Abstract
Alkyltransferase-like (ATL) proteins in Schizosaccharomyces pombe (Atl1) and Thermus thermophilus (TTHA1564) protect against the adverse effects of DNA alkylation damage by flagging O6-alkylguanine lesions for nucleotide excision repair (NER). We show that both ATL proteins bind with high affinity to oligodeoxyribonucleotides containing O6-alkylguanines differing in size, polarity, and charge of the alkyl group. However, Atl1 shows a greater ability than TTHA1564 to distinguish between O6-alkylguanine and guanine and in an unprecedented mechanism uses Arg69 to probe the electrostatic potential surface of O6-alkylguanine, as determined using molecular mechanics calculations. An unexpected consequence of this feature is the recognition of 2,6-diaminopurine and 2-aminopurine, as confirmed in crystal structures of respective Atl1-DNA complexes. O6-Alkylguanine and guanine discrimination is diminished for Atl1 R69A and R69F mutants, and S. pombe R69A and R69F mutants are more sensitive toward alkylating agent toxicity, revealing the key role of Arg69 in identifying O6-alkylguanines critical for NER recognition.
The exposure of DNA to alkylating agents can lead to the formation of mutagenic and toxic O6-alkylguanine lesions (Fig. 1A) (reviewed in ref. 1). Mutagenicity arises from mispairing with T during DNA replication, whereas toxicity is associated with mismatch repair processing of O6-alkylguanine:T sites. O6-Alkylguanine-DNA alkyltransferases (AGTs) repair many different O6-alkylguanine lesions by transferring the alkyl group to an active site Cys (1–4), although some are reported to be poorer substrates (5–10). Alkyltransferase-like (ATL) proteins (for reviews see refs. 3, 11, and 12) are highly homologous to AGTs but have a different amino acid (typically Trp or Ala) in place of the nucleophilic active site Cys. ATL proteins retain the ability to bind single-stranded or duplex DNA containing O6-alkylguanine, and although unable to undertake the de-alkylative repair reaction, they protect against the adverse effects of DNA alkylation damage by downstream recruitment of nucleotide excision repair (NER) (13–22).
Fig. 1.
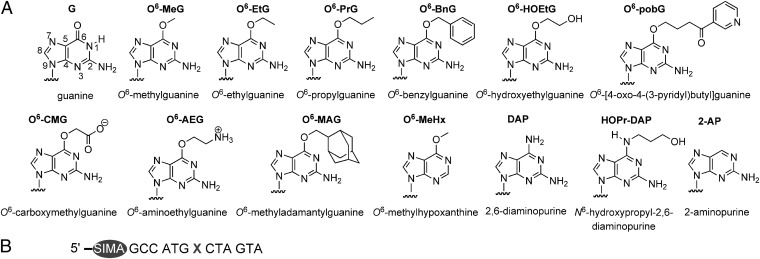
Structures of guanine, modified purines, and ODNs used in study. (A) Guanine (numbered), O6-alkylguanine, and modified purines incorporated into 13-mer ODN substrate (B) (X = guanine, O6-alkylguanine, or modified purine).
In common with other proteins (including AGTs) that repair or modify DNA bases, ATL proteins use nucleotide flipping (also referred to as base flipping) to insert the target base into a tight-fitting binding pocket and thereby recognize its unique characteristics (for reviews see refs. 23–25). For ATL proteins the fidelity of this process and the formation of high-affinity complexes with cognate substrates is critical before subsequent processing by NER. Nucleotide-flipping proteins typically distinguish correct from incorrect base by using mechanisms involving steric exclusion, in which only the cognate base can access the binding site, or processes that probe the hydrogen bonding characteristics of the base. For example, in the human DNA glycosylase UDG, Tyr147 blocks access of the methyl group of T such that only U fits in the active site before its excision (26). In contrast, the human alkyladenine DNA glycosylase distinguishes between the ethenobases of A and C (Fig. S1) and the natural bases by forming a specific hydrogen bond between His136 main chain NH and the imino nitrogens (N6 and N4) of the respective ethenobases (27, 28). The bases A and C have amino groups at these positions that are hydrogen bond donors rather than acceptors, and this key interaction with His136 is therefore disrupted. Human DNA glycosylase hOGG1 distinguishes 8-oxoguanine (8-oxoG) (Fig. S1) from G by forming a hydrogen bond between the main chain carbonyl oxygen of Gly42 and the N7 proton of 8-oxoG, which is absent in G (29). The corresponding interaction with G is that of repulsion between the lone pairs on the respective positions.
Somewhat paradoxically Atl1 does not have a tight-fitting pocket into which an O6-alkylguanine is bound. Indeed the O6-alkylguanine binding site is substantially larger than that found in AGT proteins (2, 16, 30, 31). This permits Atl1 to bind O6-alkylguanines with much bulkier alkyl groups, for example O6-pobG (Fig. 1A) (16). However, the mechanism by which Atl1 distinguishes between O6-alkylguanine and G has not been investigated, nor indeed has a comprehensive assessment of the substrate characteristics for ATL proteins been undertaken. Here we describe recognition by ATL proteins from Schizosaccharomyces pombe (Atl1) and Thermus thermophilus (TTHA1564) of oligodeoxyribonucleotides (ODNs) containing several different O6-alkylguanine analogs and related purines that derive from functional group modifications to the O6-alkylguanine structure. Both ATL proteins derive from organisms that lack an AGT protein. In comparison with processes used for base recognition by other proteins (25), our study supports an unprecedented and exquisite mechanism involving molecular readout of the electrostatic potential surface of the target base by Atl1 that permits O6-alkylguanine to be distinguished from G.
Results
To determine equilibrium dissociation constants (KD values) of ATL–DNA complexes we prepared 5′-SIMA(HEX)-labeled ODNs containing O6-alkylguanines and related purine bases (Fig. 1A) by using post-DNA synthesis methods (Fig. S2) using “convertible bases” 2-amino-6-methylsulfonylpurine (8, 32) or 6-chloropurine (33). The SIMA(HEX) (dichlorodiphenylfluorescein) fluorophore shows similar luminescence properties to hexachlorofluorescein (HEX), but its higher chemical stability was found to be fully compatible with DNA modification chemistry. The O6-alkylguanines chosen contain a wide variety of alkyl side chains that differ in charge, size, and polarity.
Atl1 and TTHA1564 Bind with High Affinities to DNA Containing Any O6-Alkylguanine.
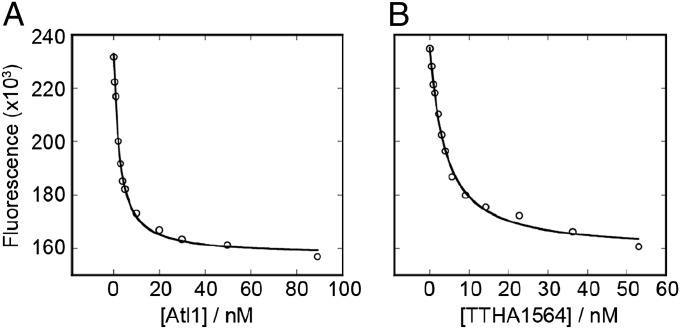
The addition of either native Atl1 (17) or maltose binding protein (MBP)-TTHA1564 (19) to a solution of fluorescently labeled O6-alkylguanine-containing ODN resulted in a concentration-dependent decrease in fluorescence intensity (Fig. 2) from which we derived KD values (Table S1) for ODNs containing the bases shown in Fig. 1A. Comparable values were obtained using fluorescence anisotropy. Because native TTHA1564 is unstable and prone to aggregation in vitro, its MBP fusion protein (19) was used. In a control experiment MBP had no discernable binding interaction with a 5′-SIMA(HEX)-labeled O6-methylguanine (O6-MeG)-containing ODN.
Fig. 2.
Analysis of binding of ATL proteins to DNA monitored using fluorescence emission intensity. Atl1 (A) or MBP-TTHA1564 (B) were added to 1 nM ODN 5′-SIMA(HEX)-GCCATGXCTAGTA (X = O6-carboxymethylguanine) in buffer (50 mM Tris.HCl (pH 7.5), 50 mM NaCl, 1mM EDTA) and fluorescence emission measured at 560 nm with excitation at 530 nm.
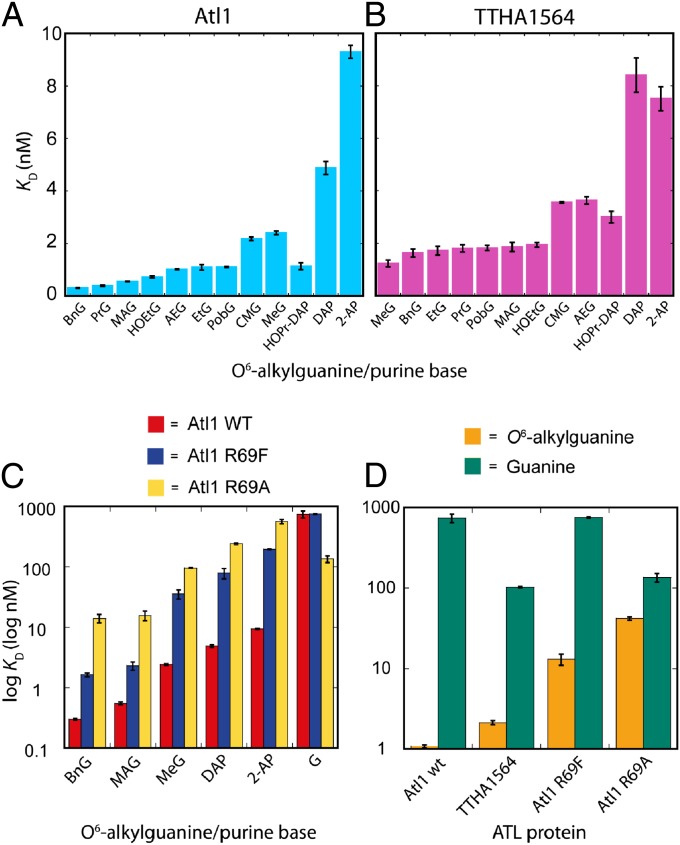
Both Atl1 and MBP-TTHA1564 bind with high affinities to ODNs containing all O6-alkylguanines (Fig. 3 A and B and Table S1). However, the T. thermophilus protein shows markedly less discrimination between damaged and natural (G-containing) DNA than that displayed by Atl1. Furthermore, Atl1 shows a preference for binding O6-alkylguanines with side chains that are larger and more hydrophobic than methyl. Both ATL proteins recognize the very bulky O6-methyladamantylguanine (Fig. 1A) that would be too large to fit in an AGT active site (2). In addition, other O6-alkylguanines such as O6-HOEtG, O6-pobG, O6-CMG, and O6-AEG (Fig. 1A) that are reportedly poorer substrates for some AGTs (6–10) are recognized with high affinity by both ATL proteins.
Fig. 3.
Equilibrium dissociation constants (KD values) determined for ATL–DNA complexes using ss ODNs containing O6-alkylguanine and other purine bases (structures, analog abbreviations, and the ODN sequence are shown in Fig. 1) with the following proteins: (A) Atl1 from S. pombe, (B) MBP-TTHA1564 from T. thermophilus, (C) Atl1 wild type (red), Atl1 mutant proteins R69F (blue) and R69A (yellow), (D) Atl1 wild type, Atl1 R69F and R69A mutants with ss ODNs containing O6-alkylguanine (mean KD values) (gold) or guanine (green). Log KD values are displayed in C and D.
Atl1 Shows Specific Recognition of the N2-Amino Group of O6-Alkylguanine but Not the O6-Alkyl Group.
The crystal structures (16) of Atl1–DNA substrate complexes suggest hydrogen bonding interactions between the N3 atom of O6-MeG and the side chain of Tyr25 and between its N2-amino group and the main chain carbonyl groups of Trp56 and Val59. Analogous interactions are also observed between O6-MeG and the corresponding residues in complexes involving human AGT (also known as O6-methylguanine-DNA methyltransferase, MGMT) (2). As expected, DNA containing O6-methylhypoxanthine (O6-MeHx; Fig. 1A), which possesses a hydrogen atom at the C2 position of the purine rather than the amino group of O6-MeG, was a poor substrate for Atl1 (Table S1). However, somewhat surprisingly, the ODN containing 2-aminopurine (2-AP; Fig. 1A), which has a hydrogen atom at the 6-position of the purine ring rather than the methoxy group of O6-MeG, was bound with comparable affinity to that of the O6-alkylguanine-containing sequences (Fig. 3A and Table S1).
ATL Proteins Recognize DNA Containing 2,6-Diaminopurine.
2,6-Diaminopurine (DAP; Fig. 1A) has a hydrogen-bond donor (amino group) replacing the hydrogen-bond acceptor (6-methoxy group) of O6-MeG. Despite this, both ATL proteins showed unexpectedly tight binding to the sequence containing DAP (Fig. 3 A and B and Table S1) that was significantly stronger than with unmodified (G-containing) DNA (Table S1) and only slightly less than with O6-alkylguanine-containing sequences. Appending an alkyl group to the 6-amino group of DAP [N6-hydroxypropyl-2,6-diaminopurine (HOPr-DAP); Fig. 1A] increased the binding affinity of both proteins for the ODN substrate, giving comparable values to those observed for O6-alkylguanine-containing ODNs (Fig. 3 A and B and Table S1).
Arg69 in Atl1 Probes the Molecular Electrostatic Potential of the Flipped Base to Distinguish Between O6-Alkylguanine and Guanine.
The free energy changes associated with formation of Atl1 complexes with the modified ODNs relative to the natural sequence (ΔΔG) were determined using Eq. 1 (34). These values vary between −11 and −19 kJ/mol (Table S1)
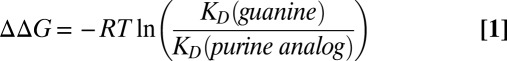 |
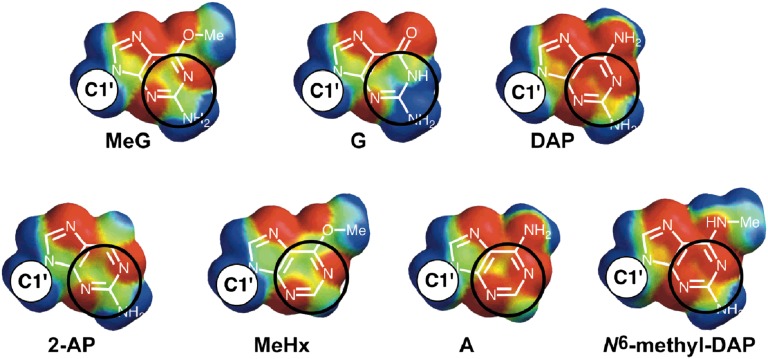
Structures of Atl1–DNA complexes (16) reveal the close proximity of the guanidinium side chain of Arg69 and the pyrimidine ring of the O6-alkylguanine, which would allow a cation–π interaction (35). The T. thermophilus ATL protein has Phe in the equivalent amino acid position (Fig. S3) and shows an approximate 10-fold decrease in its ability to discriminate between O6-alkylguanine-containing and unmodified DNA (Fig. 3 B and D and Table S1). Using coordinates from the published (16) complex of Atl1 with O6-MeG-containing DNA (Protein Data Bank accession code 3GX4) we found using molecular mechanics calculations that the interaction of Arg69 with O6-MeG is more favorable than that with G by −14 kJ/mol, in good agreement with the experimentally determined ΔΔG values for the complexes (Table S1). There are strong substituent effects on the strength of the cation–π interaction (36), and the origin of the discrimination between O6-MeG and G can be found by examining the molecular electrostatic potential (MEP) surfaces of the bases. Fig. 4 shows the calculated MEP on the van der Waals surface of the π face of selected purine analogs; areas shaded red represent regions of negative electrostatic potential and are therefore attractive to a positively charged side chain, whereas areas shaded blue correspond to regions of positive electrostatic potential, which would lead to repulsive interactions with an Arg side chain (37). Areas of the purine bases where MEP values are zero (neutral) are green. The MEP surfaces differ most markedly between O6-MeG and guanine on the N1, C2, and N3 ring atoms (numbering shown in Fig. 1A). It is this region of π surfaces of the bases that makes a close contact with the Arg69 side chain of Atl1 in crystal structures of Atl1–DNA complexes (16): the circles on Fig. 4 highlight the regions on the purine bases that contact Arg69 (interatomic separations less than 3.5 Å). Furthermore, the MEP surfaces of O6-MeG, DAP, and 2-AP are remarkably similar and in turn are very different to that of G. Although the MEP surface of A is more similar to O6-MeG than G, it lacks the 2-amino group essential for Atl1 binding.
Fig. 4.
Calculated MEP surfaces for guanine, O6-methylguanine, and other purines. Black circles highlight the regions on the purine bases that contact Arg69 (interatomic separations less than 3.5 Å). Colors correspond to: blue +50 kJ/mol, green neutral, red −50 kJ/mol. The site to which the anomeric carbon (C1') of the nucleoside is attached to the purine is indicated in each figure.
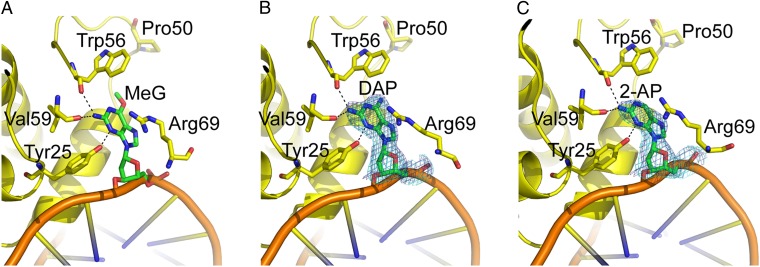
To further investigate the interaction of Atl1 with substrates containing DAP and 2-AP, we solved the crystal structures of Atl1–DNA complexes containing these bases to 2.7-Å and 2.85-Å resolution, respectively (Fig. 5 and Table S2). Both structures display minor groove binding, significant bending of the DNA duplex, and nucleotide flipping in common with Atl1–O6-MeG and Atl1–pobG complexes (16). In addition, hydrogen bonding interactions are observed between main chain carbonyls and the N2-amino groups of the bases and between the Tyr25 side chain and the N3-ring atoms. These structures differ from those containing O6-MeG in that the indole side chain of Trp56 is rotated slightly and tilted somewhat closer toward the pyrimidine rings of DAP and 2-AP, possibly resulting from slight differences in the MEP surfaces of these purines and O6-MeG (Fig. 4). Of significance is the positioning of the binding site loop (residues 65–73), which switches between open and closed conformations for free and DNA-bound Atl1, respectively (16). This loop is in the closed conformation for the DAP and 2-AP complex structures, as observed for the O6-MeG complex structure (16), positioning Arg69 near the modified purine to allow its side chain to interact with the N1, C2, and N3 ring atoms. The repulsive electrostatic interactions between G and Arg69 may help promote opening of the binding site loop to aid release of nonsubstrate DNA.
Fig. 5.
Crystal structures of (A) Atl1:O6-MeG-DNA duplex, (B) Atl1:DAP-DNA duplex, and (C) Atl1:2-AP-DNA duplex ODN complexes. 2Fo-Fc (1 σ) and Fo-Fc (+3 σ) electron density maps with DAP (B) or 2-AP (C) omitted from the model are shown in blue and cyan, respectively.
Atl1 Mutants R69A and R69F Display Weaker Binding to ODNs Containing O6-Alkylguanines and a Diminished Ability to Discriminate Between These and the Natural Sequence.
To further probe the role of Atl1 Arg69 in O6-alkylguanine recognition, mutants R69A and R69F were prepared by site-directed mutagenesis. The latter mutant was made because in TTHA1564 the residue corresponding to R69 in Atl1 is Phe130 (Fig. S3). The replacement of Arg69 with Ala significantly disrupts the ability of Atl1 to bind ODNs containing any of the modified bases, an ability that was partially restored for the R69F mutant (Fig. 3C and Table S3). Discrimination between ODNs containing O6-alkylguanines and G were reduced by ∼10-fold for the R69F mutant and almost 100-fold for Atl1 R69A.
S. pombe Strains Harboring R69A and R69F Mutations Show Enhanced Sensitivity to the Alkylating Agent MNNG.
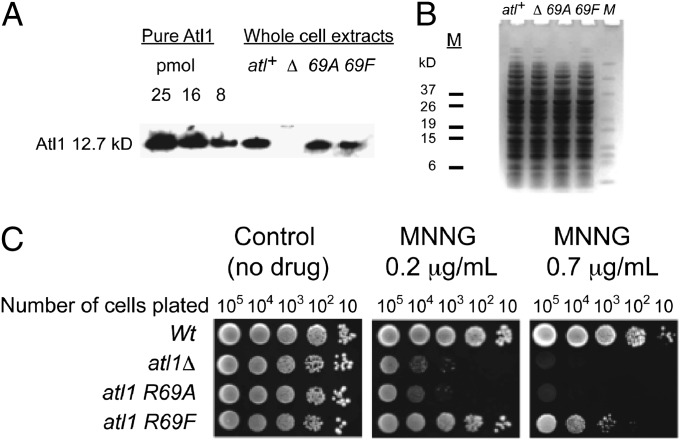
To examine the role of the Arg69 residue in the function of Atl1 we introduced mutated atl1-R69A and atl1-R69F alleles into the S. pombe genome by gene replacement. Genotype analysis and Western blotting demonstrated the presence and expression of the mutant genes (Fig. 6 A and B). We then assessed the sensitivity of the mutants to the DNA methylating agent N-methyl-N-nitroso-N′-nitroguanidine (MNNG) by agar plate (“spot”) assay. The atl1-R69A point mutant strain was found to be as sensitive to killing by MNNG as the full atl1 deletion strain, whereas the atl1-R69F mutant had intermediate sensitivity between these and the resistant wild-type strain (Fig. 6C).
Fig. 6.
Growth inhibition assays with S. pombe and MNNG. (A) Analysis of atl1 expression by SDS/PAGE of whole-cell extracts followed by Western blot with anti-Atl1 antibody. Purified recombinant Atl1 protein was used as a positive control. (B) Loading control for the Western blot analysis. Equal amounts of proteins (15 µg per lane) were loaded onto two 4–12% NuPAGE Bis-Tris Pre-Cast Gels. One gel was probed with anti-Atl1 antibody, whereas a second gel was stained in 0.05% (wt/vol) Coomassie Brilliant Blue R250. Wild type or atl+–wild type (GM1); atl1Δ or Δ - full deletant of the atl1 gene (GM3); atl1-R69A or 69A (GM177); atl1-R69F or 69F (GM178). M, BenchMark Pre-Stained Protein Ladder (Invitrogen). (C) Agar plate (“spot”) assay for atl1-R69A and atl1-R69F mutants after treatment with MNNG. Aliquots of 10-fold serial dilutions were spotted on plates with YEA media (growth medium containing yeast extract, glucose, and adenine sulfate) containing MNNG under indicated concentrations and photographed after 5 d.
Discussion
Both ATL proteins examined here bound with high affinities to DNA containing all of O6-alkylguanines regardless of the nature of the alkyl side chain. The preference of Atl1 for binding bulkier lesions (Figs. 1A and 3A and Table S1) likely derives from increased hydrophobic interactions of the O6-alkyl side chain with the Trp residue (W56) that is Ala in TTHA1564 (16). The same trend is observed upon adding an alkyl chain to the 6-amino group of DAP (HOPr-DAP) (Figs. 1A and 3 A and B and Table S1) and is related to both the size and hydrophobicity of the alkyl group. Thus, CMG, which has a polar carboxylate group, although larger than Me is bound with a similar affinity consistent with our previous data obtained using surface plasmon resonance (SPR) (4, 21). In contrast, TTHA1564 binds all O6-alkylguanine-containing ODNs with similar affinities except for those with charged side chains, such as CMG and AEG (Fig. 3 A and B and Table S1). Interestingly, both ATL proteins recognize ODNs containing AEG, pobG, CMG, and HOEtG (Fig. 1A), all of which are reported to be repaired less efficiently by AGT proteins (6–10). This result, along with the observation that the AGT proteins Ogt and Ada from Escherichia coli are even less capable than the human protein of repair of large O6-alkylguanine lesions (38–40) is consistent with ATLs having a much broader substrate specificity than AGT proteins (16, 17), as would be required to highlight subsequent processing by NER.
ATL proteins must efficiently distinguish between O6-alkylguanine and G that differ in the size of the group at purine C6 position and the absence of an N1 proton in the damaged base (Fig. 1A). Because G is smaller this precludes a steric exclusion mechanism in which only the damaged base can access the protein’s binding pocket. Furthermore, hydrogen bonding interactions to the N1 atom of O6-MeG that could distinguish this base from G are not apparent in the crystal structures of Atl1–DNA complexes (16). In contrast, hydrogen bonding interactions to the N3 atom and N2-amino group of O6-MeG (16) do exist, as supported by the poor binding of Atl1 to the ODN containing O6-methylhypoxanthine (O6-MeHx) (Figs. 1A and 3A and Table S1). Recognition of the N2-amino group seems to be critical in locating O6-alkylguanine within Atl1 binding site to allow other interactions with the base and would support binding of O6-alkylguanine and G but not A, C, and T.
Atl1 distinguishes between O6-alkylguanines and G with a difference in KD values of approximately three orders of magnitude (Fig. 3 C and D and Table S1). A major component of this selectivity is the Arg69 cation–π interaction that reads the MEP surfaces of the flipped base (16) at the N1, C2, and N3 ring atoms (MEP surfaces at the Arg69 contact point are circled in Fig. 4). In contrast, AGT proteins that use analogous H-bonding interactions to the flipped base but lack Arg69 show little binding discrimination between O6-MeG and G, with dissociation constants in the μM range (41, 42). The need to distinguish G from O6-alkylguanine and the tight binding of substrate are clearly not required by AGT proteins because these proteins, unlike ATL proteins, undertake direct substrate repair.
The Atl1 R69A mutant cannot distinguish effectively between DNA containing O6-alkylguanine and G owing to its lower affinity for the modified sequences and its much higher affinity for the natural (G-containing) sequence (Fig. 3C and Table S3). For wild-type Atl1 the critical repulsive interaction between the Arg69 side chain and the C2 atom of G is removed by mutation of this residue to Ala, and hence its ability to discriminate between O6-alkylguanines and G is diminished. The sensitivity to MNNG of mutated S. pombe expressing the R69A Atl1 protein is comparable to that of the full deletant ΔAtl1, in agreement with that predicted from in vitro binding data.
Both DAP and 2-AP have similar patterns of MEP to O6-alkylguanines (Fig. 4), and DNA containing these bases is therefore a good substrate for both ATL proteins (Fig. 3 A, B, and D and Table S1). This presumably is a consequence of the very similar electronic characteristics of these two purines and of O6-alkylguanines rather than being of biological significance because these bases are not found in S. pombe or T. thermophilus DNA (Fig. 4). This is supported by very similar structures for Atl1 complexes with DAP, 2-AP, or O6-MeG (16).
TTHA1564 displays approximately a 50-fold difference in its affinity for DNA containing O6-alkylguanine compared with the unmodified sequence (Fig. 3D). In TTHA1564 Phe replaces Arg69 in Atl1 (Fig. S3). Relative to the Atl1 R69A mutant, mutation of Atl1 Arg69 to Phe restores some ability of the protein to distinguish between O6-alkylguanine lesions and G (Fig. 3C and Table S3). The Phe69 residue is unable to make the cation–π interaction but can make hydrophobic contacts with the alkoxy substituents to exercise this discrimination, leading to tighter binding to DNA containing larger hydrophobic O6-alkylguanines. Several other DNA repair proteins have an active site Phe residue that makes hydrophobic interactions with the substrate (26, 29). In MGMT, Tyr158 corresponds to Atl1 Arg69, and this amino acid is invariantly Tyr or Phe in all known AGT sequences. Indeed, mutation of MGMT Tyr158 to Ala causes an 800-fold decrease in the ability of the protein to repair O6-MeG-containing DNA, whereas the Y158F mutant has comparable activity to wild-type MGMT (43).
Analysis of the sequences of ATL proteins from a variety of prokaryotic and lower eukaryotic organisms (139 in total) reveals that the Arg69 of Atl1 is conserved across all fungal species, whereas Lys, Pro, or Ser exist in most prokaryotes. Therefore, it may be that the unique qualities of this Arg residue are confined to eukaryotic proteins and provide an enhanced ability to distinguish O6-alkylguanines in DNA.
In summary, we have demonstrated the ability of ATL proteins to recognize a broad spectrum of alkylated guanine and other purine modifications. Furthermore, we have highlighted an unprecedented mechanism for base damage recognition in which a cation–π interaction is used to probe the molecular electrostatic potential surface of a flipped DNA base.
Methods
ODN Synthesis.
ODNs were synthesized using standard protocols (0.1 M phosphoramdite solutions) using mild/fast deprotection phosphoramidites, reagents, and columns (acetyl dC, phenoxyacetyl dG and dA) and 5′-SIMA(HEX) phosphoramidite and 0.15-M solutions of 2-amino-6-methylsulfonylpurine-2'-deoxyriboside phosphoramidite (8, 21, 32) or 6-chloropurine-2'-deoxyriboside phosphoramidite (33). After DNA synthesis, the CPG-bound protected ODNs were reacted with alcohol or amine and 1,8-diazabicyclo[5.4.0]undec-7-ene in dry acetonitrile followed by deprotection in aqueous sodium hydroxide solution or concentrated aqueous ammonia solution as previously described (8, 32). All ODNs were purified by reversed-phase HPLC (8, 21) and characterized by ESI–mass spectrometry (Table S4).
Expression and Purification of Wild-Type and Mutant ATL Proteins.
Atl1 was overexpressed as an MBP fusion protein using a pMAL-2c construct transformed into E. coli (DH5α) and purified over amylose resin. The MBP–Atl1 fusion protein was cleaved with Factor Xa (NEB) and purified over a Superdex 200 column (HiLoad 16/60; GE Healthcare). To introduce Arg69 to Phe and Ala point mutations into the atl1 gene of pMAL-2c-atl1 vector (17), the Phusion site-directed mutagenesis kit was used (Finnzymes, NEB) and the constructs verified by sequencing. E. coli clones harboring the pMAL-2c-atl1-R69F and pMAL-2c-atl1-R69A constructs were grown to OD260 of ∼0.6, induced for 3 h by the addition of isopropyl-β-thiogalactoside and the protein purified as above.
MBP-TTHA1564 was overexpressed and purified as described by Morita et al. (19).
SDS/PAGE and Western Blot Analysis.
Cell-free extracts were prepared by the glass bead methods as described elsewhere. Proteins (15 μg per lane) were subject to 4–12% NuPAGE then transferred onto nitrocellulose in an electroblotting apparatus. Membranes were then blocked with nonfat dried milk (Marvel) in TBST, washed with TBST, and incubated with rabbit anti-Atl1 antibody in 0.5% (wt/vol) nonfat dried milk in TBST for 1 h. After washing with TBST, the membrane was incubated for 1 h with goat anti-rabbit (1/1,000 dilution, P0448; Dako) horseradish peroxidase-linked secondary antibody, diluted in 0.5% (wt/vol) nonfat dried milk in TBST and washed with TBST. Chemiluminescence detection was carried out as described in the manufacturer’s protocol (ECL Plus; Amersham, GE Healthcare). A duplicate gel was stained by immersion in 0.05% (wt/vol) coomassie Brilliant Blue R250, 30% (vol/vol) methanol, 10% (vol/vol) glacial acetic acid for 1 h at room temperature, and subsequently destained in 30% (vol/vol) methanol, 10% (vol/vol) glacial acetic acid.
Binding Assays.
Fluorescence emission intensity measurements (determined in triplicate) used 1-nM solutions of ODNs in 1 mL of titration buffer [50 mM Tris·HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA] to which were added ATL protein in 1-μL aliquots. Excitation and emission wavelengths used were 530 and 560 nm, respectively. The binding isotherms were fitted by nonlinear least-squares regression using KaleidaGraph to the standard equation describing the equilibrium D + E ↔ DE (where D = ODN, E = enzyme, and DE = ODN–enzyme complex)
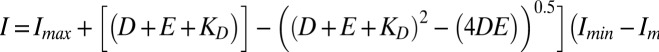 |
[where I = intensity measured at a certain concentration of enzyme, Imax = maximum intensity (i.e., before protein addition), Imin = minimum intensity (i.e., when binding is saturated), D = ODN concentration, and KD = dissociation constant].
Crystallization, X-Ray Diffraction Data Collection, and Structural Refinement.
C-terminally hexahistidine-tagged Atl1 was expressed and purified as previously described (4). A 13-nt DNA duplex with a 5′ overhang on either end was prepared by annealing single-strand ODN of sequence 5′-GCCATGXCTAGTA-3′, where X = 2,6-diaminopurine or 2-aminopurine, with equimolar complementary ODN of sequence 5′-CTACTAGCCATGG-3′. Complexes were prepared by mixing double-strand ODN with purified Atl1 at a 1.5:1 ODN:protein molar ratio. Crystals were grown by the hanging drop vapor diffusion method mixing 1 μL of protein–DNA complex with 1 μL well solution. Crystals of Atl1:DAP and Atl1:2-AP ODNs were grown from 20% mPEG 2000, 0.5 M sodium formate, 200 mM imidazole-malate (pH 5.4), and 30% xylose. Crystals in mother liquor were flash-frozen in liquid nitrogen for diffraction data collection. Diffraction data were collected at Stanford Synchrotron Radiation Lightsource beamline 11-1 at a wavelength of 0.97945 Å on a MAR325 detector. Diffraction data were processed with HKL2000 (44). Structures were solved by molecular replacement with Phaser (45), using the Atl1 structure (pdb 3GVA) as a search model for Atl1–DNA complexes. Crystallographic refinement was done with Python-based Hierarchical ENvironment for Integrated Xtallography (PHENIX) (46). Coot was used for manual model building into 2Fo-Fc and 2Fo-Fc and Fo-Fc omit electron density maps (47). DNA was built into the model after two rounds of refinement. Structural superpositions were done with Sequoia (48). Structure figures were made with PyMol (http://www.pymol.org).
Calculations.
All calculations were carried out using Spartan ’08 on an Apple Macintosh computer (Wavefunction, www.wavefun.com). Molecular mechanics calculations using the MMFF and MMFF(aq) force fields gave practically identical results for the cation–π interaction energy. The coordinates of Arg69 and O6-MeG were taken from the X-ray crystal structure of the Atl1 complex with O6-MeG-containing DNA (16), and C1' of O6-MeG and ε-CH2 of Arg69 were replaced by methyl groups. The total energy for the complex of these two fragments was compared with the energies of two fragments in isolation to estimate the interaction energy (∆E).
Then O6-MeG was converted into G by deleting the methoxy group, and the calculation was repeated. The difference between the two values of ∆E is −14.45 kJ mol−1 using the MMFF force field and −14.01 kJ mol−1 using the MMFF(aq) force field. Molecular electrostatic potential surfaces were calculated on the 0.002 Bohr Å-3 electron density isosurface using density functional theory (B3LYP with a 6-31G* basis set).
Supplementary Material
Acknowledgments
We thank Simon Thorpe for obtaining mass spectral data. This work was supported by the Biotechnology and Biological Sciences Research Council (O.J.W. and D.M.W.), the Japan Society for the Promotion of Science (O.J.W.), National Institutes of Health Grant CA097209 (to J.A.T.), and Cancer Research UK (G.P.M.). X-ray diffraction technologies at Stanford Synchrotron Radiation Lightsource are supported by the US Department of Energy and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4HDU (Atl1-2-AP-DNA) and 4HDV (Atl1-DAP-DNA)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209451109/-/DCSupplemental.
References
- 1.Margison GP, Santibáñez-Koref MF. O6-alkylguanine-DNA alkyltransferase: Role in carcinogenesis and chemotherapy. Bioessays. 2002;24(3):255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 2.Daniels DS, et al. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11(8):714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 3.Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem Res Toxicol. 2011;24(5):618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6(8):1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMurry TBH. MGMT inhibitors—The Trinity College-Paterson Institute experience, a chemist’s perception. DNA Repair (Amst) 2007;6(8):1161–1169. doi: 10.1016/j.dnarep.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Coulter R, et al. Differences in the rate of repair of O6-alkylguanines in different sequence contexts by O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 2007;20(12):1966–1971. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 7.Wang LJ, et al. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 1997;10(5):562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 8.Shibata T, et al. Novel synthesis of O6-alkylguanine containing oligodeoxyribonucleotides as substrates for the human DNA repair protein, O6-methylguanine DNA methyltransferase (MGMT) Nucleic Acids Res. 2006;34(6):1884–1891. doi: 10.1093/nar/gkl117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pletsas D, et al. Polar, functionalized guanine-O6 derivatives resistant to repair by O6-alkylguanine-DNA alkyltransferase: implications for the design of DNA-modifying drugs. Eur J Med Chem. 2006;41(3):330–339. doi: 10.1016/j.ejmech.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Shuker DEG, Margison GP. Nitrosated glycine derivatives as a potential source of O6-methylguanine in DNA. Cancer Res. 1997;57(3):366–369. [PubMed] [Google Scholar]
- 11.Margison GP, et al. Alkyltransferase-like proteins. DNA Repair (Amst) 2007;6(8):1222–1228. doi: 10.1016/j.dnarep.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Tubbs JL, Tainer JA. Alkyltransferase-like proteins: Molecular switches between DNA repair pathways. Cell Mol Life Sci. 2010;67(22):3749–3762. doi: 10.1007/s00018-010-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson SJ, Ferguson J, Santibanez-Koref M, Margison GP. Inhibition of O6-methylguanine-DNA methyltransferase by an alkyltransferase-like protein from Escherichia coli. Nucleic Acids Res. 2005;33(12):3837–3844. doi: 10.1093/nar/gki696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O6-alkylguanine adducts in E. coli. DNA Repair (Amst) 2009;8(6):697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, et al. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods. 2008;5(1):69–74. doi: 10.1038/NMETH1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tubbs JL, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459(7248):808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson SJ, et al. A novel DNA damage recognition protein in Schizosaccharomyces pombe. Nucleic Acids Res. 2006;34(8):2347–2354. doi: 10.1093/nar/gkl270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onodera TMK, et al. Role of alkyltransferase-like (ATL) protein in repair of methylated DNA lesions in Thermus thermophilus. Mutagenesis. 2011;26(2):303–308. doi: 10.1093/mutage/geq093. [DOI] [PubMed] [Google Scholar]
- 19.Morita R, Nakagawa N, Kuramitsu S, Masui R. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus interacts with a nucleotide excision repair protein. J Biochem. 2008;144(2):267–277. doi: 10.1093/jb/mvn065. [DOI] [PubMed] [Google Scholar]
- 20.Aramini JM, et al. Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein. J Biol Chem. 2010;285(18):13736–13741. doi: 10.1074/jbc.M109.093591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latypov VF, et al. Atl1 regulates choice between global genome and transcription-coupled repair of O6-alkylguanines. Mol. Cell. Cell. 2012;47:50–60. doi: 10.1016/j.molcel.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazon G, et al. Alkyltransferase-like protein (eATL) prevents mismatch repair-mediated toxicity induced by O6-alkylguanine adducts in Escherichia coli. Proc Natl Acad Sci USA. 2010;107(42):18050–18055. doi: 10.1073/pnas.1008635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 24.Yang CG, Garcia K, He C. Damage detection and base flipping in direct DNA alkylation repair. ChemBioChem. 2009;10(3):417–423. doi: 10.1002/cbic.200800580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6(4):410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Parikh SS, et al. Uracil-DNA glycosylase-DNA substrate and product structures: Conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci USA. 2000;97(10):5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA. 2000;97(25):13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingaraju GM, Davis CA, Setser JW, Samson LD, Drennan CL. Structural basis for the inhibition of human alkyladenine DNA glycosylase (AAG) by 3,N4-ethenocytosine-containing DNA. J Biol Chem. 2011;286(15):13205–13213. doi: 10.1074/jbc.M110.192435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403(6772):859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 30.Wibley JEA, Pegg AE, Moody PCE. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28(2):393–401. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore MH, Gulbis JM, Dodson EJ, Demple B, Moody PCE. Crystal structure of a suicidal DNA repair protein: The Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994;13(7):1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millington CL, et al. Convenient and efficient syntheses of oligodeoxyribonucleotides containing O6-(carboxymethyl)guanine and O6-(4-oxo-4-(3-pyridyl)butyl)guanine. Nucleosides Nucleotides Nucleic Acids. 2012;31(4):328–338. doi: 10.1080/15257770.2012.656784. [DOI] [PubMed] [Google Scholar]
- 33.Kim H-Y, et al. Synthesis and adduction of fully deprotected oligodeoxynucleotides containing 6-chloropurine. Tetrahedron Lett. 1998;39:6803–6806. [Google Scholar]
- 34.Fersht AR. In: Structure and Mechanism in Protein Science. Fersht AR, editor. New York: Freeman; 1999. pp. 340–341. [Google Scholar]
- 35.Ma JC, Dougherty DA. The cation-π interaction. Chem Rev. 1997;97(5):1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 36.Hunter CA, Low CMR, Rotger C, Vinter JG, Zonta C. Substituent effects on cation-π interactions: A quantitative study. Proc Natl Acad Sci USA. 2002;99(8):4873–4876. doi: 10.1073/pnas.072647899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter CA. Quantifying intermolecular interactions: Guidelines for the molecular recognition toolbox. Angew Chem Int Ed Engl. 2004;43(40):5310–5324. doi: 10.1002/anie.200301739. [DOI] [PubMed] [Google Scholar]
- 38.Graves RJ, Li BF, Swann PF. Repair of O6-methylguanine, O6-ethylguanine, O6-isopropylguanine and O4-methylthymine in synthetic oligodeoxynucleotides by Escherichia coli ada gene O6-alkylguanine-DNA-alkyltransferase. Carcinogenesis. 1989;10(4):661–666. doi: 10.1093/carcin/10.4.661. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson MC, et al. Purification of the E. coli ogt gene product to homogeneity and its rate of action on O6-methylguanine, O6-ethylguanine and O4-methylthymine in dodecadeoxyribonucleotides. Nucleic Acids Res. 1989;17(21):8475–8484. doi: 10.1093/nar/17.21.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodtzova K, et al. Repair of O6-benzylguanine by the Escherichia coli Ada and Ogt and the human O6-alkylguanine-DNA alkyltransferases. J Biol Chem. 1997;272(13):8332–8339. doi: 10.1074/jbc.272.13.8332. [DOI] [PubMed] [Google Scholar]
- 41.Spratt TE, Wu JD, Levy DE, Kanugula S, Pegg AE. Reaction and binding of oligodeoxynucleotides containing analogues of O6-methylguanine with wild-type and mutant human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 1999;38(21):6801–6806. doi: 10.1021/bi982908w. [DOI] [PubMed] [Google Scholar]
- 42.Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase. Effects of protein and DNA alkylation on complex stability. J Biol Chem. 2003;278(10):7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 43.Edara S, Goodtzova K, Pegg AE. The role of tyrosine-158 in O6-alkylguanine-DNA alkyltransferase activity. Carcinogenesis. 1995;16(7):1637–1642. doi: 10.1093/carcin/16.7.1637. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Bruns CM, Hubatsch I, Ridderström M, Mannervik B, Tainer JA. Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J Mol Biol. 1999;288(3):427–439. doi: 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.