Abstract
Background
Genetic variants that contribute to asthma susceptibility may be present at varying frequencies in different populations, which is an important consideration and advantage for performing genetic association studies in admixed populations.
Objective
To identify asthma-associated loci in African Americans.
Methods
We compared local African and European ancestry estimated from dense single nucleotide polymorphism (SNP) genotype data in African American adults with asthma and non-asthmatic controls. Allelic tests of association were performed within the candidate regions identified, correcting for local European admixture.
Results
We identified a significant ancestry association peak on chromosomes 6q. Allelic tests for association within this region identified a SNP (rs1361549) on 6q14.1 that was associated with asthma exclusively in African Americans with local European admixture (OR=2.2). The risk allele is common in Europe (42% in the HapMap CEU) but absent in West Africa (0% in the HapMap YRI), suggesting the allele is present in African Americans due to recent European admixture. We replicated our findings in Puerto Ricans and similarly found that the signal of association is largely specific to individuals who are heterozygous for African and non-African ancestry at 6q14.1. However, we found no evidence for association in European Americans or in Puerto Ricans in the absence of local African ancestry, suggesting that the association with asthma at rs1361549 is due to an environmental or genetic interaction.
Conclusion
We identified a novel asthma-associated locus that is relevant to admixed populations with African ancestry, and highlight the importance of considering local ancestry in genetic association studies of admixed populations.
Keywords: asthma, population structure, genome-wide association study, admixture mapping, ancestry association testing, admixed populations, African Americans, Puerto Ricans
Introduction
Asthma is a chronic respiratory disease characterized by airway inflammation, airway obstruction, and episodic respiratory symptoms. Although it is a common disease shared across diverse human populations, African Americans have higher rates of asthma, steroid dependency, and a more limited response to treatment as compared to European Americans 1, which may not be attributable to differences in access to healthcare alone 2, 3. Although there is a strong environmental component, recent estimates of the heritability of asthma are around 75% 2, 3, suggesting there are important genetic contributions to asthma susceptibility. As of 2006, polymorphisms in over 100 genes had been associated with asthma and asthma-related phenotypes, with more than 40 genes showing replication in at least one independent population 4. More recently, numerous genome-wide association studies (GWAS) have identified additional candidate genes and genetic loci that contribute to asthma susceptibility 5–11. While the majority of GWAS have focused largely on populations of European origin 6, 7, 9–11, candidate loci have also been identified through GWAS in African Americans 8, Puerto Ricans 12 and Mexicans 5.
One of the challenges of performing genetic association studies in African Americans is avoiding confounding resulting from complex population structure. Due to differences in demographic histories, populations of African origin exhibit higher levels of genetic variation, reduced linkage disequilibirum (LD), and proportionally more rare variation as compared to European populations 13. The colonization of the Americas by Europeans, followed by the African slave trade, resulted in populations with “mixed” genetic ancestry in the Americas (admixed populations), including African Americans that have on average ~ 20% European ancestry 14. Because European admixture is relatively recent, recombination has yet to break up the European and African haplotypes completely, resulting in long haplotypes specific to each ancestral background. Identifying regions whereby admixture varies between cases and controls is important to avoid confounding due to population structure in genetic association studies. However, it can also be used to perform admixture mapping to identify novel disease susceptibility loci 15.
Admixture mapping in African Americans has proven to be a valuable tool for identifying novel risk alleles for diseases such as multiple sclerosis 16, prostate cancer 17, obesity 18, and hypertension 19. In theory, admixture mapping is applicable to genetic studies of admixed populations for any disease in which risk alleles vary in frequency between in the ancestral populations, such that detecting differences in ancestry can identify regions that contain variants associated with disease. Admixture mapping has previously implicated the 5q23 locus in asthma susceptibility in Puerto Ricans 12, indicating this method of analysis is relevant to genetic studies of asthma. A potential advantage of admixture mapping is to increase the power of genetic association studies, for example by identifying candidate regions to look for allelic associations to reduce the multiple-testing burden (e.g. 20), and by increasing genomic coverage through long-range tagging of single nucleotide polymorphisms (SNPs) with local ancestry (ancestry-LD). While admixture mapping has historically utilized a set of ancestral informative markers (AIMs), methods are available to identify local ancestry using dense SNP genotype data 21, 22, enabling genome-wide ancestry association testing at a higher resolution and accuracy over more traditional admixture mapping.
To identify genetic loci that contribute to asthma susceptibility in African Americans, we performed genome-wide ancestry association testing using estimates of local ancestry inferred from ~ 1 million genotyped SNPs in 355 African American adults with asthma and 444 African American adult controls. We performed allelic tests of association within an ancestry association peak on chromosome 6q while adjusting for local ancestry, and identified a polymorphism on 6q14.1 that is associated with asthma exclusively in African American adults with local European admixture. We replicated both the ancestry and allelic association in the same direction in an independent population of Puerto Ricans. The risk allele is common in the HapMap CEU but absent in the HapMap YRI, suggesting the allele is present in African Americans due to recent European admixture.
Methods
Study Subjects
This research was approved by the Institional Review Boards at the University of Chicago and Wake Forest University. Adults and children with asthma and adult non-asthmatic controls were recruited as part of the Collaborative Study on the Genetics of Asthma (CSGA) 23 and the Severe Asthma Research Program (SARP) 24, 25. The CSGA study sample includes subjects recruited at the University of Chicago, University of Maryland, and Wake Forest University from a) families ascertained through affected sib pairs, b) trios of affected children and their parents, c) adults and children with severe persistent asthma, and d) non-asthmatic control subjects (over the age of 18 years). The multicenter SARP study sample includes mild to severe asthmatics and controls. In both studies, subjects were carefully characterized using similar procedures 23–25. Besides having a physician’s diagnosis of asthma, all asthma cases met objective criteria of bronchial hyperresponsiveness after a methacholine challenge, or reversibility to an inhaled bronchodilator. Subjects with a significant history of smoking were excluded.
Because 65% of the cases and 100% of the controls were over 17 years of age at time of recruitment, we limited our study to those subjects to reduce phenotypic heterogeneity. Among these subjects the mean age at enrollment was 36 years of age, the mean age of onset was 14 years of age, and 70% of the participants were female. All of the primary subjects included in the current study were self-reported African Americans, however we also present results from self-reported European Americans who participated in the CSGA/SARP studies (complete results will be presented elsewhere).
Genotyping and QC
Genotyping was performed on the Illumina 1Mv1 platform, with individual genotypes called using clustering algorithms as implemented in the BeadStudio software by Illumina. Following an initial round of clustering and genotype calling, 3 additional rounds of clustering was performed alternating between filtering of subjects and markers based on call rates < 95% and < 90%, respectively. SNPs were then filtered for “Cluster_Sep” values < 0.3 (score of genotype cluster separation), and for “GenTrain” scores < 0.75. The resulting number of SNPs was 1,033,467 prior to additional quality control measures.
Data quality control was performed using PLINK 26, R (http://www.R-project.org), and Eigenstrat 27, 28. SNPs were filtered based on 95% call rates, Hardy-Weinberg equilibrium p-values > 10−6, consistency in allele frequency from the HapMap ASW (chi-square p-value > 10−6), and < 5 heterozygous genotype calls in males for X-linked markers. The total number of markers following QC checks was 1,026,072. Subjects were filtered based on >95% call rates, consistency between genetic and reported sex (Fstat from X-linked markers between −0.2 and 0.3 for females, and between 0.8 and 1 for males), consistency in self-reported ethnicity (no obvious clustering with the HapMap CEU in a principal component analysis, 3 iterations of outlier removal on the first 3 principal components), and high or low heterozygosity (Fstat < 0.5 and > −0.2). Samples were also flagged for unexpected pairwise relatedness (IBD > 30%) or genetic identity (IBS > 90%). The total number of subjects remaining following QC was 355 cases and 444 controls.
Population Structure and Ancestry Association Testing
To evaluate the extent of global European admixture, genotypes were oriented to the plus strand following the removal of 5,634 A/T and C/G SNPs, and merged with the 11 phase 3 HapMap populations using PLINK. SNPs were filtered for linkage disequilibrium based on r2 values > 0.5, and a principal component analysis was conducted in Eigenstrat with no outlier iterations (outliers were removed in the QC process). Global estimates of European admixture were estimated from the first principal component that separated the HapMap CEU and YRI populations (see Figure E1 in the Online Repository). Global ancestry was compared between cases and controls using logistic regression in R.
Segments of local European ancestry were estimated using LAMP - Local Ancestry in adMixed Populations 21. Admixture in African Americans was modeled under 7 generations of admixture with a 2-population model of 81% ancestry from Africa and 19% ancestry from Europe as estimated based on the first principal component in the PCA analysis described above. Windows were offset by a factor of 0.2, the cutoff for linkage was set to 0.1, and a constant recombination rate was set to 10−8 (bp)−1. Ancestry association testing was performed using logistic regression in R by comparing the estimated number of African chromosomes in cases vs. controls at individual loci including global ancestry as a covariate (asthma ~ local ancestry + global ancestry). A total of 100 permutations were performed genome-wide by shuffling the case/control labels to maintain the haplotype structure of African and European segments. Statistical significance was evaluated based on the distribution of p-values expected at the genomic level under the null hypothesis of no significant differences in local ancestry between cases and controls. An ancestry association peak was defined as a window of unadjusted p-values < 0.01.
Association Testing
Tests of allelic associations were performed for SNPs within ancestry association peaks using logistic regression in R while controlling for local ancestry at each SNP (asthma ~ genotype + local ancestry). Using this approach we tested for allelic associations with asthma beyond that explained by local population structure. We also ran a stratified comparison to identify allelic associations specific to one ancestral background at a locus by separating cases and controls into three groups: 1) individuals with two chromosomes of European ancestry, 2) individuals with one chromosome of European ancestry and one chromosome of African ancestry, and 2) individuals with both chromosomes of African ancestry. The distribution of allelic association p-values was compared to a uniform distribution using the function qqplot in R. Tests of allelic association were performed in a similar manner for the European American samples for SNPs within the ancestry association peak identified in the African American sample. Imputation of genotypes from the 1000 Genomes Pilot Project 29 was performed using IMPUTE2 30, 31 using all of the European and African populations as a reference.
Replication Studies
The Genetics of Asthma in Latino Americans (GALA) Study included 277 asthma cases and 191 asthma controls recruited from schools, clinics and hospitals in New York City and Puerto Rico whose parents and 4 sets of grandparents self-identified as Puerto Rican. The research was approved by the Institional Review Board at the University of California San Francisco. Although the GALA study also includes individuals of Mexican ancestry, we restricted our analysis to Puerto Rican individuals because they have a higher degree of African ancestry 32. Asthma was defined as a physician diagnosis of mild to moderate asthma, having had two or more symptoms of asthma in the previous two years at time of recruitment (wheezing, coughing and/or shortness of breath), and current use of asthma medications. The mean age at enrollment was 14 years of age, the mean age of onset was 3.2 years of age, and the proportion of females was 45%. Controls were recruited from the same locations and determined to be negative for a history of asthma based on a standardized questionnaire.
Subjects were genotyped at > 900,000 SNPs using the Affymetrix 6.0 GeneChip Array and filtered using standard QC procedures including >95% call rate and consistency with Hardy-Weinberg equilibrium (p>10−5). Local ancestry was estimated using LAMP in a similar manner as for the CSGA/SARP subjects, but with 20 generations of admixture, and a 3-population model with genomic ancestral proportions of 16% African ancestry, 66% European ancestry, and 18% Native American ancestry as estimated using ADMIXTURE 33. Replication of ancestry and allelic association for the top associated SNP was performed using logistic regression in R, including global African ancestry as a covariate in tests of ancestry association, and local African and European ancestry as covariates in tests of allelic association (to account for 3 different local ancestries).
Results
Following subject and marker quality control, our study included a total of 1,026,072 genotyped SNPs in 355 African American adults with asthma, and 444 African American non-asthmatic controls. Estimates of global African ancestry among self-reported African Americans ranged from 30–100% African ancestry (mean=81%) based on the first principal component using the HapMap CEU and YRI individuals as a reference for European and African ancestry (see Figure E1 in the Online Repository). However, there was no significant difference in the distribution of global European admixture between cases and controls (logistic regression p-value=0.78, see Figure E2 in the Online Repository), suggesting that the proportion of European and African ancestry when pooled across the genome was balanced between cases and controls. Mean local ancestry as estimated using LAMP was highly correlated with global estimates of ancestry based on the first principal component in Eigenstrat (see Figure E3 in the Online Repository). Patterns of local European ancestry were variable both within and between individuals as expected under a model of more recent European admixture (see Figure E4 in the Online Repository).
We identified four ancestry association peaks suggestive of differences in local ancestry between African American cases and controls (p<0.01) (Figure 1A). Two of the peaks were on chromosome 4 (4p and 4q) in the direction of increased European ancestry in cases (p=4.0×10−3, p=4.4×10−4, Figure 1B), and two of the peaks were on chromosomes 6 (6q) and 7 (7p) in the direction of increased African ancestry in cases (p=2.5×10−4 and p=2.7×10−3 respectively) (Figure 1C). The ancestry association peak on 6q reached genome-wide significance based on 100 permutations across ~1,000,000 SNPs, (permutation p-value<0.01).
Figure 1.
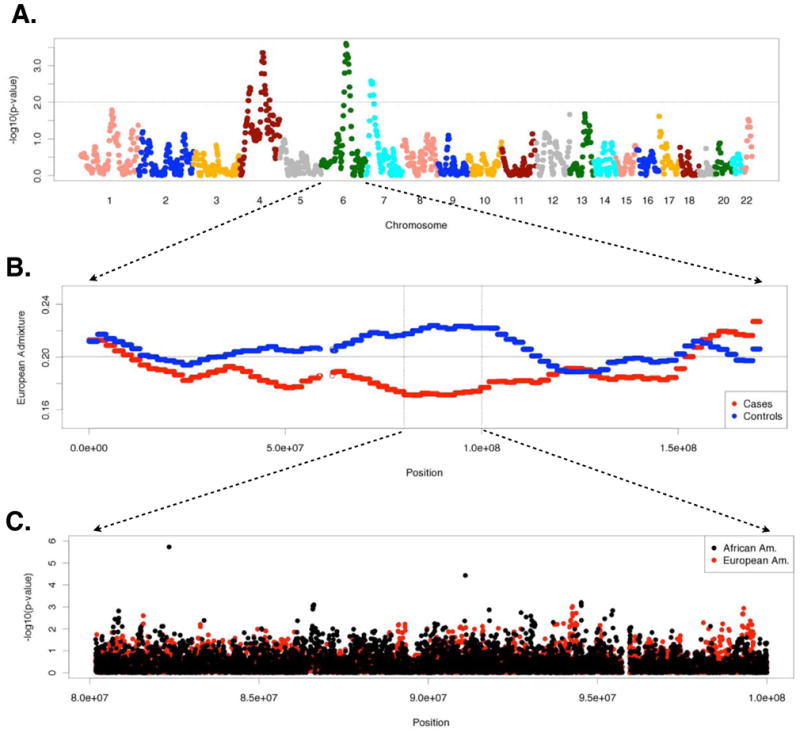
Results of genome-wide ancestry and allelic association testing in African American adults with asthma and healthy controls using logistic regression. A. −log10 (p-values) across the genome showing the location of four ancestry association peaks with p-values < 0.01 (above the dashed line). B. Average local European ancestry in the cases (red) and controls (blue) on chromosome 6; vertical lines indicate the boundaries of the ancestry association peak with p<0.01, and the horizontal line indicates local European ancestry averaged across the genome. C. Tests of allelic association within the ancestry association peak on 6q in African Americans (using local ancestry as a covariate) and European Americans in CSGA/SARP.
We then tested for allelic associations with asthma for 9,266 SNPs within the ancestry association peak on 6q while adjusting for local ancestry (Figure 1D). The most significant SNP, rs1361549, had an association p-value that passed Bonferroni correction for 9,266 tests at 6q14.1 (p=1.87×10−6, α9,266=5.4×10−6, Figure 2). Notably, rs1361549 is only weakly associated with asthma unless local ancestry is included as a covariate in the logistic regression model (p=0.047 with no covariates, p=0.053 with global ancestry from PC1 as a covariate) (Table 1). Furthermore, when we correct for both local and global ancestry the association at rs1361549 is moderately increased (p=6.6×10−7). Comparisons of allele frequencies at rs1361549 for cases and controls with and without local European admixture indicated that the signal of association is exclusive to African American individuals with at least one chromosome of European ancestry at rs1361549 (Table 2). The frequency of the minor allele in African American cases heterozygous for European ancestry at rs1361549 was 27%, as compared to only 14% in controls heterozygous for European ancestry (Odds Ratio=2.2, 95% confidence interval=1.4–3.5). Although the numbers of individuals is small, the frequency of the risk allele was 63% in 8 African American cases homozygous for European ancestry, as compared to 27% in 22 African American controls homozygous for European ancestry (Odds Ratio=4.3, 95% confidence interval=1.2–18). In individuals homozygous for local African ancestry, rs1361549 was monomorphic in both cases and controls. Stratified tests of allelic association based on local ancestry (see methods) for 287 SNPs within 500 Kb of rs1361549 identified additional SNPs that show similar, but weaker signals of association exclusively for African American subjects with local European ancestry that were not identified in the logistic model. None of the SNPs within 500 Kb of rs1361549 were significantly associated with asthma in the European American subjects (Figure 1D, and Table E1); however, we replicated both the ancestry association (p=5.3×10−3) and allelic association (p=0.025) with asthma at rs1361549 in Puerto Ricans in GALA (Table 3), which was attributable to individuals that were heterozygous for African and either Native American or European ancestry (non-African ancestry) (Table 2). Although we attempted to replicate the signal of allelic association in African Americans in the Genomic Research on Asthma in the African Diaspora (GRAAD) study and in families from Barbados 8, rs1361549 was not genotyped directly and imputed genotypes were of low quality (imputation R2=0.43).
Figure 2.
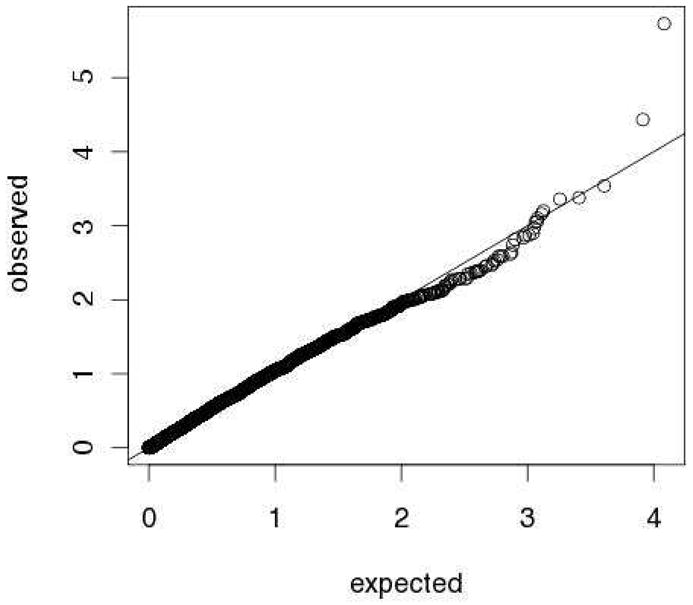
QQplot comparing the observed distribution of p-values to a uniform distribution using the function qqplot in R for SNPs within the admixture mapping peak on chr6q. Local ancestry was included as a covariate in tests of association.
Table 1.
Top signals of allelic association at SNPs within an ancestry association peak on 6q in African Americans. Included in the table are logistic regression p-values from local ancestry association testing, and allelic association testing with no correction for ancestry (no correction), a correction for global ancestry based on PC1 (global correction), and a correction for local ancestry (local correction). Results of association testing in European Americans is in the last column, the top signals of allelic association at SNPs using only a global correction for population structure are in Table E1.
| SNP (major/minor) | Closest Gene (distance) | MAF (cases, controls) | p-value local ancestry | p-value allelic, no correction | p-value allelic, global correction | p-value allelic, local correction | p-value, allelic European Americans (freq cases, controls) |
|---|---|---|---|---|---|---|---|
| rs1361549 (G/A) | FAM46A (−171661) | 0.093, 0.069 | 0.00079 | 0.0472 | 0.0527 | 1.87×10−6 | 0.428 (0.42, 0.40) |
| rs6926330 (G/A) | BACH2 (−32265) | 0.013, 0.052 | 0.00049 | 1.13×10−4 | 1.14×10−4 | 3.70×10−5 | 0.851 (0.0015, 0.0018) |
| rs576231 (A/G) | PRDM13 (−75107) | 0.028, 0.079 | 0.0090 | 3.19×10−5 | 1.67×10−5 | 4.38×10−4 | 0.15 (0.25, 0.28) |
| rs9363108 (C/T) | EPHA7 (−332718) | 0.25, 0.31 | 0.00062 | 0.013 | 0.014 | 6.32×10−4 | 0.52 (0.0066, 0.0089) |
| rs1414614 (T/C) | EPHA7 (−331225) | 0.38, 0.46 | 0.00062 | 1.65×10−3 | 1.83×10−3 | 6.93×10−4 | 0.40 (0.33, 0.31) |
Table 2.
Allele frequencies and estimated odds ratios for rs1361549 in African American, European American, and Puerto Rican cases and controls. Comparisons for African American and Puerto Rican cases and controls are shown for individuals regardless of local ancestry (all), individuals homozygous for African ancestry (African Ancestry), individuals heterozygous for African/European or African/Native American ancestry (African/European Ancestry, African/Non-African Ancestry), and individuals homozygous for non-African ancestry (Native American or European ancestry).
| Sample | Frequency in Cases (N) | Frequency in Controls (N) | Odds Ratio (95% CI) |
|---|---|---|---|
| HapMap YRI | - | 0 (120) | - |
| African Americans (all) | 0.09 (355) | 0.07 (444) | 1.4 (0.98–2.1) |
| African Ancestry | 0 (243) | 0 (257) | - |
| African/European Ancestry | 0.27 (104) | 0.14 (165) | 2.2 (1.4–3.5) |
| European/European Ancestry | 0.63 (8) | 0.27 (22) | 4.3 (1.2–18) |
| European Americans | 0.42 (678) | 0.40 (561) | 1.1 (0.9–1.3) |
| Puerto Ricans (all) | 0.31 (277) | 0.29 (191) | 1.1 (0.79–1.4) |
| African Ancestry | 0 (21) | 0 (3) | - |
| African/Non-African Ancestry | 0.20 (73) | 0.11 (47) | 2.1 (0.92–5.0) |
| Non-African Ancestry | 0.38 (183) | 0.36 (141) | 1.1 (0.78–1.5) |
| HapMap CEU | - | 0.42 (120) | - |
CI = 95% confidence interval.
Table 3.
Replication of ancestry and allelic association with asthma at rs1361549 in Puerto Rican cases and controls included in the GALA study. The sign of the regression coefficient indicates the direction of the effect, which is consistent between the two studies. The regression coefficient for tests of association at rs1361549 in European Americans in the CSGA/SARP was 0.066 (p=0.43). Also shown are stratified tests of association in African Americans based on local ancestry.
|
Primary/Discovery Sample African Americans, CSGA/SARP: |
Replication Sample Puerto Ricans, GALA: |
|||||
|---|---|---|---|---|---|---|
| Logistic Model | Regression Coefficient | Standard Error | p-value | Regression Coefficient | Standard Error | p-value |
| Local African Ancestry | 0.96 | 0.29 | 7.9×10−4 | 0.22 | 0.080 | 5.4×10−3 |
| rs1361549 (No Correction) | 0.094 | 0.047 | 0.0472 | 0.01 | 0.036 | 0.79 |
| rs1361549 (Global Ancestry Correction) | 0.093 | 0.048 | 0.0527 | 0.007 | 0.036 | 0.85 |
| rs1361549 (Local Ancestry Correction) | 0.28 | 0.058 | 1.9×10−6 | 0.099 | 0.044 | 0.025 |
| rs1361549 (2 African Chrom) | n/a | n/a | n/a | n/a | n/a | n/a |
| rs1361549 (1 African, 1 European/Native American) | 0.25 | 0.059 | 2.5×10−5 | 0.19 | 0.11 | 0.073 |
| rs1361549 (2 European/Native American) | 0.35 | 0.12 | 5.4×10−3 | 0.068 | 0.050 | 0.18 |
Lastly, we performed in silico fine mapping by imputing 7,124 SNPs from the 1000 Genomes Project within 500 Kb of rs1361549, and identified an additional variant (rs4706896) that shows a similarly strong signal of association when adjusting for local ancestry (p=2.7×10−6) (Figure 3). The SNP is 49 Kb more proximal to the centromere than rs1361549, however it is in high LD (r2=0.97 in the HapMap CEU) and shows similar patterns of allele frequencies as rs1361549 (frequency in the CEU = 41%, frequency in the YRI = 0%).
Figure 3.
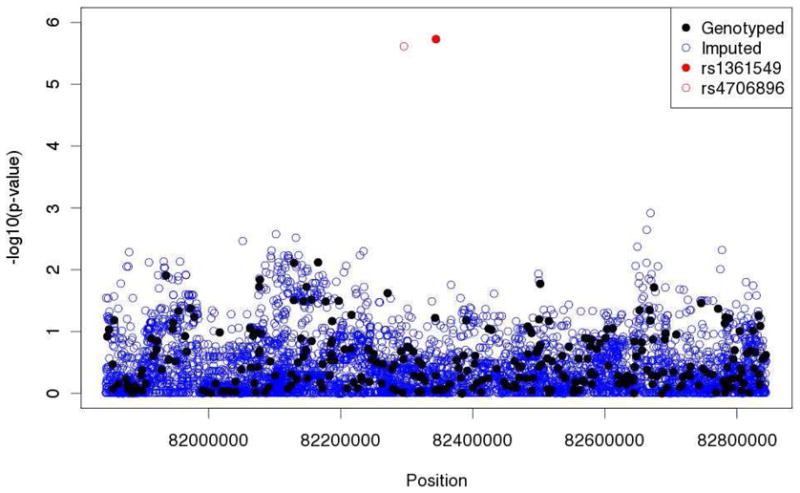
Tests of allelic association controlling for local ancestry within 500 Kb of rs1361549 in African Americans in CSGA/SARP at both genotyped and imputed SNPs from the 1000 Genomes Project.
Discussion
In this study we performed genome-wide ancestry association testing using genotypes from ~ 1 million SNPs in 344 African American adults with asthma and 444 non-asthmatic controls. Given the relatively low effect sizes of asthma-associated SNPs identified in previous GWAS 5–11, our study was underpowered for performing a traditional GWAS. We therefore utilized genome-wide ancestry association testing to identify ancestry association peaks, and then performed allelic tests of association within the associated region. This strategy significantly lowers the multiple testing burden inherent in GWAS. We identified a significant ancestry association peak at 6q14.1 that is more likely to have underlying differences in patterns of allele frequencies between cases and controls as compared to a randomly selected region of the genome. By focusing on SNPs within this peak, we identified an intergenic SNP, rs1361549, with an association p-value that surpassed Bonferroni correction for a reduced number of tests after adjusting for local ancestry.
The signal of association with asthma at rs1361549 is highly dependent on correcting for local rather than global ancestry. We observed only a minor difference in the allele frequency between cases and controls prior to including local ancestry as a covariate in the logistic model. The risk allele is common in Europe (frequency in the CEU = 42%) and either rare or absent in West Africa (frequency in the YRI = 0%), highlighting the importance of adjusting for population structure in allelic tests of association at this SNP. Admixture association testing identified increased African ancestry in the cases as compared to the controls at 6q14.1, and therefore under the null hypothesis of no association the frequency of the minor allele is expected to be higher in the controls (due to increased European ancestry, where the allele is more common). By including local ancestry in the model this prior expectation was taken into consideration, leading to the identification a signal of association with asthma at rs1361549 that would have otherwise gone unnoticed.
By comparing the frequency of rs1361549 between cases and controls after stratifying by local ancestry, we discovered that the frequency of the risk allele is only increased in African Americans cases with local European admixture. Moreover, we found additional, but weaker signals of association at SNPs more proximal to the centromere that showed a similar pattern to rs1361549 (Table E2). However, neither rs1361549 nor any of the nearby SNPs showed an association with asthma in European Americans, suggesting that the underlying causal variant (or variants) only increases the risk of asthma when present on an African genetic background. In support of this, we replicated our signal of association at rs1361549 in Puerto Ricans in the same direction (Table 3), and similarly found that the frequency of the risk allele was predominantly increased in cases that were heterozygous for local African and non-African ancestry.
Although the number of individuals was small, rs1361549 also showed a signal of association with asthma in African American individuals that were homozygous for local European ancestry, however we found no significant association in Puerto Ricans homozygous for non-African ancestry (either Native American or European). Our findings suggesting the association at rs1361549 could be due to an environmental or genetic interaction with a variant (or variants) more common in Africa. Under the latter scenario, we hypothesize that the risk of asthma may only be increased for admixed individuals who carry both the minor allele at rs1361549 and an unobserved allele that is more common on African haplotypes. However, because we observed an increase in African ancestry in cases compared to controls in both African Americans and Puerto Ricans at this locus, we cannot rule out the possibility there are additional untyped and untagged variants that increase the risk of asthma for individuals that are homozygous for African ancestry that is independent of the effect of rs1361549.
In silico fine-mapping identified one additional SNP in high LD with rs1361549 that showed a similar pattern of association (rs4706896). Both rs1361549 and rs4706896 lie within an intergenic region that is ~ 170 Kb downstream of the closest annotated gene, FAM46A. FAM46A codes for a protein of unknown function, however it is expressed in the lung and various immune-related cells including B-cells, natural killer cells, and monocytes 34 suggesting a potential avenue for involvement in asthma susceptibility. In addition, there are two reported transcripts within 36 Kb of rs1361549 in GenBank, including AF130064, a cDNA identified from fetal liver cells, and AK126547, a cDNA identified from mesenchymal cells. Patterns of linkage disequilibrium suggest the underlying causal variant(s) may be more proximal to the centromere (see Figures E5 and E6 in the Online Repository). Within this region there are multiple candidate eQTLs 35 in weak LD with rs1361549 (r2 > 0.3) that show an association p-value < 0.001 (Table E1), including eQTLs for Fc receptor-like and mucin-like 1 (FCRLA), proline rich 15 (PRR15), and cannabinoid receptor 2 (macrophage) (CNR2). The function of PRR15 is unknown, however FCRLA is involved in B-cell development, and increased levels of the FCRLA protein has been found in patients with rheumatoid arthritis 36. CNR2 is a cannabinoid/G-protein coupled receptor that is also expressed in B-cells and may function to mediate immunoglobulin class switching from IgM to IgE 37. IgE production is known to play an important role in asthma; however, additional characterization of the genetic variation within this region is required to determine the underlying mechanism behind the association of rs1361549 with asthma susceptibility.
An important consideration is whether estimates of local European admixture around rs1361549 were accurately assigned. In support of this, we found that rs1361549 was monomorphic in both African Americans and Puerto Ricans that were homozygous for African ancestry, consistent with our expectations based on the SNP being absent in the HapMap YRI. Given our estimate that African Americans in CSGA/SARP have ~19% European ancestry, if the variant were attributable solely to European admixture we would expect the frequency of the minor allele to be around 8% given a frequency of 42% in the HapMap CEU (42% × 19% European ancestry), which is similar to what we observe (7.8%). Therefore, our results suggest that rs1361549 is present in African Americans predominantly due to recent European admixture, and that estimates of local ancestry at this locus are accurately distinguishing between African and European chromosomes.
From a methodological viewpoint, our results highlight the importance of considering local ancestry in genetic association studies of admixed populations. This was accomplished by including local ancestry as a covariate in allelic tests of association. However, it is foreseeable that in some situations a stratified analysis may be necessary to identify allelic associations that are unique to a specific genetic background. For example, we identified a hidden signal of association at rs1361549 when using local ancestry as a covariate in a logistic model, in part due to the SNP being monomorphic on the African chromosomes that minimized the signal to noise ratio. Signals of association at nearby SNPs that were polymorphic on both African and European chromosomes were exclusively identified in a stratified analysis, as signals were weakened in the combined subjects due to the majority of individuals having local African ancestry (and the SNPs not being associated in these individuals). Although we did not address the issue of false positive associations in our current study, it is important to note that correcting for local ancestry is likely equally important for avoiding spurious signals of association.
In summary, we identified an ancestry association peak on 6q, and a SNP on 6q14.1 that increases the risk of asthma exclusively for individuals heterozygous for local African and non-African ancestry,. Genetic associations that are dependent on local ancestry present unique challenges at both the discovery and replication stage, highlighting the need for additional large-scale genetic studies of asthma in admixed populations. Overall our results demonstrate the utility of ancestry association testing for identifying novel genetic variants that are associated with asthma, and the importance of considering local rather than global genetic ancestry in association studies involving admixed populations.
Supplementary Material
Key Messages.
Genome-wide comparisons of local ancestry in African Americans identified an admixture mapping peak on 6q that is more likely to contain variants associated with asthma.
An allele at 6q14.1 that is common to European populations is associated with an increased risk of asthma in African Americans and Puerto Ricans, but not in European Americans.
A consideration of local genetic ancestry is essential when performing genome-wide association studies in admixed populations.
Acknowledgments
The authors wish to thank all of the participants in the CSGA, SARP, and GALA studies. SARP is a multicenter asthma research group funded by the National Heart, Lung, and Blood Institute and consisting of the following contributors (principal investigators are marked with asterisks): Brigham & Women’s Hospital—Elliot Israel,* Bruce D. Levy, Michael E. Wechsler, Shamsah Kazani, and Gautham Marigowda; Cleveland Clinic—Serpil C. Erzurum,* Raed A. Dweik, Suzy A. A. Comhair, Emmea Cleggett-Mattox, Deepa George, Marcelle Baaklini, and Daniel Laskowski; Emory University—Anne M. Fitzpatrick, Denise Whitlock, and Shanae Wakefield; Imperial College School of Medicine—Kian Fan Chung,* Mark Hew, Patricia Macedo, Sally Meah, and Florence Chow; University of Iowa—Eric Hoffman* and Janice Cook- Granroth; University of Pittsburgh—Sally E. Wenzel,* Fernando Holguin, Silvana Balzar, and Jen Chamberlin; University of Texas-Medical Branch —William J. Calhoun* and Bill T. Ameredes; University of Virginia —Benjamin Gaston,* W. Gerald Teague,* and Denise Thompson-Batt; University of Wisconsin—William W. Busse,* Nizar Jarjour, Ronald Sorkness, Sean Fain, and Gina Crisafi; Wake Forest University—Eugene R. Bleecker,* Deborah Meyers, Wendy Moore, Stephen Peters, Rodolfo M. Pascual, Annette Hastie, Gregory Hawkins, Jeffrey Krings, and Regina Smith; Washington University in St Louis—Mario Castro,* Leonard Bacharier, and Jaime Tarsi; Data Coordinating Center—Douglas Curran-Everett,* Ruthie Knowles, Maura Robinson, and Lori Silveira; and the National Heart, Lung, and Blood Institute—Patricia Noel and Robert Smith. The authors also wish to thank Vito J. Mantese, BA.
Sources of funding: This work was supported by grants from the National Institute of Health (U01 HL49596, R01 HL072414, R01 HL087665, RC2 HL101651, 2T32GM007546, 1RC2 HL101651, ES015794, U19 AI077439, HL088133, HL078885), and the Flight Attendant Medical Research Institute (FAMRI).
Abbreviations
- GWAS
Genome-wide association study
- SNP
Single nucleotide polymorphism
- OR
Odds ratio
- LD
Linkage disequilibrium
- CSGA
Collaborative Study on the Genetics of Asthma
- SARP
Severe Asthma Research Program
- CEU
Utah residents with Northern and Western European ancestry from the CEPH collection
- CEPH
Centre d’Etude du Polymorphisme Humain
- YRI
Yoruba in Ibadan, Nigeria
- GRAAD
Genomic Research on Asthma in the African Diaspora
- GALA
Genetics of Asthma in Latino Americans
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barnes KC, Grant AV, Hansel NN, Gao P, Dunston GM. African Americans with asthma: genetic insights. Proc Am Thorac Soc. 2007;4:58–68. doi: 10.1513/pats.200607-146JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40:1054–1061. doi: 10.1111/j.1365-2222.2010.03525.x. [DOI] [PubMed] [Google Scholar]
- 3.Willemsen G, van Beijsterveldt TC, van Baal CG, Postma D, Boomsma DI. Heritability of self-reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res Hum Genet. 2008;11:132–142. doi: 10.1375/twin.11.2.132. [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 5.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owen SA, Weiss ST, Raby BA, Gao H, Eng C, Chapela R, Burchard EG, Tang H, Sullivan PF, London SJ. Genome-wide association study implicates chromosome 9q21. 31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, Lazarus R, Murphy AJ, Soto-Quiros ME, Avila L, Beaty T, Mathias RA, Ruczinski I, Barnes KC, Celedon JC, Cookson WO, Gauderman WJ, Gilliland FD, Hakonarson H, Lange C, Moffatt MF, O’Connor GT, Raby BA, Silverman EK, Weiss ST. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335e11. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, Gao P, Togias A, Hansel NN, Diette G, Adkinson NF, Liu MC, Faruque M, Dunston GM, Watson HR, Bracken MB, Hoh J, Maul P, Maul T, Jedlicka AE, Murray T, Hetmanski JB, Ashworth R, Ongaco CM, Hetrick KN, Doheny KF, Pugh EW, Rotimi CN, Ford J, Eng C, Burchard EG, Sleiman PM, Hakonarson H, Forno E, Raby BA, Weiss ST, Scott AF, Kabesch M, Liang L, Abecasis G, Moffatt MF, Cookson WO, Ruczinski I, Beaty TH, Barnes KC. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346.e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 11.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, Wang K, Rafaels NM, Michel S, Bonnelykke K, Zhang H, Kim CE, Frackelton EC, Glessner JT, Hou C, Otieno FG, Santa E, Thomas K, Smith RM, Glaberson WR, Garris M, Chiavacci RM, Beaty TH, Ruczinski I, Orange JM, Allen J, Spergel JM, Grundmeier R, Mathias RA, Christie JD, von Mutius E, Cookson WO, Kabesch M, Moffatt MF, Grunstein MM, Barnes KC, Devoto M, Magnusson M, Li H, Grant SF, Bisgaard H, Hakonarson H. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 12.Choudhry S, Taub M, Mei R, Rodriguez-Santana J, Rodriguez-Cintron W, Shriver MD, Ziv E, Risch NJ, Burchard EG. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123:455–468. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, White TJ, Nielsen R, Clark AG, Bustamante CD. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray T, Beaty TH, Mathias RA, Rafaels N, Grant AV, Faruque MU, Watson HR, Ruczinski I, Dunston GM, Barnes KC. African and non-African admixture components in African Americans and an African Caribbean population. Genet Epidemiol. 2010;34:561–568. doi: 10.1002/gepi.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 17.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng CY, Kao WH, Patterson N, Tandon A, Haiman CA, Harris TB, Xing C, John EM, Ambrosone CB, Brancati FL, Coresh J, Press MF, Parekh RS, Klag MJ, Meoni LA, Hsueh WC, Fejerman L, Pawlikowska L, Freedman ML, Jandorf LH, Bandera EV, Ciupak GL, Nalls MA, Akylbekova EL, Orwoll ES, Leak TS, Miljkovic I, Li R, Ursin G, Bernstein L, Ardlie K, Taylor HA, Boerwinckle E, Zmuda JM, Henderson BE, Wilson JG, Reich D. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5:e1000490. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng CY, Reich D, Wong TY, Klein R, Klein BE, Patterson N, Tandon A, Li M, Boerwinkle E, Sharrett AR, Kao WH. Admixture mapping scans identify a locus affecting retinal vascular caliber in hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) study. PLoS Genet. 2010;6:e1000908. doi: 10.1371/journal.pgen.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, Tayo B, Adeyemo A, Sun YV, Li Y, Morrison A, Newton-Cheh C, Liu K, Ganesh SK, Kutlar A, Vasan RS, Dreisbach A, Wyatt S, Polak J, Palmas W, Musani S, Taylor H, Fabsitz R, Townsend RR, Dries D, Glessner J, Chiang CW, Mosley T, Kardia S, Curb D, Hirschhorn JN, Rotimi C, Reiner A, Eaton C, Rotter JI, Cooper RS, Redline S, Chakravarti A, Levy D. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: Contributions from the CARe consortium. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, Miller ME, Dunston GM, Solway J, Wolf RL, Samet JM, Marsh DG, Meyers DA, Ober C, Bleecker ER. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108:357–362. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 24.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, Dweik RA, Fitzpatrick AM, Gaston B, Hew M, Hussain I, Jarjour NN, Israel E, Levy BD, Murphy JR, Peters SP, Teague WG, Meyers DA, Busse WW, Wenzel SE. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D’Agostino RJ, Castro M, Curran-Everett D, Fitzpatrick AM, Gaston B, Jarjour NN, Sorkness R, Calhoun WJ, Chung KF, Comhair SA, Dweik RA, Israel E, Peters SP, Busse WW, Erzurum SC, Bleecker ER. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 33.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, Dolan ME, Cox NJ. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huizinga TW, Ioan-Facsinay A. Soluble FcgammaRIIIa as a marker for rheumatoid arthritis: the use of genetics in selected populations to study pathogenetic pathways. J Rheumatol. 2003;30:1904–1906. [PubMed] [Google Scholar]
- 37.Agudelo M, Newton C, Widen R, Sherwood T, Nong L, Friedman H, Klein TW. Cannabinoid receptor 2 (CB2) mediates immunoglobulin class switching from IgM to IgE in cultures of murine-purified B lymphocytes. J Neuroimmune Pharmacol. 2008;3:35–42. doi: 10.1007/s11481-007-9088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


