Abstract
Vitamin D signaling regulates cell proliferation and differentiation, and epidemiological data suggest that it functions as a cancer chemopreventive agent, although the underlying mechanisms are poorly understood. Vitamin D signaling can suppress expression of genes regulated by c-MYC, a transcription factor that controls epidermal differentiation and cell proliferation and whose activity is frequently elevated in cancer. We show through cell- and animal-based studies and mathematical modeling that hormonal 1,25-dihydroxyvitamin D (1,25D) and the vitamin D receptor (VDR) profoundly alter, through multiple mechanisms, the balance in function of c-MYC and its antagonist the transcriptional repressor MAD1/MXD1. 1,25D inhibited transcription of c-MYC–regulated genes in vitro, and topical 1,25D suppressed expression of c-MYC and its target setd8 in mouse skin, whereas MXD1 levels increased. 1,25D inhibited MYC gene expression and accelerated its protein turnover. In contrast, it enhanced MXD1 expression and stability, dramatically altering ratios of DNA-bound c-MYC and MXD1. Remarkably, F-box protein FBW7, an E3-ubiquitin ligase, controlled stability of both arms of the c-MYC/MXD1 push–pull network, and FBW7 ablation attenuated 1,25D regulation of c-MYC and MXD1 turnover. Additionally, c-MYC expression increased upon VDR knockdown, an effect abrogated by ablation of MYC regulator β-catenin. c-MYC levels were widely elevated in vdr−/− mice, including in intestinal epithelium, where hyperproliferation has been reported, and in skin epithelia, where phenotypes of VDR-deficient mice and those overexpressing epidermal c-MYC are similar. Thus, 1,25D and the VDR regulate the c-MYC/MXD1 network to suppress c-MYC function, providing a molecular basis for cancer preventive actions of vitamin D.
Vitamin D is obtained naturally from limited dietary sources. It is also generated by cutaneous conversion of 7-dehydrocholesterol in the presence of adequate surface solar UV-B radiation, which varies with latitude and time of year (1). Vitamin D has attracted broad clinical interest because insufficiency or deficiency is widespread in several populations worldwide (2–4). Although initially identified as a regulator of calcium homeostasis, vitamin D is now known to have a broad spectrum of actions, driven by the virtually ubiquitous expression of the vitamin D receptor (VDR), a nuclear receptor and hormone-regulated transcription factor. For example, it acts as a chemopreventive agent in several animal models of cancer and induces cell-cycle arrest and nonmalignant and malignant cell differentiation (5–11). Epidemiological data have provided associations between lack of UV-B exposure, vitamin D insufficiency, and the prevalence of certain cancers (12). A large prospective study associated vitamin D sufficiency with reduced total cancer incidence and mortality, particularly in digestive cancers [head and neck squamous cell carcinoma (HNSCC), esophageal, pancreatic, stomach, and colorectal cancers] and leukemias (13). VDR gene polymorphisms also correlate with protection against different malignancies, including HNSCC (12, 14). However, results of epidemiological studies on the protective effects of vitamin D are not unanimous, and uncertainties as to the potential benefits persist (15, 16), underlining the need for not only more clinical studies, but also a better understanding of potential molecular mechanisms of the protective effects of vitamin D.
The VDR is bound by hormonal 1,25-dihydroxyvitamin D (1,25D), which is produced from vitamin D by largely hepatic 25-hydroxylation, followed by 1α-hydroxylation by widely expressed CYP27B1 (17, 18). Potential cancer preventive actions of 1,25D signaling through the VDR can be explained in part by the direct interaction of the VDR with FoxO transcription factors, leading to 1,25D-stimulated FoxO DNA-binding and target gene regulation (19). FoxO proteins regulate cell proliferation, differentiation, and metabolism and control longevity (20–23). Serial ablation of foxo genes in mice revealed that they are bona fide tumor suppressors (24–26). Conversely, although FoxO expression is often suppressed in cancer, elevated or deregulated expression of transcription factor c-MYC is widespread (27, 28). c-MYC is a critical regulator of cell-cycle progression and, like the VDR (29), controls epidermal differentiation (30). Inducible epidermal expression of c-MYC rapidly induced actinic keratosis, a squamous cell carcinoma precursor (31). Heterodimers of c-MYC and its cofactor MAX bind E-box motifs (CACGTG) to induce expression of cell-cycle regulatory genes such as CCND2 and CDK4. c-MYC is highly regulated posttranslationally and is rapidly turned over by proteasomal degradation controlled by the SCF ubiquitin ligase complex containing F-box protein Fbw7 (32). c-MYC and FoxO proteins are often regulated by common mechanisms with opposing effects. For example, both are ubiquitinated by p45SKP2 (33, 34), which induces FoxO protein turnover, but coactivates c-MYC.
Recent studies have shown that 1,25D gradually reduces MYC RNA levels in cancer cells (35, 36). Because signaling through the VDR enhances FoxO protein function (19), we investigated potential mechanisms of cross-talk between c-MYC and VDR signaling. We found that signaling through the VDR controls expression and FBW7-dependent turnover of both c-MYC and its antagonist MAD1/MXD1, leading to dramatic changes in the ratio of c-MYC and MXD1 in vitro and in vivo and strongly favoring repression of c-MYC target genes by MXD1. These findings provide a compelling mechanism for the cancer chemopreventive actions of vitamin D and implicate VDR-dependent regulation of c-MYC in control of epidermal differentiation. Moreover, c-MYC is critical for normal epidermal differentiation, and its deregulated expression in skin depletes epidermal stem cells (30, 37, 38), disrupting hair follicle development and increasing sebaceous activity, which is very similar to vdr knockout mice (29). Our results are thus consistent with c-MYC overexpression contributing to the alopecia observed in vdr−/− mice.
Results and Discussion
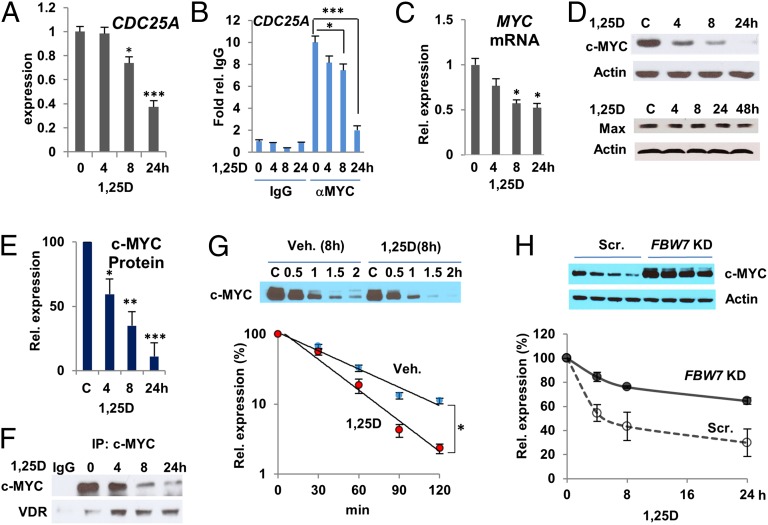
We have been using human SCC25 HNSCC cells as a model to investigate mechanisms of 1,25D-induced cell-cycle arrest (19, 39). c-MYC overexpression has been implicated in the development of HNSCC (40), and inducible overexpression of c-MYC induces actinic keratosis, a squamous cell cancer precursor (31). In SCC25 cells, 1,25D inhibited expression of c-MYC target genes implicated in cell-cycle progression, CDC25A, CCND2, and CDK4 (Fig. 1A and SI Appendix, Fig. S1A). This coincided with loss of c-MYC binding to E-box regions of the corresponding promoters (Fig. 1B and SI Appendix, Fig. S1B), consistent with repression of these genes by 1,25D occurring at least in part by suppression of c-MYC function. We also observed a gradual diminution of MYC mRNA over 24 h (Fig. 1C), in agreement with other studies (35, 36). In contrast, there was an almost total loss of c-MYC protein over the same period, whereas expression of c-MYC heterodimeric partner MAX was unaffected (Fig. 1 D and E). A similar decline in c-MYC target gene expression, and MYC mRNA, along with a dramatic loss of c-MYC protein, was observed in 1,25D-treated primary cultures of keratinocytes (SI Appendix, Fig. S2 A–C). Similarly, treatment of promyelocytic human HL60 cells with 1,25D also suppressed c-MYC protein expression (SI Appendix, Fig. S2D), indicating that the effect of 1,25D on c-MYC expression is not limited to epithelial cells.
Fig. 1.
1,25D signaling suppresses c-MYC expression and function. (A) 1,25D treatment represses expression of c-MYC target gene CDC25A in SCC25 cells. Cells were treated with 100 nM 1,25D for the times indicated. (B) 1,25D treatment blocks binding of c-MYC to E boxes target gene promoters in SCC25 cells as assessed by ChIP assay followed by quantitative PCR. (C) Gradual inhibition of MYC mRNA expression in SCC25 cells treated with 100 nM 1,25D. (D) c-MYC protein expression but not that of cofactor MAX is strongly suppressed by 1,25D (100 nM) in SCC25 cells. (E) Quantification of Western blots of c-MYC expression in 1,25D-treated cells from four different experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 as determined by one-way ANOVAs followed by Tukey’s post hoc test for multiple comparisons. (F) The VDR coimmunoprecipitates with c-MYC from extracts of vehicle or 1,25D-treated SCC25 cells. (G) 1,25D treatment enhances turnover of c-MYC protein expression in SCC25 cells in the presence of protein synthesis inhibitor cycloheximide. (Upper) Western blot of a single experiment. (Lower) Quantification of results of three independent experiments. Statistical analysis was performed by two-way ANOVA with Bonferroni correction for multiple comparisons. *P = 0.05. (H) Ablation of FBW7 expression attenuates 1,25D-induced turnover of c-MYC expression. SCC25 cells were transfected with control scrambled (Scr.) or FBW7-specific siRNAs, as indicated.
Importantly, coimmunoprecipitation studies showed that 1,25D treatment induced a rapid association (<4 h) of the VDR with c-MYC, suggesting that 1,25D may control c-MYC stability (Fig. 1F). Indeed, when protein synthesis was blocked with cycloheximide, c-MYC turnover was accelerated in 1,25D-treated cells (Fig. 1G). 1,25D-induced turnover was abolished or largely attenuated by ablation of expression of either the VDR (SI Appendix, Fig. S3) or FBW7 (also known as FBXW7, CDC4, AGO, and SEL10; Fig. 1H and SI Appendix, Fig. S4A), the F-box component of the SCFFbw7 E3 ubiquitin ligase that recognizes c-MYC (32). Ablation of p45SKP2 or Elav1/HuR, which can also regulate c-MYC expression (41), had no substantial effect on c-MYC expression in the absence or presence of 1,25D (SI Appendix, Fig. S4 B and C).
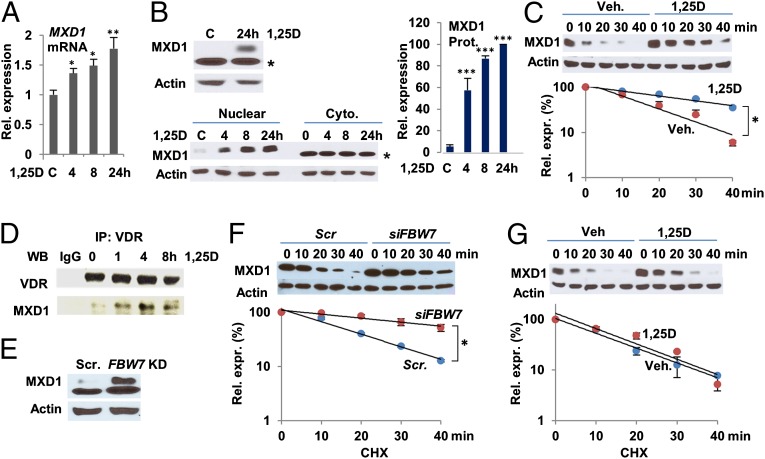
c-MYC is antagonized by transcriptional repressor MAD1/MXD1, which heterodimerizes with MAX to repress c-MYC target genes (42). MXD1 levels correlate with epithelial differentiation (43, 44), and MXD1’s expression in SCCs is associated with the capacity of cells to differentiate (45, 46). The change in MXD1 mRNA in 1,25D-treated SCC25 cells was opposite to that of MYC transcripts, increasing modestly over 24 h (Fig. 2A). In contrast, MXD1 protein levels increased dramatically over the same period (Fig. 2B). Note that MXD1 corresponds to the upper nuclear protein (Fig. 2B, Lower), because siRNA-mediated knockdown of MXD1 expression eliminated the upper band, but had no effect on the lower cytoplasmic protein (SI Appendix, Fig. S5). Treatment of primary human keratinocytes with 1,25D led to changes in MXD1 mRNA and MXD1 protein parallel to those seen in SCC25 cells (SI Appendix, Fig. S6 A and B). Similarly, MXD1 expression was strongly enhanced by 1,25D treatment of HL60 cells (SI Appendix, Fig. S6C). Consistent with these findings, short-term (6 h) 1,25D pretreatment of SCC25 cells reduced MXD1 turnover (Fig. 2C). Similar to the rapid association of the hormone-bound VDR with c-MYC, we observed 1,25D-dependent coimmunoprecipitation of MXD1 and the receptor (Fig. 2D), with MXD1 recruitment detectable by 1 h of 1,25D treatment. The rapid association of c-MYC and MXD1 with the VDR in the presence of 1,25D strongly suggests that regulation of their function contributes to, rather than is a consequence of, cell-cycle arrest by 1,25D.
Fig. 2.
1,25D signaling enhances stability and expression of MXD1. (A) Gradual rise in MXD1 mRNA levels in SCC25 cells treated with 100 nM 1,25D. (B) 1,25D treatment induces expression of MXD1 protein. (Upper) Western blots for MXD1 and actin from total cell extracts. (Lower) Western blots of nuclear and cytoplasmic extracts of cells treated with 1,25D as indicated. The band marked by an asterisk in the top and bottom Western blots is a nonspecific cytoplasmic protein. (Right) Quantification of the results of four independent analyses of MXD1 protein expression. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 as determined by one-way ANOVAs followed by Tukey’s post hoc test for multiple comparisons. (C) Effect of short-term (6 h) treatment with 1,25D on turnover of MXD1 protein cycloheximide-treated cells. A quantification of the results of three different experiments is shown. (D) MXD1 coimmunoprecipitates in a 1,25D-dependent manner with the VDR from extracts of SCC25 cells treated with vehicle or 1,25D as indicated. (E) Knockdown of FBW7 enhances expression of MXD1. (F) Ablation of FBW7 expression reduces MXD1 turnover in cycloheximide-treated cells (Scr; scrambled control siRNA). A quantification of four independent experiments is shown. Statistical analysis was performed by two-way ANOVA with Bonferroni correction for multiple comparisons; *P = 0.001. (G) Ablation of FBW7 abolishes the effect of 1,25D treatment on MXD1 turnover. A quantification of five independent experiments is shown.
Although control of c-MYC turnover has been extensively studied, little is known about regulation of MXD1. Remarkably, ablation of FBW7 expression also led to increased MXD1 protein levels (Fig. 2E), similar to 1,25D treatment. MXD1 turnover was also reduced, although not abolished, in FBW7-deficient cells (Fig. 2F), revealing that FBW7 regulates both the activator and repressor arms of the c-MYC/MXD1/MAX network. Notably, 1,25D-treatment had no substantial effect on MXD1 turnover in FBW7-deficient cells (Fig. 2G), consistent with the hormone-bound VDR protecting MXD1 from FBW7-mediated turnover. Regulation of MXD1 turnover by FBW7 was unexpected because it lacks a CDC4 phospho-degron recognized by FBW7 present in c-MYC, and other targets such as cyclin E, characterized by a TPxxS/E core (47). The control of MXD1 stability by FBW7 may thus be indirect (i.e., by regulating turnover or function of another unknown protein critical for direct regulation of MXD1 degradation).
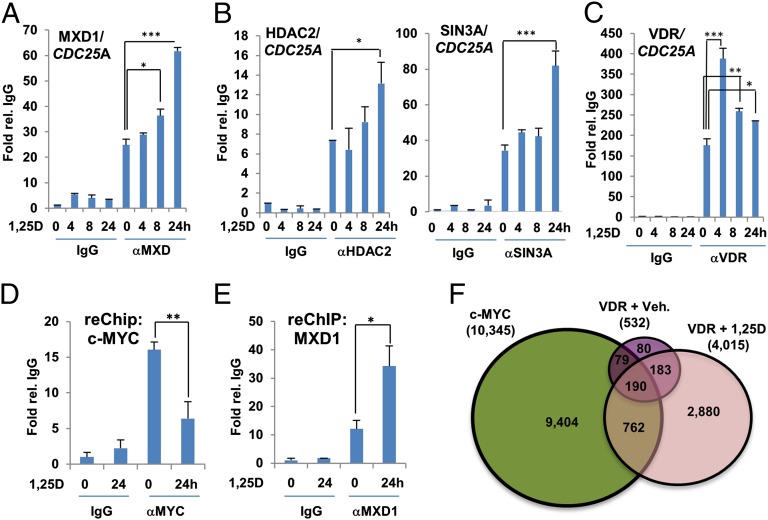
1,25D induced binding of MXD1 along with MXD1-associated corepressors HDAC2 and mSIN3A to E-box regions of CDC25A, CDK4, and CCND2 promoters (Fig. 3 A and B and SI Appendix, Fig. S7). Analysis of the interaction of the VDR with the same promoters revealed a pattern intermediate between that of c-MYC and MXD1 (Fig. 3C and SI Appendix, Fig. S8), consistent with the association of the VDR with both c-MYC and MXD1 observed by coimmunoprecipitation. The direct interactions of the VDR with c-MYC and MXD1 on the CDC25A promoter were confirmed by re-ChIP analysis (Fig. 3 D and E). The degree of association of the VDR with c-MYC or MXD1 after 24 h in the presence of 1,25D as detected by re-ChIP was very consistent with the effects of hormone on c-MYC and MXD1 DNA binding (Figs. 1B and 3A and SI Appendix, Figs. S1B and S7). To gain a broader insight into the potential association between the VDR and c-MYC binding sites on the genome, we reanalyzed with equally stringent parameters VDR and c-MYC peaks identified by ChIP sequencing from related lymphoblastoid cell lines (48, 49). This revealed an overlap between approximately one half of the VDR-binding sites detected in the absence of 1,25D (269/532), and almost one quarter of those (952/4,015) in the presence of 1,25D, with high-fidelity c-MYC sites (Fig. 3F). Although this type of comparison is not as definitive as ChIP sequencing studies performed in parallel in the same cell line, it nonetheless supports our experimental findings that DNA-bound c-MYC and the VDR do associate in vivo.
Fig. 3.
Analysis of the 1,25D-dependent association of MXD1 (A), its cofactors HDAC2 (B, Left), and SIN3a (B, Right), and the VDR (C) with the E box region of the promoter of the CDC25A gene by ChIP assay. ReChIP analysis of association of c-MYC (D) or MXD1 (E) with the VDR on the CDC25A promoter. Samples were immunoprecipitated with and anti-VDR antibody followed by immunoprecipitation of c-MYC or MXD1. (F) Results of a comparative analysis of overlap of genomic binding sites from ChIP sequencing studies of c-MYC and the VDR (from cells treated with vehicle [Veh] or 100 nM 1,25D for 36 h) performed in related lymphoblastoid cell lines. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 as determined by one-way ANOVAs followed by Tukey’s post hoc test for multiple comparisons.
Taken together, these results reveal that 1,25D treatment has opposing effects on MYC and MXD1 mRNA expression and FBW7-dependent turnover of the corresponding proteins. This ultimately leads to elevated DNA binding of MXD1 and its associated corepressors and repression of c-MYC target gene transcription. To determine whether the opposing but modest changes in both MYC and MXD1 mRNA levels and protein turnover could account for the relatively dramatic changes in protein content observed over 24 h of 1,25D treatment (Figs. 1E and 2B), we mathematically modeled the effects of 1,25D signaling on mRNA expression and protein turnover of c-MYC and MXD1 (details of derivations appear in Supplemental Experimental Procedures). Parameters for exponential changes in mRNA levels and changes in protein turnover were derived from experimental data, which led to predicted changes in total protein over 24 h that fit very well with those observed by Western blotting (R2 = 0.89 [c-MYC], and 0.97 [MXD1]; SI Appendix, Fig. S9). Thus, the combined effects of 1,25D signaling on mRNA levels and protein turnover can largely account for the dramatically opposing effects on c-MYC and MXD1 expression over 24 h.
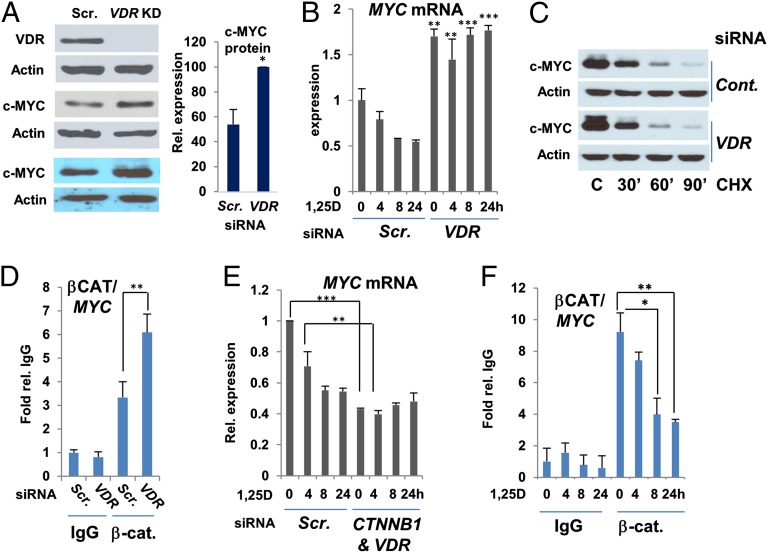
In addition to the effects of 1,25D on c-MYC and MXD1 expression and function, we noted that c-MYC protein levels were elevated in VDR-deficient cells (Fig. 4A and SI Appendix, Fig. S3). This effect was not limited to SCC25 cells, as c-MYC expression was also increased in VDR-depleted human LNCaP prostate carcinoma cells (SI Appendix, Fig. S10). c-MYC target gene CDC25A transcription was enhanced in VDR-deficient cells (SI Appendix, Fig. S11A), as was MYC gene expression (Fig. 4B). Ablation of VDR expression had little effect on c-MYC turnover (Fig. 4C), suggesting that higher MYC gene expression rather than increased protein stability led to elevated c-MYC levels and function in the absence of the VDR. The canonical Wnt signaling pathway stimulates MYC transcription through β-catenin (50), and the vitamin D signaling suppresses β-catenin function (51, 52). Notably, association of β-catenin with the MYC promoter was higher in VDR-deficient cells (Fig. 4D). Therefore, we simultaneously ablated β-catenin (CTNNB1) and VDR expression, which reduced MYC and CDC25A transcript levels to those seen in VDR-replete cells treated with 1,25D (Fig. 4E and SI Appendix, Fig. S11B). Consistent with these findings, association of β-catenin with the MYC promoter in VDR-replete cells was reduced by 1,25D treatment (Fig. 4F).
Fig. 4.
MYC gene transcription is elevated in VDR-deficient cells. (A) SiRNA-mediated knockdown of VDR expression increases levels of c-MYC protein. (Left) Western blotting of VDR and c-MYC protein levels in cells transfected with control or VDR-specific siRNAs. (Right) Quantification of c-MYC levels in VDR-replete and -deficient cells from three independent experiments. (B) MYC transcript levels are elevated in VDR-deficient cells and resistant to 1,25D-mediated repression. SCC25 cells were transfected with scrambled (Scr.) or VDR-specific siRNAs and incubated with 1,25D, as indicated, before measurement of MYC mRNA levels by RT/quantitative PCR. (C) Effect of VDR ablation on c-MYC turnover in cycloheximide-treated cells. (D) Ablation of VDR expression leads to elevated β-catenin binding to the MYC promoter as measured by ChIP assay. (E) MYC mRNA levels are reduced upon simultaneous ablation of β-catenin (CTNNB1) and VDR expression. (F) 1,25D treatment inhibits association of β-catenin with the MYC gene promoter. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 as determined by one-way ANOVAs followed by Tukey’s post hoc test for multiple comparisons.
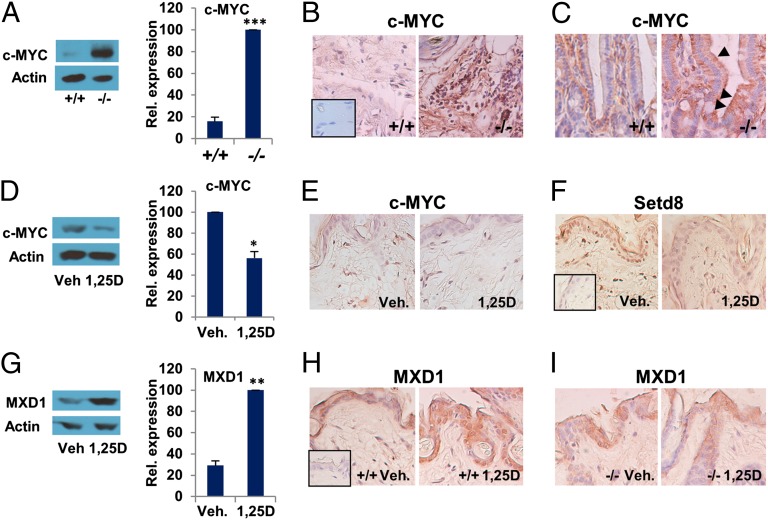
These observations are striking because vdr null mice or patients with vitamin d–resistant rickets resulting from inactivating mutations in the VDR gene (29, 53) develop alopecia because of dysregulated epidermal differentiation, and studies of vdr null mice have revealed that loss of the VDR in skin leads to elevated β-catenin signaling (29). In addition, vdr−/− mice display a hyperproliferative phenotype in colonic epithelia (29). Therefore, we compared c-MYC levels by Western blotting and immunohistochemistry in skin and colonic epithelia of wild-type and vdr null mice (Fig. 5 and SI Appendix, Fig. S12 and S13), using normal and malignant breast tissue as a control for c-MYC overexpression (SI Appendix, Fig. S12A). This analysis revealed substantially elevated c-MYC in both skin and colon of null mice (Fig. 5 A–C and SI Appendix, Fig. S12 C and D), with colonic overexpression visible in epithelial crypt cells (arrowheads; Fig. 5C) as well as in other tissues including heart, muscle and brain (SI Appendix, Fig. S12B). Consistent with observed effects on c-MYC in human cells, topical application of 1,25D to wild-type mouse skin suppressed c-MYC levels (Fig. 5 D and E). Because c-MYC expression was modest in wild-type skin, we also analyzed the product of one of its epidermal target genes, setd8 (54), whose expression was suppressed by 1,25D (Fig. 5F). Finally, in agreement with the induction of MXD1 expression observed in vitro, topical 1,25D increased MXD1 levels in the skin of wild-type mice (Fig. 5 G and H) but not vdr−/− mice (Fig. 5I).
Fig. 5.
1,25D and the VDR control c-MYC and MXD1 expression in vivo. (A) Western blotting of c-MYC expression in extracts of skin from wild-type (+/+) or vdr null (−/−) mice. Immunocytochemistry of c-MYC expression in the skin (B) and colon (C) of vdr wild-type (+/+) and null (−/−) mice. Arrowheads point to elevated c-MYC expression in enterocytes of null animals. (D) Western blotting of c-MYC expression in extracts of skin from wild-type mice treated topically with vehicle (Veh.) or 1,25D (15 ng/100 µL⋅cm2) for 24 h. Topical application (24 h) of vehicle or 1,25D to the skin of wild-type animals suppresses expression of c-MYC (E) and the product of epidermal c-MYC target gene setd8 (F). (G) Western blotting of MXD1 expression in extracts of skin from wild-type mice treated topically with Veh. or 1,25D (15 ng/100 µL⋅cm2) for 24 h. Topical 1,25D enhances expression of MXD1 in skin of wild-type (+/+; H) but not vdr null (−/−; I) mice. (Insets) Negative controls (no primary antibody). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 as determined by one-way ANOVAs followed by Tukey’s post hoc test for multiple comparisons.
In summary, we have found that the VDR suppresses c-MYC expression and function in vitro and in vivo in the absence and presence of 1,25D by multiple mechanisms. The VDR is not unique among the nuclear receptors in its regulation of c-MYC. Estrogen receptor α is a well-established enhancer of MYC gene transcription, and induction of c-MYC expression is a key component of the mitogenic actions of estrogens in breast cancer. Moreover, elevated expression of c-MYC in cancer cells can lead to resistance to antiestrogen therapy (55, 56). In contrast, recent work has shown that signaling through liver X receptors inhibit MYC gene expression through regulation of β-catenin (57). Our results indicate that the suppression of β-catenin function largely accounts for the inhibition of MYC mRNA expression by the VDR. Importantly, the relatively modest and opposing effects on MYC and MXD1 expression and protein turnover in the presence of 1,25D combine to dramatically alter the balance in c-MYC and MXD1 and levels of DNA-bound c-MYC/MAX and MXD1/MAX heterodimers, resulting in repression of c-MYC target gene transcription. Given that c-MYC controls normal epidermal differentiation and that its deregulated expression depletes epidermal stem cells (30, 37, 38), our findings strongly suggest that overexpression of c-MYC in the absence of the VDR contributes to alopecia observed in humans and mice. In addition, the regulation of c-MYC expression and function by the VDR provides a compelling molecular basis for its potential cancer preventive actions. In particular, β-catenin function and c-MYC expression are often elevated in colon cancer (50, 58). Moreover, the elevated c-MYC levels observed in several tissues of knockout mice suggest that the VDR, which is very broadly expressed, regulates c-MYC expression in widely different tissue types.
Experimental Procedures
All experiments presented are representative of three to five biological replicates. Experimental procedures for siRNA knockdowns, RT-quantitative PCR, immunoprecipitations and Western blotting, ChIP assays, and animal experiments including genotyping, Western blotting, immunohistochemistry, and topical 1,25D treatments are provided in SI Appendix. A complete description of derivations used for mathematical modeling of c-MYC and MXD1 expression and protein turnover is also provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by operating grants from the Canadian Institutes of Health Research (to J.H.W. and D.G.), the Canadian Cancer Society Research Institute (to J.H.W.), and the Natural Sciences and Engineering Research Council (to L.G.). R.S.-T. was funded by a J. P. Collip Fellowship in Medical Research and a McGill Integrated Cancer Research Training Program (MICRTP) fellowship. B.-S.A. was funded by a postdoctoral fellowship from the Fonds de Recherche en Santé de Québec.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210037109/-/DCSupplemental.
References
- 1.Tavera-Mendoza LE, White JH. Cell defenses and the sunshine vitamin. Sci Am. 2007;297(5):62–72. doi: 10.1038/scientificamerican1107-62. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357(3):266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 3.White JH. Vitamin D signaling, infectious diseases, and regulation of innate immunity. Infect Immun. 2008;76(9):3837–3843. doi: 10.1128/IAI.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arabi A, El Rassi R, El-Hajj Fuleihan G. Hypovitaminosis D in developing countries—prevalence, risk factors and outcomes. Nat Rev Endocrinol. 2010;6(10):550–561. doi: 10.1038/nrendo.2010.146. [DOI] [PubMed] [Google Scholar]
- 5.Gurlek A, Pittelkow MR, Kumar R. Modulation of growth factor/cytokine synthesis and signaling by 1alpha,25-dihydroxyvitamin D(3): Implications in cell growth and differentiation. Endocr Rev. 2002;23(6):763–786. doi: 10.1210/er.2001-0044. [DOI] [PubMed] [Google Scholar]
- 6.Lin R, et al. Expression profiling in squamous carcinoma cells reveals pleiotropic effects of vitamin D3 analog EB1089 signaling on cell proliferation, differentiation, and immune system regulation. Mol Endocrinol. 2002;16(6):1243–1256. doi: 10.1210/mend.16.6.0874. [DOI] [PubMed] [Google Scholar]
- 7.Hosomi J, Hosoi J, Abe E, Suda T, Kuroki T. Regulation of terminal differentiation of cultured mouse epidermal cells by 1 alpha,25-dihydroxyvitamin D3. Endocrinology. 1983;113(6):1950–1957. doi: 10.1210/endo-113-6-1950. [DOI] [PubMed] [Google Scholar]
- 8.Pálmer HG, et al. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63(22):7799–7806. [PubMed] [Google Scholar]
- 9.Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10(2):142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 10.Munker R, et al. A new series of vitamin D analogs is highly active for clonal inhibition, differentiation, and induction of WAF1 in myeloid leukemia. Blood. 1996;88(6):2201–2209. [PubMed] [Google Scholar]
- 11.Lin R, Wang TT, Miller WH, Jr, White JH. Inhibition of F-Box protein p45(SKP2) expression and stabilization of cyclin-dependent kinase inhibitor p27(KIP1) in vitamin D analog-treated cancer cells. Endocrinology. 2003;144(3):749–753. doi: 10.1210/en.2002-0026. [DOI] [PubMed] [Google Scholar]
- 12.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 13.Giovannucci E, et al. Prospective study of predictors of vitamin D status and cancer incidence and mortality in men. J Natl Cancer Inst. 2006;98(7):451–459. doi: 10.1093/jnci/djj101. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, et al. Polymorphisms of vitamin D receptor gene protect against the risk of head and neck cancer. Pharmacogenet Genomics. 2005;15(3):159–165. doi: 10.1097/01213011-200503000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Manson JE, Mayne ST, Clinton SK. Vitamin D and prevention of cancer—ready for prime time? N Engl J Med. 2011;364(15):1385–1387. doi: 10.1056/NEJMp1102022. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas J. Vitamin D and cancer: Uncertainty persists; research continues. J Natl Cancer Inst. 2011;103(11):851–852. doi: 10.1093/jnci/djr215. [DOI] [PubMed] [Google Scholar]
- 17.Zehnder D, et al. Extrarenal expression of 25-hydroxyvitamin d(3)-1 α-hydroxylase. J Clin Endocrinol Metab. 2001;86(2):888–894. doi: 10.1210/jcem.86.2.7220. [DOI] [PubMed] [Google Scholar]
- 18.Hewison M, et al. Extra-renal 25-hydroxyvitamin D3-1alpha-hydroxylase in human health and disease. J Steroid Biochem Mol Biol. 2007;103(3-5):316–321. doi: 10.1016/j.jsbmb.2006.12.078. [DOI] [PubMed] [Google Scholar]
- 19.An B-S, et al. Stimulation of Sirt1-regulated FoxO protein function by the ligand-bound vitamin D receptor. Mol Cell Biol. 2010;30(20):4890–4900. doi: 10.1128/MCB.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchard C, Marquardt J, Brás A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23(14):2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117(4):421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 22.Ho KK, Myatt SS, Lam EW. Many forks in the path: Cycling with FoxO. Oncogene. 2008;27(16):2300–2311. doi: 10.1038/onc.2008.23. [DOI] [PubMed] [Google Scholar]
- 23.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27(16):2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci USA. 2005;102(5):1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dehan E, Pagano M. Skp2, the FoxO1 hunter. Cancer Cell. 2005;7(3):209–210. doi: 10.1016/j.ccr.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Barreyro FJ, et al. Transcriptional regulation of Bim by FoxO3A mediates hepatocyte lipoapoptosis. J Biol Chem. 2007;282(37):27141–27154. doi: 10.1074/jbc.M704391200. [DOI] [PubMed] [Google Scholar]
- 27.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28(27):2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouillon R, et al. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocr Rev. 2008;29(6):726–776. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang LL, Peng RY. Unveiling hair follicle stem cells. Stem Cell Rev. 2010;6(4):658–664. doi: 10.1007/s12015-010-9172-z. [DOI] [PubMed] [Google Scholar]
- 31.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3(5):565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 32.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: The role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29(35):4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 34.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11(5):1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 35.Rohan JNP, Weigel NL. 1Alpha,25-dihydroxyvitamin D3 reduces c-Myc expression, inhibiting proliferation and causing G1 accumulation in C4-2 prostate cancer cells. Endocrinology. 2009;150(5):2046–2054. doi: 10.1210/en.2008-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toropainen S, Väisänen S, Heikkinen S, Carlberg C. The down-regulation of the human MYC gene by the nuclear hormone 1alpha,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J Mol Biol. 2010;400(3):284–294. doi: 10.1016/j.jmb.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28(2):165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 38.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11(8):558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 39.Akutsu N, et al. Regulation of gene expression by 1alpha,25-dihydroxyvitamin D3 and its analog EB1089 under growth-inhibitory conditions in squamous carcinoma cells. Mol Endocrinol. 2001;15(7):1127–1139. doi: 10.1210/mend.15.7.0655. [DOI] [PubMed] [Google Scholar]
- 40.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 41.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23(15):1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 43.Chin L, et al. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci USA. 1995;92(18):8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gandarillas A, Watt FM. Changes in expression of members of the fos and jun families and myc network during terminal differentiation of human keratinocytes. Oncogene. 1995;11(7):1403–1407. [PubMed] [Google Scholar]
- 45.Lymboussaki A, et al. Expression of Mad, an antagonist of Myc oncoprotein function, in differentiating keratinocytes during tumorigenesis of the skin. Br J Cancer. 1996;73(11):1347–1355. doi: 10.1038/bjc.1996.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurlin PJ, et al. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11(12):2487–2501. [PubMed] [Google Scholar]
- 47.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 48.Ramagopalan SV, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rozowsky J, et al. AlleleSeq: Analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 51.Beildeck ME, Gelmann EP, Byers SW. Cross-regulation of signaling pathways: An example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res. 2010;316(11):1763–1772. doi: 10.1016/j.yexcr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larriba MJ, et al. Vitamin D receptor deficiency enhances Wnt/β-catenin signaling and tumor burden in colon cancer. PLoS ONE. 2011;6(8):e23524. doi: 10.1371/journal.pone.0023524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes MR, Malloy PJ, O’Malley BW, Pike JW, Feldman D. Genetic defects of the 1,25-dihydroxyvitamin D3 receptor. J Recept Res. 1991;11(1-4):699–716. doi: 10.3109/10799899109066437. [DOI] [PubMed] [Google Scholar]
- 54.Driskell I, et al. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J. 2012;31(3):616–629. doi: 10.1038/emboj.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carroll JS, Brown M. Estrogen receptor target gene: An evolving concept. Mol Endocrinol. 2006;20(8):1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 56.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 57.Uno S, et al. Suppression of beta-catenin signaling by liver X receptor ligands. Biochem Pharmacol. 2009;77(2):186–195. doi: 10.1016/j.bcp.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25(57):7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.