Abstract
Here we report a microfluidics method to enrich physically deformable cells by mechanical manipulation through artificial microbarriers. Driven by hydrodynamic forces, flexible cells or cells with high metastatic propensity change shape to pass through the microbarriers and exit the separation device, whereas stiff cells remain trapped. We demonstrate the separation of (i) a mixture of two breast cancer cell types (MDA-MB-436 and MCF-7) with distinct deformabilities and metastatic potentials, and (ii) a heterogeneous breast cancer cell line (SUM149), into enriched flexible and stiff subpopulations. We show that the flexible phenotype is associated with overexpression of multiple genes involved in cancer cell motility and metastasis, and greater mammosphere formation efficiency. Our observations support the relationship between tumor-initiating capacity and cell deformability, and demonstrate that tumor-initiating cells are less differentiated in terms of cell biomechanics.
Keywords: cell mechanics, cytoskeleton, genomic profiling
Cell deformability is commonly measured using magnetic twisting cytometry, particle tracking rheometry, optical tweezers, micropipette aspiration, atomic force microscope, and other derivative cell stretching or poking methods (1–4). Applications of these methods to stem cells have revealed the greater deformability of the cytoskeleton and nucleoskeleton in less differentiated cells, whereby deformability generally decreases during differentiation to mature cells (5–8). Research on cancer cell deformability has also consistently revealed that increased deformability is correlated with increased metastatic potential (9–15).
Despite the success achieved using cell deformability measurements, isolation of cells with differential deformabilities remains a great challenge (10, 16). Microfabrication-assisted technology, using microscale arrays of round or rectangular posts, channels, or other simple patterns, has the potential to solve this problem (17–27). Here, we focused on the mechanical properties of cancer cells in designing a unique cell purification system for the purpose of generating subpopulations enriched in highly deformable cells. We used microfabrication technology and obtained a subpopulation of SUM149 breast cancer cells with stem-cell–like deformability and mammosphere formation capability.
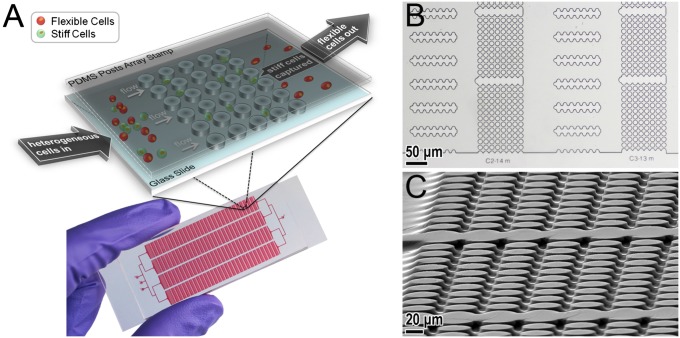
The separation device, a mechanical separation chip (MS-chip), employs artificial microbarriers in combination with hydrodynamic force to separate deformable from stiff cells (Fig. 1A). Both the microbarrier structures and the fluidic parameters are essential to the cell-enrichment process. The most notable feature of the device is the precise placement of the microbarriers to impede the passage of stiff cells. Published in vivo observations suggest that the minimum crossable barrier for cancer cells is ∼8 μm or less (28, 29). Here, we took those barrier dimensions into account in designing the MS-chip to separate cells based on perfusion through constrictions. As depicted in Fig. 1, the gaps between posts range in size from 15 to 7 μm, following the direction of fluid flow; this allows continuous optimization of separation efficiency. The rectangular, matrix-like arrangement of the microbarriers ensures that the most flexible cells are able to seek alternate routes in the event of a blockage. The second important feature is the placement of wide channels between post arrays (Fig. 1B). The main benefits of those channels are to prevent obstruction that could easily occur in continuously repeating arrays of posts and regulate and equalize hydrodynamic pressure throughout the chip. Thus, the cells actually have a chance to reorient themselves before continuing and adopt an alternative path before passing through another set of post arrays.
Fig. 1.
(A) Scheme (Upper) and photograph (Lower) of MS-chip. (B) Optical microscopic images of the MS-chip. (C) Scanning electron microscopic image of the post array taken at an angle. The post height is ∼13 μm. The microscale geometry of the flow channels and post arrays ensures that fluid flow is laminar, resulting in continuous cell movement and deformation in this device.
Using the MS-chip, we demonstrate the separation of: (i) an artificial mixture of two breast cancer cell types (MDA-MB-436 and MCF-7) with distinct deformabilities and metastatic potentials, and (ii) a heterogeneous breast cancer cell line (SUM149), into enriched flexible and stiff subpopulations. We have demonstrated that the flexible phenotype is associated with overexpression of multiple genes involved in cancer cell motility and metastasis. We have also assayed the tumorigenicity of the flexible and stiff subpopulations of SUM149 cells using the mammosphere formation efficiency assay. Our study provides an example of application of a microfluidics-based approach to cancer cell metastasis research.
Results and Discussion
Chip Design and Operation.
In a typical experiment, the MS-chip was fabricated using standard polydimethylsiloxane (PDMS) microfluidics technology (30). Suspended cells were then applied to the MS-chip via a Tygon tube connected to the chip inlet. The flow of cells was controlled by nitrogen pressure, normally at 5–10 psi (34–69 × 103 Pa), for a total flow rate of 1–2 mL/h. Cells applied to the MS-chip were imaged by fluorescence microscopy. The separated subpopulations were then subjected to gene-expression analysis, flow cytometry, or mammosphere formation assays.
On-Chip Cell Separation.
To demonstrate the capacity of the MS-chip to separate cells based on flexibility, we tested the separation of a mixture containing two different breast cancer cell types. We used a less flexible, nonmetastatic breast cancer cell line, MCF-7/GFP, which expresses GFP, and a more flexible metastatic breast cancer cell line, MDA-MB-436/RFP, which expresses red fluorescent protein (RFP); fluorescent proteins had been introduced into the cells with lentivirus. Mesenchymal-like breast cancer cell lines [such as MDA-MB-231 (9) and MDA-MB-436] and epithelium-like breast cancer cell lines (such as MCF-7) were previously shown to have significantly different deformabilities, quantified by optical deformability assays. Typically, the mesenchymal-like cells show greater deformability. As a proof-of-concept study, MCF-7/GFP and MDA-MB-436/RFP cells were cultured separately and then mixed in equal amounts at a final density of 1 × 106 cells/mL. These two cell types express different levels of cytoskeletal proteins, with MCF-7 having the stiffer cytoskeleton (31). We characterized the higher deformability of MDA-MB-436/RFP cells compared with MCF-7/GFP cell using atomic force microscopy indentation and examined cytoskeletal organization after staining of cytoskeletal protein (Figs. S1–S4 and SI Materials and Methods).
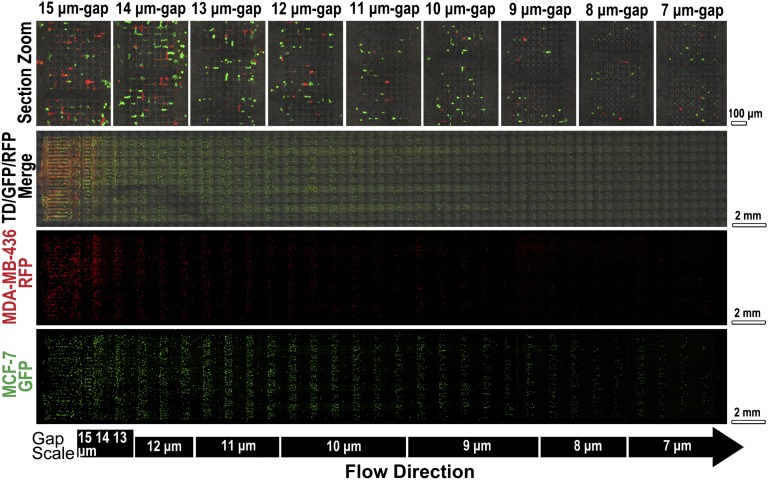
Cells trapped on MS-chip were imaged by fluorescence microscopy (Fig. 2 and Fig. S5). The proportion of the two cell lines varied along the length of the chip. At the inlet, equal amounts of green MCF-7 and red MDA-MB-436 fluorescent cells were observed. In this region, channels were much wider than the diameter of the cells, resulting in no significant separation of flexible and stiff cells. In the direction of flow, MCF-7/GFP cells were consistently trapped but MDA-MB-436/RFP cells passed through the gaps more frequently, as seen in Fig. 2. The smallest relative numbers of MDA-MB-436/RFP cells were in the center of the chip, where the gap sizes ranged from 11 to 9 μm at the flow direction. The results of statistical analysis of data derived from three independent experiments are presented in Fig. S6. These data demonstrate the overall efficiency of separation of the two cell lines in the MS-chip. The efficiency of separation was lower in the rear part of the chip. Two factors may aid in the interpretation of this observation. First, sorting in the front and middle parts of the chip resulted in an increased proportion of MDA-MB-436 cells, and therefore a higher proportion of MDA-MB-436 cells to MCF-7 cells presented when the mixture reached the rear part of the chip. Second, after sorting in the front and middle parts of the chip, the cells that were able to reach the rear part of the chip showed similar deformability, regardless of cell type.
Fig. 2.
Fluorescence images of cells retained on MS-chip after separation of MCF-7/GFP and MDA-MB-436/RFP cells. Gap sizes are shown on the left. Green, red, and merged channels are shown in the first three columns to demonstrate cell types trapped on the MS-chip. Higher magnification images of the merged image are on the right, indicating the efficiency of separation through gaps ranging in size from 15 to 7 μm.
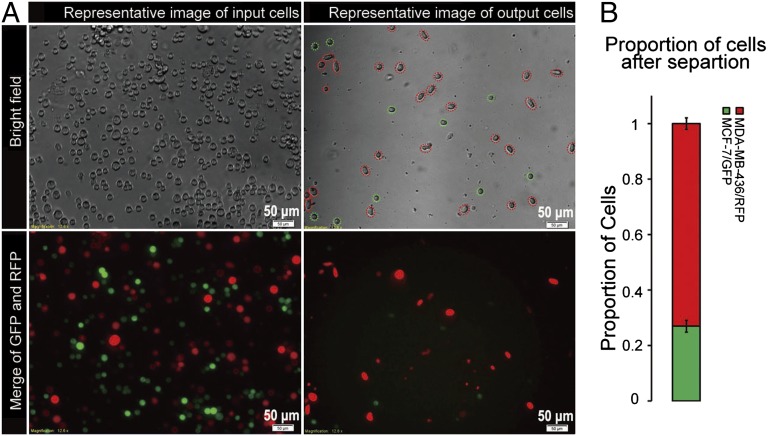
Fig. 3A shows representative images of cells input at the inlet and collected at the outlet, and Fig. 3B shows the proportion of red to green cells after the enrichment process. The fraction of MDA-MB-436/RFP cells in the mixture was increased from 50% to 73% (mean value from three experiments). It should be noted that cellular heterogeneity does exist within the cell types and affects the quality of the separation; specifically, a proportion of MCF-7/GFP cells were released from the chip, and some MDA-MB-436/RFP were retained. MDA-MB-436/RFP cells had a wider size distribution, with a mean size of 282 μm2, compared with MCF-7/GFP cells, which had a mean size of 184 μm2. These two cell types showed slightly decreased size after separation, but still preserved distinct mean sizes relative to each other (Fig. S7). These data indicate that cell size has little effect in the mechanical separation of cancer cells in MS-chip.
Fig. 3.
(A) Comparison of input and output cells in a typical MCF-7/GFP and MDA-MB-436/RFP separation. Both bright-field and fluorescence images are presented. The representative image of input cells (cell sample before loading into the MS-chip) shows approximately equal numbers of green and red cells (Lower, Left), and the representative image of output cells (cells that passed through the entire MS-chip and were collected at the outlet) shows a significantly enriched population of red cells (Lower Right). (B) The bar graph displays the proportion of MCF-7/GFP and MDA-MB-436/RFP cells after separation of an equal mixture. Values represent mean ± SD from three independent experiments analyzing 250–300 cells each.
Analyses of the Enriched Cell Subpopulation.
Given that the MS-chip can enrich cells with high deformability and metastatic potential from a complex cell mixture, we next explored the mechanical subtype of SUM149 cells in detail. The SUM149 cell line is known for its intrinsic heterogeneity (32), and it is a well-established in vitro model of inflammatory breast cancer that we have studied previously (33, 34).
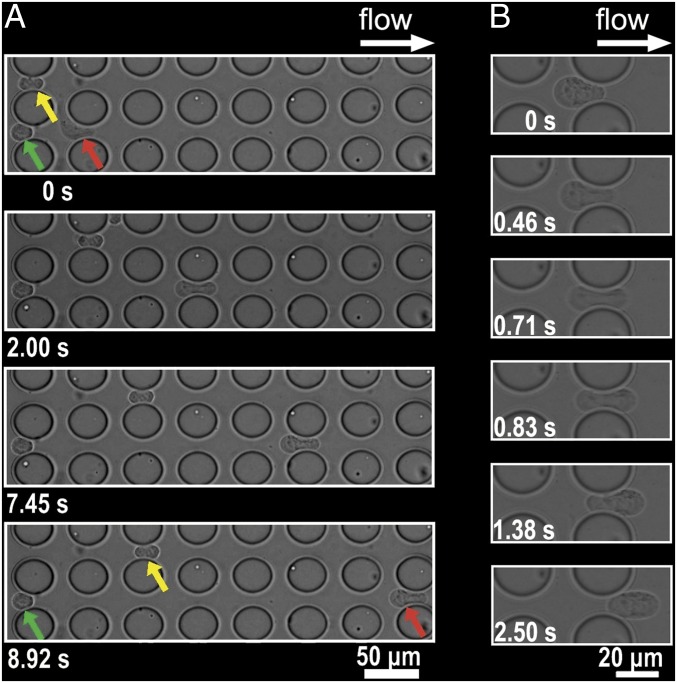
The evaluation of the heterogeneous deformability of SUM149 cells is shown in Movies S1 and S2. These two movie clips record the cancer cells as they cross MS-chip post arrays. Fig. 4 is a magnified field showing the deformed cells. The green arrow points to a stiff cell with limited deformability trapped in a gap between posts, despite the hydrodynamic force of fluid flow. The red arrow points to a malleable cell that changes shape and is highlighted in a time sequence analysis on the right (L-shaped in the top frame, dumbbell-shaped in the second, and bowling pin-shaped in the two lower frames). This cell was able to pass through the post arrays because of its high degree of deformability. The cell indicated by the yellow arrow passed halfway through the device, demonstrating intermediate deformability. During this experiment, cell size played a less important role, as indicated by the observation that a smaller stiff cell was trapped (green arrow), but a much larger, but flexible, cell passed through (red arrow). Fig. 4B indicates the deformation and partial recovery of a cell in the MS-chip. Multiple examples of cell deformation are shown in Movie S3.
Fig. 4.
(A) Images of SUM149 cells crossing the MS-chip gaps. Red, green, and yellow arrows indicate different levels of cell deformability. The most flexible cell type (red) crosses the entire barrier array; the less flexible one (yellow) crosses halfway; and the least flexible one (green) stops at the initial intersection. (B) High-magnification images show cell deformation in the right six frames.
After flexible and stiff SUM149 cells were separated based on deformability (Movie S3), the cells were immediately lysed (Movie S4), RNA was extracted, and genome-wide gene-expression analysis was conducted using the Affymetrix HG-U133 plus 2.0 GeneChip with 54,675 probe sets. Based on cell number and quantitation of RNA from duplicate samples in two experiments, we estimated that ∼10% of applied cells successfully flowed through the MS-chip and 90% were retained. In total, seven sets of gene-expression data were collected and analyzed. Samples from the first of three independent cell-sorting experiments (Exp. 1) were performed on three technical replicates and those from others (Exps. 2 and 3) were performed on two technical replicates. For bioinformatic analysis of gene-expression data, all seven sets of data were processed together.
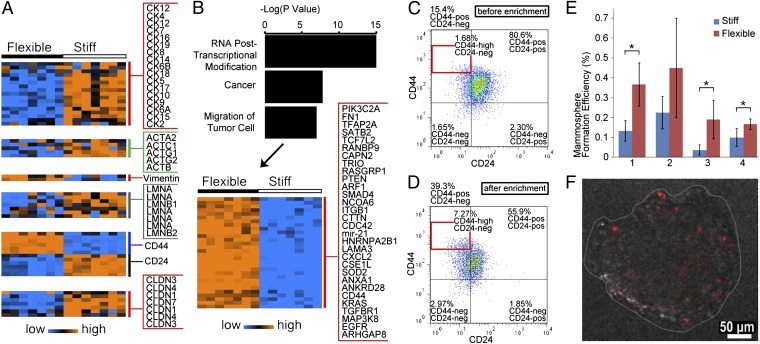
To further evaluate whether the enriched flexible subpopulation exhibits stem cell-like deformability, we examined expression of cytoskeletal and nucleoskeletal filament protein genes (Fig. 5A). The expression profile of the enriched flexible SUM149 cell subpopulation shows underexpression of epithelial cytokeratins, actins, and lamins, as well as overexpression of vimentin. These properties could be indicators of greater flexibility and a less differentiated state (5–8, 35), providing support for the proposed mechanism of MS-chip separation.
Fig. 5.
(A) Heat map shows expression levels of cytokeratins, actins, vimentin, lamins, CD44, CD24, and claudins in flexible and stiff cells. The genechip contains more than 54,000 probe sets, including 38,500 for human genes. Some genes have multiple probe sets. (B) Bar graph of the top three cell functions over the –log(p). The heat map shows 30 significantly overexpressed genes in the cell movement category. (C and D) Scatterplot of CD44/CD24 expression for SUM149 cells before (C) and after (D) MS-chip separation. (E) Mammosphere formation efficiency of flexible vs. stiff subpopulations. All four replicates show consistently greater efficiency in flexible cells. *P < 0.05. (F) Representative fluorescence image of a mammosphere derived from PKH26-labeled flexible SUM149 cells. PKH26 is significantly diluted during mammosphere formation.
The profiles of genes differentially expressed in flexible and stiff cells also demonstrate greater cell motility in the flexible cells. The 288 most highly overexpressed genes in flexible cells (>threefold change, P < 1 × 10−4, and false-discovery rate <0.1%) were mapped by Ingenuity Pathway Analysis (IPA) (Datasets S1 and S2). The results indicate that 53 highly expressed genes are relevant to cell motility and 30 regulate the migration of tumor cells, as shown in the heat map in Fig. 5B. The upper gene panel contains molecules involved in cytoskeletal reorganization, which is critical in cell motility and tumor progression; examples include Cdc42, CD44, c-Raf, and PIK3C2A (35–38). This panel in Fig. 5 also includes ITGβ1, TGFβR1, SMAD4, and EGFR, which are known to drive tumor invasion and metastasis (39, 40).
To further explore the relationship between stem cell-like deformability and tumor-initiating features, we next analyzed expression of the tumor-initiating cell (TIC) markers CD44, CD24, and claudins, and assessed TIC function by examining colony-forming capacity in mammosphere culture. Gene-expression analysis (Fig. 5A) revealed that the cell subtype with CD44+/CD24−/claudin-low expression was enriched by separation on the MS-chip, suggesting higher tumor-initiating capacity in these flexible breast cancer cells (41, 42). In agreement with this observation, flow cytometric analysis confirmed a significant enrichment of the CD44+/CD24− (or CD44high/CD24−) population in the flexible cells (Fig. 5 C and D, Figs. S8–10, and SI Results). Here, we have used mammosphere formation efficiency under conditions of serum-free growth in suspension culture as an assay of tumorigenicity. Less than 1% of tumor cells are able to survive under these culture conditions and the surviving cells are capable of self-renewal, differentiation, and tumor formation upon transplantation (41). The mammosphere formation assay demonstrated that flexible cells show higher growth efficiency (Fig. 5E) and generate larger mammospheres (Fig. S11), providing functional evidence of TIC enrichment. Under mammosphere culture conditions, rare quiescent and slowly dividing cells retain PKH26 fluorescence, but rapidly growing cells lose fluorescence by dilution with each proliferative cycle (Fig. 5F).
Conclusions
Overall, the enriched population of flexible SUM149 cells has a gene-expression profile indicative of greater cytoskeletal and nucleoskeletal deformability and motility (Figs. 4 and 5 A and B), as well as multiple characteristics that define TICs (Fig. 5 A, and C–E). Using cell separation, gene-expression analysis, and tumorigenicity assay, we have demonstrated the relationship between mechanical deformability and efficiency of mammosphere formation in breast cancer cells, which suggests that TICs preserve the mechanical deformability of normal stem cells. Our TIC-enrichment approach using a unique microfluidic cell-sorting technology will contribute to the advance of TIC research and the development of TIC-targeted therapy in a variety of cancer cell types. The MS-chip could be used to purify stem cells and TICs based on their unique mechanical characteristics and thereby identify molecular signatures and genes essential for tumor initiation. MS-chip separation will be particularly useful in other types of cancer for which TIC biomarkers have not yet been discovered.
Materials and Methods
Materials and Reagents.
SPR 220–7 photoresist was purchased from Rohm and Haas Electronic Materials. PDMS (GE 615 RTV), lyophilized BSA, FBS, trypsin, and penicillin-streptomycin were purchased from Fisher Scientific. DMEM and Ham’s F-12 medium, insulin, hydrocortisone, blasticidin, and PBS were purchased from Life Technologies. Basal membrane extracts (BME) was purchased from Trevigen. RNeasy micro kit was purchased from Qiagen. Human genechip U133 plus 2.0 chips were purchased from Affymetrix. The WT-Ovation Pico RNA amplification system and Encore Biotin module were purchased from Nugen. Tygon tubing (inner diameter, 0.02 in) was purchased from Saint-Gobain. Flat steel pins were purchased from New England Small Tube.
MS-Chip Design and Fabrication.
The microchip pattern was designed with AutoCAD (Autodesk). Two sizes of MS-chip were produced to fit either a 75 × 25-mm or 75 × 50-mm glass slides. The 75 × 25-mm MS-chip design consists of four independent working rows running in parallel between the same inlet and outlet through 200-μm channels. Each working row (∼41 mm in length and 5.3 mm in width) consists of two types of regions: post-array regions (n = 30) and wide-channel regions (n = 30). These two types of regions alternate on the chip. Each post-array region spans the entire width of the working row consisting of ∼800 posts (40 μm in diameter) that are sectioned into eight or nine sets of posts (depending on gap sizes); each of these sets consist of 9 × 10 arrays of posts. The gaps between posts decrease from a maximum of 15 μm to a minimum of 7 μm from left to right across the chip (see the gap size distribution in Fig. 2: column 1 of post arrays, 15 μm; column 2, 14 μm; column 3, 13 μm; columns 4–6, 12 μm; columns 7–10, 11 μm; columns 11–16, 10 μm; columns 17–22, 9 μm; columns 23–26, 8 μm; columns 27–30, 7 μm. Each channel region consists of 24 channels arranged vertically (420 μm in length and 160 μm in width). All patterns on the 75 × 25-mm chip, excluding the last three 7-μm-gap post arrays, are duplicated on the 75 × 50-mm chip. A total of eight independent working rows are arranged in parallel between the same inlet and outlet. Three generations of MS-chip have been designed and tested in this study. The design and examples of use of early generations are shown in Fig. S12 and Movie S5.
PDMS elastomer devices were fabricated using standard photolithography and molding technology (30). A high-resolution chrome mask with the design pattern purchased from Photo Sciences was used for photolithography. The photomask pattern was first translated into a positive structure on a 4-in silicon wafer using SPR 220-7 photoresist, which is a negative mold for casting PDMS materials. The SPR 220-7 mold was spin-cast at ∼1,500 rpm for 40 s, and was ∼13 μm in height after exposure and development. The height of the mold was designed to be slightly smaller than the average diameter of breast cancer cells (e.g., MCF-7, MDA-MB-436, and SUM149) for these experiments. Changes in the mold specifications are easily made by photolithography. A lower spin-coating rate will result in increased photoresist thickness and, thus, the height of the mold. The positive structure on the silicon wafer was then used to fabricate the PDMS layer. The mold and PDMS layer were then baked at 80 °C for 2 h and the cured PDMS was cut and removed from the mold. The holes for the inlets and outlets were punched using needle sizes compatible with the size of the fluid input/output pins. The PDMS layer was then cleaned by briefly rinsing with isopropyl alcohol and deionized water and dried with nitrogen gas. After treatment with oxygen plasma, the PDMS layer was bonded immediately to a glass slide. Finally, the bonded device was baked for 2 h at 80 °C.
Cell Culture and Synchronization.
The SUM149 cell line (estrogen receptor- and progesterone receptor-negative; EGFR- and HER2-low) was purchased from Asterand. The cells were grown in Ham’s F-12 medium supplemented with 5% (vol/vol) FBS, 1% (wt/vol) penicillin-streptomycin, 5 μg/mL insulin, and 1 μg/mL hydrocortisone in a humidified atmosphere of 5% CO2/95% air at 37 °C. The cell lines MCF-7/GFP and MDA-MB-436/RFP, which stably express GFP and RFP, respectively, were purchased from Cell Biolabs; the GFP and RFP genes had been introduced using lentivirus. Fluorescent proteins are widely used to visualize cancer cells in vivo and in vitro and have minimal effect on the characteristics of the cells (43, 44). These cells were grown in DMEM supplemented with 10% (vol/vol) FBS, 1% penicillin-streptomycin, and 2 μg/mL blasticidin under the same conditions as above.
Cell cultures were grown in serum-free medium for 48 h to synchronize the cells. Nutrition supply was cut off by serum-free medium for minimizing RNA synthesis in cells during separation experiment. This process minimizes sampling errors between flexible and stiff cells for downstream microarray analyses. After synchronization, the medium was aspirated and cells were washed twice with PBS and treated with trypsin for ∼5 min. Immediately before the on-chip experiment, cells were collected by centrifugation at 200 × g for 5 min, counted, and resuspended in serum-free DMEM at a concentration of 106 cells/mL. The mixture of MCF-7/GFP and MDA-MB-436/RFP cells contained 5 × 105 cells/mL of each cell type.
Cell Sorting on MS-Chip.
MS-chip pretreatment.
The channels in the MS-chip were first wetted with PBS and then treated with a 10% (vol/vol) BME solution in PBS for 1 h. BME represents a more physiological microenvironment than the native PDMS surface. The channels were then washed with 0.5% BSA in PBS for 1 h and loaded with 0.1% BSA in PBS. BSA blocks the surface and further prevents the nonspecific adhesion of cells to PDMS. The chip was kept on ice during pretreatment.
Cell-sorting process.
The mixture of MCF-7/GFP and MDA-MB-436/RFP cells was separated on a 75 × 25-mm MS-chip. Cell suspension (0.25 mL) was loaded into plastic Tygon tubing (inner diameter 0.02 in) with a 1-mL syringe and the tubing was connected to the MS-chip by a flat steel pin (outer diameter of 0.025 in). During the flow experiment, compressed nitrogen gas was applied to the cell suspension at a pressure of ∼5 psi. In this discussion on this experiment, “input cells” and “output cells” refer to the samples that were loaded into the MS-chip and that were collected at outlets. A typical separation lasted ∼15 min, and the average flow rate was controlled at ∼1 mL/h. The fluorescence images of cell mixtures before and after sorting were taken with an Olympus IX81 inverted fluorescence microscope (Fig. 3A). Paraformalin solution [4% (wt/vol)] was injected into the MS-chip with a 3 psi pressure. The whole chip was then scanned on a Nikon A1 confocal microscope with an image stitching function. The stitching function allowed the generation of large images covering the entire device, permitting continuous cell counts based on fluorescence signal.
The sorting of SUM149 cells was performed on a 75 × 50-mm MS-chip (Movie S3). The chip size was enlarged from 75 × 25 to 75 × 50 mm to increase throughput and generate a sufficient amount of cells for downstream analyses, including microarray, FACS, and mammosphere formation assay. The larger chip can process approximately a half million cells, at least twice as many as the 75 × 25-mm chip. The flow at the inlet is split evenly so that the flow pressure is uniformly distributed. The design change did not appear to affect cell separation efficiency. The 75 × 50-mm chip requires more space in both inlet and outlet area to distribute flow pressure. To fit the device into the 75-mm length, three sets of post arrays were killed for a 3-mm space. The rear arrays were removed because their separation efficiency is negligible comparing to front ones, as shown in the proof of concept experiment (Fig. S5). Because the flow pressure was carefully distributed to all functional areas of the chip, the separation efficiency was consistent.
The cell suspension (0.5 mL) was applied to the chip as described above. A mean flow rate was controlled at 2 mL/h by adjusting applied nitrogen pressure. The duration of the separation was roughly 15 min. The flexible cells were collected from the outlet and the stiff cells remained inside the MS-chip. The stiff cells were collected for mammosphere culture by back-flushing cells to the inlet.
Cell lysis.
Flexible and stiff cells were lysed for mRNA analysis immediately after separation. Flexible cells were immediately centrifuged and lysed with RNeasy lysis buffer (Qiagen). Stiff cells inside the device were lysed by perfusing lysis buffer through the chip (Movie S4). The time difference between lysis of the two sets of cells was less than 5 min. Serum-free culture medium was used to deplete the nutrient supply and minimize RNA synthesis. This strategy minimizes error because of differences in sampling the two populations of cells.
Empirical evidence indicates that 12–13 μg RNA can be extracted from 1 million breast cancer cells using the Qiagen RNeasy Micro Kit. In this study, 5 μg RNA were obtained from the lysate of stiff cells and 0.5 μg RNA from flexible cells. Thus, we estimated that 380,000–410,000 stiff cells and 38,000–41,000 flexible cells had been lysed.
Image Acquisition and Analysis.
Whole-slide images (Fig. 2A and Fig. S5) were scanned and stitched using a Nikon A1 laser confocal microscope equipped with NIS-Elements microscope imaging software. The fluorescence images were taken with an Olympus IX81 inverted fluorescence microscope.
Analytical Flow Cytometry.
Cells were suspended in cold HBSS containing 2% FBS (HBSS+) at 50,000 cells/mL for antibody staining. Antibodies CD44-APC and CD24-PeCy7 (both from BD Bioscience) were diluted 1:20 and 1:100, respectively. After staining on ice, the cells were washed twice with cold HBSS+ and kept in HBSS+ on ice until FACS analysis. Isotype control antibodies were used as the gating control.
Mammosphere Formation Assay.
After the separation experiments, cells were maintained in mammary epithelial cell growth medium containing 2% FBS (MEGM+; Lonza) on ice. Flexible and stiff cells showed similar viability in four groups of independent experiments, with respective values of 89%, 89%, 84%, and 75%, and 84%, 85%, 83%, and 78%. For each group of experiments, six pairs of mammosphere cultures were characterized based on number and size, and the means and SDs were calculated.
For the mammosphere assay, cells were seeded at 2,000 viable cells per well in 24-well plates (six wells per group) and incubated at 37 °C in an atmosphere of 5% CO2/95% air. After 2 wk, the mammospheres were counted using GelCount (Oxford Optronix). Mammoshpere size was evaluated by optical images.
Gene Expression Array and Analysis.
Total RNA was extracted from lysates of the flexible and stiff SUM149 subpopulations (0.5 μg and 5 μg, respectively) using the Qiagen RNeasy Micro kit. cDNA was prepared from 50 ng total RNA with the Nugen WT-Ovation Pico RNA amplification system, generating 250-fold amounts of cDNA. The OD260/OD280 ratios were ∼2, indicating a pure preparation of DNA. Five micrograms of cDNA from each sample were labeled with biotin using the Nugen FL-Ovation cDNA Biotin Module V2. The labeled, single-stranded cDNA probes were hybridized to Affymetrix HG-U133 plus 2.0 Genechips.
Gene-expression data were normalized with the MAS5 algorithm using BRB ArrayTool software (v4.2.0, http://linus.nci.nih.gov/BRB-ArrayTools.html) and R software (http://www.r-project.org v2.7.2) and log2-transformed before further analysis. Data from flexible and stiff cells were compared using two-class unpaired analysis. The restrictions on the proportion of false discoveries were 0.01 for a maximum proportion of false discovery and 90% for a confidence level. With a probability of 90%, the total set of differentially expressed genes contains no more than 1% false discoveries. Core analysis was used to interpret the gene expression data in the context of biological pathways, functions, and networks using IPA (Ingenuity Systems, www.ingenuity.com). The stricter criteria of >threefold change, 0.001 maximum allowed proportion of false-positive genes, and 90% confidence level of false-discovery rate assessment were applied to reduce the number of up-regulated genes for analysis; 688 genes satisfied the criteria and were submitted to the IPA software. Of these, 288 genes were mapped by IPA pathways, functions, and networks analysis (Dataset S1). Significance of the biofunctions and the canonical pathways was assessed by P values obtained using the Fisher Exact test.
Supplementary Material
Acknowledgments
We thank Ms. Stephanie Mena and Mr. Wenborui Zhong for experimental assistance; Drs. Kemi Cui and James Gu at the Methodist Hospital Research Institute Advanced Tissue and Cellular Imaging Core for atomic force microscopy assistance; and Dr. Ana Maria Zaske at the University of Texas-Health Science Center Atomic Force Microscopy core. Array analyses were performed using Biometric Research Branch-ArrayTools developed by Dr. Richard Simon and the Biometric Research Branch-ArrayTools Development Team. This study is funded by the Cancer Prevention and Research Institute of Texas (CPRIT-R1007), the U54-CA149196-Pilot Project, the Emily Herman Research Fund, and the Golfers Against Cancer Foundation; and a grant from the State of Texas Rare and Aggressive Breast Cancer Research Program (to N.T.U. and K.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209893109/-/DCSupplemental.
References
- 1.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: The force journey of a tumor cell. Cancer Metastasis Rev. 2009;28(1–2):113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2(11):715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 3.Wirtz D. Particle-tracking microrheology of living cells: Principles and applications. Annu Rev Biophys. 2009;38(1):301–326. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 4.Khismatullin DB. 2009. The cytoskeleton and deformability of white blood cells. Leukocyte Adhesion, ed Ley K, Current Topics in Membranes (Elsevier Academic, San Diego), Vol 64, pp 47–111.
- 5.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104(40):15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titushkin I, Cho M. Modulation of cellular mechanics during osteogenic differentiation of human mesenchymal stem cells. Biophys J. 2007;93(10):3693–3702. doi: 10.1529/biophysj.107.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury F, et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater. 2010;9(1):82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12(11):695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 9.Guck J, et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J. 2005;88(5):3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11(7):512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beil M, et al. Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nat Cell Biol. 2003;5(9):803–811. doi: 10.1038/ncb1037. [DOI] [PubMed] [Google Scholar]
- 12.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2(12):780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 13.Swaminathan V, et al. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011;71(15):5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remmerbach TW, et al. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009;69(5):1728–1732. doi: 10.1158/0008-5472.CAN-08-4073. [DOI] [PubMed] [Google Scholar]
- 15.Darling EM, Zauscher S, Block JA, Guilak F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: do cell properties reflect metastatic potential? Biophys J. 2007;92(5):1784–1791. doi: 10.1529/biophysj.106.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossett DR, et al. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc Natl Acad Sci USA. 2012;109(20):7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 19.Melin J, Quake SR. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct. 2007;36(1):213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 20.Heath JR, Davis ME, Hood L. Nanomedicine targets cancer. Sci Am. 2009;300(2):44–51. doi: 10.1038/scientificamerican0209-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craighead H. Future lab-on-a-chip technologies for interrogating individual molecules. Nature. 2006;442(7101):387–393. doi: 10.1038/nature05061. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50(13):3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26(12):1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson RH, et al. Self-sorting of white blood cells in a lattice. Phys Rev Lett. 1997;79(11):2149–2152. [Google Scholar]
- 25.Carlson RH, Gabel C, Chan S, Austin RH. Activation and sorting of human white blood cells. Biomed Microdevices. 1998;1(1):39–47. [Google Scholar]
- 26.Mohamed H, Murray M, Turner JN, Caggana M. Isolation of tumor cells using size and deformation. J Chromatogr A. 2009;1216(47):8289–8295. doi: 10.1016/j.chroma.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 27.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. Deformability-based cell classification and enrichment using inertial microfluidics. Lab Chip. 2011;11(5):912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi K, et al. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005;65(10):4246–4252. doi: 10.1158/0008-5472.CAN-05-0069. [DOI] [PubMed] [Google Scholar]
- 29.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane) Anal Chem. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 31.Sommers CL, et al. Vimentin rather than keratin expression in some hormone-independent breast cancer cell lines and in oncogene-transformed mammary epithelial cells. Cancer Res. 1989;49(15):4258–4263. [PubMed] [Google Scholar]
- 32.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, et al. Epidermal growth factor receptor tyrosine kinase inhibitor reverses mesenchymal to epithelial phenotype and inhibits metastasis in inflammatory breast cancer. Clin Cancer Res. 2009;15(21):6639–6648. doi: 10.1158/1078-0432.CCR-09-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwamoto T, et al. Different gene expressions are associated with the different molecular subtypes of inflammatory breast cancer. Breast Cancer Res Treat. 2011;125(3):785–795. doi: 10.1007/s10549-010-1280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Mouw JK, Weaver VM. Forcing form and function: Biomechanical regulation of tumor evolution. Trends Cell Biol. 2011;21(1):47–56. doi: 10.1016/j.tcb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Föger N, Marhaba R, Zöller M. Involvement of CD44 in cytoskeleton rearrangement and raft reorganization in T cells. J Cell Sci. 2001;114(Pt 6):1169–1178. doi: 10.1242/jcs.114.6.1169. [DOI] [PubMed] [Google Scholar]
- 38.Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390(6660):632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 39.Dunning AM, et al. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63(10):2610–2615. [PubMed] [Google Scholar]
- 40.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 41.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 42.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J-Y, et al. Establishment of red fluorescent protein-tagged HeLa tumor metastasis models: Determination of DsRed2 insertion effects and comparison of metastatic patterns after subcutaneous, intraperitoneal, or intravenous injection. Clin Exp Metastasis. 2003;20(2):121–133. doi: 10.1023/a:1022645116030. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer. 2005;5(10):796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.