent. Although nitrite was also detected in the depth profile, its concentrations were much lower (maximum of 4.2 μM, mostly around the detection limit of the colorimetric method) than those of nitrate. There was no depth-related pattern, and values were not constant over time. The nitrite needed by n-damo bacteria active in the soil might be supplied by other microorganisms (e.g., denitrifying bacteria) or “Ca. Methylomirabilis” itself converting nitrate to nitrite using organic carbon compounds other than methane. This would explain why nitrate is sufficient as an electron acceptor for methane oxidation in situ and in the initial batch incubations, whereas after enrichment, concomitant with a relative loss of other bacteria and a degradation of labile organic carbon, this supply path is insufficient and nitrite addition becomes mandatory for methane oxidation.
The present study shows an additional, so far hardly investigated pathway linking the biogeochemical cycling of nitrogen and methane in peatlands. Given the worldwide increasing groundwater nitrate and atmospheric nitrogen loads (32, 45), this methane sink may become more relevant for mitigating the mobilization of carbon in the form of methane from wetlands in the future.
ACKNOWLEDGMENTS
We thank Jelle Eygensteyn, Paul van der Ven, and Jeroen Graafland for technical assistance with chemical analyses and Nicko Straathof, Linda Wortel, and Maurice Mouthaan from Natuurmonumenten for their cooperation in access to the Brunssummerheide. We also thank Boran Kartal and Huub op den Camp for helpful discussions.
The research was funded by a CAS-KNAW grant (09PhD02) to B. Zhu, a Darwin Center for Biogeosciences grant to K. F. Ettwig (project 142.16.3071, publication number DW-2012-1007), and an ERC Advanced Grant (232937) to M. S. M. Jetten.
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Bastviken D, Tranvik LJ, Downing JA, Crill PM, Enrich-Prast A. 2011. Freshwater methane emissions offset the continental carbon sink. Science 331:50. [DOI] [PubMed] [Google Scholar]
- 2. Bodelier PLE. 2011. Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr. Opin. Environ. Sustainability 3:379–388 [Google Scholar]
- 3. Bodelier PLE, Laanbroek HJ. 2004. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47:265–277 [DOI] [PubMed] [Google Scholar]
- 4. Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 5. Cicerone RJ, Oremland RS. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 2:299–327 [Google Scholar]
- 6. Clymo RS, Bryant CL. 2008. Diffusion and mass flow of dissolved carbon dioxide, methane, and dissolved organic carbon in a 7-m deep raised peat bog. Geochim. Cosmochim. Acta 72:2048–2066 [Google Scholar]
- 7. Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 8. Denman KL, et al. 2007. Couplings between changes in the climate system and biogeochemistry. In Solomon S, et al. (ed), Climate change 2007: the physical science basis. Contribution of Working Group I to the fourth assessment report of the Intergovernmental Panel on Climate Change Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 9. Deutzmann JS, Schink B. 2011. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl. Environ. Microbiol. 77:4429–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Downes J, et al. 2009. Pyramidobacter piscolens gen. nov., sp nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 59:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eller G, Kanel L, Krüger M. 2005. Cooccurrence of aerobic and anaerobic methane oxidation in the water column of Lake Plusssee. Appl. Environ. Microbiol. 71:8925–8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ettwig KF, et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 13. Ettwig KF, et al. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164–3173 [DOI] [PubMed] [Google Scholar]
- 14. Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fritz C, et al. 2011. Zero methane emission bogs: extreme rhizosphere oxygenation by cushion plants in Patagonia. New Phytol. 190:398–408 [DOI] [PubMed] [Google Scholar]
- 16. Gärdenäs AI, et al. 2011. Knowledge gaps in soil carbon and nitrogen interactions–from molecular to global scale. Soil Biol. Biochem. 43:702–717 [Google Scholar]
- 17. Gauci V, Dise N, Fowler D. 2002. Controls on suppression of methane flux from a peat bog subjected to simulated acid rain sulfate deposition. Global Biogeochem. Cycles 16:1004 [Google Scholar]
- 18. Gauci V, et al. 2004. Sulfur pollution suppression of the wetland methane source in the 20th and 21st centuries. Proc. Natl. Acad. Sci. U. S. A. 101:12583–12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu S, et al. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ. Microbiol. Rep. 1:377–384 [DOI] [PubMed] [Google Scholar]
- 21. Juretschko S, et al. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kampman C, et al. 2012. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment. J. Hazard. Mater. 227-228:164–171 [DOI] [PubMed] [Google Scholar]
- 23. Kartal B, et al. 2006. Adaptation of a freshwater anammox population to high salinity wastewater. J. Biotechnol. 126:546–553 [DOI] [PubMed] [Google Scholar]
- 24. Kip N, et al. 2010. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3:617–621 [Google Scholar]
- 24a. Kool DM, et al. 2012. Rare branched fatty acids characterize the lipid composition of the intra-aerobic methane oxidizer Candidatus Methylomirabilis oxyfera.” Appl. Environ. Microbiol. 78:8650–8656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334 [DOI] [PubMed] [Google Scholar]
- 26. Lai DYF. 2009. Methane dynamics in northern peatlands: a review. Pedosphere 19:409–421 [Google Scholar]
- 27. Le Mer J, Roger P. 2001. Production, oxidation, emission and consumption of methane by soils: a review. Eur. J. Soil Biol. 37:25–50 [Google Scholar]
- 28. Leser TD, et al. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu LL, Greaver TL. 2009. A review of nitrogen enrichment effects on three biogenic GHGs: the CO2 sink may be largely offset by stimulated N2O and CH4 emission. Ecol. Lett. 12:1103–1117 [DOI] [PubMed] [Google Scholar]
- 30. Lovley DR, Klug MJ. 1983. Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol. 45:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lovley DR, Phillips EJ. 1987. Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl. Environ. Microbiol. 53:2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lucassen ECHET, Smolders AJP, Van der Salm AL, Roelofs JGM. 2004. High groundwater nitrate concentrations inhibit eutrophication of sulphate-rich freshwater wetlands. Biogeochemistry 67:249–267 [Google Scholar]
- 33. Luesken FA, et al. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. 92:845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luesken FA, et al. 2011. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lüke C, Frenzel P. 2011. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 77:6305–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDonald IR, Bodrossy L, Chen Y, Murrell JC. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pester M, Knorr KH, Friedrich MW, Wagner M, Loy A. 2012. Sulfate-reducing microorganisms in wetlands-fameless actors in carbon cycling and climate change. Front. Microbiol. 3:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raghoebarsing AA, et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 39. Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem. Rev. 107:486–513 [DOI] [PubMed] [Google Scholar]
- 40. Schubert CJ, et al. 2011. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol. Ecol. 76:26–38 [DOI] [PubMed] [Google Scholar]
- 41. Segers R. 1998. Methane production and methane consumption: a review of processes underlying wetland methane fluxes. Biogeochemistry 41:23–51 [Google Scholar]
- 42. Sivan O, et al. 2011. Geochemical evidence for iron-mediated anaerobic oxidation of methane. Limnol. Oceanogr. 56:1536–1544 [Google Scholar]
- 43. Smemo KA, Yavitt JB. 2011. Anaerobic oxidation of methane: an underappreciated aspect of methane cycling in peatland ecosystems? Biogeosciences 8:779–793 [Google Scholar]
- 44. Smith RL, Howes BL, Garabedian SP. 1991. In situ measurement of methane oxidation in groundwater by using natural-gradient tracer tests. Appl. Environ. Microbiol. 57:1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smolders AJP, Lucassen ECHET, Bobbink R, Roelofs JGM, Lamers LPM. 2010. How nitrate leaching from agricultural lands provokes phosphate eutrophication in groundwater fed wetlands: the sulphur bridge. Biogeochemistry 98:1–7 [Google Scholar]
- 46. Stevens CJ, et al. 2011. Ecosystem responses to reduced and oxidised nitrogen inputs in European terrestrial habitats. Environ. Pollut. 159:665–676 [DOI] [PubMed] [Google Scholar]
- 47. Strous M, Jetten MS. 2004. Anaerobic oxidation of methane and ammonium. Annu. Rev. Microbiol. 58:99–117 [DOI] [PubMed] [Google Scholar]
- 48. Tomassen HBM, Smolders AJP, Leon PML, Roelofs JGM. 2003. Stimulated growth of Betula pubescens and Molinia caerulea on ombrotrophic bogs: role of high levels of atmospheric nitrogen deposition. J. Ecol. 91:357–370 [Google Scholar]
- 49. Turner S, Pryer KM, Miao VPW, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small submit rRNA sequence analysis. J. Eukaryot. Microbiol. 46:327–338 [DOI] [PubMed] [Google Scholar]
- 50. van Breukelen BM, Griffioen J. 2004. Biogeochemical processes at the fringe of a landfill leachate pollution plume: potential for dissolved organic carbon, Fe(II), Mn(II), NH4, and CH4 oxidation. J. Contam. Hydrol. 73:181–205 [DOI] [PubMed] [Google Scholar]
- 51. Vile MA, Bridgham SD, Wieder RK, Novák M. 2003. Atmospheric sulfur deposition alters pathways of gaseous carbon production in peatlands. Global Biogeochem. Cycles 17:1058 [Google Scholar]
- 52. Waughman GJ. 1980. Chemical aspects of the ecology of some south German peatlands. J. Ecol. 68:1025–1046 [Google Scholar]
- 53. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu G, Jetten MS, Kuschk P, Ettwig KF, Yin C. 2010. Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl. Microbiol. Biotechnol. 86:1043–1055 [DOI] [PubMed] [Google Scholar]