Abstract
Klebsiella pneumoniae carbapenemases (KPCs) are considered a serious threat to antibiotic therapy, as they confer resistance to carbapenems, which are used to treat extended-spectrum beta-lactamase (ESBL)-producing bacteria. Here, we describe the development and evaluation of a DNA microarray for the detection and genotyping of KPC genes (blaKPC) within a 5-h period. To test the whole assay procedure (DNA extraction plus a DNA microarray assay) directly from clinical specimens, we compared two commercial DNA extraction kits (the QIAprep Spin miniprep kit [Qiagen] and the urine bacterial DNA isolation kit [Norgen]) for the direct DNA extraction from urine samples (dilution series spiked in human urine). Reliable single nucleotide polymorphism (SNP) typing was demonstrated using 1 × 105 CFU/ml urine for Escherichia coli (Qiagen and Norgen) and 80 CFU/ml urine, on average, for K. pneumoniae (Norgen). This study presents, for the first time, the combination of a new KPC microarray with commercial sample preparation for detecting and genotyping microbial pathogens directly from clinical specimens; this paves the way toward tests providing epidemiological and diagnostic data, enabling better antimicrobial stewardship.
INTRODUCTION
The increasing prevalence of carbapenem-resistant Enterobacteriaceae (CRE) is a growing public health concern (24, 30, 33). Resistance conferred by the Klebsiella pneumoniae carbapenemases (KPCs) in particular is an emerging problem of significant clinical importance (25). KPC enzymes are class A beta-lactamases, which confer resistance to penicillins, cephalosporins, monobactams, and carbapenems (17). KPCs were first identified in a multidrug-resistant Klebsiella pneumoniae isolate from a hospital in the United States in 1996 but have since then spread worldwide and to other Gram-negative species, like Escherichia coli and Acinetobacter baumannii (17, 35). Due to a lack of new antibiotics, there is only a limited number of treatment options left against carbapenemase-producing bacteria, such as the last-line drugs polymyxin B and colistin, which have been associated with high rates of nephrotoxicity (16). In addition, recent reports have even shown the appearance of a KPC-producing K. pneumoniae isolate that is also resistant to colistin (2, 20, 41).
In order to reduce and control the further spread of carbapenem resistance, rapid identification is crucial so that appropriate treatment can be applied (18). Classical microbiological methods are often slow and give results only after additional cultivation for 24 or even 48 h (24, 34). To address this problem, a variety of molecular methods have been developed, such as PCR or real-time PCR assays detecting carbapenemase genes (3, 6, 11, 13, 19, 32). PCR-based methods are a lot quicker than microbiological methods and can give results within a few hours. However, these methods lack the ability to detect single nucleotide polymorphisms (SNPs), which are helpful for detailed outbreak investigation and epidemiological studies. Currently, there are 11 published KPC variants (KPC-2 to KPC-12), which differ from each other only in single point mutations. For each variant, slightly different carbapenem MIC values and efficacies of beta-lactam inhibitors, like clavulanic acid, have been observed. KPC-2 and KPC-6 especially seem to confer resistance to all carbapenems, whereas other variants show less activity against imipenem or meropenem (36, 45). To identify all the different variants, sequencing is the gold standard, but this method is very time consuming and too demanding for routine clinical diagnostics. An alternative method is the use of a DNA microarray, which allows for rapid identification of SNPs and parallel detection of several resistance genes (4, 5, 7, 14, 15, 22, 23, 44, 47). However, the currently described methods for KPC gene detection (the Check-MDR CT101, CT102, and CT103 assays and Check-KPC extended-spectrum beta-lactamase [ESBL] microarray [Check-Points Health BV, Wageningen, Netherlands] and the hyplex SuperBug ID test system [Amplex BioSystems GmbH, Gießen, Germany]) do not allow for differentiation between the KPC variants. Here, we report the development and evaluation of a new DNA microarray, which is capable of SNP detection, allowing an identification of all variants from KPC-2 to KPC-11 directly from urine samples and without prior growth in culture.
MATERIALS AND METHODS
The new KPC microarray was designed to run under the same conditions as our previously developed ESBL microarray (15). We evaluated the performance of this new microarray on characterized reference strains and analyzed its detection limits. We further tested the performance of the microarray to identify KPC variants directly from urine samples without further cultivation. For this, we used two different DNA extraction kits, the QIAprep Spin miniprep kit (Qiagen) and the urine bacterial DNA isolation kit (Norgen), and validated their performances in combination with the KPC microarray. Urine samples that were spiked with different dilutions of E. coli or K. pneumoniae reference strains, carrying different variants of the blaKPC gene, were used as the testing material.
Reference strains.
Twelve well-characterized reference strains carrying blaKPC-type genes were used for the development and validation of the microarray probes and primers. E. coli (producing KPC-2) and K. pneumoniae (KPC-2 and KPC-3) were from the Robert Koch Institute, Wernigerode, Germany (31), and K. pneumoniae (KPC-3) was from the Health Protection Agency, United Kingdom (46). Three strains of K. pneumoniae—VIN, AUB, and GOU (KPC-2)—were provided by the Hopital Paul Brousse, France (12), and another five strains—VA 367 (KPC-3), VA 375 (KPC-3), VA 361 (KPC-2), VA 184 (KPC-2), and VA 406 (KPC-2)—were provided by Robert Bonomo from the Louis Stokes Cleveland VA Medical Center (8). All isolates were cultivated at 37°C in Luria-Bertani (LB) medium.
Spiking of urine samples and DNA extraction.
Noninfected urine samples (tested by routine microbiological culture) from several patients (New Royal Infirmary, Edinburgh, United Kingdom) were pooled and subsequently spiked with reference strains carrying variants of blaKPC. For an accurate determination of the limit of detection (LOD), dilution series of bacteria were produced in urine, covering a range from 1 to 109 CFU/ml urine in 11 dilution steps. The number of bacteria in each dilution step was determined via the counting of colonies on LB agar plates in duplicate. CFU numbers that were too large to be counted were extrapolated from the lower concentrations. Dilution series tests were carried out for all three strains received from the Robert Koch Institute (E. coli [KPC-2], K. pneumoniae [KPC-2], and K. pneumoniae [KPC-3]). After spiking of the urine samples, each tube was mixed and set aside at room temperature for 30 min. Before the DNA extraction procedures were applied, 100 μl of each dilution step was used to determine the exact number of CFU/ml in the urine by plating onto LB agar.
During DNA microarray development, plasmid DNA from each clinical isolate was extracted from 2 ml overnight culture using the QIAprep Spin miniprep kit (Qiagen, Hilden, Germany). For the detection study of clinical specimens, plasmid DNA from spiked urine samples was extracted from 1.7 ml of urine using the QIAprep Spin miniprep kit (Qiagen) or the urine bacterial DNA isolation kit (Norgen, Thorold, Canada), both of which were applied according to the manufacturer's instructions.
Target DNA preparation.
The target DNA used for the hybridization onto the oligonucleotide microarrays was synthesized via PCR. The primers used for the amplification of the blaKPC gene were the forward primer KPC_PR_F1 (5′-TGTCACTGTATCGCCGTG-3′) (48) and the reverse primer KPC_PR_R2 (5′-TTGACGCCCAATCCCT-3′), developed as part of this study. The amplicon was expected to be 871 bp in length. The amplification and labeling of blaKPC took place in a total reaction mixture volume of 30 μl using the following reagents: 0.4 μM each primer, 1× Taq buffer, 1 mM MgCl2, 3 U of HotStar Taq polymerase (Qiagen, Hilden, Germany), 0.1 mM each dATP, dGTP, and dTTP, 0.06 mM dCTP, and 0.04 mM cyanine 3 (Cy3)-dCTP (Fisher Scientific, Leicestershire, United Kingdom). The reactions were carried out on a Techne TC-512 thermocycler (Keison Products, Essex, United Kingdom) using an initial denaturing and activation step at 95°C for 15 min followed by 40 cycles consisting of 30 s of denaturing at 94°C, 30 s of annealing at 54°C, and 1 min of elongation at 72°C, followed by a final extension step at 72°C for 10 min. The PCR product was purified using the QIAquick Spin PCR purification kit (Qiagen) following the standard instructions and a final elution in 30 μl double-distilled water (ddH2O). The DNA yield and rate of Cy3-dCTP incorporation, expressed as the quotient of the number of nucleotides and the number of incorporated fluorescent dyes (NT/F), were determined by measuring the absorption at 260 and 550 nm (ND-1000 spectrophotometer; Nanodrop Technologies, Rockland, DE). Directly before hybridization, the labeled target DNA was fragmented for 5 min at room temperature using 0.8 mU DNase I (Promega, Mannheim, Germany) for each ng DNA in a total reaction mixture volume of 40 μl containing 1× DNase buffer. The reaction was stopped through the addition of 3 mM EGTA and incubation at 65°C for 10 min. The fragmentation efficiency was estimated by capillary gel electrophoresis using a DNA 1000 LabChip kit (Bioanalyzer 2100; Agilent, Böblingen, Germany).
Oligonucleotide microarray fabrication.
The following protocol is based on our previously published array production methods (15). All oligonucleotide capture probes were purchased from Metabion (Martinsried, Germany) and diluted to a final concentration of 20 μM in spotting buffer (Nexterion Spot I and Spot III, in a 1:3 ratio). Each probe had an 11-thymidine spacer and an amino modification at the 5′ end. Using a contact printer (MicroGrid II; BioRobotics, Cambridge, United Kingdom) with split pins (MicroSpot 2500; BioRobotics), each probe was spotted in triplicate onto epoxy-coated slides (Nexterion Slide E; Schott, Jena, Germany). A total of 4 arrays were printed per slide. In order to immobilize the probes after spotting, the slides were incubated for 30 min at 60°C in a drying oven (Memmert, Schwabach, Germany). At this stage, the slides could be stored for several months. Before hybridization, the slides were rinsed for 5 min in 0.1% (vol/vol) Triton X-100, for 4 min in 0.5 μl of concentrated HCl per ml of ddH2O, for 10 min in 100 mM KCl, and finally for 1 min in ddH2O. Subsequently, the slides were blocked for 15 min at 50°C in blocking solution containing 0.3% (vol/vol) ethanolamine in 100 mM Trizma base adjusted to pH 9 with HCl. Finally, they were rinsed for 1 min in ddH2O and spun dry at 1,300 rpm for 2 min in an Eppendorf 5810 R centrifuge (Eppendorf AG, Hamburg, Germany) equipped with swing-bucket rotor adapters for 96-well plates using a metal slide rack (Lipshaw, Detroit, MI). In addition to blaKPC-specific probes, several control probes were included on each array: a prelabeled spotting control (5′-TTTTTTTTTTTTCTAGACAGCCACTCATA-Cy3-3′), a positive hybridization control (5′-TTTTTTTTTTTGATTGGACGAGTCAGGAGC-3′) complementary to a labeled oligonucleotide target (5′-Cy3-GCTCCTGACTCGTCCAATC-3′), which was spiked during hybridization, and a negative control (5′-TTTTTTTTTTTTCTAGACAGCCACTCATA-3′). All control sequences were derived from Arabidopsis thaliana and are very distant from any target sequences found in bacteria. Spotting controls were spotted at every corner of each subarray (10 μM), whereas positive and negative controls were distributed alternately along the sides of each subarray.
Hybridization and washing.
For the analysis of KPC strains, 100 ng target DNA was used for hybridization onto each microarray. In the case of the dilution series, the total amount of target DNA received from the labeling PCR was used for hybridization (28 μl) and ranged from 1 to 1,600 ng DNA. For hybridization, the target DNA was supplemented with 0.2 pmol of oligonucleotide complementary to the positive hybridization control in 100 μl with 2× SSPE (20× SSPE: 3 M NaCl, 200 mM NaPO4, 20 mM EDTA [pH 7.4]) and 0.01% SDS. The hybridization was performed in an Agilent microarray hybridization chamber using gasket slides to cover the microarray, with incubation for 1 h at 47°C in an Agilent hybridization oven at 6 rpm (Agilent Technologies). After hybridization, the slides were washed at room temperature for 10 min each in 2× SSC (20× SSC: 3 M NaCl and 0.3 M sodium citrate) with 0.2% SDS, 2× SSC, and 0.2× SSC. Subsequently, the slides were dipped in ddH2O for less than 2 s and spun dry at 1,300 rpm for 2 min in an Eppendorf 5810 R centrifuge. At this point, the slides could be stored at room temperature until scanning.
Image acquisition and data analysis.
After hybridization, the fluorescent signals were acquired with a Tecan LS reloaded laser scanner (Tecan Austria GmbH, Grödig, Austria) at 532 nm and a 575-nm Cy3 filter. Each slide was scanned with 3 different photomultiplier tube (PMT) gain settings (150, 180, and 200), using a resolution of 10 μm. The quantification of signal intensities was performed using QuantArray Analysis Software (Packard BioChip Technologies, Billerica, MA) followed by data analysis and processing in Microsoft Excel (Microsoft, Redmond, WA). First, the local background of each spot was subtracted from the raw spot intensity value, followed by the calculation of the mean net signal intensity (NI) and standard deviation (SD) of the three replicates. Within each probe set (probes interrogating one mutation site), the probe with the highest signal intensity was termed the perfect match (PM), whereas the remaining probes were marked as mismatches (MM). In order to evaluate the performance of each probe set, the ratios between the MM and PM signal intensities were calculated. The larger the relative difference between the MM and PM signals, the better the discriminative power of the probe set. The MM probe with the highest signal intensity was used for the calculation of the relative signal intensity [RImax(MM) = NImax(MM)/NIPM]. Only the probe sets that showed performances with RImax(MM) values of <0.7 were used for analysis. The use of this threshold has been proven to result in high-quality discriminations (10, 15). In addition to the RI value, the limit of detection (LOD) was used to evaluate performance of the probe sets. The LOD was calculated based on the maximum signal intensity (NImax) obtained within each probe set, based on a no-template control (NTC) hybridization plus 3 times the highest standard deviation [LOD = NImax + (3 × SDmax)]. Only probe sets with perfect-match signal intensities above the limit of detection (NIPM > LOD) were used for analysis. In addition, the coefficient of variation (CV) was calculated for each set of replicate probes (CV = SD/NIPM). Probe sets with CV values of >30% were flagged and excluded from analysis to ensure that only the probe signals with high reproducibility were used for the analysis. The correct blaKPC variant was then identified based on the combination of all valid perfect-match signals. The KPC variants and their single nucleotide polymorphisms (SNPs) used for identification corresponded to recently published data (3). The mathematics described above were applied automatically using Excel (Microsoft) using the input of the raw quantification files obtained from QuantArray to identify the correct KPC variant.
RESULTS
Construction of the KPC microarray.
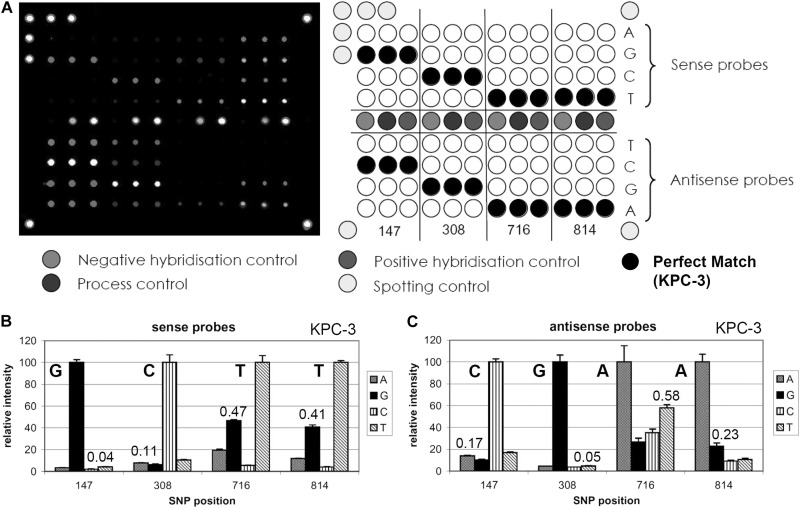
In this study, we developed a DNA microarray for the rapid detection of KPC beta-lactamase (blaKPC)-positive bacteria, which is capable of distinguishing between the different KPC variants. The probes used for the DNA microarray were designed to identify single nucleotide changes in the four mutation hot spots (positions 147, 308, 716, and 814) of the blaKPC gene, allowing an identification of all known KPC variants (3). For each position of interest, two sets of probes were designed, sense and antisense probes, resulting in a very robust detection system. Each probe consisted of a 16- to 19-bp oligonucleotide with a 13-thymidine spacer and a C6-amino modification at the 5′ end. All 32 oligonucleotide probes and the two primers that were used are listed in Table 1. The array layout as shown in Fig. 1 can easily be expanded in the future by the addition of new probes to cover potential KPC variants with different mutation hot spots.
TABLE 1.
blaKPC primer and oligonucleotide probe sequences
| Oligonucleotide namea | 5′–3′ sequenceb,c | Position/SNP in blaKPC | Tm (°C)d | Reference |
|---|---|---|---|---|
| KPC_SNP1_s | TGTACGCGATNGATACCGG | 147 | 55.4 | This study |
| KPC_SNP1_as | CCGGTATCNATCGCGTACA | 147 | 55.4 | This study |
| KPC_SNP2_s | GCTGGTTCNGTGGTCAC | 308 | 54.9 | This study |
| KPC_SNP2_as | GTGACCACNGAACCAGC | 308 | 54.9 | This study |
| KPC_SNP3_s | TGCGGAGNGTATGGCA | 716 | 55.2 | This study |
| KPC_SNP3_as | TGCCATACNCTCCGCA | 716 | 55.2 | This study |
| KPC_SNP4_s | GATGACAAGNACAGCGAGG | 814 | 54.5 | This study |
| KPC_SNP3_as | CCTCGCTGTNCTTGTCATC | 814 | 54.5 | This study |
| KPC_PR_F1 | TGTCACTGTATCGCCGTC | 2–20 | 54.5 | 48 |
| KPC_PR_R2 | AGGGATTGGGCGTCAA | 857–872 | 53.8 | This study |
Every probe was spotted as sense (s) or antisense (as).
For each single nucleotide polymorphism (SNP) position, four probes were designed that differ only at their central bases (N = A, G, C, or T). The relevant nucleotide triplets are underlined.
Every probe was modified with a 13-thymidine spacer and a C6-amino modification at the 5′ end.
The melting temperatures (Tm) were calculated with the OligoAnalyzer (Integrated DNA Technologies) using default parameters.
Fig 1.
(A, left panel) Typical fluorescent image of a DNA microarray hybridized with blaKPC target DNA from Klebsiella pneumoniae (Health Protection Agency [HPA] isolate) carrying variant KPC-3. (A, right panel) KPC DNA microarray layout. All relevant SNP positions are covered by a set of 8 probes (all four bases as sense and antisense) spotted in triplicate. The perfect-match positions are marked with black circles corresponding to the blaKPC-3 variant. The bottom images represent the resulting relative fluorescent signal intensities of sense (A) and antisense (B) probes hybridized. The corresponding perfect-match signal patterns from the sense (GCTT) and antisense (CGAA) probes identified variant KPC-3 correctly. The combined analysis of sense and antisense strands increases the robustness of the system. The numbers represent the mismatch-to-perfect-match ratios (MM/PM), a measure for the discriminative power of each probe set. In general, the probe sets with MM/PM ratios larger than 0.7 were omitted from the analysis. The respective SNP was then covered by the corresponding sense/antisense probe set.
Validation of the DNA microarray using reference strains.
The performance of the KPC microarray was validated using 12 well-characterized KPC-producing reference strains, which were all identified correctly. In all cases, the Cy3 labeling PCR amplification yielded, as expected, an 871-bp product in a concentration range of 15 to 25 ng/μl. The rate of label incorporation, the number of nucleotides per number of incorporated fluorescent dyes (NT/F), varied between 34 and 76, depending on the quality of the template DNA. The best results were obtained using 200 ng labeled DNA product per microarray (2 ng/μl), but as little as 50 ng (0.5 ng/μl) was sufficient in all cases for the correct identification of each variant (equivalent to 870 pmol/liter). The performance of each probe set was measured using the maximum mismatch-to-perfect-match ratio (MMmax/PM). With only one exception, these values were always below 0.7 for all tested reference strains, defining a high level of discrimination for each probe set. In the single exception, the antisense probe for position 716 had a MMmax/PM of 0.711, in which case the sense probe was used for discrimination instead with a MMmax/PM value of 0.54. Based on all reference strain hybridizations, the best discrimination for the sense probes was achieved with the probe set SNP-147, which had a median relative intensity value (MMmax/PM) of 0.037, followed by SNP-308 (0.055), SNP-814 (0.377), and SNP-716 (0.526). For the antisense probes, the best discrimination was achieved with probe set SNP-308, which had a median relative intensity value (MMmax/PM) of 0.041, followed by SNP-814 (0.09), SNP-147 (0.133), and SNP-716 (0.347). Figure 1B and C shows, as an example, the relative fluorescent signal intensities of all sense and antisense probes obtained through hybridization with target DNA from K. pneumoniae carrying the blaKPC-3 variant. The relative intensity values between the maximum mismatch and perfect-match signal (MMmax/PM) are also included in Fig. 1. Both the sense and antisense probes identified variant KPC-3 correctly. The results of all other strains are shown in Fig. S1 (KPC-2) and S2 (KPC-3) in the supplemental material.
Microarray limit of detection.
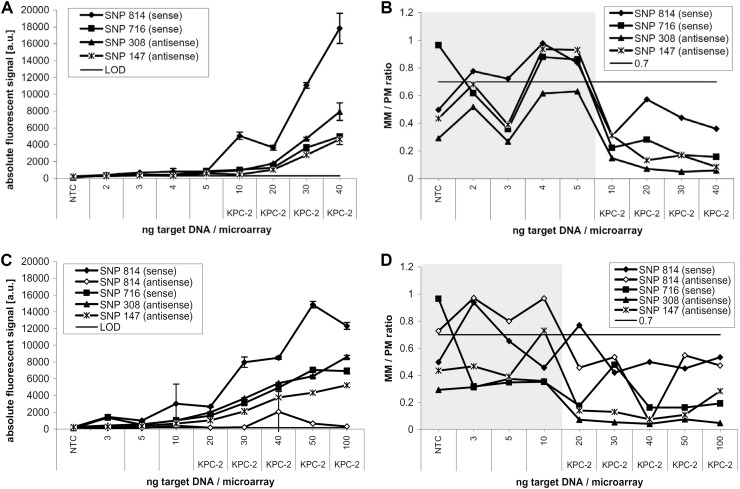
Before the limit of detection (LOD) of the whole assay was analyzed, the LOD of the microarray to labeled target DNA itself was tested. For this purpose, a dilution series of labeled target DNA (2 to 100 ng) was made, amplified from E. coli (KPC-2). Before hybridization, two different methods of target DNA treatment were applied, one using a DNase concentration adjusted to the actual amount of target DNA and the other using a fixed concentration independent of the amount of target DNA (resembling the clinical test situation, where the amount of DNA would be unknown). For the adjusted protocol, a DNase concentration of 0.8 mU DNase for each ng DNA was found to be most efficient, whereas for the fixed concentration experiment, 16 mU DNase was used, optimized to an average amount of 20 ng target DNA. The first method was more accurate but also more time consuming, due to the additional purification and measuring steps, which are necessary to acquire the exact concentration of the target DNA. The second method using a fixed amount of DNase would be the more practical solution in terms of developing an automated diagnostic tool, which contributes to a significant reduction in assay time. A comparison of the microarray results using both methods is shown in Fig. 2. With the adjusted method (Fig. 2A), the correct KPC variant was detected using as little as 10 ng of target DNA (equivalent to 170 pmol/liter), whereas when using a fixed amount of DNase (Fig. 2B), the correct KPC variant was identified using as little as 20 ng target DNA (350 pmol/liter). Hybridizations using the adjusted method resulted generally in higher absolute fluorescent signals as well as better (lower) MM/PM ratios. Therefore, this method was applied to all the following experiments.
Fig 2.
Limit of detection of the DNA microarray using dilutions of target DNA. A dilution series of labeled target DNA (2 to 100 ng) was hybridized onto the KPC microarray to identify its LOD. (A) Absolute fluorescent signal intensities obtained after the hybridization of target DNA, which was digested with 0.8 mU DNase/ng DNA. The identification of KPC-2 was possible with as little as 10 ng target DNA. a.u. indicates arbitrary units. (C) The DNA was digested with a fixed amount of DNase (16 mU), which is equivalent to the amount used for 20 ng (panel A). Here, as little as 20 ng target DNA/microarray was still correctly identified. (B and D) Corresponding MM/PM ratios. At 20 ng target DNA/microarray, the SNP-814 sense probe was out of range (MM/PM > 0.7); therefore the antisense probe was used for discrimination instead, which still correctly identified variant KPC-2. For all following experiments, the method presented in panels A and B (0.8 mU DNase/ng DNA) was used due to its higher reproducibility and sensitivity.
Limit of detection estimated directly from spiked urine samples.
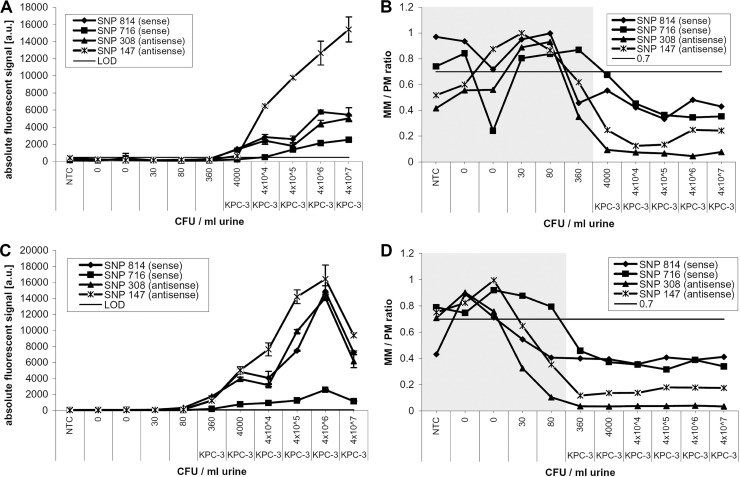
In order to determine the limit of detection (LOD) of the whole assay, uninfected urine samples were pooled and subsequently spiked with bacteria carrying variants of the blaKPC gene. These samples were diluted in 11 steps, resulting in dilution series covering a range of 1 to 109 CFU/ml urine, confirmed via colony counts on LB agar plates. All dilution series were counted at least in duplicate. Several dilution series were produced using E. coli blaKPC-2, K. pneumoniae blaKPC-2, and K. pneumoniae blaKPC-3. Subsequently, plasmid DNA was extracted from each dilution step using the QIAprep Spin miniprep kit (Qiagen) and the urine bacterial DNA isolation kit (Norgen, Thorold, Canada) in duplicate. Nonspiked urine samples were extracted as well and were used as no-template controls (NTC). The extracted DNA was amplified and analyzed using the DNA microarray. As an example, Fig. 3A and B shows the data obtained from analyzing a dilution series of K. pneumoniae (KPC-3) extracted with the QIAprep Spin miniprep kit. KPC-3 was correctly identified at a concentration of 4 × 103 CFU/ml urine. One dilution step further (360 CFU/ml), the criteria for a correct identification were not fulfilled anymore. At this dilution step, the mismatch-to-perfect-match ratio (MM/PM) for one SNP position (SNP-716) was below the threshold of 0.7 for both probe sets (sense/antisense), and in addition, the limit of detection for more than one probe set was reached. Figure 3C and D shows the corresponding data obtained with the microarray after extraction using the urine bacterial DNA isolation kit from Norgen. The correct identification of the variant KPC-3 using this method was still possible from a dilution containing 360 CFU/ml urine.
Fig 3.
Analysis of limit of detection (LOD) directly from urine samples. Overnight cultures of Klebsiella pneumoniae carrying blaKPC-3 were spiked into urine samples in a dilution series from 4 × 107 to 1 CFU/ml urine. The samples were then left for 30 min at room temperature before the DNA was extracted. In this example, the QIAprep Spin miniprep kit (Qiagen) was used for extraction. (A) Absolute fluorescent signal intensities of 2 sense and 2 antisense perfect-match probes obtained after DNA microarray analysis of the extracts from each dilution. (B) The mismatch-to-perfect-match ratios of the same probes are presented, showing data up to the dilution step at which a good discrimination (MM/PM of <0.7) was possible. In this case, KPC-3 was still identified correctly to a dilution step of 4,000 CFU/ml urine. The identified variant is shown underneath the concentration. (C and D) Data obtained from the same dilution series after extraction using the urine bacterial DNA isolation kit (Norgen). With this method, the correct KPC variant was still identified from a dilution containing 360 CFU/ml urine.
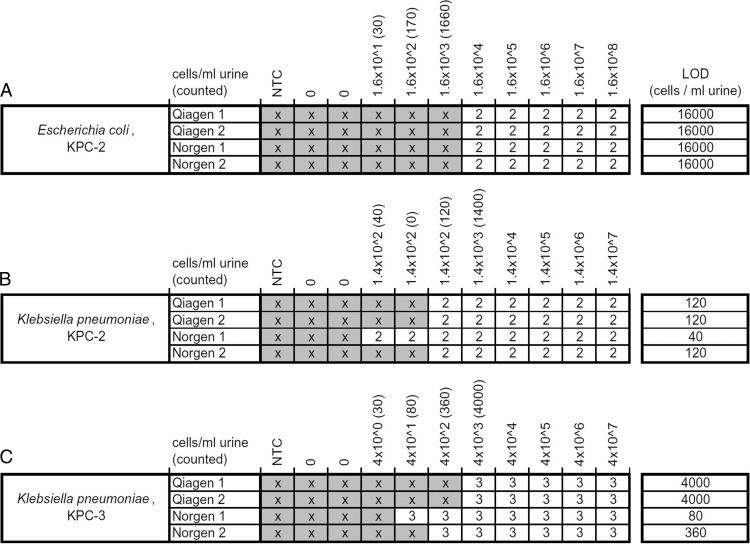
Figure 4 shows a summary of all 132 DNA microarray experiments carried out to determine the limit of detection for the whole assay. E. coli (KPC-2), which was spiked into urine samples, was still identified correctly at a concentration of 1.6 × 104 CFU/ml urine in all cases. For E. coli (KPC-2), the LOD results were the same for all replicates carried out with the Qiagen extraction kit as well as the urine extraction kit from Norgen (Fig. 4A). The cells of K. pneumoniae (KPC-2) were still identified correctly to a concentration of 120 CFU/ml urine with the Norgen and Qiagen kits. One of the Norgen extractions allowed the correct identification of the KPC-2 variant from as little as 40 CFU/ml urine (Fig. 4B). The third tested strain, K. pneumoniae (KPC-3), was identified correctly at a concentration of 4 × 103 CFU/ml urine using the Qiagen extraction kit for both replicates, whereas the Norgen kit allowed an identification at 360 CFU/ml urine (80 CFU/ml in one of the replicates) (Fig. 4C). Over all experiments, the LOD seemed to be higher for urine samples spiked with E. coli (1.6 × 104 CFU/ml urine) and lower for those spiked with K. pneumoniae (40 to 4,000 CFU/ml urine). In addition, we observed that the Norgen kit gave slightly higher yields than the Qiagen kit when extracting DNA from K. pneumoniae, resulting in a lower LOD. A more-detailed table containing all absolute PM signal intensities and the corresponding MMmax/PM ratios can be found in Fig. S3, S4, and S5 in the supplemental material. When processing 20 urine samples in parallel, the extraction using the urine bacterial DNA isolation kit (Norgen) took 2 h, on average, resulting in a total time to results of 6 h after urine sampling. When applying the QIAprep Spin miniprep kit (Qiagen), the extraction took 1 h, on average, for 20 urine samples, resulting in a total time to results of 5 h.
Fig 4.
Limit of detection (LOD) from urine samples. Here, the results obtained from 132 microarray hybridizations carried out to determine the limit of detection of the whole assay are summarized. Two extraction kits (from Norgen and Qiagen) were used to isolate DNA from urine samples spiked with 3 different strains carrying variants of the blaKPC gene. The fields marked with an “x” represent array experiments that did not fulfill all mathematical criteria for a correct analysis and therefore were beyond the limit of detection (MM/PM < 0.7, PM > LOD). The numbers represents the KPC variants, which are identified here.
DISCUSSION
The rapid detection of antibiotic resistance in clinical samples is crucial in order to provide appropriate treatment for patients in a timely manner. ESBLs and carbapenemases especially have become worldwide threats to successful antibiotic therapy. In particular, KPC carbapenem resistance has been reported increasingly in recent years, resulting in a need for new and rapid detection methods. Conventional routine methods are mostly based on phenotypical detection procedures. An example is the modified Hodge test, which can confirm the presence of carbapenemases but cannot distinguish between KPC and other carbapenemases (25, 29). To distinguish KPCs from other carbapenem producers, boronic disk tests can be used (42, 43), but still, the identification of single KPC variants is not possible. In general, all phenotypic methods are very time consuming, delivering results often only after 1 or 2 days (24). Faster are the molecular tests, such as real-time PCR assays, allowing for quick identification of KPC genes (3, 6, 11, 13, 18, 21, 24). Nevertheless, these assays often have only a limited multiplexing capability and also cannot distinguish single KPC variants from each other.
Therefore, DNA microarrays are a good alternative, offering a high multiplexing capability, and furthermore allow for the identification of SNPs, which is necessary to distinguish between single variants. The possibility of identifying single variants from each other using a DNA microarray has been demonstrated for the ESBL-relevant genes blaTEM, blaSHV, and blaCTX-M (10, 15). The commercially available microarray assays from Check-Points enable only the identification of genes and mutation hot spots relevant to resistance caused by ESBLs and carbapenemases, including the detection of blaKPC (4, 5, 7, 9, 22, 23, 44, 47). However, the Check-Points system can be used as a reliable screening tool to guide PCR sequencing, allowing in this way an identification of single variants (14).
The capability to identify single variants of the KPC gene might not have been a requirement in the past, as there were only a very limited number of KPC variants reported showing very similar phenotypes. However, recent studies have suggested that an increasing number of different KPC variants confer different resistance profiles. Knowing which variant is present might open new treatment options in the future, especially under strict antibiotic stewardship. The difference in resistance profiles and their effects on beta-lactam inhibitors were demonstrated directly in clinical samples and transformants with KPC variants and through comparisons of hydrolytic activities (1, 27, 28, 37, 45). Robledo et al. (36) reported a variation of antimicrobial susceptibility to carbapenems depending on the KPC variant during a 1-year study based on KPC-producing isolates taken from 6 Puerto Rico Medical Center hospitals. All isolates were resistant to ertapenem irrespective of the KPC variant. Isolates with KPC-2 and KPC-6 were resistant to all carbapenems tested. Isolates with KPC-4 were susceptible to imipenem and meropenem, while those with KPC-3 demonstrated variable susceptibility (36). Therefore, knowing the exact KPC variant might allow for a more target-driven use of individual carbapenems or beta-lactam inhibitors. However, the greatest benefit of SNP detection in blaKPC genes is the application in epidemiological studies to examine if the resistance found is a single case or a pandemic (10).
The KPC microarray described here was able to identify and distinguish all KPC variants that were published at the time of design (KPC-2 to KPC-11). These variants differ from each other in 4 SNP positions (nucleotides 147, 308, 716, and 814). The recently reported variant KPC-12 (see www.lahey.org/Studies/) differs from KPC-2 by a single mutation at SNP position 502, a new position, which is not covered in the current version but could easily be added to future versions of this microarray. Therefore, KPC-12 would currently be identified as KPC-2 using the microarray. Due to the selected melting temperatures of the probes and primers, the KPC microarray could be used together with our existing ESBL microarray (15), or both could be spotted onto one new microarray in the future by applying the same reaction conditions.
The KPC microarray was tested successfully on 12 different reference strains carrying either variant KPC-2 or KPC-3. These are the most frequently found KPC variants. During the course of the project, we had no access to any other KPC variants. Nevertheless, all probe sets could still be validated due to the fact that each probe set is covered by the amplicon used. Each probe set gave a clear positive hybridization signal with a high level of discrimination between perfect-match and mismatch probes when being tested with KPC-2 or KPC-3. Consequently, there are no untested probes on the array. We would consider this to be sufficient at this stage, as the method of allele-specific hybridization for SNP detection using microarrays is well established, and all probe sets were tested positive in over 160 separate hybridization experiments. Although theoretically possible, we did not design synthetic targets to test all possible hybridization patterns (all variants for each position), as this would have gone beyond the scope of the study while giving only a limited scientific benefit due to the differences in PCR amplicons and synthetic targets. The limit of detection for labeled target DNA was found to be 10 ng per assay when a DNase amount that was adjusted to the target DNA concentration was used. When a fixed DNase amount, optimized for 20 ng target DNA, was used, the limit of detection also turned out to be 20 ng. Smaller DNA amounts were probably overdigested and could therefore not be detected anymore. Higher fluorescent signals and better discrimination values (MM/PM) were obtained using the adjusted method. With the adjusted method, a total assay time of 3.5 h after DNA extraction was possible, which is significantly shorter than conventional PCRs followed by sequencing or phenotypical methods that require 1- or 2-day overnight cultivation (26, 39, 40). By using a fixed amount of DNase before hybridization, this assay time could be reduced by at least 30 min, which would otherwise be necessary for DNA purification, concentration measurements, and final digestions. Therefore, this microarray has the potential to be used as a rapid KPC resistance test.
Disregarding the much faster time to results, the introduction of molecular assays into routine diagnostics depends on the cost. In general, molecular assays are still more costly than culture-based tests. Commercially available molecular assays currently have prices of approximately $19 (real-time PCR [RT-PCR]) to $40 (microarrays) per sample. For our KPC microarray, we calculated a price of $38 per sample, which includes array production, DNA extraction from urine samples, and consumables for running the assay. The sequencing is already cheaper, with prices around $6 per sample, but prior overnight cultivation and DNA extraction are still necessary additions. Therefore, sequencing is still too demanding for routine clinical diagnostics.
Most importantly, this study demonstrates, possibly for the first time, the direct identification of KPC variants from urine samples without prior cultivation. Two different DNA extraction kits (from Qiagen and Norgen) were tested for the extraction of bacterial DNA from urine, followed directly by the microarray analysis. Urine samples spiked with dilution series of different reference strains were used as the testing material. In total, 132 extractions and microarray experiments were carried out to determine the limit of detection (LOD). In all experiments, the correct KPC variant was identified from urine samples with as little as 1.6 × 104 CFU/ml. This LOD was obtained analyzing urine samples spiked with E. coli, whereas for K. pneumoniae, an even lower limit of detection was observed (4 × 103 CFU/ml for the Qiagen kit and 360 CFU/ml for the Norgen kit). For the DNA extraction of K. pneumoniae, the Norgen kit seemed to be slightly more sensitive than the Qiagen kit. On average, only 80 CFU/ml urine was needed when using the Norgen kit. If such a level of sensitivity is not required, the Qiagen kit seems to be a lot more practicable for routine extractions, with a much shorter handling time. Phenotypic tests have a lower detection limit (e.g., 4 × 101 to 9 × 102 CFU/ml for the CHROMagar KPC test), but the results can be obtained only after 24 to 48 h or even later (26, 39). Bacterial loads of more than 106 CFU/ml in urine are considered to be a clear indication of a urinary tract infection (UTI) (38). Therefore, the KPC microarray test presented in our study would be sensitive enough to identify bacteria with KPC resistance from patients with UTIs. Since only 1.7 ml urine was used for the analyses, the limit of detection for both extraction methods could still be improved further by increasing the amount of urine used for DNA extraction. This would be especially interesting for the analysis of symptomatic patients, where the presence of 100 CFU/ml is enough to diagnose bacteriuria (38). The technology is, in principle, suitable for the direct testing of patient samples. However, the performance ability, in terms of sensitivity and specificity, needs to be further investigated in a separate study.
This study demonstrates the possibility of identifying single KPC variants directly from urine samples, without prior cultivation, using a new DNA microarray. The total assay times of 5 h (Qiagen extraction plus a DNA microarray) and 6 h (Norgen extraction plus a DNA microarray) are a lot shorter than those of classical methods of analyzing antimicrobial susceptibilities in urine samples. The bacteria could be analyzed directly from urine samples without further cultivation, and the exact KPC variant could be identified, allowing for direct information toward possible treatment options and epidemiology. A larger study on urine samples carrying KPC variants would further confirm the performance of this test.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part within the Era-Net PathoGenoMics project “Deciphering the Intersection of Commensal and Extraintestinal Pathogenic E. coli” and was financially supported by the German Federal Ministry of Education and Research.
We thank the following people for providing us with reference strains: David Livermore (Antibiotic Resistance Monitoring and Reference Laboratory, Health Protection Agency, United Kingdom), Najiby Kassis-Chikhani (Hopital Paul Brousse, France), and Robert Bonomo (Louis Stokes Cleveland Department of Veterans Affairs Medical Center, USA).
Footnotes
Published ahead of print 3 October 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 49:4760–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogdanovich T, et al. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 53:373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen LA, et al. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen Stuart J, et al. 2010. Rapid detection of TEM, SHV and CTX-M extended-spectrum beta-lactamases in Enterobacteriaceae using ligation-mediated amplification with microarray analysis. J. Antimicrob. Chemother. 65:1377–1381 [DOI] [PubMed] [Google Scholar]
- 5. Cohen Stuart J, Voets G, Scharringa J, Fluit A, Leverstein-Van HM. 2012. Detection of carbapenemase producing Enterobacteriaceae with a commercial DNA microarray. J. Med. Microbiol. 61:809–812 [DOI] [PubMed] [Google Scholar]
- 6. Cole JM, Schuetz AN, Hill CE, Nolte FS. 2009. Development and evaluation of a real-time PCR assay for detection of Klebsiella pneumoniae carbapenemase genes. J. Clin. Microbiol. 47:322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Endimiani A, et al. 2010. Evaluation of a commercial microarray system for detection of SHV-, TEM-, CTX-M-, and KPC-type beta-lactamase genes in Gram-negative isolates. J. Clin. Microbiol. 48:2618–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Endimiani A, et al. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J. Antimicrob. Chemother. 63:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gazin M, Paasch F, Goossens H, Malhotra-Kumar S, MOSAR WP2 and SATURN WP1 Study Teams 2012. Current trends in culture-based and molecular detection of extended-spectrum-beta-lactamase-harboring and carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 50:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grimm V, et al. 2004. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J. Clin. Microbiol. 42:3766–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hindiyeh M, et al. 2008. Rapid detection of bla(KPC) carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kassis-Chikhani N, et al. 2010. Outbreak of Klebsiella pneumoniae producing KPC-2 and SHV-12 in a French hospital. J. Antimicrob. Chemother. 65:1539–1540 [DOI] [PubMed] [Google Scholar]
- 13. Krafft CA, et al. 2009. Development of a real-time PCR assay to detect Klebsiella pneumoniae that produce carbapenemase (KPC) in clinical specimens. J. Mol. Diagn. 11:646–647 [Google Scholar]
- 14. Lascols C, et al. 2012. Using nucleic acid microarrays to perform molecular epidemiology and detect novel beta-lactamases: a snapshot of ESBLs throughout the world. J. Clin. Microbiol. 50:1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leinberger DM, et al. 2010. Integrated detection of extended-spectrum-beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J. Clin. Microbiol. 48:460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim LM, et al. 2010. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy 30:1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lolans K, Calvert K, Won S, Clark J, Hayden MK. 2010. Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J. Clin. Microbiol. 48:836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mangold KA, et al. 2011. Real-time detection of bla(KPC) in clinical samples and surveillance specimens. J. Clin. Microbiol. 49:3338–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendes RE, et al. 2007. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mezzatesta ML, et al. 2011. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin. Microbiol. Infect. 17:1444–1447 [DOI] [PubMed] [Google Scholar]
- 21. Monteiro J, Widen RH, Pignatari ACC, Kubasek C, Silbert S. 2012. Rapid detection of carbapenemase genes by multiplex real-time PCR. J. Antimicrob. Chemother. 67:906–909 [DOI] [PubMed] [Google Scholar]
- 22. Naas T, Cuzon G, Bogaerts P, Glupczynski Y, Nordmann P. 2011. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. J. Clin. Microbiol. 49:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naas T, Cuzon G, Truong H, Bernabeu S, Nordmann P. 2010. Evaluation of a DNA microarray, the check-points ESBL/KPC array, for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and KPC carbapenemases. Antimicrob. Agents Chemother. 54:3086–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nordmann P. 2010. Gram-negative bacteriae with resistance to carbapenems. Med Sci (Paris). 26:950–959 (In French) [DOI] [PubMed] [Google Scholar]
- 25. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 26. Panagea T, et al. 2011. Evaluation of CHROMagar KPC for the detection of carbapenemase-producing Enterobacteriaceae in rectal surveillance cultures. Int. J. Antimicrob. Agents 37:124–128 [DOI] [PubMed] [Google Scholar]
- 27. Papp-Wallace KM, et al. 2010. Inhibitor resistance in the KPC-2 beta-lactamase, a preeminent property of this class A beta-lactamase. Antimicrob. Agents Chemother. 54:890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papp-Wallace KM, et al. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 beta-lactamase. Antimicrob. Agents Chemother. 54:2867–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pasteran F, et al. 2011. A simple test for the detection of KPC and metallo-beta-lactamase carbapenemase-producing Pseudomonas aeruginosa isolates with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 17:1438–1441 [DOI] [PubMed] [Google Scholar]
- 30. Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfeifer Y. 2010. ESBL, AmpC and carbapenemases: emergence, dissemination and diagnostics of beta-lactamase-producing Gram-negative pathogens. LaboratoriumsMedizin/J. Lab. Med. 34:205–215 [Google Scholar]
- 32. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 33. Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reglier-Poupet H, et al. 2008. Performance of chromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum beta-lactamases. J. Med. Microbiol. 57:310–315 [DOI] [PubMed] [Google Scholar]
- 35. Robledo IE, et al. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robledo IE, et al. 2011. Dissemination and molecular epidemiology of KPC-producing Klebsiella pneumoniae collected in Puerto Rico Medical Center hospitals during a 1-year period. Epidemiol. Res. Int. 2011:1–8 [Google Scholar]
- 37. Sacha P, et al. 2009. The KPC type beta-lactamases: new enzymes that confer resistance to carbapenems in Gram-negative bacilli. Folia Histochem. Cytobiol. 47:537–543 [DOI] [PubMed] [Google Scholar]
- 38. Salvatore S, et al. 2011. Urinary tract infections in women. Eur. J. Obstet. Gynecol. Reprod. Biol. 156:131–136 [DOI] [PubMed] [Google Scholar]
- 39. Samra Z, et al. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schechner V, et al. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toth A, et al. 2010. Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 29:765–769 [DOI] [PubMed] [Google Scholar]
- 42. Tsakris A, et al. 2009. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J. Clin. Microbiol. 47:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsakris A, et al. 2011. Comparative evaluation of combined-disk tests using different boronic acid compounds for detection of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae clinical isolates. J. Clin. Microbiol. 49:2804–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willemsen I, et al. 2011. New diagnostic microarray (Check-KPC ESBL) for detection and identification of extended-spectrum beta-lactamases in highly resistant Enterobacteriaceae. J. Clin. Microbiol. 49:2985–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wolter DJ, et al. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woodford N, et al. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 48:4793–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woodford N, Warner M, Pike R, Zhang J. 2011. Evaluation of a commercial microarray to detect carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 66:2887–2888 [DOI] [PubMed] [Google Scholar]
- 48. Woodford N, et al. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62:1261–1264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.