Abstract
Whole HIV-1 genome sequences are pivotal for large-scale studies of inter- and intrahost evolution, including the acquisition of drug resistance mutations. The ability to rapidly and cost-effectively generate large numbers of HIV-1 genome sequences from different populations and geographical locations and determine the effect of minority genetic variants is, however, a limiting factor. Next-generation sequencing promises to bridge this gap but is hindered by the lack of methods for the enrichment of virus genomes across the phylogenetic breadth of HIV-1 and methods for the robust assembly of the virus genomes from short-read data. Here we report a method for the amplification, next-generation sequencing, and unbiased de novo assembly of HIV-1 genomes of groups M, N, and O, as well as recombinants, that does not require prior knowledge of the sequence or subtype. A sensitivity of at least 3,000 copies/ml was determined by using plasma virus samples of known copy numbers. We applied our novel method to compare the genome diversities of HIV-1 groups, subtypes, and genes. The highest level of diversity was found in the env, nef, vpr, tat, and rev genes and parts of the gag gene. Furthermore, we used our method to investigate mutations associated with HIV-1 drug resistance in clinical samples at the level of the complete genome. Drug resistance mutations were detected as both major variant and minor species. In conclusion, we demonstrate the feasibility of our method for large-scale HIV-1 genome sequencing. This will enable the phylogenetic and phylodynamic resolution of the ongoing pandemic and efficient monitoring of complex HIV-1 drug resistance genotypes.
INTRODUCTION
Next-generation sequencing (NGS) provides unprecedented possibilities for the large-scale sequencing of virus genomes. The sequencing of RNA viruses such as human immunodeficiency virus type 1 (HIV-1) depends on reverse transcription (RT) and amplification to enrich virus genomes to the amounts of DNA required for NGS. Furthermore, short-read sequence assemblies of RNA virus and HIV-1 genomes are complicated by the high level of population diversity (17) due to error-prone RNA polymerases and reverse transcriptases, respectively, and high rates of virus genome replication.
HIV-1 is one of the most genetically diverse viruses known. Four genetic groups have been described: the major group M, which causes ∼85% (7) of the ∼34 million infections worldwide (13a) and is further divided into nine subtypes (subtypes A to D, F to H, J, and K) (15); the outlier group O (3, 6, 31); the nonmajor and nonoutlier group N (28); and another recently designated group, group P (23). Up to 35% amino acid differences between subtypes are found, and strains belonging to the same subtype can vary by up to 20% (7). In addition, intersubtype recombination is common (25). Circulating recombinant forms (CRFs), found in three or more epidemiologically unlinked individuals, and unique recombinant forms (URFs), identified in fewer than three individuals, consist of mosaic genomes with sections of two or more subtypes. To date, 51 CRFs have been identified.
The ability to generate HIV-1 genome sequences is crucial for an understanding of the dynamics of the pandemic at the population level and viral diversification, including the acquisition of drug resistance mutations, in individual patients. As the phenotype of a virus is the compound effect of polymorphisms present in a genome and as HIV therapy now targets proteins encoded by genes dispersed throughout the 9.7-kb genome, there is a need for high-performance HIV-1 whole-genome sequencing. A method for NGS of HIV-1 genomes of subtype B and analysis of minor variants was described recently (8), but due to the remarkable degree of sequence heterogeneity, a universal method for the generation of large numbers of HIV-1 genome sequences has remained elusive.
Here we present a novel method for rapid and reliable amplification, NGS, and the assembly of HIV-1 genomes that does not necessitate prior knowledge of the HIV-1 group or subtype. We apply the method to investigate the genetic diversity of HIV-1 genes, groups, and subtypes as well as mutations associated with HIV-1 drug resistance in clinical samples.
MATERIALS AND METHODS
Samples and RNA extraction.
Virus strains and isolates were obtained from the National Institute for Biological Standards and Control at the Health Protection Agency or University College London. Residual ETDA plasma samples sent to the Department of Clinical Microbiology and Virology, University College London Hospitals NHS Foundation Trust, for routine genotypic analysis and samples provided by Imperial College London were anonymized before analysis (see Table S1 in the supplemental material). Viral RNA was purified with a QIAamp viral RNA minikit (Qiagen).
Primer design and one-step RT-PCR.
A “pan”-HIV-1 primer set for the amplification of HIV-1 genomes of all groups and subtypes was designed based on 1,496 sequences of the 2009 “Web alignment” from the Los Alamos HIV Sequence Database (Table 1). One-step RT-PCRs generating overlapping amplicons of 1.9 kb, 3.6 kb, 3 kb, and 3.5 kb were performed by using a SuperScriptIII One-Step RT-PCR system with Platinum Taq DNA High Fidelity polymerase (Invitrogen). Each 25-μl reaction mixture contained 12.5 μl reaction mix (2×), 4.5 μl RNase-free water, 1 μl each of each primer (20 pmol/μl), 1 μl SuperScriptIII RT/Platinum Taq High Fidelity mix, and 5 μl of template RNA. Cycling conditions were 50°C for 30 min; 94°C for 2 min; 35 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 4 min 30 s; and, finally, 68°C for 10 min. Amplicons were verified by agarose gel electrophoresis and quantified by using Quant-iT PicoGreen dsDNA reagent (Invitrogen).
TABLE 1.
Primers used in this study
| Set and primer | Sequence (5′–3′) | Positions (nt)a | Product size (bp)a |
|---|---|---|---|
| 1 | |||
| Pan-HIV-1_1F | AGC CYG GGA GCT CTC TG | 26–42 | 1,928 |
| Pan-HIV-1_1R | CCT CCA ATT CCY CCT ATC ATT TT | 1953–1931 | |
| 2 | |||
| Pan-HIV-1_2F | GGG AAG TGA YAT AGC WGG AAC | 1031–1051 | 3,574 |
| Pan-HIV-1_2R | CTG CCA TCT GTT TTC CAT ART C | 4604–4583 | |
| 3 | |||
| Pan-HIV-1_3F | TTA AAA GAA AAG GGG GGA TTG GG | 4329–4351 | 3,066 |
| Pan-HIV-1_3R | TGG CYT GTA CCG TCA GCG | 7394–7377 | |
| 4 | |||
| Pan-HIV-1_4F | CCT ATG GCA GGA AGA AGC G | 5513–5531 | 3,551 |
| Pan-HIV-1_4R | CTT WTA TGC AGC WTC TGA GGG | 9063–9043 |
According to HIV-1 reference strain HXB2 (GenBank accession number NC001802). nt, nucleotides.
Roche/454 sequencing, quality control, and read assembly.
Amplicons were pooled in equimolar amounts, and 500 ng of DNA was sequenced by using a Genome Sequencer FLX Titanium XL+ instrument (Roche/454 Life Sciences) (19). Up to 17 samples were sequenced on one-quarter of a PicoTiterPlate using multiplex identifier (MID) adaptors, as previously described (4). SFF files were de-multiplexed and converted to FASTQ files, primer sequences were removed, and quality control (removing reads of <200 bp and trimming low-quality bases from the 3′ end of the reads until the median quality of the read was >30) was performed by using QUASR (http://sourceforge.net/projects/quasr/). A de novo assembly was constructed by using GS De Novo Assembler, version 2.6 (Roche/454 Life Sciences). Overlapping contiguous sequences were aligned by using CAP3 (11) and visually inspected with Se-Al, version 2.0a11 (http://tree.bio.ed.ac.uk/software/seal/), to derive a consensus sequence.
Phylogenetic analysis.
A reference set of 29 full-genome sequences was obtained. A total of 25 representative HIV-1 sequences from each group and subtype, 2 simian immunodeficiency virus (SIV) SIVcpz sequences, and 2 SIVgor sequences were selected. The sequences have the following GenBank/EMBL/DDBJ accession numbers: AB253421, AB287379, K03455, AY423387, EF469243, AB254141, K03454, DQ054367, U54771, AB253423, FJ771010, AB485658, AF084936, AY612637, AF190127, FJ711703, GU237072, AF082394, AJ249235, AJ249239, L20571, AJ302647, AJ006022, AJ271370, GU111555, DQ373066, AF103818, FJ424866, and FJ424863. Sequences derived in this study were aligned with the reference set by using MAFFT, version 6.857b (14), and the alignment was manually curated. A Bayesian phylogeny was reconstructed by using MrBayes, version 3.2 (12), under the general time-reversible model of nucleotide substitution with the proportion of invariable sites and gamma-distributed rate heterogeneity, as determined by jModelTest, version 0.1.1 (24). The Markov chain Monte Carlo search was set to 50,000,000 iterations, with trees being sampled every 2,500th generation and a 20% burn-in being discarded. Multiple chains were run to check chain convergence. The tree was edited with FigTree, version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Detection of HIV-1 subtypes and recombination.
The Rega HIV-1 Subtyping Tool, version 2.0 (http://dbpartners.stanford.edu/RegaSubtyping/), and the Recombinant Identification Program, version 3.0 (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html), were used to identify the subtypes of sequences and the presence of recombinants. Recombination was verified with the manual bootscan method (27), implemented in the Recombination Detection program, version 3.42 (20, 21).
Visualization of HIV-1 genome diversity.
Circos software (16) was used to visualize the diversity of HIV-1 genes, groups, and subtypes.
Drug resistance mutations.
The consensus sequence for each sample was used as a sample-specific reference sequence for the mapping of reads using Burrows-Wheeler Aligner (18). To analyze minor species, read depth and frequencies of mutations associated with drug resistance, as outlined in the Stanford HIV drug resistance database (http://hivdb.stanford.edu/), were calculated by using custom Python scripts.
Nucleotide sequence accession number.
The Roche/454 Life Sciences sequencing data obtained in this study are available from the EMBL/GenBank/DDBJ Sequence Read Archive under study accession number ERP001257.
RESULTS AND DISCUSSION
Design of a protocol for pan-HIV-1 RT-PCR, NGS, and assembly of HIV-1 genomes.
We designed a “pan”-HIV-1 primer set targeting semiconserved regions of the genome, based on ∼1,500 HIV-1 genome sequences (Table 1; see also Fig. S1 in the supplemental material). This novel primer set reverse transcribes and amplifies the amino acid coding region and partial long terminal repeats (LTRs) of the HIV-1 genome in four overlapping products. We chose a one-step RT-PCR protocol and a MID-tagged library preparation to minimize sample handling and to enable the processing and potential automation of large numbers of samples. We did not apply the primer ID approach that addresses the problems of resampling, PCR error and recombination, differential amplification, and sequencing errors (13), as it cannot be adapted easily to a one-step RT-PCR protocol using long amplicons. It is important to sequence the viral RNA genome and not the proviral DNA, as the former represents the current replication pool contributing to HIV-1 diversity and evolution. We combined our pan-HIV-1 RT-PCR method with Roche/454 sequencing and de novo virus genome assembly, as the reference genome-based assembly introduces biases (2). Therefore, our new method can be adapted readily to other RNA viruses as primer sets (34) or algorithms to design them (33) become available. We estimate the cost of sequencing reagents to be $270 per sample, if 100 samples are sequenced on a PicoTiterPlate to achieve a minimum coverage of 500-fold to identify minor species, and Roche/454 list prices are applied.
Broad application of the protocol.
As a first step, we used the WHO HIV-1 genotype reference panel (10) to validate our primer set. The panel comprises 10 RNA samples of HIV-1 groups M, N, and O and various subtypes, and we successfully amplified all four products from each sample (see Fig. S2 in the supplemental material). An in silico PCR was performed by using the 468 HIV-1 genome sequences in GenBank of sufficient length to cover all primer sites (≥9,300 bp) to investigate computationally the function of the primer set across a broad range of HIV-1 diversities (see Fig. S3 in the supplemental material). This virtual PCR showed that the primer set could successfully amplify full genomes from 394 (84%) of these viruses, while 74 (16%) failed to bind at least one primer and would not be expected to yield full genome coverage.
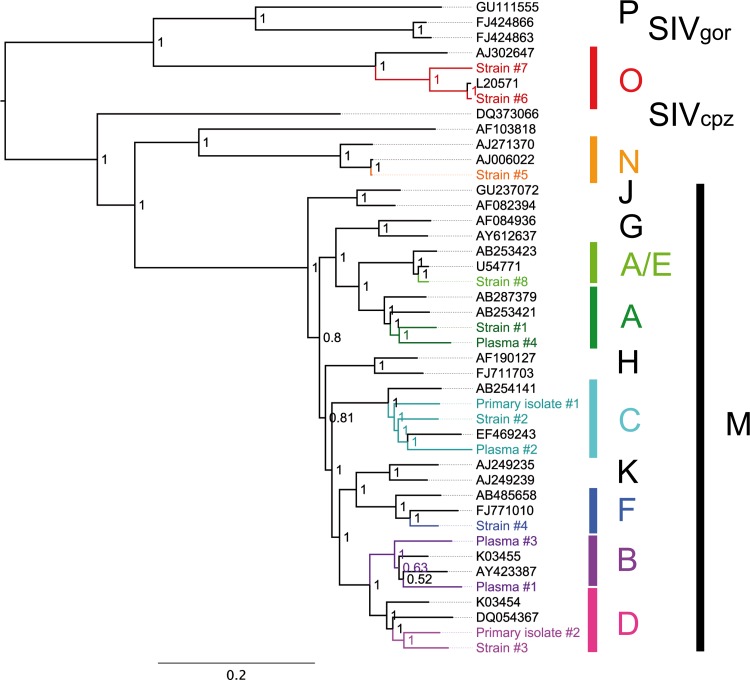
As a second step, we reverse transcribed, amplified, and sequenced 15 RNA samples of various HIV-1 groups and subtypes as well as CRFs, ranging from cell culture reference subtype virus strains to primary clinical isolates to uncultured EDTA plasma virus samples. Virus strains were selected to be representative of the HIV-1 diversity spectrum, and rare subtypes/groups were also included, while primary isolates and plasma virus samples were randomly selected. Short-read data were preprocessed by using QUASR (http://sourceforge.net/projects/quasr/), and de novo genome assemblies were performed. For all samples, the complete amino acid coding region of the HIV-1 genome was covered. Sequences of all major subtypes of group M, as well as groups N and O, were generated by NGS. Bayesian phylogenetic analysis of 14 assembled HIV-1 genomes (not including CRF14_BG) together with a reference set of complete HIV-1 and SIV genomes showed that NGS and assembly were successful across the range of HIV-1 diversities (Fig. 1). Previously sequenced HIV-1 strains were included in the analysis. GenBank sequences and sequences obtained by NGS showed nucleotide identities of 98.8 to 99.4% (see Table S1 in the supplemental material). As expected, a clear phylogenetic clustering of GenBank sequences and sequences obtained by NGS was found for both strain 5 (group N) and strain 6 (group O). The minimal differences reflect accumulated genetic variations from cultures of these HIV-1 strains since their original isolation. We also sequenced two CRFs, CRF01_AE and CRF14_BG and used the derived HIV-1 genomes to verify recombination (see Fig. S4 in the supplemental material). The ability to amplify and sequence CRFs using one set of pan-HIV-1 primers is of central importance, as CRFs play a significant role in regional epidemics and new CRFs continue to emerge (7, 30).
Fig 1.
Bayesian phylogeny of HIV-1 genome sequences derived in this study. As a reference set, 25 HIV-1 full-genome sequences representing all groups and subtypes, 2 SIVcpz sequences, and 2 SIVgor sequences were included. New NGS sequences produced in this study are shown in color. Bayesian posterior probabilities are indicated on the corresponding nodes. The tree is midpoint rooted. The scale bar represents the number of nucleotide substitutions per site.
Specificity.
For specificity tests, we used RNA or DNA from plasma samples of patients infected with other blood-borne viruses important for the differential diagnosis of HIV-1 and samples from healthy individuals. All samples, including nucleic acid obtained from plasma samples positive for HIV-2, cytomegalovirus, and hepatitis C virus, with various viral loads, failed to amplify the HIV-1 PCR products (see Table S1 in the supplemental material).
Sensitivity.
The sensitivity of the RT-PCR was determined by using a total of 90 plasma virus samples of known copy numbers, ranging from 3,000 to 2,800,000 copies/ml (see Fig. S5 in the supplemental material). We were able to amplify all four products from 84/90 samples (93.33%) and determined a sensitivity of at least 3,000 copies/ml. For the remaining 6 samples (6.66%) that did not have specifically low viral loads, we successfully amplified three products. This sensitivity of 3,000 copies/ml is sufficient for many samples, but a higher sensitivity would be preferred for clinical utility. Nevertheless, there is the possibility of extracting RNA from a larger volume of plasma if the detection limit of the protocol described here is reached.
Reproducibility and accuracy.
To investigate the reproducibility and accuracy of our method, we repeated both emulsion PCR and sequencing runs for the complete set of 15 samples, using the Roche/454 libraries as starting material. The consensus sequences that were derived were 100% identical to the consensus sequences generated by the first sequencing run (see Table S1 in the supplemental material). The frequency of mutations associated with drug resistance in clinical samples differed by a maximum of 1.1% (Table 2). The clinical impact of these minor species is as yet undetermined, but they can represent a reservoir for the emergence of resistant viruses (32). While the exact frequencies of minor species are clearly of research interest, mutations that imply drug resistance should be considered, regardless of their frequencies.
TABLE 2.
Mutations associated with HIV-1 drug resistance
| Inhibitor and codona | Amino acid of wild-type sequence | Mutation(s) associated with drug resistanceb | Frequency of mutation in run 1/frequency of mutation in run 2 (%)c |
||||
|---|---|---|---|---|---|---|---|
| Plasma sample 1 | Plasma sample 2 | Plasma sample 3 | Plasma sample 4 | Strain 92UG037 | |||
| Protease inhibitor | |||||||
| 46 | M | I, L | 1.4/0.3 | 1.0/0.7 | |||
| 47 | I | V, A | 0.9/1.1 | 1.0/0.6 | |||
| 50 | I | V | 1.1/0.8 | 1.0/0.7 | |||
| 82 | V | A, T, F, S, L | 1.2/2.1 | ||||
| Nucleoside reverse transcriptase inhibitor | |||||||
| 65 | K | R | 46.9/47.0 | ||||
| 67 | D | N | 8.2/8.0 | 1.5/1.4 | 7.2/7.6 | 9.7/10.2 | 4.6/4.1 |
| 115 | Y | F | 1.2/0.7 | 1.3/1.7 | |||
| 184 | M | V, I | 0.7/1.2 | 66.2/65.8 | |||
| 215 | T | Y, F | 1.0/0.9 | ||||
| 219 | K | Q, E | 2.2/1.7 | 2.8/2.9 | 5.7/6.1 | 5.5/4.9 | |
| Nonnucleoside reverse transcriptase inhibitor | |||||||
| 101 | K | E, P | 1.9/2.4 | ||||
| 103 | K | N, S | 38.1/38.5 | ||||
| 181 | Y | C, I, V | 38.9/38.4 | 91.4/92.1 | |||
| 190 | G | A, S, E | 59.9/59.2 | 91.2/92.2 | |||
| Integrase inhibitor | No mutations associated with drug resistance found | ||||||
| Fusion inhibitor (enfuvirtide) | |||||||
| 42 | N | T | 1.2 | ||||
According to HIV-1 reference strain HXB2 (GenBank accession number NC001802).
Only mutations that were found at frequencies of ≥1% in at least one sample are shown.
Frequencies determined by two independent sequencing runs are shown.
Frequency of in vitro recombination.
To determine the frequency of in vitro recombination and the accuracy of frequencies of minor species, we prepared a 1:1 mix of RNA from two plasma samples containing HIV-1 subtype B (viral load, 360,000 copies/ml) and subtype C (viral load, 120,000 copies/ml), respectively. We amplified one amplicon from this mixed population using primer set 1, endpoint diluted the amplicon, and reamplified it in 96-well plates so that fewer than 20% of the wells yielded a product (26). We sequenced 64 reamplified products separately and included amplicons derived from the nonmixed RNA of subtypes B and C as controls. For each sequence, we determined the subtype and controlled for recombination. We found 46/64 (71.88%) products of subtype B, 17/64 (26.56%) products of subtype C, and a single product with an intersubtype B-C recombinant sequence corresponding to an in vitro recombination rate of 1.56% (see Table S2 in the supplemental material). Therefore, both in vitro recombination and differential amplification did not represent a major problem when this protocol was applied. However, in vitro recombination can occur during PCR amplification and depends on various factors such as the amount of template DNA and the specific PCR conditions (22). NGS deep-sequencing studies need to take into account and aim to minimize in vitro recombination, as it can be mistaken for viral recombination or lead to an overestimation of the viral diversity.
Genetic diversity of HIV-1 genes, groups, and subtypes.
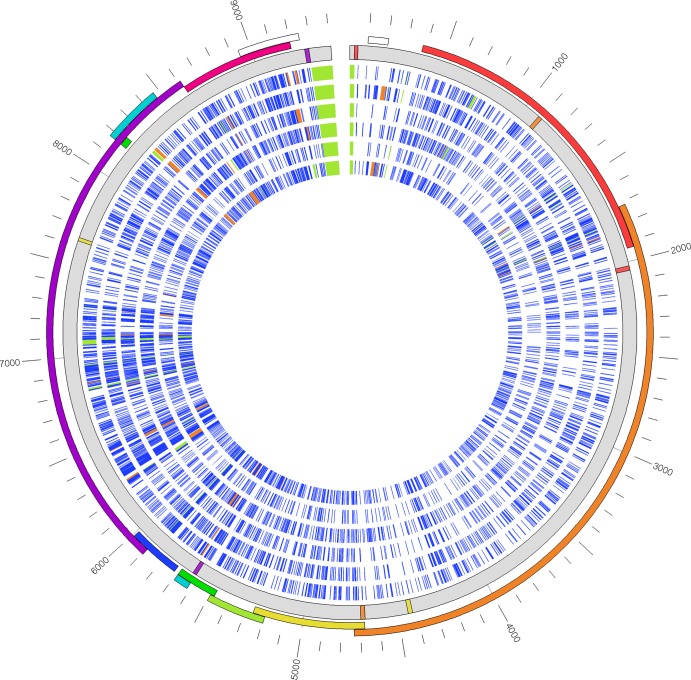
To further validate our method, we compared and visualized the genetic diversities of HIV-1 in different genes and between groups and subtypes (Fig. 2; see also Fig. S6 in the supplemental material). We found variation across the genome sequences where we expected it, and all open reading frames were maintained. The sequence similarities of different HIV-1 groups and subtypes to subtype B reference strain HXB2 were particularly low in the env, nef, vpr, tat, and rev genes and parts of the gag gene, while the pol gene showed the highest level of intersubtype similarity. These results are consistent with a previous study that obtained a comprehensive map of positive selection as well as functional and structural constraints of the HIV-1 subtype B genome (29). The group O and N sequences differed most from subtype B reference strain HXB2 and are shown relative to the relevant reference sequence in Fig. S6 in the supplemental material. Large-scale HIV-1 genome sequencing applying our new method will enable the combined analysis of host and viral genetic information for the range of HIV-1 subtypes.
Fig 2.
Genetic diversity of HIV-1 genes, groups, and subtypes. Six representative HIV-1 genomes sequenced in this study are shown in a circular format. They are displayed in the inner tracks and represent subtypes A, B, C, D, A/E, and F (from the inside to the outside of the plot). Sequences are compared to the sequence of subtype B reference strain HXB2 (GenBank accession number NC001802), shown in gray. For each sequence, every nucleotide differing from the reference strain is shown as a blue line, an insertion is shown in orange, and a deletion is shown in green. The positions of primer sets 1 to 4 for the pan-HIV-1 RT-PCR are shown as colored lines in the reference strain. The genomes contain the complete coding sequence of HIV-1, a complete U5 and a partial R region of the 5′ LTR, and a partial U3 region of the 3′ LTR. The outer track shows the open reading frames in rainbow colors and a scale bar. Genome sequences of groups N and O are shown in Fig. S6 in the supplemental material.
Mutations associated with HIV-1 drug resistance.
Highly active antiretroviral therapy is one of the most potent selective pressures on the HIV-1 genome. We used our method to investigate mutations associated with drug resistance in clinical samples (Table 2). Rigorous quality control was performed so that the median quality score of each read was >30, corresponding to a base call accuracy of 99.9%. We suggest that a minimum average coverage of 500-fold is necessary for the reliable identification of minor species, as a cutoff of 1% (5/500 reads) was used for the lowest frequency of a variant to be considered genuine. This level is conservatively well in excess of the next-generation sequencing error rate of 0.1% (1, 4). Between 40 (88.8%; plasma sample 4) and 44 (97.7%; plasma sample 1) of the 45 positions of interest were sequenced at >500-fold coverage. While drug resistance mutations were present in two plasma virus samples as the major variant (i.e., the consensus sequence), additional minor species with frequencies of between 1% and 46.9% were also identified at various positions. In contrast, HIV-1 strain 92UG037, which was included as a control, exhibited only minority D67N or K219Q/E mutations. These are secondary thymidine analogue mutations that occur in untreated populations (5). We expect that the cost-effective monitoring of complex drug resistance genotypes will be most efficiently achieved by NGS of complete HIV-1 genomes, as current antivirals target the HIV-1 protease, reverse transcriptase, integrase, and envelope (gp41), and CCR5/CXCR4 tropism needs to be considered as well.
In conclusion, we demonstrate the feasibility and the potential for large-scale HIV-1 genome sequencing from the range of HIV-1 genetic groups and subtypes prevalent globally. We expect that this new method will be pivotal for large-scale studies of the ongoing inter- and intrahost evolution of HIV-1. It has the potential to set a new standard for the clinical management of HIV infection by combining the detection of minor drug resistance mutations of clinical significance as well as covering gene targets of all present and future drug classes across the entire HIV-1 genome.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust Sanger Institute (grant S0753) and the NIHR University College London Hospitals Biomedical Research Centre.
We thank Richard Rance, Vicky Murray, and Tarryn Porter for the generation of Roche/454 sequencing data and Adele McCormick for viral RNA extractions.
A patent, “HIV-1 detection” (U.S. patent application no. 61/696,508), has been filed with the U.S. Patent Office. It will be used to facilitate the nonexclusive licensing of this method. We declare no other conflict of interest.
Footnotes
Published ahead of print 19 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Archer J, et al. 2012. Analysis of high-depth sequence data for studying viral diversity: a comparison of next generation sequencing platforms using Segminator II. BMC Bioinformatics 13:47 doi:10.1186/1471-2105-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker M. 2012. Structural variation: the genome's hidden architecture. Nat. Methods 9:133–137 [DOI] [PubMed] [Google Scholar]
- 3. Charneau P, et al. 1994. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247–253 [DOI] [PubMed] [Google Scholar]
- 4. Gall A, et al. Restriction of sequence diversity in the V3 region of the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters. Retrovirology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garcia-Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. 2004. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J. Virol. 78:7545–7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gurtler LG, et al. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23 doi:10.1097/01.aids.0000247564.73009.bc [DOI] [PubMed] [Google Scholar]
- 8. Henn MR, et al. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 8:e1002529 doi:10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10. Holmes H, Davis C, Heath A. 2008. Development of the 1st international reference panel for HIV-1 RNA genotypes for use in nucleic acid-based techniques. J. Virol. Methods 154:86–91 [DOI] [PubMed] [Google Scholar]
- 11. Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 13. Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a primer ID. Proc. Natl. Acad. Sci. U. S. A. 108:20166–20171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a. Joint United Nations Programme on HIV/AIDS 2011. UNAIDS World AIDS Day report. Joint United Nations Programme on HIV/AIDS, Geneva, Switzerland [Google Scholar]
- 14. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korber B, et al. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796 [DOI] [PubMed] [Google Scholar]
- 16. Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lauring AS, Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6:e1001005 doi:10.1371/journal.ppat.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21:98–102 [DOI] [PubMed] [Google Scholar]
- 21. Martin DP, Williamson C, Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 22. Meyerhans A, Vartanian JP, Wainhobson S. 1990. DNA recombination during PCR. Nucleic Acids Res. 18:1687–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plantier JC, et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 [DOI] [PubMed] [Google Scholar]
- 24. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 25. Robertson DL, Sharp PM, McCutchan FE, Hahn BH. 1995. Recombination in HIV-1. Nature 374:124–126 [DOI] [PubMed] [Google Scholar]
- 26. Salazar-Gonzalez JF, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salminen MO, Carr JK, Burke DS, Mccutchan FE. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type-1 by bootscanning. AIDS Res. Hum. Retroviruses 11:1423–1425 [DOI] [PubMed] [Google Scholar]
- 28. Simon F, et al. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032–1037 [DOI] [PubMed] [Google Scholar]
- 29. Snoeck J, Fellay J, Bartha I, Douek DC, Telenti A. 2011. Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology 8:87 doi:10.1186/1742-4690-8-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 358:1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanden Haesevelde M, et al. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westby M, et al. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Q, et al. 2011. PriSM: a primer selection and matching tool for amplification and sequencing of viral genomes. Bioinformatics 27:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou B, et al. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J. Virol. 83:10309–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.