Abstract
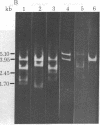
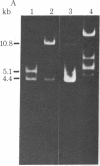
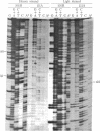
Restriction enzyme analysis of mtDNAs for the purpose of determining sequence divergence rests on the assumption that variant recognition sites differ with respect to sequence and not methylation. This assumption was tested on two mtDNAs, A and B, which are distributed throughout the laboratory rat population and which can be distinguished by a number of restriction enzymes. The mtDNAs were cloned and the nucleotide sequences of corresponding small HindIII fragments, in which a variant EcoRI site occurs, were determined. Evidence that the fragments differ in sequence and not methylation is as follows: (i) The cloned mtDNA yielded the same fragment pattern as did native mtDNA when treated with EcoRI, Hha I, HinfI, and Hae III; (ii) three nucleotide replacements were found in the 169-base pair fragment, A.T in equilibrium G.C, A.T in equilibrium G.C, T.A in equilibrium G.C; (iii) one of these replacements, A.T in equilibrium G.C at position 80, accounts for the presence of the EcoRI site in the type A and its absence in the type B mtDNA. Examination of the sequence leads to the suggestion that these three nucleotide replacements are silent; i.e., they would not lead to amino acid substitutions in a possible encoded protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Lansman R. A., Shade R. O. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979 May;92(1):279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Clayton D. A. The transcription map of mouse mitochondrial DNA. Cell. 1978 May;14(1):143–156. doi: 10.1016/0092-8674(78)90309-4. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton W. G., Grabowy C. T., Sager R. Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1390–1394. doi: 10.1073/pnas.76.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzo K., Fouts D. L., Wolstenholme D. R. EcoRI cleavage site variants of mitochondrial DNA molecules from rats. Proc Natl Acad Sci U S A. 1978 Feb;75(2):909–913. doi: 10.1073/pnas.75.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D. J., Tait A., Goddard J. M. Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta. 1974 Nov 20;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Dawid I. B. 5-methylcytidylic acid: absence from mitochondrial DNA of frogs and HeLa cells. Science. 1974 Apr 5;184(4132):80–81. doi: 10.1126/science.184.4132.80. [DOI] [PubMed] [Google Scholar]
- Dawid I. B. Evolution of mitochondrial DNA sequences in Xenopus. Dev Biol. 1972 Oct;29(2):139–151. doi: 10.1016/0012-1606(72)90051-6. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Five TGA "stop" codons occur within the translated sequence of the yeast mitochondrial gene for cytochrome c oxidase subunit II. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6534–6538. doi: 10.1073/pnas.76.12.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco J. F., Brown G. G., Simpson M. V. Further studies on types A and B rat mtDNAs: cleavage maps and evidence for cytoplasmic inheritance in mammals. Plasmid. 1979 Jul;2(3):426–436. doi: 10.1016/0147-619x(79)90026-x. [DOI] [PubMed] [Google Scholar]
- Francisco J. F., Simpson M. V. The occurrence of two types of mitochondrial DNA in rat populations as detected by EcoRI endonuclease analysis. FEBS Lett. 1977 Jul 15;79(2):290–294. doi: 10.1016/0014-5793(77)80805-3. [DOI] [PubMed] [Google Scholar]
- Groot G. S., Kroon A. M. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in -CCGG- sequences. Biochim Biophys Acta. 1979 Sep 27;564(2):355–357. doi: 10.1016/0005-2787(79)90233-8. [DOI] [PubMed] [Google Scholar]
- Hayashi J. I., Yonekawa H., Gotoh O., Tagashira Y., Moriwaki K., Yosida T. H. Evolutionary aspects of variant types of rat mitochondrial DNA'S. Biochim Biophys Acta. 1979 Sep 27;564(2):202–211. doi: 10.1016/0005-2787(79)90219-3. [DOI] [PubMed] [Google Scholar]
- Hayashi J. I., Yonekawa H., Gotoh O., Watanabe J., Tagashira Y. Strictly maternal inheritance of rat mitochondrial DNA. Biochem Biophys Res Commun. 1978 Aug 14;83(3):1032–1038. doi: 10.1016/0006-291x(78)91499-7. [DOI] [PubMed] [Google Scholar]
- Hayashi J., Yonekawa H., Gotoh O., Motohashi J., Tagashira Y. Two different molecular types of rat mitochondrial DNAs. Biochem Biophys Res Commun. 1978 Apr 14;81(3):871–877. doi: 10.1016/0006-291x(78)91432-8. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon A. M., de Vos W. M., Bakker H. The heterogeneity of rat-liver mitochondrial DNA. Biochim Biophys Acta. 1978 Jun 22;519(1):269–273. doi: 10.1016/0005-2787(78)90079-5. [DOI] [PubMed] [Google Scholar]
- Macino G., Coruzzi G., Nobrega F. G., Li M., Tzagoloff A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3784–3785. doi: 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass M. M. Differential methylation of mitochondrial and nuclear DNA in cultured mouse, hamster and virus-transformed hamster cells. In vivo and in vitro methylation. J Mol Biol. 1973 Oct 15;80(1):155–175. doi: 10.1016/0022-2836(73)90239-8. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Attardi G. Identification and partial characterization of multiple discrete polyadenylic acid containing RNA components coded for by HeLa cell mitochondrial DNA. J Mol Biol. 1974 Sep 5;88(1):205–219. doi: 10.1016/0022-2836(74)90305-2. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M. Restriction endonuclease cleavage maps of rat and mouse mitochondrial DNAs. Nucleic Acids Res. 1977;4(5):1291–1299. doi: 10.1093/nar/4.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S., Newbold J. E., Hutchison C. A., 3rd, Edgell M. H. Specific cleavage analysis of mammalian mitochondrial DNA. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4496–4500. doi: 10.1073/pnas.72.11.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastl E., Dawid I. B. Expression of the mitochondrial genome in Xenopus laevis: a map of transcripts. Cell. 1979 Oct;18(2):501–510. doi: 10.1016/0092-8674(79)90067-9. [DOI] [PubMed] [Google Scholar]
- Royer H. D., Sager R. Methylation of chloroplast DNAs in the life cycle of Chlamydomonas. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5794–5798. doi: 10.1073/pnas.76.11.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens G. J., Buse G. Studies on cytochrome c oxidase, IV[1--3]. Primary structure and function of subunit II. Hoppe Seylers Z Physiol Chem. 1979 Apr;360(4):613–619. [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Van Tuyle G. C., Hamilton F. D., Vissering F. F., Simpson M. V. Biosynthesis of mitochondrial DNA. Is 8 S DNA an artifact? J Biol Chem. 1977 May 10;252(9):2984–2991. [PubMed] [Google Scholar]
- Vanyushin B. F., Kirnos M. D. Structure of animal mitochondrial DNA (base composition, pyrimidine clusters, character of methylation). Biochim Biophys Acta. 1977 Mar 18;475(2):323–336. doi: 10.1016/0005-2787(77)90023-5. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Bakker H., Saccone C., Kroon A. M. Further analysis of the type differences of rat-liver mitochondrial DNA. Biochim Biophys Acta. 1980 Mar 28;607(1):1–9. doi: 10.1016/0005-2787(80)90215-4. [DOI] [PubMed] [Google Scholar]