Abstract
Th17 cells have critical roles in mucosal defense and are major contributors to inflammatory disease. Their differentiation requires the nuclear hormone receptor RORγt working with multiple other essential transcription factors (TFs). We have used an iterative systems approach, combining genome-wide TF occupancy, expression profiling of TF mutants, and expression time series to delineate the Th17 global transcriptional regulatory network. We find that cooperatively-bound BATF and IRF4 contribute to initial chromatin accessibility, and with STAT3 initiate a transcriptional program that is then globally tuned by the lineage-specifying TF RORγt, which plays a focal deterministic role at key loci. Integration of multiple datasets allowed inference of an accurate predictive model that we computationally and experimentally validated, identifying multiple new Th17 regulators, including Fosl2, a key determinant of cellular plasticity. This interconnected network can be used to investigate new therapeutic approaches to manipulate Th17 functions in the setting of inflammatory disease.
INTRODUCTION
The vertebrate immune system, composed of numerous phenotypically well-defined cell types, is ideally suited for studying the combinatorial action of transcription factors (TFs) and epigenetic regulators whose target gene products confer unique cellular functions. TFs that are selectively expressed in subsets of myeloid and lymphoid lineage have been designated “master regulators” if they are both essential and sufficient to induce defined cell fates. It is becoming clear, however, that networks of multiple TFs are required to achieve the full differentiation programs (Mattick et al., 2010; Novershtern et al., 2011). How such factors cooperate to determine specific programs remains poorly understood.
CD4-expressing T lymphocytes are among the best-characterized immune system cells (Zhu et al., 2010). They develop in the thymus and acquire the potential to become T-helper cells that guide B lymphocytes to produce distinct classes of antibody and carry out multiple other effector functions; or they up-regulate the TF Foxp3 and become anti-inflammatory regulatory T cells (Treg). T-helper cells differentiate further in the periphery following induction or activation of TFs in response to signals from the T cell antigen receptor (TCR), cytokines, and other ligands in the microenvironment. T-helper effector subsets include Th1 cells, which produce interferon-γ and control infections with intracellular microbes, Th2 cells, which secrete IL-4, IL-5, and IL-13 and are required for clearance of helminths, and Th17 cells, producers of IL-17A, IL-17F, and IL-22 that protect mucosa from bacterial and fungal infection (Korn et al., 2009). In addition, follicular helper T cells (TFH) provide B cells with signals for immunoglobulin class switching and affinity maturation (Crotty, 2011). CD4+ T cell subsets exhibit plasticity, but are considered distinct lineages based on expression of TFs with properties of “master regulators”. Th1 cells are defined by their expression of T-bet (Tbx21), Th2 cells by GATA3, Th17 cells by RORγt, and TFH cells by Bcl6. Effector T cells expressing distinct subset-specific cytokines are common in vivo, although cells with combinations of such cytokines are often observed. Differentiation of naïve CD4+ T cells into Th1, Th2, Th17, or Treg cells can be mimicked in vitro by TCR stimulation and combinations of defined cytokines. Genome-wide histone modifications, chromatin accessibility, and occupancy by lineage-specifying TFs have been studied in such models (Durant et al., 2010; Kwon et al., 2009; Wei et al., 2009).
Th17 cells have critical functions in many autoimmune diseases and in cancer (Korn et al., 2009). The orphan nuclear receptor RORγt is required for the differentiation of Th17 cells and for inflammatory diseases in mice. Its forced expression in mouse and human T cells induces transcripts present in Th17 cells, including those coding for the key cytokines, for the IL-23 receptor, and for the chemokine receptor CCR6 (Ivanov et al., 2006; Manel et al., 2008). However, RORγt is not sufficient to specify the full Th17 program, and other TFs, including STAT3, IRF4, BATF, and IκBδ, are required for induction of RORγt and IL-17A in vivo and upon polarization in vitro with IL-6, TGF-β, with or without IL-1β and IL-23 (Brustle et al., 2007; Okamoto et al., 2010; Schraml et al., 2009; Yang et al., 2007). Multiple other TFs are also involved in Th17 cell differentiation, including c-Maf, Runx1, and Ahr (Bauquet et al., 2009; Veldhoen et al., 2008; Zhang et al., 2008). RORα, which is closely related to RORγt, can also contribute to IL-17 expression in the absence of RORγt (Yang et al., 2008).
Investigation of TF functions in Th17 cell differentiation has been limited to how single factors affect expression of a limited number of targets (e.g. IL-17A). However, the Th17 differentiation program extends beyond functions of individual cytokines, as highlighted by studies showing Th17-mediated pathogenesis in the absence of IL-17A and IL-17F (Codarri et al., 2011; Leppkes et al., 2009). We therefore wished to examine how multiple TFs regulate each other and their targets in order to model a transcriptional network and identify novel critical factors in Th17 cell differentiation. New regulatory interactions can be found using genome-wide methods to learn networks from time series and genetic and environmental perturbations (Bonneau et al., 2007; Ernst et al., 2007; Faith et al., 2007; Greenfield et al., 2010). Integrating multiple data types and analytical methods for network inference using meta-analyses (averaging over several data types and computational approaches) can leverage the complementary weaknesses and strengths of each component to produce higher accuracy networks (Marbach et al., 2012b). We have applied an integrative approach, with meta-analysis of genome occupancy of multiple TFs, RNA-seq of TF-deficient T cells and immune cell transcriptome data, to build a network model for Th17 cells. We find in Th17 cell differentiation early cooperative binding of BATF and IRF4 that governs chromatin accessibility and subsequent recruitment of RORγt to regulate a select set of Th17-relevant genes. We used the network model to identify additional candidate genes that were in turn incorporated through an iterative process, and validated several genes critical in Th17 cell differentiation, including TFs that influence the expression of > 2,000 genes. We found that the AP-1 family member Fosl2 (confidently predicted to be a core Th17 factor) has a key role in a mouse model of autoimmune disease, limiting plasticity of T helper cell differentiation. In addition, loci implicated in genome-wide association studies (GWAS) to have roles in autoimmune disease were enriched in the Th17 network. This analysis can therefore identify candidate genes that serve as cogs in functional specialization of Th17 cells and that have potential to be valuable for new therapeutic approaches.
RESULTS
TF co-occupancy enriches for functional cis regulatory modules
We studied early Th17 cell specification events in a largely synchronized population of naïve CD4+ T cells that produce IL-17 upon stimulation of the TCR in the presence of IL-6 and low levels of TGF-β (Figure S1A). In this model, cells receiving TCR stimulation alone serve as a non-polarized (Th0) control.
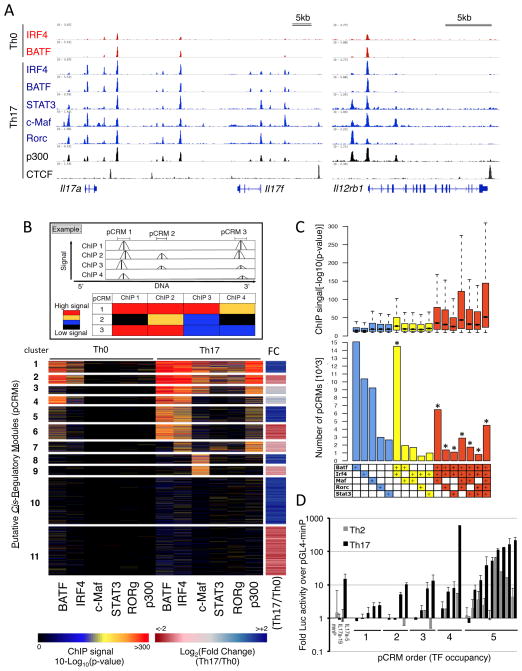
To assemble a high resolution map of TF-DNA interactions in Th17 differentiation, ChIP-seq was performed with antibodies directed against STAT3, IRF4, BATF, c-Maf, RORγ, and p300 with cells cultured in Th0 and Th17 conditions for 48h, when Th17-specifying TFs are simultaneously expressed or active (Figure S1B). Many high confidence bound regions were observed for each TF in Th17 conditions and for BATF and IRF4 in Th0 cells with the cognate consensus binding motif recovered for each (Figure S1C). TFs binding at key lineage-associated loci (Il17a, Il17f, Il12rb1, Il1r1, Rorc) revealed a high degree of co-localization in Th17 cells (Figure 1A, S1D), indicating that these TFs occupy common cis regulatory regions and highlighting their roles in integrating cytokine and TCR-derived signals. In contrast, insulator-binding factor CTCF displayed a distinct occupancy pattern.
Figure 1. Genome-wide co-occupancy of Th17 lineage TFs.
A. ChIP-seq binding tracks for core TFs, p300, and CTCF at selected Th17 loci in Th0 and Th17 cells at 48h (visualized with IGV, Broad Institute).
B. A clustered heat map of pCRM regions (rows) based on TF ChIP signals and the associated gene expression fold changes (FC) in Th17 vs. Th0 cells. A schematic illustration of the clustering approach is shown (top panel).
C. Numbers of pCRMs with > 500 occurrences in Th17 cells (bottom) with associated distributions of TF ChIP p-values (top). Boxplots (top) show median (line), 25th to 75th percentile (box) ± 1.5 interquartile range. *denotes that we observed significantly more pCRMs than expected by chance based on 10000 simulations (P value <0.001).
D. Luciferase reporter assay of enhancer activity for selected pCRM DNA regions in CD4 T cells cultured under Th2 and Th17 conditions. Y-axis: luciferase activity relative to that of pGL4-minP with a minimal promoter. X-axis: TF occupancy order of tested pCRMs (1 to 5 TFs); Il17a-5 and Il17a-19 serve as positive and negative controls, respectively. Error bars = SD of two experiments.
To examine genome-wide TF binding patterns in Th0 and Th17 cells, we merged TF-binding peaks with summits that clustered in close proximity to each other (within 100 bp) to define putative Cis Regulatory Modules (pCRM; Figure 1B). In this manner, 162,113 significant TF peaks clustered into 83,138 unique pCRMs with distributions provided in Figure 1C. The clustered heat map in Figure 1B displays TF binding significance for TSS-proximal (+/−5kb) pCRMs and the associated fold change in gene expression in Th17 versus Th0 cells for the nearest gene (right panel).
The pCRM clustering revealed that the most prominent signature was co-occupancy by all five TFs in Th17 cells and by IRF4 and BATF in Th0 cells (clusters 1 and 2, Figure 1B). This was not a result of TF binding enrichment expected near the TSSs, as approximately 70% of the 5-TF pCRMs localized to distal sites (>5 kb from TSS; Figure S1E). Notably, BATF and IRF4 showed a striking binding overlap regardless of occupancy by other Th17 TFs (clusters 3–6, Figure 1B). TF average binding significance increased with pCRM order and was the highest at 5-TF pCRMs (Figure 1C), possibly reflecting enhanced accessibility and/or cooperativity between factors at these regions. Moreover, strong RORγt binding was almost exclusively restricted to five-factor pCRMs (clusters 1 and 2), suggesting that these elements represent important regulatory domains for the integration of specification signals. Accordingly, these pCMRs were proximal to loci showing differential expression in Th17 versus Th0 cells (Figure 1B, right column). Nearly all (>99%) 5TF-pCRMs co-localized with the histone acetyltransferase p300 (Figure 1B), a factor for which occupancy can be predictive of tissue-specific regulatory activity (Visel et al., 2009). Notably, p300 binding in Th17 cells, while induced relative to Th0, was not restricted to up-regulated loci, which may reflect the contribution of inhibitory gene regulation by distal pCRMs.
To relate pCRM occupancy to cis regulatory function, genomic regions corresponding to pCRMs occupied by different numbers of TFs were cloned upstream of a minimal promoter driving a luciferase reporter and assayed for activity (Figure 1D). The most active regions coincided with high order occupancy that were also significantly more active in Th17 versus control Th2 cells. Thus, cis regions that integrate Th17 signals (4 and 5 TF pCRMs) display lineage selectivity and their combinatorial occupancy increases the likelihood of activity. Together, these findings support the view that core Th17 TFs function synergistically and that shared regulatory targets of STAT3, IRF4, BATF, c-Maf and RORγt are likely enriched with key targets for Th17 cell specification.
Cooperative binding of BATF/IRF4 complexes pre-patterns chromatin for specification
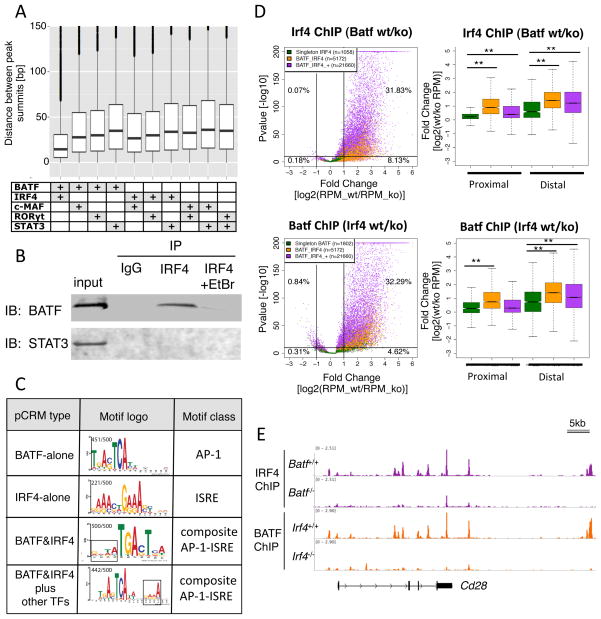
The strong association between IRF4 and BATF occupancy in both Th17 and Th0 cells suggested a regulatory interaction between these factors. Indeed, in five-TF pCRMs, the binding summits for IRF4 and BATF were more proximal than any other pair of Th17 TFs (Figure 2A), and BATF was uniquely co-immunoprecipitated with IRF4 in a DNA-dependent manner in Th17-polarized cells (Figure 2B), indicating that they form a complex on DNA. Consistent with this, pCRMs co-occupied by BATF and IRF4 lacked the interferon stimulated response element (ISRE) consensus, but were enriched with two types of dominant AP1-ISRE composite elements with AP-1 motifs adjacent to ISRE half sites (Figure 2C). As a similar motif structure underlies cooperative binding of IRF4 and PU.1 (Eisenbeis et al., 1995), we tested functional cooperativity of BATF and IRF4 binding in a cellular context. We thus performed ChIP-seq for each TF in Th0 and Th17-polarized cells genetically deficient for the other TF. IRF4 and BATF occupancy was markedly reduced in Batf and Irf4 mutant cells, respectively. The effect was most significant at pCRMs occupied by IRF4 and BATF in combination or with additional TFs when compared to regions harboring single IRF4 or BATF peaks, where cooperativity is not expected (Figure 2D). The dependence between IRF4 and BATF was stronger at distal pCRMs, suggesting that additional factors binding near promoters compensate for the loss of either TF. Similar results were obtained in Th0 cells in the absence of cytokine-induced factors (Figure S2A). Several examples of this mutual binding dependency are shown (Figures 2E and S2B).
Figure 2. Cooperative occupancy by BATF and IRF4.
A. Proximal binding of BATF and IRF4. Distribution of the distance between ChIP peak summits for pairs of TFs in 5-TF pCRMs. Boxplot: as in Figure 1; points=outliers.
B. Co-immunoprecipitation of BATF, but not STAT3, with IRF4 in 48h Th17-polarized cultures. Ethidium bromide (EtBr) disrupts DNA-protein interactions. IP: immunoprecipitating antibody; IB: immunoblotting antibody. Representative of two experiments.
C. MEME-ChIP motifs identified in 4 sub-types of pCRMs as indicated. The AP-1 and ISRE consensus is recovered in regions singly occupied by BATF, and IRF4, respectively. A new AP-1-ISRE composite motif comprising an AP-1 site (TGA(C/G)TCA) adjacent to an ISRE half site (GAAA; boxed region) is only recovered at pCRMs occupied by BATF and IRF4. ISRE half site orientation differs according to whether or not there is a 4bp interval. The fraction of pCRMs for which the motif is found is indicated.
D. Genome-wide interdependence of IRF4 and BATF co-occupancy in Th17-polarized cells. Scatter plots display the fold change in ChIP-seq reads vs. significance. Top: IRF4 in Batf wt vs. KO. Bottom: BATF in Irf4 wt vs. KO. Differences in ChIP-Seq reads displayed for 3 relevant pCRM sub-types: BATF or IRF4 alone (green); BATF and IRF4 (orange); and BATF, IRF4, plus additional TFs (purple). Distribution of fold changes of wt vs. KO occupancy are displayed for proximal and distal pCRMs; boxplots as in Figure 1C. RPM; reads per million. ** Significant at P-value<0.001, Kolmogorov-Smirnov test.
E. Reciprocally reduced occupancy of BATF and IRF4 at the Cd28 locus in Th17-polarized cells deficient for IRF4 and BATF, respectively. ChIP-seq tracks were normalized for library size.
The cooperativity of IRF4 and BATF, paired with our finding that cis regions bound by both TFs in TCR stimulation conditions (Th0) exclusively acquire additional strong binding by STAT3, RORγt, c-Maf, and p300 in Th17 cells (Figure 1B), suggests that IRF4 and BATF function as pioneer factors in nucleating binding of Th17 TFs upon cytokine-stimulated differentiation. Consistent with this, occupancy of IRF4 and BATF is enriched in the center of five-TF pCRMs (Figure S3A). To investigate this hypothesis directly, we examined chromatin accessibility at regions co-occupied by all five TFs, using Formaldehyde Assisted Isolation of Regulatory Elements sequencing (FAIRE-seq). Deletion of IRF4 or BATF in Th0 or Th17 cells had little effect on regions already accessible in naïve cells, but most regions with inducible FAIRE-seq signal exhibited marked reductions in Irf4−/− and Batf−/− compared to WT cells in both Th17 and Th0 conditions (Fig. S3B). Thus, in the absence of IRF4 and BATF, regions normally bound by all five Th17 TFs are less accessible, providing further evidence that IRF4 and BATF remodel the chromatin landscape, potentially facilitating subsequent recruitment of TFs involved in regulating expression of adjacent genes.
Consistent with IRF4 and BATF mediating accessibility, these TFs also globally affect p300 occupancy, which was reduced in IRF4- or BATF-null Th17 cells (Figure S3C). STAT3 deficiency also reduced p300 binding, but loss of RORγt resulted in a much smaller genome-wide effect (Figure S3C). The focal influence of RORγt was also reflected in occupancy of IRF4 and STAT3 and the presence of H3K4me2 and H3Kme3, histone marks associated with active transcription, in RORγt-deficient Th17 cells (Figure S3D). Strikingly, few genes were dependent on RORγt, as measured by more than 2-fold reduction (and p-value<0.01) in both H3K4me3 at locus-linked pCRMs and expression of the respective genes, namely Il17a, Il17f, Il23r, Ccl20, Il1r1, Ltb4r1 that were known and novel targets (2310007L2Rik, Furin, Fam124b, Tmem176a, Tmem176b). These findings are consistent with RORγt having a highly specific regulatory footprint relative to initiator TFs that establish broader changes in chromatin remodeling.
The Th17 network reveals lineage specification by combinatorial regulation
While providing mechanistic insight, TF occupancy does not sufficiently explain target gene regulation. We thus complemented TF ChIP-seq (that finds direct targets) with RNA-seq of TF wild-type vs. knock-out (KO) Th17 cells (that finds functional targets). ChIP p-values for peaks falling within 10kb of the gene body were consolidated to a gene-wide p-value, and KO-RNA-seq p-values were calculated based on target gene differential expression. For each data set, p-values were mapped to rank-based scores from 0 (least significant) to 1 (most significant) and combined such that the integrated ChIP+KO scores ranged from 0 to 2, with the highest scoring genes likely direct and functional targets. Rank-based approaches have proved successful in similar settings (Madar et al., 2010; Marbach et al., 2012a; Marbach et al., 2012b; Prill et al., 2010) (see Supplementary Methods). High confidence TF-target interactions (score >1.5; FDR<10%) are visualized in Cytoscape.
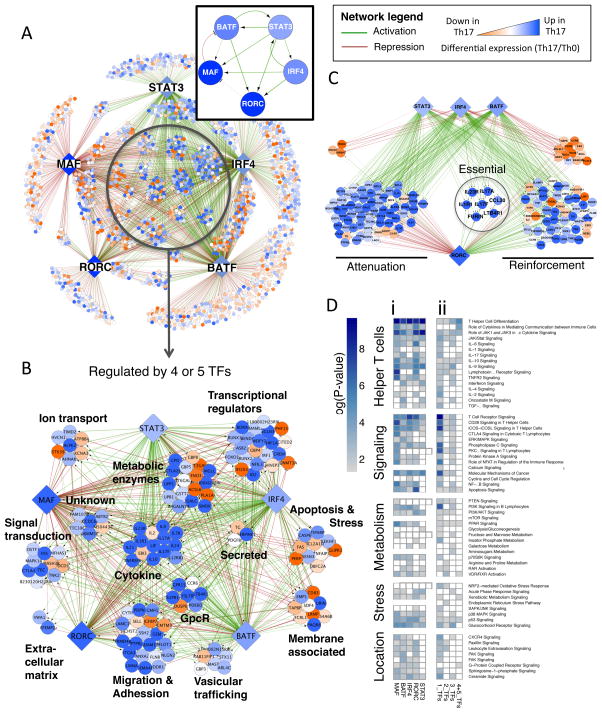
The resulting causal network captures known relationships between core TFs, including Rorc activation by STAT3, IRF4, and BATF (Brustle et al., 2007; Schraml et al., 2009; Yang et al., 2007), and further reveals feed-forward loops that reinforce Rorc expression in response to TCR and cytokine signals (Figure 3A, box). Notably, there is high interconnectivity among TFs, including positive feedback loops reinforcing expression of initiator TFs BATF, IRF4 and STAT3, and a negative feedback loop (c-Maf to BATF) that limits response. Conversely, RORγt does not participate in stabilizing positive feedback relationships with inducing TFs, thus rendering its expression sensitive to changing environmental signals. This is consistent with the need for continuous STAT3 activation (McGeachy et al., 2009) and the plasticity of the Th17 subset when cytokine conditions are altered (Hirota et al., 2011; Lee et al., 2009).
Figure 3. TF combinatorial interactions specify the Th17 lineage.
A. High confidence regulatory edges (FDR<10%; based on 10,000 simulations) focused on five core TFs identify direct (ChIP-seq) and functional (KO RNA-seq) regulatory targets (visualized using Cytoscape). Boxed inset displays the regulatory interactions between core Th17 TFs. See network legend for visualization scheme.
B. Expanded view of highly regulated nodes with four to five core regulatory inputs, grouped based on general functional categories.
C. Regulatory interactions shared by STAT3, IRF4, BATF, and RORγt highlighting different aspects of RORγt transcriptional function. Attenuation: RORγt repression targets that are up-regulated in Th17 cells; Reinforcement: Activation targets that are up-regulated in Th17 cells; Essential: targets having a two-fold change in RORγt KO differential expression and KO H3K4me3 ChIP.
D. Targets for single TFs are enriched for pathways in multiple functional categories (i). Targets of multiple TFs (increasingly regulated by 2, 3, or 4+5 TFs) are selectively enriched for pathways related to T helper differentiation and effector function (ii). Analysis performed using the Ingenuity analysis tool (IPA) is presented as a heat map of enrichment p-values.
Highly regulated nodes, defined by combinatorial regulation by 4 or 5 core TFs, comprise many genes with critical lineage modulatory and effector activity. These include key signature genes (Il17a, Il17f, IL23r), other relevant cytokines and receptors (Il2, Il9, Lif, Il10, Il1r1, Il21, Il12rb1, Ebi3 (IL-27) Ltb4r1, and Ccr6), and TFs (Rora, Hif1a, Runx1, and Foxo1) (Korn et al., 2009). This indicates that other highly regulated genes in diverse categories (e.g. ion transport, migration, metabolism, and stress response) likely include novel Th17 regulators or effectors (Figure 3B).
To assess the regulatory relationships between Th17 TFs, we summarized the activating and repressing regulatory inputs for each network gene in a clustered heat map (Figure S4A). Notably, initiator TFs BATF, IRF4, and STAT3 regulate the largest number of genes and impose complementary control of shared targets, particularly in activation. This is most striking for highly regulated Th17 genes (Figure 3C, S4B), and is mirrored by the regulation of similar pathways (Figure 3D(i)), including helper T cell differentiation and activation, cytokine signaling, metabolism, and oxidative/xenobiotic stress response (some previously attributed to these TFs (Durant et al., 2010; Kwon et al., 2009; Schraml et al., 2009)). Thus, together initiator TFs establish a broad, coherent transcriptional program in Th17 cells.
c-Maf is generally appreciated as an activator of cytokine loci (Ho et al., 1999). Unexpectedly, in Th17 cells it functions mainly as a negative regulator (Figure 3A, S4A), attenuating the expression of pro-inflammatory loci (e.g. Rora, Runx1, Il1r1, Ccr6, Tnf) and globally repressing genes in pathways regulated by other core TFs (Figure 3D(i)). Notably c-Maf does positively regulate a few loci, several linked to attenuating inflammation (e.g. Il9, Il10, Lif, Ctla4). Together with the recent description of c-Maf as an Il22 repressor (Rutz et al., 2011), the global c-Maf target repertoire identifies an underappreciated general anti-inflammatory role for this TF.
RORγt, the key lineage-specifier, functions as an activator and a repressor within the network (Figure S4A). Notably, it either reinforces or antagonizes the coherent activation program initiated by IRF4, BATF, and STAT3 (Figure 3C, S4B). While RORγt positively regulates many genes, it has a strong role at only a small number of key Th17 loci (as defined above; Figure 3C, S3D; see Figure S4C for the magnitude of RORγt dependency). As a repressor, RORγt limits target expression, including regulators of metabolism and quiescence (e.g. Il10, Hif1a, Egln3, Foxo1, and Il7r) and alternative lineage fates (ll4ra and Il12rb2) promoted by initiator TFs. In this regard, RORγt acts as a modulator; its repressive activity is poorly correlated with actual expression changes, with its repressed targets often up-regulated in Th17 relative to Th0 cells (Figure 3C, S4D). Thus, RORγt is essential in licensing the expression of a select few loci and elsewhere it functions as a rheostat to tune mRNA levels to those of a Th17-specifying program.
Individual Th17 TFs regulate broad cellular functions (Fig. 3D(i)). When network targets were subdivided according to the complexity of inputs (either 1, 2, 3, or 4/5 TF edges), pathway analysis revealed a selective enrichment for genes involved in helper T cell differentiation with increasing number of TF inputs (Fig. 3D, (ii)). Accordingly, nodes regulated by 4 or 5 TFs were most enriched for genes highly differentially expressed in Th17 cells (Figure 3A, compare node color in center vs. periphery). Thus, although lineage TFs orchestrate the expression of genes in similar pathways, lineage specificity is a product of high order combinatorial regulation.
Data integration allows for discovery of new Th17-relevant genes
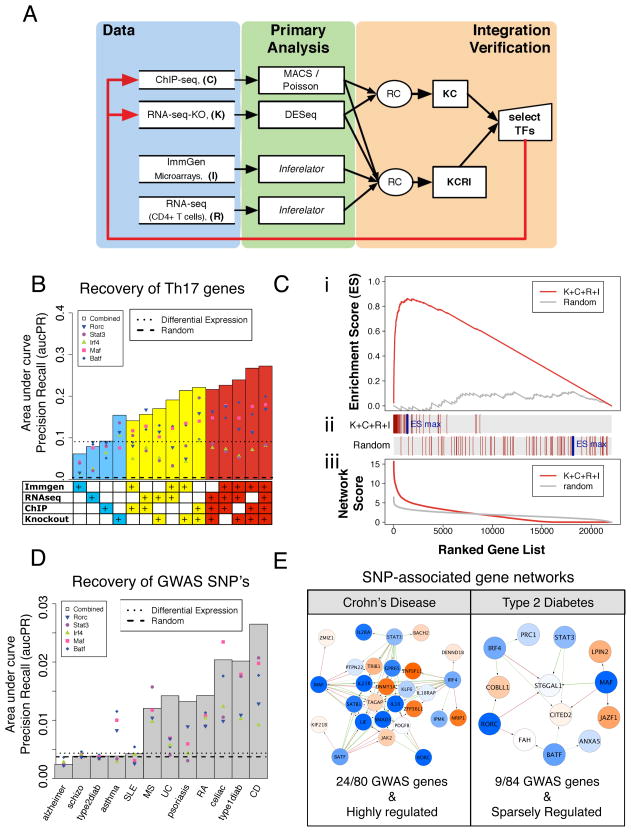
The ChIP and KO based network is accurate but lacks regulatory information for other TFs with roles in Th17 cells. To learn interactions for those TFs, we integrated into our pipeline two other datasets (Figure 4A). The four data types are designated: [C] ChIP-seq; [K] RNA-seq KO; [R] 155 helper T cell RNA-seq experiments; and [I] public microarray data spanning 167 immune cell types and conditions (Immunological Genome Project; ImmGen) (Heng and Painter, 2008). In addition to providing regulatory information for new TFs, R and I can provide further support for interactions identified by the other two data types (C, K). Z-scores for putative TF-target regulatory interactions were individually assigned for R and I using the Inferelator (Greenfield et al., 2010), and converted to rank-based scores, as above (ranging from 0–1), to facilitate their integration with K and C (Supplementary Methods).
Figure 4. Network model performance and validation.
A. Schematic for integration of four genomics and systems data types (K, C, R, and I) using a rank combined (RC) approach, resulting in the KC and KCRI networks.
B. Performance measured as aucPR values indicating enrichment of literature-curated Th17 genes in networks derived from all possible data combinations. Points indicate single TF predictions (e.g. BATF → target) and bars indicate TF sum predictions (i.e. [BATF+IRF4+STAT3+MAF+RORγt]→target). Dotted line, reference performance for targets prioritized by differential expression (Th17 vs. Th0) and, dashed line, for random.
C. Gene Set Enrichment Analysis (GSEA, Broad Institute) for the top-performing KCRI network. (i) The ranked list of TF sum targets recovers Th17-relevant genes with a maximal enrichment score (ES) of 0.86 out of 1 (red line); random (gray line). (ii) Vertical red lines indicate where in the ranked list literature-derived genes were recovered. (iii) Summed TF score distributions for KCRI (red line) and random (gray line).
D. The KCRI network selectively recovers GWAS SNP-linked genes for Th17-implicated inflammatory diseases. Recovery of SNP-associated disease genes is measured in terms of aucPR for TF sum predictions (gray bars) and single TF predictions (points).
E. Core TF networks for genes associated with GWAS of Crohn’s disease and Type 2 Diabetes. The KCRI network recovered 24 out of 80 Crohn’s disease genes (p-val=10−7), and 9 out of 84 Type 2 Diabetes genes (p-val=0.29). Network display is as in Figure 3. P-values calculated by Fisher’s exact test.
To validate our approach and evaluate complementarity of data sets, we estimated the predictive power of the resulting Th17 networks at recovering 74 literature-curated Th17-relevant genes (Table S1). As key Th17 genes were strongly regulated by multiple core TFs (Fig. 3B), we reasoned that ranking genes based on their summed scores over all five core TFs would enrich for relevant target genes. We used two performance metrics, the areas under precision recall (aucPR) and under receiver operator curves (aucROC) that together provide a balance between estimating the sensitivity (aucROC) and accuracy (aucPR) of top ranked predictions. Regardless of the metric used, for each data combination, summed TF target predictions (Fig. 4B; bar) better identified Th17-relevant genes than targets for individual TFs (Fig. 4B; points in bar), highlighting the predictive power of leveraging combinatorial regulation (also Fig. S5A). Moreover, combining across data types resulted in higher quality networks (note the performance increase with data integration, Figure 4B, S5A). This is true for both K+C that produces a smaller (core-TF centered) but accurate network, and for R+I that generates a more comprehensive (more than 170 differentially expressed TFs) but less accurate model. Thus, combining data as in K+C+R+I provides additional network information without compromising the K+C network accuracy, leading to a three-fold performance boost at finding relevant Th17 genes over the conventional differential expression list (Figure 4B, dotted line). Moreover, the non-parametric ranked-based scheme is better suited for combining data than an alternative parametric Fisher method for combining p-values (Figure S5C, D).
We further characterized the predictions of the top performing KCRI network using gene set enrichment analysis (GSEA; Figure 4C). Literature-curated genes were highly enriched as top predictions (60 of 74 total), ranking many more putative Th17-relevant genes with comparable KCRI scores (1,328 of ~22,000 total).
The wealth of GWAS information for diseases in which Th17 cells are implicated allowed us to judge the relevance of our mouse Th17 network for human disease (Hindorff et al. ). Loci linked to SNPs associated with ulcerative colitis (UC), Crohn’s disease (CD), multiple sclerosis, psoriasis and rheumatoid arthritis were significantly enriched in the core (KCRI) Th17 network (Figures 4D, E, S5B). This is in striking contrast to diseases with enrichment scores expected by chance (Alzheimer’s disease and schizophrenia) and other inflammatory diseases in which Th17 cell involvement is not established (e.g. type 2 diabetes and systemic lupus erythematosus (SLE)). Notably, for those diseases with strong links to Th17 function, GWAS associated genes are more highly regulated by the core Th17 TFs (Figure 4E), and TF sum scores (Figure 4D; bars) better recovered GWAS loci relative to individual TFs (Figure 4D; points). This strongly supports the notion that synergy between the five core Th17 TFs is also relevant for function of the Th17 lineage in human disease.
Network analysis identifies novel modulators of the Th17 program
Our validations revealed that highly regulated targets of the KCRI network can be exploited to identify new Th17-relevant effector genes. However, early response regulators that function upstream of or in parallel with core TFs may not be captured (Figure 3A). To address this, we used an independent Inferelator-derived ImmGen network (I) to predict new TFs that demonstrate significant target overlap with the five core TFs (KC network; see Supplementary Methods); a similar method was proven successful (Carro et al., 2010). Accordingly, 26 new candidate Th17 TFs were prioritized that either 1) were top scoring KCRI network-regulated genes (90th percentile), or 2) showed significant overlap between their predicted targets and targets of the five core TFs. The latter TF enrichment analysis identified all core TFs as top hits and several known Th17 regulators (see full list Table S2), including RORα, AHR, RBPJ, and TBX21 (Alam et al., 2010; Veldhoen et al., 2008; Yang et al., 2008).
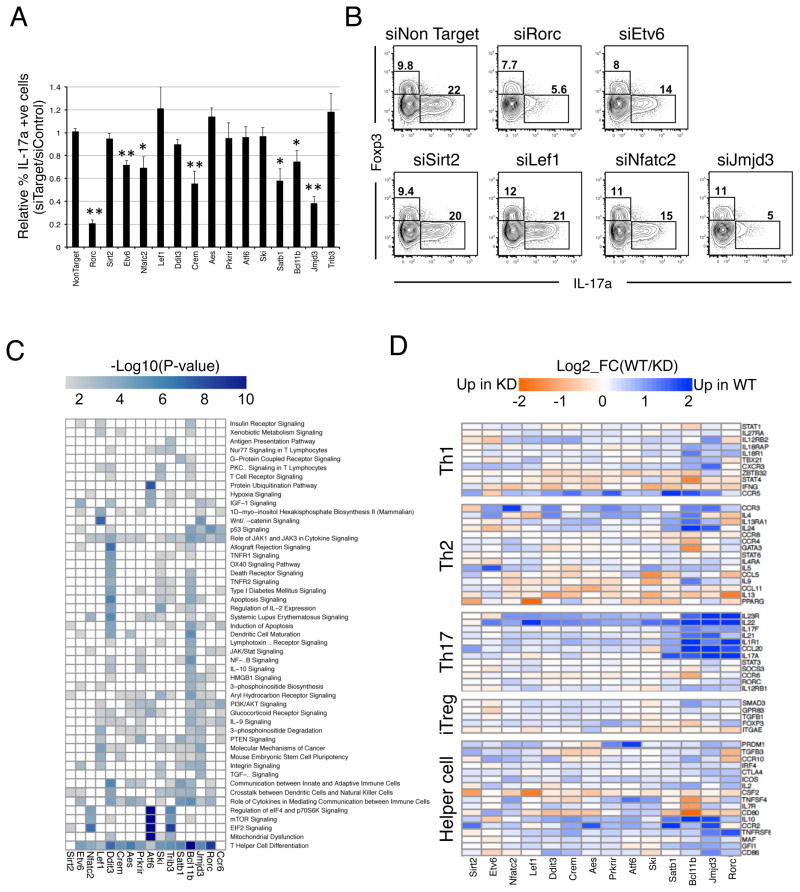
To assess the effects of prioritized TFs on Th17 cell differentiation, we performed gain- and loss-of-function experiments. Among the 16 TFs overexpressed in CD4+ T cells by retroviral transduction, several had a significant effect on the percentage of IL-17A+ cells generated, including Ets factor Etv6, Ncoa2, Smad3, Hif1a, Skil, and Trib3 (Figure S6A, B). Most striking was the AP-1 family member Fosl2, the top predicted factor in the TF enrichment analysis (Table S2), whose overexpression significantly reduced the number of IL-17A-producing cells.
In a complementary approach, we performed siRNA-mediated knock-down (KD) experiments for 14 candidate TFs with siRNA pools electroporated into activated CD4+ T cells subjected to Th17 polarizing conditions. Reductions in target TF mRNAs were similar to those observed with siRorc, which effectively reduced RORγt (Figure S6C, D), its targets (Il17a/f, Il23r, Il22), and Th17 differentiation (Figure 5A,B, S6C). Strikingly, six TF KDs significantly altered IL-17A production relative to a non-targeting control, without affecting the concomitant generation of Foxp3+ iTreg cells. These included Etv6, Nfatc2, Bcl11b, Crem, and regulators of chromatin remodeling (Satb1, Kdm6b (Jmjd3)) (Figures 5A, 5B). Thus, the network method prioritized and made accurate predictions about genes that influence expression of IL-17A, a key component of the Th17 phenotype.
Figure 5. Identification of novel Th17 regulators.
A. siRNA knock-down screen of candidate TF function in Th17 differentiation. Percent of IL17A-producing cells relative to the control siRNA condition for knock-down cultures analyzed at 24h of Th17 polarization. Error bars = SD of two experiments conducted in triplicate. *P<0.05 and **P<0.01, T-test.
B. Flow cytometric analysis for Th17-polarized cells transfected with siRNAs for the indicated gene targets.
C. Shared and unique functions of novel Th17 TF regulators. Heat map of Ingenuity pathway enrichment (IPA, p-value<0.01) for candidate TF-dependent genes.
D. TF candidates influence the expression of immune-modulatory genes. Heat map of log2 fold change in expression of T helper signature genes in the siRNA knock-downs relative to non-targeting control for 24h Th17 polarization cultures.
To assess factor influence on the broad Th17 program, we performed RNA-seq of TF KD cultures. Global pathway analysis for TF-dependent genes identified distinct factor-related pathways, yet showed a striking convergence in enrichment for genes involved in T helper cell differentiation/function for all factors, except Sirt2 and a non-TF control, Ccr6 (Figure 5C). Indeed, TF-dependent loci comprise a variety of helper T cell effector or lineage-specializing genes (Il4, Ifng, Gata3, Foxp3, Tbx21, Il22, Il1r1, Il23r, Il10, Il24, Il9, Ccl20) (Figure 5D). Of particular interest, Bcl11b influenced the expression of a broad set of helper T cell-modulatory genes, suggesting a key regulatory role in subset diversification. Similarly, Jmjd3, a lysine K27 demethylase and known T-bet partner (Miller et al., 2010), displayed a marked influence on the activation of multiple Th17-expressed cytokines, suggesting that it may partner with core Th17 TFs. Indeed, Jmjd3 shares many direct targets (KD+C) with RORγt and STAT3 (Figure S6E). Taken together, the results show that the network model identifies many novel candidate modulators of the Th17 program.
Fosl2 is a core component of the Th17 specification program
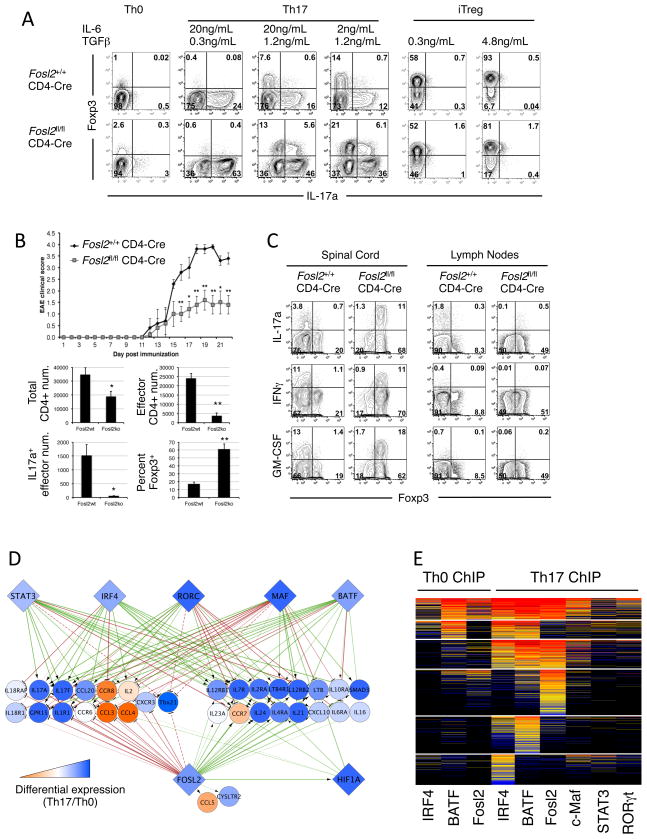
The AP-1 family TF Fosl2 was the highest-ranking candidate to co-regulate targets with core TFs (Table S2). We interrogated its role in helper T cell differentiation and function using mice with conditional deletion of Fosl2 in T cells (Fosl2fl/fl CD4-Cre). Fosl2-deficient CD4+ T cells could be polarized in vitro into Th1, Th2, Th17 or iTreg cells, but, notably, cytokine production was dysregulated (Figure 6A, S7A). Fosl2-null Th17 cultures were markedly increased for IL-17A-producing and atypical Foxp3+IL-17A+ cells. They also had low-level derepression of Il17a in Th1 and Th2 cells, consistent with Fosl2 function as an Il17a repressor (Figure S6A,B, S7A). Fosl2-deficiency also enabled IFNγ production in Th17 and Th2 cultures, particularly when Th17 cells were subsequently exposed to Th1-skewing conditions (Figure S7A, B).
Figure 6. Fosl2 regulates loci critical to lineage identity and function.
A. Fosl2 negatively regulates IL-17A expression. Flow cytometric analysis for naïve Fosl2 wt and KO CD4 T cells cultured as indicated for 3 days. Representative of 4 experiments.
B. Conditional deficiency of Fosl2 in CD4 T cells reduces the severity of EAE. Top panel shows clinical scores (mean ± s.e.m.) for CD4-cre mice with wt or conditional Fosl2 alleles (*P<0.05 and **P<0.01, T-test). Bottom panel displays total numbers or percentages of CD4+ T cell populations isolated from spinal cord (mean ± s.e.m.). Effector CD4+ T cells are defined as Foxp3−. Representative of two experiments.
C. Predominance of cytokine-producing Foxp3+ cells in mice during EAE. Flow cytometric analysis of re-stimulated CD4+ T cells isolated from the spinal cord and lymph nodes of mice on day 22 post EAE induction. Plots gated CD4+CD45+ cells.
D. Combinatorial core TF targets involved in T cell specification are highly regulated by Fosl2. Network depiction is as in Figure 3. Dashed lines: edges added manually to account for Tbx21 with ChIP peaks outside of the set boundaries (10kb flanking the transcribed region).
E. The clustered heat map of Fosl2 and core TF occupancy reveals that the binding domain of Fosl2 largely overlaps with that of BATF.
We next examined the role of Fosl2 in the Th17-dependent disease model experimental autoimmune encephalomyelitis (EAE), which mimics the CNS pathology in multiple sclerosis. Fosl2fl/fl CD4-Cre mice had significantly attenuated disease severity compared to wild-type controls (Figure 6B). Analysis of spinal cord infiltrates at 21 days post immunization revealed reduced CD4+ T cells, but similar percentages of IL-17A, IFNγ, and GM-CSF producers, in mutant mice. Strikingly, the Fosl2-deficient cytokine-producing T-helper cells also expressed the TF Foxp3, which specifies the Treg program (Figure 6B, C). Consistent with our in vitro observations (Figure 6A), these findings suggest Fosl2 as a key regulator of T-helper lineage plasticity, particularly under inflammatory conditions.
To gain insight into Fosl2 function we identified its direct targets using K+C analysis. Consistent with previous studies, Fosl2 both activates and represses target loci (Wagner and Eferl, 2005). It attenuates expression of Th17 signature genes in addition to Il17a (Il17f, Ccl20, Ccr6, Il1r1, Batf) and of Th1- regulatory loci (Tbx21, Il18r1, Il18rap, and Il2), suggesting a role in controlling inflammatory responses and preventing Th1 specification (Figure 6D). Conversely, Fosl2 also promotes the expression of genes that drive Th17 maintenance and survival (Il6ra, Il-23r, Il12rb1, il7r, Il21) and helper T cell diversification or function (Il4ra, Il12rb2, Il2ra, Il10ra, Ltb4r1, Smad3, Hif1a) (Figure 6D). These loci are also targeted by the five core TFs, indicating that Fosl2 modulates the lineage identity and functional programs regulated by core Th17 TFs.
In light of the dominant regulation by AP-1 factors in the Th17 lineage, it was interesting to observe a high degree of overlap in occupancy by Fosl2 and BATF (Figure 6E), suggestive of an antagonistic relationship between them. This may be mediated by direct competition for the same binding sites (Figure S7C), and enhanced by direct transcriptional repression of Batf by Fosl2 (Figure S7D). Thus, as predicted, Fosl2 is a highly interconnected component of the core Th17 specification program. This is in contrast to Hif1α, a recently identified regulator of Th17 cells (Dang et al., 2011), that was not predicted to share a significant number of targets with core TFs (Table S3) and is not as interconnected as Fosl2 (Figure 6D).
Visualization and exploration of the extended KCRI Th17 network
The final KCRI network comprises 173 differentially expressed TFs controlling 3,679 genes with approximately 19,000 interactions. The subset of this network surrounding regulators based on ChIP-seq and KO data is more accurate but limited (this network has 7 TFs controlling 2218 genes, with 4237 edges). To facilitate interrogation of the Th17 transcriptional network by the scientific community, we provide access to the primary data (see Table S3, Supplemental Data), the networks (KC and KCRI), and analysis tools at http://th17.bio.nyu.edu.
DISCUSSION
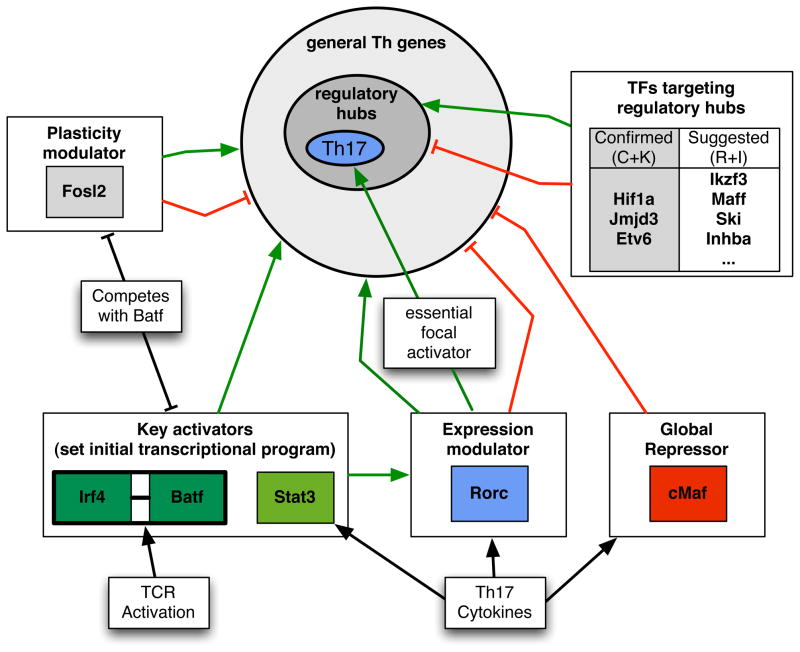
Th17 cells exert critical functions in immune defense at mucosal barriers and are implicated as contributors to multiple autoimmune diseases (Korn et al., 2009). Since the discovery of Th17 cells, multiple TFs involved in the production of IL-17A and in inflammation were described, but little was known of how they collaborate in the global transcriptional program governing Th17 specification and function. Here we aimed to accurately define how Th17 TFs integrate functionally to execute this program, and created a useful network model that can be exploited to uncover novel lineage regulators, effectors, and potential therapeutic targets (Figure 7). To achieve this, we used a culture system for Th17 differentiation to build a transcriptional network model based on combinations of datasets and analytical approaches, and determined its performance using both computational and experimental validations, testing the role of predicted regulators in vitro and in a disease model of inflammation. Our combined computational and experimental approach allowed for iteration between the generation of a data-integrative network and follow-up investigation of individual genes, and, hence, for continuous network refinement (Figure 4A). This work provides a clear experimental design and analysis framework that can be adopted for other cell lineages in the immune system and elsewhere.
Figure 7. Model for Th17 TF functions during lineage specification.
Functions of the core TFs and a selection of newly-identified TFs in regulating expression of general T-helper cell- and Th17 cell-associated genes. BATF/IRF4 complexes, transcriptionally induced following TCR signaling, mutually activate the expression of a large set of target genes, together with STAT3. RORγt drives expression of a small subset of key Th17 genes and modulates the expression of genes activated by initiator TFs, BATF/IRF4/STAT3. Fosl2 restricts the expression of genes required for alternate CD4+ differentiation programs. c-Maf functions as a general repressor. Regulatory hubs include loci that receive a high level of input from Th17 TFs and are enriched for genes that are critical for Th17 differentiation and function.
Dynamics of TF function in the specification of CD4+ T cells
Transcription initiation at sites occluded by nucleosomes and high-order chromatin structure requires mechanisms for making specific regions accessible to appropriate regulators (Zaret and Carroll, 2011). In TCR-activated CD4+ T cells, BATF and IRF4 bind cooperatively to sites throughout the genome. In the presence of Th17-polarizing cytokines, STAT3, c-Maf, and RORγt are recruited to many of the same sites. Chromatin accessibility analysis suggests that BATF/IRF4 complexes pioneer the access of other TFs that further specify functional subsets. Indeed, BATF and IRF4 have critical roles in multiple Th cells (Brustle et al., 2007; Ise et al., 2011; Rengarajan et al., 2002; Schraml et al., 2009). As these TFs are up-regulated in Th0 cells, it is interesting to speculate that pioneering function provides the T cell with plasticity to differentiate in multiple directions, depending on the cytokine environment. Thus, while TGF-β and STAT3-activating signals would recruit STAT3/RORγt to a subset of BATF/IRF4 binding sites, Th1 or Th2 signals may recruit STAT1/T-bet or STAT6/GATA-3 to others. It will be of interest to compare the global distribution of BATF, IRF4, and lineage-specifying TFs in Th1 and Th2 cells.
Fosl2 is a negative regulator of IL-17A. Thus, the finding that Fosl2-deficient mice had a reduced inflammatory response in the EAE model was unexpected. This may reflect the requirement for Fosl2 for expression of key loci supporting Th17 cell maintenance. The result may also be explained by derepression of Foxp3 in inflammatory T cells producing IL-17A, IFNγ, and GM-CSF, which may be mediated, in part, by reduced expression of Hif1α—a Foxp3 inhibitor—in Fosl2-deficient T cells (Dang et al., 2011). Foxp3+ T helper cells that produce effector cytokines have been described in humans and have been shown to have regulatory activity (Voo et al., 2009). Hence, reduced disease scores in Fosl2 deficiency may be due to increased activity of Treg-like cells infiltrating the CNS. Fosl2-deficient T cells also derepress T-bet and IFNγ, suggesting that Fosl2 serves as a brake, binding to sites otherwise occupied by BATF and IRF4 to prevent expression. Indeed, Fosl2 occupancy overlaps with that of BATF and IRF4 in Th0 and Th17 cells, where it likely competes for AP-1 sites (Figure 7). Thus, Fosl2 is a highly integrated regulator of T helper cell lineage identity, functioning to limit plasticity of Th17 cells by repressing Th1 and Treg transcriptional programs, potentially by balancing the activity of BATF/IRF4 at key loci. Our analysis also highlights that regulation of a single cytokine, i.e. IL-17A, does not reflect broad functions of the controlling TFs. A global perspective in the context of a multi-TF causal regulatory network aids in deciphering the role of individual factors.
RORγt has been described as a “master regulator” for the Th17 program, yet it has a surprisingly small regulatory footprint. RORγt deficiency had limited effects on p300 recruitment and H3K4 methylation, suggesting that it lacks a major role in remodeling its regulated loci. However, a handful of loci were highly dependent on RORγt for these early inductive events (Figure 7); what distinguishes this selectivity remains to be uncovered. This focal mode of regulation, coupled with a generalized program upon which RORγt functions to tune expression, is consistent with the plasticity of Th17 cells, suggesting that expression and chromatin state at key Th17 loci might be amenable to rapid change depending on cytokine environment. The lack of stabilizing positive feedback of RORγt to initiators may permit such T-helper program switching. Moreover, while RORγt attenuates the expression of regulators of alternative Th subsets (il4ra, il12rb, Tbx21), these loci are nevertheless expressed. Thus, RORγt is not a prototypical “master” regulator that functions to “lock-in” lineage programs. This renders RORγt an exceptional drug target, as therapeutic intervention would not be expected to perturb the generic regulatory programs shared by other cell types.
A highly predictive Th17 cell network model
The iterative approach applied here was successful in uncovering important aspects of Th17 biology, generating a model that captures most of the previously identified Th17-relevant genes among the top candidates and predicting many more with equal confidence. Among these, several TFs and chromatin modifiers were shown to affect Th17 differentiation or the expression of immune-modulatory genes, including Bcl11b, Etv6, and Jmjd3. Although we focused on the top predicted TF Fosl2, we expect that many more candidates will be pertinent to Th17 biology. We anticipate the network model presented herein to be a highly useful tool for exploration and for generation of new hypotheses.
Although the Th17 network largely models in vitro differentiation, it is nonetheless likely to be relevant for in vivo Th17 cell functions. Indeed, Fosl2-mutant T cells were compromised in effector function in an autoimmunity model and similar phenotypes were reported in mice deficient for many top scoring network genes. Moreover, the network is selectively enriched for genes with orthologs that harbor SNPs associated with human inflammatory diseases linked to Th17 cell-mediated pathology, such as Crohn’s disease and psoriasis. GWAS studies, while facilitating the identification of genes involved in complex diseases involving multiple cell types, are often difficult to translate into biological hypotheses amenable to investigation. However, our analysis identified several GWAS-implicated genes as candidate Th17-specific mediators of pathogenesis (e.g. PTPN22, LIF, KLF6) and may be used to implicate Th17 cells in the etiology of particular conditions. Deconvoluting GWAS data by leveraging the information from accurate and comprehensive transcriptional regulatory networks to provide cellular context, reveal functional epistasis, and prioritize genes of potential medical importance will likely prove to be a powerful approach in uncovering disease mechanisms and developing new diagnostic and therapeutic tools (Califano et al., 2012). Taken together, this body of work is an excellent example of how the power of systems biology can be harnessed to answer a specific large-scale biological question, thus providing a validated paradigm for similar undertakings.
EXPERIMENTAL PROCEDURES
For full details of experimental and computational approaches, see Extended Experimental Procedures in the Supplemental Information. Briefly, naïve CD4+ T cells were purified from lymph nodes and spleen of wild-type mice or TF KO mice. The cells were cultured under Th0 and Th17 conditions, and processed for ChIP-, RNA- and FAIRE-seq. The analysis pipeline included dedicated methods for learning from individual data sources.
Supplementary Material
Acknowledgments
We are grateful to Andrew Sczesnak for generating TDF files and for multiple other contributions to data analysis during the course of the study. We thank R. Dalla Favera for providing IRF4 mutant mice; O. Uluckan and S. Wurm for coordinating shipping of AP-1 mutant mice; K. Ward for system administration; and K. Gunsalus and M. Dustin for helpful discussions. Support was provided by NIH grants RC1 AI087266 and RC4 AI092765 (A.A, D.R.L, R.B, and R.M.M.), PN2 EY016586 (A.G, A.M, and R.B), and IU54CA143907-01, EY016586-06 (A.G and R.B); NSF grant IOS-1126971 (R.B); and fellowships from the Leukemia and Lymphoma Society and Crohn’s and Colitis Foundation of America (M.C.), the National Arthritis Research Foundation (Y.Y.), and Irvington Institute Fellowships from the Cancer Research Institute (M.S. and W.H.).
Footnotes
The Supplemental Information includes the Extended Experimental Procedures, seven supplemental figures, and three tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam MS, Maekawa Y, Kitamura A, Tanigaki K, Yoshimoto T, Kishihara K, Yasutomo K. Notch signaling drives IL-22 secretion in CD4+ T cells by stimulating the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2010;107:5943–5948. doi: 10.1073/pnas.0911755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. Reconstructing dynamic regulatory maps. Mol Syst Biol. 2007;3:74. doi: 10.1038/msb4100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield A, Madar A, Ostrer H, Bonneau R. DREAM4: Combining genetic and dynamic information to identify biological networks and dynamical models. PLoS One. 2010;5:e13397. doi: 10.1371/journal.pone.0013397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, MacArthur J, Wise A, Junkins HA, Hall PN, Klemm AK, Manolio TA. [Accessed Feb 29, 2012];A Catalog of Published Genome-Wide Association Studies. In Available at: wwwgenomegov/gwastudies.
- Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho IC, Kim JI, Szabo SJ, Glimcher LH. Tissue-specific regulation of cytokine gene expression. Cold Spring Harb Symp Quant Biol. 1999;64:573–584. doi: 10.1101/sqb.1999.64.573. [DOI] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009 doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Madar A, Greenfield A, Vanden-Eijnden E, Bonneau R. DREAM3: network inference using dynamic context likelihood of relatedness and the inferelator. PLoS One. 2010;5:e9803. doi: 10.1371/journal.pone.0009803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Aderhold A, Allison KR, Bonneau R, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012a doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 2012b doi: 10.1101/gr.127191.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information--moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, Ohya K, Jetten AM, Akira S, Muta T, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- Prill RJ, Marbach D, Saez-Rodriguez J, Sorger PK, Alexopoulos LG, Xue X, Clarke ND, Altan-Bonnet G, Stolovitzky G. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS One. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Noubade R, Eidenschenk C, Ota N, Zeng W, Zheng Y, Hackney J, Ding J, Singh H, Ouyang W. Transcription factor c-Maf mediates the TGF-beta-dependent suppression of IL-22 production in T(H)17 cells. Nat Immunol. 2011;12:1238–1245. doi: 10.1038/ni.2134. [DOI] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH, Arima K, Bover L, Hanabuchi S, Khalili J, Marinova E, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106:4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.