Abstract
PP1 (protein phosphatase 1) is an essential serine/threonine phosphatase that plays a critical role in a broad range of biological processes, from muscle contraction to memory formation. PP1 achieves its biological specificity by forming holoenzymes with more than 200 known regulatory proteins. Interestingly, most of these regulatory proteins (≥70%) belong to the class of IDPs (intrinsically disordered proteins). Thus structural studies highlighting the interaction of these IDP regulatory proteins with PP1 are an attractive model system because it allows general parameters for a group of diverse IDPs that interact with the same binding partner to be identified, while also providing fundamental insights into PP1 biology. The present review provides a brief overview of our current understanding of IDP–PP1 interactions, including the importance of pre-formed secondary and tertiary structures for PP1 binding, as well as changes of IDP dynamics upon interacting with PP1.
Keywords: intrinsically disordered protein, nuclear magnetic resonance (NMR), protein phosphatase 1 (PP1), small-angle X-ray scattering (SAXS)
Introduction
Phosphorylation is a key mechanism for signal transduction and the regulation of a broad range of physiological processes, including cell proliferation, differentiation, survival, migration and death. Thus dysregulation of phosphorylation signalling pathways is directly correlated with a plethora of diseases such as cancer and diabetes. The phosphorylation state of a protein is controlled by the action of kinases and phosphatases on both serine/threonine residues, which account for 98.2% of all phosphorylation reactions, and tyrosine residues, which account for only 1.8%. Yet, although the human genome has more than 420 genes that encode serine/threonine kinases, only ~40 genes encode serine/threonine phosphatases [1–4].
The metalloenzyme PP1 (protein phosphatase 1) (~38.5 kDa, 2 Mn2 + are bound in the active site when expressed in bacteria) is a particularly well-characterized serine/threonine phosphatase [4,5]. PP1 is exceptionally conserved in eukaryotes from fungi to humans, and mammalian genomes contain three different genes that encode four distinct catalytic PP1 subunits: PP1α, PP1β/δ and the splice variants PP1γ1 and PP1γ2. The differences in the amino acid composition between the PP1 isoforms are primarily localized to the N- and C-termini. Free PP1 has essentially no specificity for substrates. Rather, its specificity is tightly controlled by its interaction with >200 known targeting proteins [5,6]. These targeting proteins fulfil two major functions. First, they localize PP1 to distinct regions of the cell and, secondly, they modulate substrate specificity of PP1. This modulation can be accomplished by PP1-specific inhibitor proteins, e.g. I-2 (inhibitor-2) [7,8] and DARPP-32 (dopamine- and cAMP-regulated phosphoprotein of 32 kDa) [9–11], which bind and block the PP1 active site, rendering PP1 inactive to all substrates. Alternatively, regulatory proteins such as spinophilin and MYPT1 (myosin phosphatase target subunit 1) bind PP1 to form a highly stable holoenzyme, which can regulate PP1 specificity via a variety of ways.
The catalytic site of PP1 is at the intersection of three substrate-binding regions: the hydrophobic, acidic and C-terminal grooves. Most PP1 regulators interact with PP1 via a primary PP1-binding motif, the RVXF motif, which conforms to the consensus sequence (K/R)(K/R)(V/I)X(F/W), where X is any residue other than phenylalanine, isoleucine, methionine, tyrosine, aspartate or proline [12–14]. For regulatory proteins to interact strongly with PP1, they must bind at the RVXF interaction site. Nevertheless, this interaction, which is ~20 Å(1 Å= 0.1 nm) from the catalytic site, does not alter the PP1 catalytic site structure or function [4]. Finally, more than 70% of PP1-regulatory proteins are predicted to be IDPs (intrinsically disordered proteins) [5]. Thus PP1 interacts with a large number of distinct IDPs and is thus an excellent model for addressing outstanding questions about IDP proteins and their functions in the cell.
Characterization of IDPs
The best tool for understanding the structures and dynamics of IDPs in their free states at atomic resolution is NMR spectroscopy (Figure 1A). NMR spectra of IDPs lack the typical hallmarks of NMR spectra of folded proteins, as both hydrogen bonds as well as hydrophobic interactions are largely absent from IDPs. Nevertheless, 15N and 13C carbonyl chemical shifts are still significantly different for each amino acid in an IDP. This provides an opportunity to interpret NMR spectra for IDPs and, in turn, obtain atomic-resolution information about IDP function.
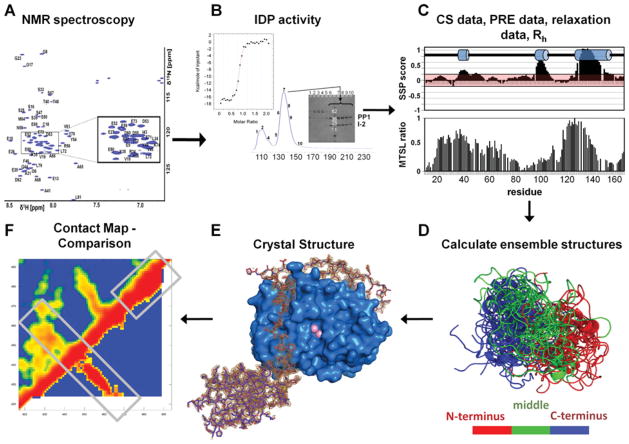
Figure 1. Typical IDP analysis workflow.
(A) NMR spectroscopy can be readily used to identify IDPs. Two-dimensional 1H,15N HSQC (heteronuclear single-quantum coherence) spectrum of IDPs have a limited peak dispersion in the 1HN dimension due to a lack of hydrogen-bonding network typical in secondary-structural elements of well-folded proteins. (B) Necessary experiments to verify protein function. For this system, IDP function is tested by IDP–PP1 complex formation, either by co-elution from a size-exclusion chromatography column or by isothermal titration calorimetry. (C) Measurement and evaluation of chemical shift (CS), PRE, auto-correlated short-timescale relaxation NMR data as well as global parameters, based on dynamic light scattering or SAXS. MTSL, S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate spin label; Rh, hydrodynamic radius; SSP, secondary-structure propensity. (D) Calculation of the IDP ensemble structure. (E) Determination of the complex structure between the IDP and the targeting protein (spinophilin–PP1 structure is shown; PDB code 3EGG) using X-ray crystallography or NMR spectroscopy. (F) Comparison of unbound and bound IDP conformations via a distance correlation map, to evaluate the mode of interaction, as well as differential and common structural features.
The analysis of NMR parameters of IDPs is challenging, as they reflect an average taken over the ensemble of conformers populated in the unstructured state of a protein. Nevertheless, numerous NMR parameters, especially chemical shifts, PREs (paramagnetic relaxation enhancements), RDCs (residual dipolar couplings) and 15N relaxation rates, among others, can be used to characterize the conformational space of IDPs [15] (Figure 1C). Recently, additional tools, such as single-molecule fluorescence spectroscopy [16] and SAXS (small-angle X-ray scattering) [17–19] have also been successfully used to gain structural insights into IDPs.
Critically, interpretation of these measured parameters is often very different for IDPs than it is for folded proteins. Specifically, IDPs interconvert rapidly between many conformations. This increased flexibility requires careful consideration when NMR data of IDPs are analysed and makes structure (or ensemble) determination of IDPs challenging. NMR structure determination of folded proteins is carried out as follows. First, all atoms in a biomacromolecule are assigned. Secondly, these assignments are used to directly measure distance constraints, i.e. the collection of a large number of 1H-1H NOEs (nuclear Overhauser effects). Thirdly, these unambiguous distance constraints are used to determine the three-dimensional structure of the protein in solution [20]. Unfortunately, only a small number of 1H-1H NOEs can be detected in IDPs, and these NOEs are mainly sequential. Thus the methods developed for the structure determination of folded proteins cannot be applied to IDPs.
To overcome this problem, multiple approaches that enable three-dimensional IDP structure ensemble determination have been developed in the last few years, including those by the groups of Martin Blackledge [21–23], Julie Forman-Kay [24] and Michele Vendruscolo [25]. In our studies, the program ENSEMBLE [24,26] was used to investigate the unbound ensemble structure(s) of a number of IDPs, all of which bind and influence the specificity of PP1. ENSEMBLE assigns populations to a large pool of pre-generated conformers that satisfy experimentally determined restraints. As a complete sampling of the conformational space is impossible, ENSEMBLE uses an iterative approach, in which an experimental data-based conformer selection method is used to increase the pool of available ensemble structures for subsequent ENSEMBLE runs (Figures 1D and 1F).
The biophysical behaviour of IDPs
Clearly, folded proteins have fundamentally different structures and stabilities compared with IDPs. Importantly, the three-dimensional structures of folded proteins are directly correlated with their functions. In the last 15 years, it has been established that this principle is also true for IDPs (Figure 1). Despite lacking a three-dimensional structure, IDPs are functional proteins that can readily bind to their binding partner, e.g. folded proteins, and carry out biological functions. Nevertheless, the modes of interaction that IDPs use to bind proteins are significantly different from those used by folded proteins. The increased flexibility in IDPs allows for a number of different binding processes, including conformational selection, folding-upon-binding and ‘fly casting’ [27–29]. The interaction mechanism that is used for complex formation often depends on the structure of the IDP in its unbound form. Whereas some IDPs are true random coil polymers, most contain elements of secondary or tertiary structure that are only transiently populated. These secondary- and tertiary-structural elements can lead to a structural organization of IDPs. Furthermore, this organization, or ‘preferred conformation’, results in a spatial restriction in IDPs that can expose primary protein interaction sites to enable faster and more effective binding, as well as folding-upon-binding, to target molecules [30–32].
Whereas numerous IDP protein–protein interactions have been reported and analysed in the last 15 years, in the present review, we focus on three distinct IDPs that bind the same well-folded enzyme, PP1: the PP1-targeting proteins spinophilin [19,33,34] and MYPT1 [35,36], and the PP1 inhibitor I-2 [17,19,37,38]. When these proteins bind PP1 to form a PP1 holoenzyme, PP1 becomes highly specific for a subset of substrates (Figures 1 and 2).
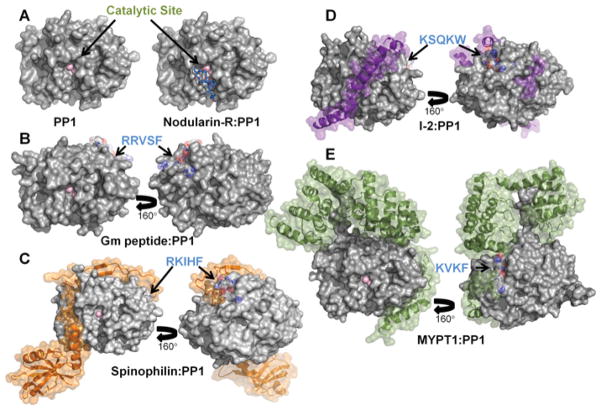
Figure 2. PP1 regulators are IDPs that adopt distinct conformations in their PP1-bound forms.
(A) PP1 is a metalloenzyme with two Mn2 + ions bound at the catalytic site. The catalytic site is also the target of molecular toxins such as Nodularin-R. (B) Gm peptide–PP1 complex; electron density for the peptide was only detected in the RVXF-binding site. (C) Spinophilin–PP1 complex (PDB code 3EGG). (D) I-2–PP1 complex (PDB code 2O8G). (E) MYPT1–PP1 complex (PDB code 1S70).
Regulation of PP1 by IDPs
Spinophilin, MYPT1 and I-2 all modulate the activity of PP1. They do so by either inhibiting the catalytic site (I-2) or by modifying the PP1 surface so specific substrates can be efficiently dephosphorylated (spinophilin and MYPT1). Despite their related functions and common binding partner, a comparison of the unbound structures of these three IDPs shows that they have different structural and dynamic characteristics in both the free and PP1-bound states.
First, spinophilin is highly dynamic in its unbound form; e.g. nearly no positive 15N{1H}-NOEs are observed [19,34]. In contrast, I-2 possess a nearly 100% populated α-helix, in which many strong (i, i + 1) HN-HN NOEs from residues in this helix are readily measured in a three-dimensional 15N-resolved 1H,1H NOESY spectrum [38]. In addition, residues near this α-helix also have reduced backbone flexibility. Finally, at the other end of the IDP spectrum, only the 40 N-terminal residues are unstructured in MYPT1 and even these residues have restricted short-timescale backbone dynamics [35].
Secondly, all three IDPs have pre-formed secondary-structural elements that are used to bind PP1. However, the number of pre-formed structures varies significantly between the three IDPs. The most dynamic protein, spinophilin, has the least amount of pre-formed secondary structure. Conversely, I-2 has three pre-formed α-helices. However, whereas some of these pre-formed secondary-structure elements in I-2 have a significant role in PP1 binding and biological function (e.g. the 100% populated α-helix in the free state binds PP1 and blocks its catalytic site, explaining the inhibitory mechanism of I-2), some pre-formed secondary-structural elements in I-2 are not used for interacting with PP1. Lastly, MYPT1 also has a pre-formed α-helix that is important for PP1 binding.
Thirdly, and as might be expected, spinophilin, I-2 and MYPT1 have different structural ensembles in their unbound forms [19,35]. However, they also adopt significantly different conformations in their bound forms, i.e. when they are bound to PP1 to form PP1 holoenzymes (spinophilin–PP1, I-2–PP1 and MYPT1–PP1) (Figure 2). Indeed, these structures reveal that, beyond the RVXF binding motif, there are no common PP1-binding motifs among the IDPs. Instead, these structures have revealed that PP1 is a protein interaction hub and that many PP1 surfaces are potential protein–protein interaction sites. In fact, using the average SASA (solvent-accessible surface area) of the RVXF motif and more recently identified PP1-binding pockets as a measure of the SASA needed for a single PP1-binding site, and comparing it with the SASA of the entire PP1 protein, we predict that PP1 may have up to 30 non-overlapping regulatory protein-binding sites [5]. IDPs, because of their increased flexibility and extended structures, have a significant advantage compared with folded proteins when binding PP1 because they can easily interact with a single or multiple PP1-binding pockets using a minimal number of residues. In contrast, if PP1 holoenzymes were formed by folded regulatory proteins, these proteins would need to be much larger (~4-fold) to engage the equivalent PP1-binding surfaces. The diversity of binding sites available on PP1, and the inherent flexibility of IDPs, explains why, for this system, the conformations of IDPs bound to PP1 are distinct.
Fourthly, spinophilin, I-2 and MYPT1 use different mechanisms to bind PP1. Two major processes can lead to the selection of a single folded conformation when IDPs bind to a folded protein: conformational selection and/or induced fit. In the first case, the IDP has an intrinsic preference for its binding conformer and therefore the interacting protein is only a scaffold, i.e. it does not direct the formation of the bound conformation. This conformational selection model of binding requires that a limited population of the IDP adopts the bound-state conformation in its free state [39]. This behaviour is observed in spinophilin and MYPT1. Both unbound states have pre-formed structures that are similar to the structures they adopt in their bound forms. This is not the case for I-2, which probably follows an ‘induced-fit’ process. Here, I-2 conformations in the unbound form are different from the PP1-bound conformer. In this case, the bound conformation is energetically accessible only in the presence of its binding partner [40], PP1, and is either not detected in the unbound state or is so minimally populated that its presence is undetectable using the techniques that we have used in our studies. Clearly, the highly populated α-helix in I-2 might play a significant role in initiating the induced-fit events that define the interaction of I-2 with PP1.
Fifthly, there are significant differences in the residual flexibility of these three IDPs in their PP1-bound states. Spinophilin, the most dynamic IDP in its unbound form, becomes completely rigid when bound to PP1. It does so by forming both a β-sheet, which extends a β-sheet in PP1, and an α-helix, which was minimally populated in the unbound state. All residues have excellent electron density in the spinophilin–PP1 holoenzyme crystal structure. This behaviour is similar to that observed for the MYPT1–PP1 complex [36], where MYPT1 becomes entirely structured when bound to PP1. This is interesting, as the most flexible (spinophilin) and most rigid (MYPT1) IDPs in their unbound states adopt fully rigid structures when bound to PP1. In contrast, I-2 is well-structured and more rigid than spinophilin in its unbound form. However, only ~25% of the I-2 residues adopt a rigid structure in the I-2–PP1 crystal structure [37]; 75% of all I-2 residues stay flexible. Using NMR spectroscopy and SAXS, we were able to determine the ensemble structure of the I-2–PP1 holoenzyme in solution, which showed that the I-2 residues that stay flexible in the I-2–PP1 complex form two extended loop structures [19]. Interestingly, using chemical shift data, we identified a ~60% populated α-helix in unbound I-2, which is located in one of these extended loops. It is possible that this α-helix forms an additional protein–protein interaction site, enabling this IDP to interact with other proteins while bound to PP1 [41].
Two IDPs fight for one folded protein
For many years, it was thought that only one PP1-regulatory protein was able to interact with PP1 at a time, as nearly every regulatory protein binds PP1 in its RVXF-binding pocket. However, it has been shown that pairs of targeting and inhibitor proteins can bind to PP1 simultaneously [42–44]. This leads to an additional layer of complexity in PP1 regulation. To understand how, at a molecular level, multiple regulatory proteins interact with PP1 simultaneously, and to establish which of the two proteins bind the PP1 RVXF-binding pocket, we investigated such a heterotrimeric PP1 complex. Specifically, we studied the triple complex formed by the interaction of two IDPs with PP1, spinophilin and I-2, which together form the PP1–spinophilin–I-2, or PSI, complex.
Using biochemical, NMR spectroscopy and SAXS analyses, we showed that, in the PSI complex, I-2 releases from the RVXF-binding site while spinophilin engages this site [17]. This release of one of the key PP1-binding pockets by I-2 significantly increases the length of the flexible loop in I-2 in the PSI complex. This leads to two interesting observations. First, IDPs can be exceedingly flexible when bound to their targeting proteins, such as I-2 in the PSI triple complex, where ~80% of all residues stay flexible. Thus contributions of enthalpy and entropy in IDP–protein interactions are likely to be weighted differently from how they are for interactions between two folded proteins, as entropy can play a larger role in an IDP-binding event due to increased flexibility in the unbound, as well as the bound, state. Clearly, spinophilin reduces its entropy upon binding, as it loses nearly all of its free-state flexibility. In contrast, I-2 stays nearly entirely flexible in the holoenzyme as it only binds PP1 via very short motifs and a long α-helix that is pre-formed in the unbound state. Secondly, it is interesting to note that the secondary-structure elements that were most highly populated in the unbound forms of spinophilin and I-2 are present and bound in the PSI complex, indicating that these pre-formed secondary-structural elements might reflect the protein–protein-binding strength.
Conclusions
During the last decade, it has become apparent that IDPs have critical roles in a significant number of regulatory processes [5,18,19,45–47]. One of these roles is the regulation of the serine/threonine phosphatase PP1, an enzyme responsible for ~30% of all dephosphorylation reactions in eukaryotic cells. Approximately 200 biochemically confirmed interaction partners bind to PP1 to regulate its activity and most of these regulatory proteins are IDPs. Thus PP1, together with its regulatory proteins, is a unique system for investigating the similarities and differences in a family of IDPs, the PP1-regulatory proteins, that bind a single protein: PP1.
Using this system, it has become apparent that PP1-regulatory IDPs use a variety of processes to interact with PP1. In all cases, pre-formed secondary structure has been shown to play a central role in the interaction of these IDPs with PP1. However, the importance of these pre-formed structures for PP1 binding varies between the IDPs. In addition, not only are their unbound structures highly variable, but also all three IDPs described in the present paper also adopt distinct conformations when bound to PP1. The similarities between the unbound and bound conformations and dynamics of a single IDP are also different for these three PP1 IDPs. For example, spinophilin and MYPT1 have unbound tertiary structures that are similar to their PP1 bound conformations, whereas I-2 does not. Furthermore, both spinophilin and MYPT1 become entirely rigid upon PP1 binding, whereas only ~25% of the I-2 residues become rigid. This shows an interesting variability of the flexibility that IDPs retain when binding PP1. Thus, although it is possible to identify common features for the interaction of IDPs with PP1, it is too early to make any predictions about how other regulatory IDPs will bind PP1. As we are currently pursuing a comprehensive structural understanding of multiple PP1 regulators, both in their bound and unbound conformations, it will be interesting to see whether these additional data enable us to make detailed and precise predictions for IDP–PP1 interactions.
Acknowledgments
We thank all current and previous members of the Page and Peti laboratories for their contribution to this ongoing research over the last 8 years. We thank Dr Angus Nairn, Dr Mathieu Bollen and Dr Shirish Shenolikar for stimulating input into the extensive biology of PP1, and Dr Julie Forman-Kay for support with the ENSEMBLE method.
Funding
The work was supported by the National Institute of Neurological Disorders and Stroke [grant number R01NS056128 (to W.P.)], the National Institute of General Medicine [grant number R01GM098482 (to R.P.)] and an American Cancer Society Research Scholar Grant [grant number RSG-08-067-01-LIB (to R.P.)].
Abbreviations used
- I-2
inhibitor-2
- IDP
intrinsically disordered protein
- MYPT1
myosin phosphatase target subunit 1
- NOE
nuclear Overhauser effect
- PP1
protein phosphatase 1
- PRE
paramagnetic relaxation enhancement
- PSI
PP1–spinophilin–I-2
- SASA
solvent-accessible surface area
- SAXS
small-angle X-ray scattering
References
- 1.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signaling. 2009;2:re9. doi: 10.1126/scisignal.2100re9. [DOI] [PubMed] [Google Scholar]
- 4.Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2012 doi: 10.1111/ j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Huang HB, Horiuchi A, da Cruze Silva EF, Hsieh-Wilson L, Allen PB, Shenolikar S, Greengard P, Nairn AC. Protein phosphatase 1 regulation by inhibitors and targeting subunits. Proc Natl Acad Sci USA. 2001;98:3080–3085. doi: 10.1073/pnas.051003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang HB, Horiuchi A, Watanabe T, Shih SR, Tsay HJ, Li HC, Greengard P, Nairn AC. Characterization of the inhibition of protein phosphatase-1 by DARPP-32 and inhibitor-2. J Biol Chem. 1999;274:7870–7878. doi: 10.1074/jbc.274.12.7870. [DOI] [PubMed] [Google Scholar]
- 8.Alessi DR, Street AJ, Cohen P, Cohen PT. Inhibitor-2 functions like a chaperone to fold three expressed isoforms of mammalian protein phosphatase-1 into a conformation with the specificity and regulatory properties of the native enzyme. Eur J Biochem. 1993;213:1055–1066. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 9.Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 10.Kwon YG, Huang HB, Desdouits F, Girault JA, Greengard P, Nairn AC. Characterization of the interaction between DARPP-32 and protein phosphatase 1 (PP-1): DARPP-32 peptides antagonize the interaction of PP-1 with binding proteins. Proc Natl Acad Sci USA. 1997;94:3536–3541. doi: 10.1073/pnas.94.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmings HC, Jr, Nairn AC, Aswad DW, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II Purification and characterization of the phosphoprotein from bovine caudate nucleus. J Neurosci. 1984;4:99–110. doi: 10.1523/JNEUROSCI.04-01-00099.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 13.Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Eliezer D. Characterizing residual structure in disordered protein states using nuclear magnetic resonance. Methods Mol Biol. 2007;350:49–67. doi: 10.1385/1-59745-189-4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dancheck B, Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry. 2011;50:1238–1246. doi: 10.1021/bi101774g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh JA, Dancheck B, Ragusa MJ, Allaire M, Forman-Kay JD, Peti W. Structural diversity in free and bound states of intrinsically disordered protein phosphatase 1 regulators. Structure. 2010;18:1094–1103. doi: 10.1016/j.str.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons; Chichester: 1986. [Google Scholar]
- 21.Bernado P, Blanchard L, Timmins P, Marion D, Ruigrok RW, Blackledge M. A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc Natl Acad Sci USA. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen MR, Markwick PR, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernado P, Blackledge M. Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure. 2009;17:1169–1185. doi: 10.1016/j.str.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Jensen MR, Salmon L, Nodet G, Blackledge M. Defining conformational ensembles of intrinsically disordered and partially folded proteins directly from chemical shifts. J Am Chem Soc. 2010;132:1270–1272. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 24.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308:1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 25.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 26.Marsh JA, Forman-Kay JD. Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J Mol Biol. 2009;391:359–374. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 28.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csizmok V, Bokor M, Banki P, Klement E, Medzihradszky KF, Friedrich P, Tompa K, Tompa P. Primary contact sites in intrinsically unstructured proteins: the case of calpastatin and microtubule-associated protein 2. Biochemistry. 2005;44:3955–3964. doi: 10.1021/bi047817f. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 32.Hoang TX, Marsella L, Trovato A, Seno F, Banavar JR, Maritan A. Common attributes of native-state structures of proteins, disordered proteins, and amyloid. Proc Natl Acad Sci USA. 2006;103:6883–6888. doi: 10.1073/pnas.0601824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. Flexibility in the PP1:spinophilin holoenzyme. FEBS Lett. 2011;585:36–40. doi: 10.1016/j.febslet.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinheiro AS, Marsh JA, Forman-Kay JD, Peti W. Structural signature of the MYPT1–PP1 interaction. J Am Chem Soc. 2011;133:73–80. doi: 10.1021/ja107810r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 37.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 38.Dancheck B, Nairn AC, Peti W. Detailed structural characterization of unbound protein phosphatase 1 inhibitors. Biochemistry. 2008;47:12346–12356. doi: 10.1021/bi801308y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 40.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sami F, Smet-Nocca C, Khan M, Landrieu I, Lippens G, Brautigan DL. Molecular basis for an ancient partnership between prolyl isomerase Pin1 and phosphatase inhibitor-2. Biochemistry. 2011;50:6567–6578. doi: 10.1021/bi200553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lesage B, Beullens M, Pedelini L, Garcia-Gimeno MA, Waelkens E, Sanz P, Bollen M. A complex of catalytically inactive protein phosphatase-1 sandwiched between Sds22 and inhibitor-3. Biochemistry. 2007;46:8909–8919. doi: 10.1021/bi7003119. [DOI] [PubMed] [Google Scholar]
- 44.Terry-Lorenzo RT, Elliot E, Weiser DC, Prickett TD, Brautigan DL, Shenolikar S. Neurabins recruit protein phosphatase-1 and inhibitor-2 to the actin cytoskeleton. J Biol Chem. 2002;277:46535–46543. doi: 10.1074/jbc.M206960200. [DOI] [PubMed] [Google Scholar]
- 45.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 46.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graphics Modell. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 47.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]