Abstract
As a promising biomaterial with numerous potential applications, various types of synthetic spider silk fibers have been produced and studied in an effort to produce manmade fibers with mechanical and physical properties comparable to those of native spider silk. In this study, two recombinant proteins based on Nephila clavipes Major ampullate Spidroin 1 (MaSp1) consensus repeat sequence were expressed and spun into fibers. Mechanical test results showed that fiber spun from the higher molecular weight protein had better overall mechanical properties (70 KD versus 46 KD), whereas postspin stretch treatment in water helped increase fiber tensile strength significantly. Carbon-13 solid-state NMR studies of those fibers further revealed that the postspin stretch in water promoted protein molecule rearrangement and the formation of β-sheets in the polyalanine region of the silk. The rearrangement correlated with improved fiber mechanical properties and indicated that postspin stretch is key to helping the spider silk proteins in the fiber form correct secondary structures, leading to better quality fibers.
INTRODUCTION
The extraordinary mechanical properties of orb-weaving spider silks have served spiders for over 400 million years.1 However, only in the late 20th century has scientific research started to shed light on the molecular nature of spider silk that contributes to its incredible properties as biomaterials.2 Despite the fact that spider silk is a promising biomaterial with various potential commercial, medical, and military applications,3,4 the spiders’ territorial nature prevents people from farming spiders like silkworms. Thus, production of synthetic spider silk fibers with mechanical properties comparable to those of native spider silk is a current focus of spider silk research.5,6
Among all six types of solid fiber that an orb-weaving spider produces,1 major ampullate silk (also known as dragline silk) is the strongest. It is more than three times tougher than the manmade fiber, Kevlar.7,8 Spiders use this silk to build the main framework of their web, a belaying line during movement, or safety line to escape from predators. Major ampullate silk consists of two main proteins: Major ampullate Spidroin 1 (MaSp1)9 and Major ampullate Spidroin 2 (MaSp2).10 Each protein has distinct sequence features that contribute to the impressive mechanical properties that dragline silk possesses. Early studies on major ampullate silk revealed that the amino acids are organized as hundreds of tandemly repeated motifs in the silk protein.11,12 These motifs are highly conserved and have been retained in the Araneidae family with little change for at least 150 million years.13,14 Different motifs have been hypothesized to control different structural and functional aspects of the silk, which balance the strength and elasticity of the silk. The MaSp1 protein generally contains two motifs, polyalanine and GGX (X = L, Y, Q, A), whereas MaSp2 protein is composed of GPGXX (X = G, Q, Y) and the polyalanine motif. One of the surprising features about spider silk is that the polyalanine motif, which is commonly found to have helical structure,15,16 is primarily in antiparallel β-sheet structure in native spider silks. The large fraction of β-sheet nanostructures in spider silk was first observed using X-ray diffraction;17,18 later, NMR, FTIR, AFM were used to verify further the nature of β-sheets and the amino acid motifs that contribute to the β-sheet component in spider silks.19–22 This self-assembled antiparallel β-sheet forms crystalline segments in the fiber and is believed to be the key element in native silk responsible for the high tensile strength of the silk.12,19 Because the β-sheet is a tightly knit structure with a large number of hydrogen bonds, the absence of water in this structure also provides large hydrophobic forces, preventing the silk molecules from separating. Notably, the conserved polyalanine motif of spider silk is also found in the structural proteins of many other organisms that possess exceptional mechanical properties, such as oyster shell matrix protein23 and mussel byssus proteins.24,25 The GPGXX motif has been implicated as an elasticity module because it forms an elastic, spring-like β-spiral,26 which is similar to the β-turn spiral in elastin.27–29 The role of GGX motifs is currently not clear, but it appears to be a Gly-II helix and likely contributes to the intermolecular interactions in the silk.21,30 The proposed structure model for spider silk fiber is crystalline β-sheet domains embedded in a glycine-rich matrix.2 These previous studies indicate that by manipulating the amount and ratio of different motifs, one could build synthetic protein with tunable mechanical properties.
In Nephila clavipes, a species that has been used to establish the fundamental understanding of spider silk, 81% of its major ampullate silk is MaSp1 with the remaining 19% MaSp2.31,32 To date, only a few synthetic proteins have been produced based on Nephila clavipes MaSp1 sequences.33,34 In this study, we expressed two synthetic silk proteins based on Nephila clavipes MaSp1 consensus sequence with only a size difference. Fibers were spun in vitro using a wet spinning technique with postspin stretch treatment performed on half of the resultant fibers. Our results suggest that postspin stretching improves the mechanical properties of synthetic silk fiber by helping the silk fiber protein increase its β-sheet content. The mechanical properties of our fibers confirm this theory by showing superior properties after postspin stretching. Carbon-13 cross-polarization magic-angle spinning (CP-MAS) NMR studies of the silk protein molecular structure further support this point.
MATERIALS AND METHODS
Gene Cloning
Two constructs were made based on the Nephila clavipes MaSp1 conserved amino acids sequence (Table 1). The basic MaSp1 repeating unit contains multiple GGX motifs, followed by a polyalanine motif. This monomer gene was synthesized by Bio S&T (Montreal, Quebec). Duplication of the monomer was achieved by compatible but nonregenerable cloning strategy developed in our lab,35 with 5′-AvrII and 3′-NheI flanking the monomer. Gene manipulation was completed in pBluescript II SK (+) vector (Ampr) in E. coli strain GM2163 (Cmr). The final constructs were ligated into pET19k (a modified pET-19b vector where the ampicillin resistance gene was replaced with the kanamycin resistance gene from the pET-26b vector)35 through 5′-NdeI and 3′-BamHI and then transformed into E. coli strain BL21(DE3) for protein expression. A deca-histidine-tag was included at the N-terminal of the silk protein for purification purposes. Restriction enzymes used in the gene cloning were purchased from New England Biolabs (Ipswich, Massachusetts).
Table 1.
Synthetic Silk Protein Sequences with Gene and Protein Sizesa
| protein sequence | DNA size (bp) | protein size (kD) | |
|---|---|---|---|
| (MaSp1)16 | (GGAGQGGYGGLGSQGAGRGGLGGQGAGA6)16 | 1584 | 46 |
| (MaSp1)24 | (GGAGQGGYGGLGSQGAGRGGLGGQGAGA6)24 | 2376 | 70 |
Note that the His-tag sequence at N-terminal of the proteins is not shown.
Protein Expression
Before large-scale protein production, positive colonies were first tested by small-scale (5–20 mL) protein expression in LB media to identify the colony with the highest synthetic silk protein yield. This was done by growing 12 selected colonies in LB media in small culture tubes (10–50 mL) until an OD600 of 0.8; follow-up processes like protein induction, bacteria collection, and silk protein detection by Western blots were similar to the large-scale expression process described below.
Selected colonies with high silk protein yields were then grown in a 19.5 L BioFlo 415 sterilizable-in-place fermentor (New Brunswick Scientific, Edison, NJ) with fermentor-recommended media (provided in the New Brunswick fermentor instructions) overnight until it reached an OD600 of 15–20, upon which IPTG (isopropyl-β-d-thiogalactopyr-anoside; Biosynth AG, Switzerland) was added to a final concentration of 1 mM to induce silk protein synthesis from lac promoter. Induced cells were allowed to grow for 4 h before harvesting. Bacteria culture was centrifuged at 3500 rpm for 20 min (Allegra 6KR, Beckman, Brea, CA). Cell pellets were resuspended in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 8.0) at a weight to volume ratio of 1:3; 0.25 mg/mL lysozyme was also added. The resuspended cell mixtures were immediately frozen in an −80 °C freezer for at least 24 h.
Protein Purification
Each 800 mL of resuspended fermentation products was defrosted and sonicated to lyse for two 4 min bursts at 90 W to ensure complete lysis. Cell debris were separated and discarded by centrifugation at 13 000g for 25 min (Beckman J2-21M). Supernatants were then heat-treated at 80 °C in a water bath to precipitate most heat-instable E. coli proteins. Spider silk proteins of interest were known to withstand this temperature without precipitation.35 The supernatant containing synthetic silk protein after heat treatment was collected by centrifuge as previously at 13 000g (25 min). The silk protein with 10× N-terminal his-tag was captured and purified by immobilized metal (Nickel) ion affinity chromatography (IMAC) using an ÄKTAexplorer (GE Healthcare, Piscataway, NJ). The protein binding column was washed with binding buffer, then 100 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 7.9 washing buffer. Silk protein was eluted by elution buffer at 250 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 7.9. Silk protein eluates were dialyzed against ddH2O in dialysis tubing with 12 kDa pore size (Spectrum Laboratories, Rancho Dominguez, CA) for 2 days, followed by lyophilization to produce dry protein powders.
Electrophoretic Analysis
DNA electrophoresis was performed on each step of cloning using 8% agarose gels at 100 V in TAE buffer, stained by ethidium bromide, and visualized under UV light. Protein electrophoresis was done on precast 4–20% SDS-PAGE gel (Precise Protein Gel, Pierce Biotechnology, Rockford, IL) at 90 V with Pierce-recommended Tris-Hepes-SDS buffer. Silk proteins of interest were detected by either Western blot using His-tag antibody (Novagen) following a standard protocol36 or His-tag In-gel stain (In Vision His-Tag In-gel Stain, Invitrogen, Carlsbad, CA). Coomassie staining with Bio-Safe Coomassie (Bio-Rad, Hercules, CA) was used to visualize all proteins on the gel.
Fiber Spinning and Postspin Treatment
We made spinning dopes by dissolving lyophilized protein powder into 1,1,1,3,3,3-hexa-fluoro-2-propanol (HFIP; TCI America, Portland, OR). Protein concentration in the spinning dope was ~30% (w/v). The glass vial containing the dope was put on a slowly spinning rotor for at least 2 days and vortexed regularly to dissolve completely the protein. Fiber spinning was performed on a spinning machine specially made at DACA Instruments (Santa Barbara, CA). Spinning dopes were loaded in a 1 mL Hamilton syringe (Hamilton, Reno, NV) and extruded through 0.005 in. PEEK tubing (SUPELCO, Bellefonte, PA) at a syringe plunger speed of 0.7 mm/min into a coagulation bath filled with 100% isopropanol. Fibers formed in coagulation bath were picked up carefully by tweezers, passed through two sets of spinning godets (speed 0.5 to 1 m/min), and finally collected on a winder. Usually, at these settings, 2 to 3 m of fibers could be collected without breakage.
As-spun fiber without postspin treatment was examined by eye for visible defects. Good fibers were then cut into 2 cm pieces. Fibers were soaked in 75% isopropanol/water solution by volume for 1 min to achieve maximum water penetration. Then, the fibers were stretched in the solution from one end slowly by tweezers until it reached a total length of 6 cm. Stretched fibers were taken out of the solution but maintained the length by tweezers until dried.
Mechanical Testing
Post-spin-stretch-treated fibers were air-dried, cut, and mounted onto 30 mm × 20 mm rectangular mechanical testing card with 15 mm square gap in the middle. Microscope pictures of each fiber were taken under 10× ocular lens plus 40× objective lens (Nikon Eclipse E200 microscope). Fiber diameters were measured at nine different places using ImageJ 1.42q (National Institute of Health, USA). The average was calculated to determine the diameter of each fiber sample. Mechanical tests were carried out at room temperature with ~15% humidity on an MTS Synergie 100 (MTS corporation, Eden Prairie, MN) using a custom-built 10 g load cell (Transducer Techniques, Temecula, CA) at a pulling rate of 10 mm/min with 30 Hz frequency data collection. The data were plotted in Matlab 7.6.0 (R2008a) Mac, and a polynomial regression curve was fitted to the data points at an order of seventh degree polynomial.
Solid-State Nuclear Magnetic Resonance Studies
Solid-state 13C CP-MAS NMR spectra were collected on a Varian VNMRS wide-bore 400 MHz spectrometer equipped with a 3.2 mm triple resonance MAS probe. The experimental parameters for CP were a 4 µs 1Hπ/2 pulse, a 1 ms ramped (~20%) 1H spin-lock pulse with a radio frequency (rf) field strength of 62.5 kHz at the ramp maximum and a square 13C contact pulse. The MAS frequency was 10 kHz, and the 1H→13C CP condition was matched to the −1 spinning sideband in the Hartmann–Hahn profile on the 13C channel. Two-pulse phase-modulated (TPPM) 1H decoupling with 100 kHz rf field strength was applied during acquisition of the free induction decay (FID) with a 6° phase shift. Typical acquisition parameters were 1024 points, 16 384 scans, a 4 s recycle delay, and 50 kHz sweep width. The spectra were processed by zero filling to 4096 points and applying 100 Hz of exponential line broadening, followed by Fourier transformation (FT). Additionally, all reported 13C CP-MAS spectra were background-subtracted to minimize contributions from probe background. The 13C chemical shift was referenced indirectly to TMS by setting the downfield (high ppm) chemical shift of adamantane to 38.56 ppm.37
RESULTS
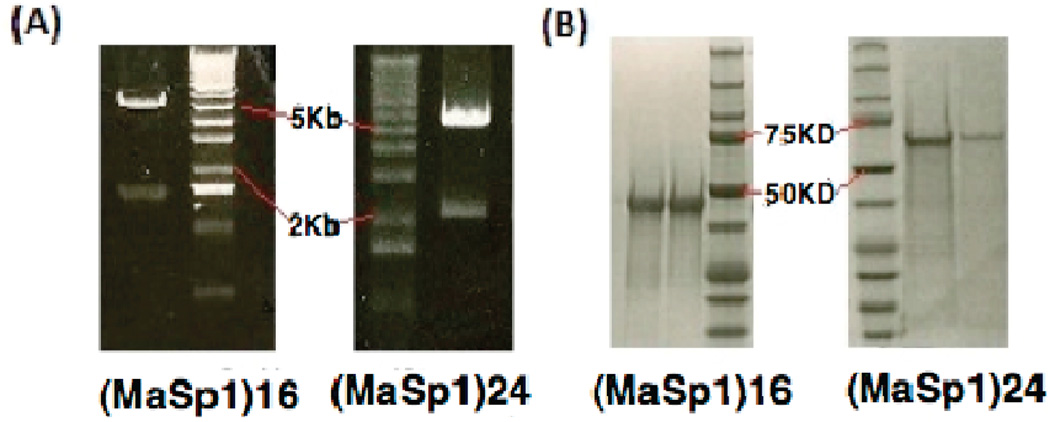
Each step of cloning was checked by restriction enzyme digest (Figure 1A) and further confirmed by DNA sequencing (Macromolecular Core Equipment Facility, University of Wyoming). T7 and T3 promoter primers were used for pBlue-script II (SK+) sequencing; T7 promoter and terminator primers were used for sequencing in pET19k. Electrophoresis confirmed the size of the protein produced by fermentation (Figure 1B). Those proteins were then dialyzed, lyophilized, and dissolved in HFIP for fiber spinning. Fibers were spun at a piston extrusion rate of 0.7 mm/min; collection starting when the fiber extrusion rate became stable. Environment temperature was 25 °C with 13–15% humidity.
Figure 1.
(A) Agarose gels show BamHI and NdeI restriction enzyme double digest result of the final synthetic silk gene in pET19k. Lower bands are synthetic silk gene insertions, upper bands are Linearized pET19k vector (5.8kb). (B) SDS-PAGE (polyacrylamide gel electrophoresis) of proteins purified from IMAC column and stained by Biosafe Coomassie.
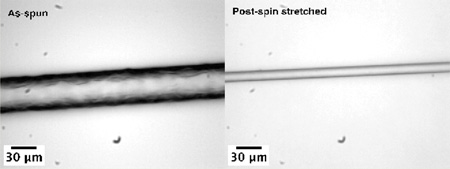
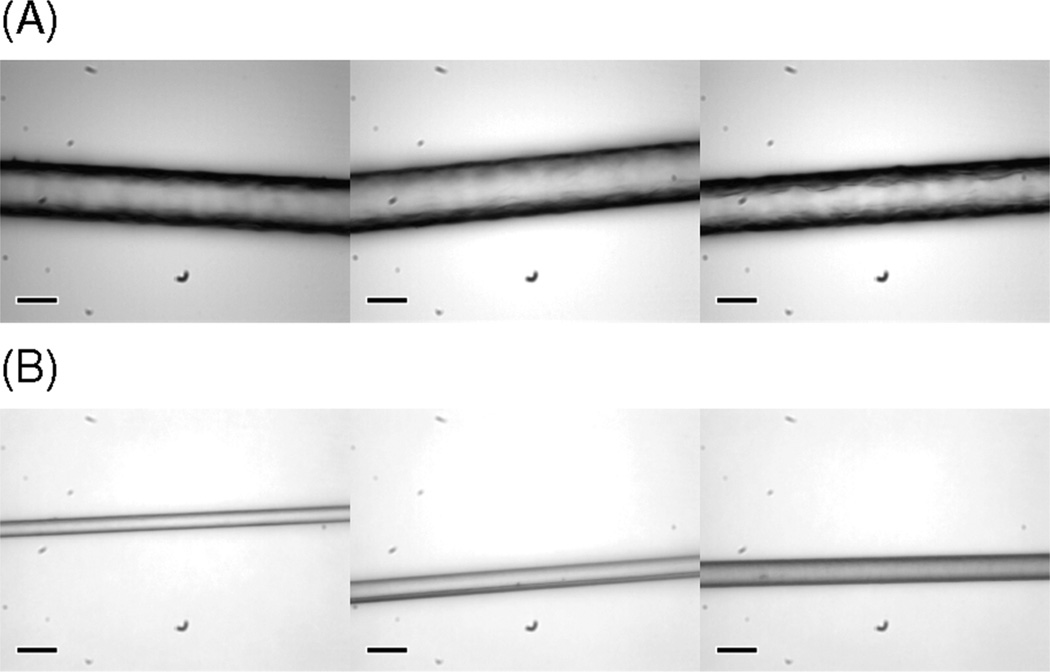
As-spun fibers were post-spin-stretched in 75% aqueous isopropanol because these synthetic fibers become partially soluble and form a gel-like phase in water. This interaction between as-spun fiber and water seems to have little relation to the polymer size but may largely depend on protein primary structure, dope microenvironments, and spinning conditions. Synthetic as-spun fibers produced in our lab based onMaSp1 and MaSp2 sequences are at least partially soluble in water. Some of our fibers spun from the same protein at different times (with different protein concentration in spinning dope and extrusion speed, etc.) showed that they withstand different degrees of water penetration, ranging from 20 to 50% water in the isopropanol solution. All fibers in this study were treated in 75% aqueous isopropanol solution by volume and post-spin-stretched three times their original length for the purpose of reducing potential variation generated by the fiber production process. It is worth noting that during stretching visible tiny air bubbles were ejected from the fiber, which indicates that the as-spun fibers have hollow pockets inside the fiber structure that likely lower mechanical properties. Microscopic images in Figure 2 show the diameters and surfaces of as-spun and post-spin-stretched (MaSp1)24 fibers.
Figure 2.
Light microscopic images of (A) as-spun (MaSp1)24 and (B) post-spin-stretched (MaSp1)24 fibers. Scale bars in lower left corner indicate 30 µm.
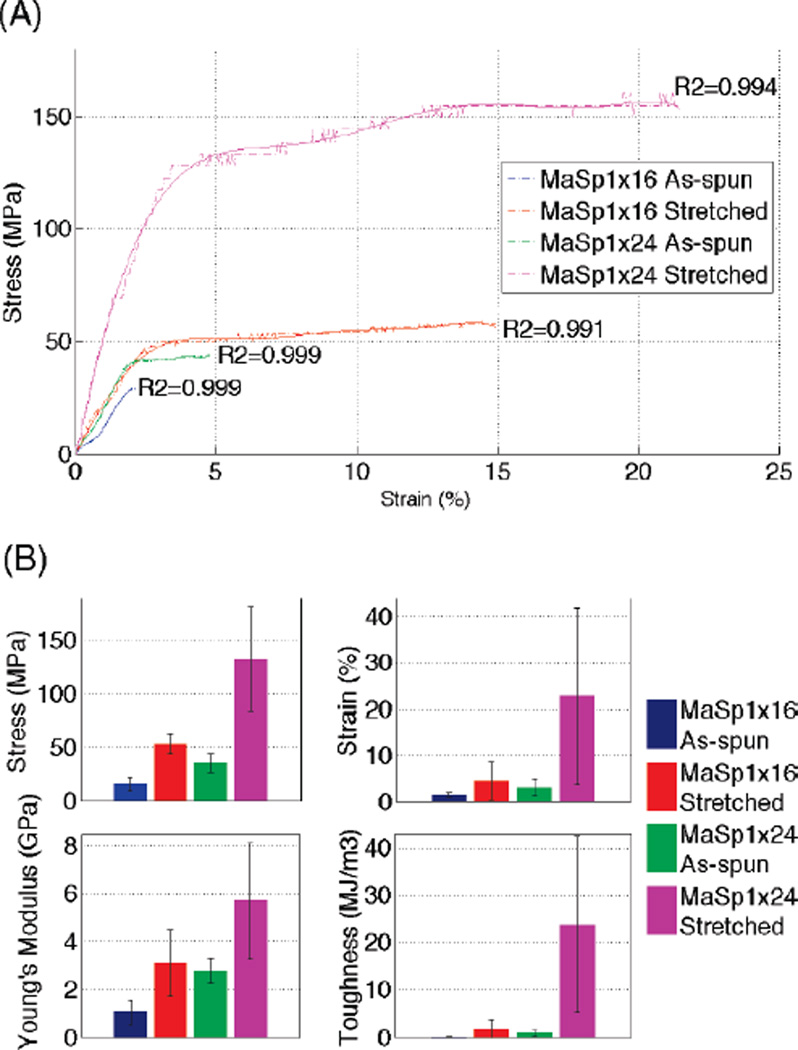
Mechanical properties of both as-spun and post-spin-stretch-treated synthetic silk fibers were evaluated and are summarized in Figure 3 and Table 2. Table 3 lists several better performers from each group to represent the best single fiber we have acquired so far. Ten or more fibers in each group were tested, including 11 (MaSp1)16 as-spun fibers with an average diameter of 41.23 ± 2.93 µm, 10 (MaSp1)16 stretched fibers with diameter of 24.12 ± 5.44 µm, 10 (MaSp1)24 as-spun fibers at 40.90 ± 3.99 µm, and 12 (MaSp1)24 stretched fibers at 17.44 ± 4.97 µm. The error in these measurements increased after post-spin-stretch, which indicates that hand stretching is a potential source of variation.
Figure 3.
Mechanical testing data analysis of synthetic silk fibers. (A) Typical stress–strain curves (dashed-dotted line) of (MaSp1)16 as-spun (blue), (MaSp1)16 stretched (red), (MaSp1)24 as-spun (green), (MaSp1)24 stretched (magenta) with trendlines (solid line with corresponding colors) fitted to an polynomial order of the seventh degree. (B) Column charts present the average performance in stress, strain, Young’s modulus, and toughness of all of the fibers tested in each group. All error bars represent standard deviation with n ≥ 10.
Table 2.
Summary of the Average Mechanical Test Performance of All the Fibers Tested in the Study
| materials |
strength |
extension |
Young’s modulus |
toughness |
|
|---|---|---|---|---|---|
| σmax (MPa) | εmax(%) | Einitial (GPa) | (MJ/m3) | ||
| (MaSp1)16 | as-spun | 16.32 ± 6.68 | 1.46 ± 0.43 | 1.06 ± 0.50 | 0.11 ± 0.07 |
| stretched | 53.93 ± 9.52 | 4.50 ± 4.07 | 3.11 ± 1.40 | 1.70 ± 2.04 | |
| (MaSp1)24 | as-spun | 35.65 ± 8.42 | 3.13 ± 1.84 | 2.78 ± 0.53 | 0.87 ± 0.63 |
| stretched | 132.53 ± 49.20 | 22.78 ± 19.06 | 5.70 ± 2.43 | 23.73 ± 18.46 | |
Table 3.
Selected Better Performers from Each Fiber Group
| strength |
extension |
Young’s modulus |
toughness |
|
|---|---|---|---|---|
| materials | σmax (MPa) | εmax (%) | Einitial (GPa) | (MJ/m3) |
| (MaSp1)16 stretched | 59.21 | 14.93 | 1.82 | 7.30 |
| 59.91 | 3.72 | 4.07 | 1.82 | |
| (MaSp1)24 stretched | 233.54 | 31.29 | 6.19 | 46.82 |
| 99.06 | 55.67 | 2.85 | 48.40 |
Stress–strain curves of all fibers showed good polynomial fit at a polynomial order of seven, as reflected by R2 value of the polynomial regression in each trendline (R2 > 0.99). Comparing the stress–strain curves of (MaSp1)16 versus (MaSp1)24, (MaSp1)24 fibers clearly showed superior mechanical properties, in tenacity, extensibility, and Young’s modulus in both as-spun and stretched fibers. Comparing the data of as-spun fiber versus stretched fiber, the tenacity and Young’s modulus of stretched fiber increased significantly, roughly 3.5-fold in tenacity and 2-fold in Young’s modulus for both (MaSp1)16 and (MaSp1)24 fiber. Extensibility also increased noticeably for most stretched fibers. However, large variations (as showed by error bars in Figure 3) for stretched fibers indicate the inconsistency of this increase, preventing useful predictions quantitatively or scientifically.
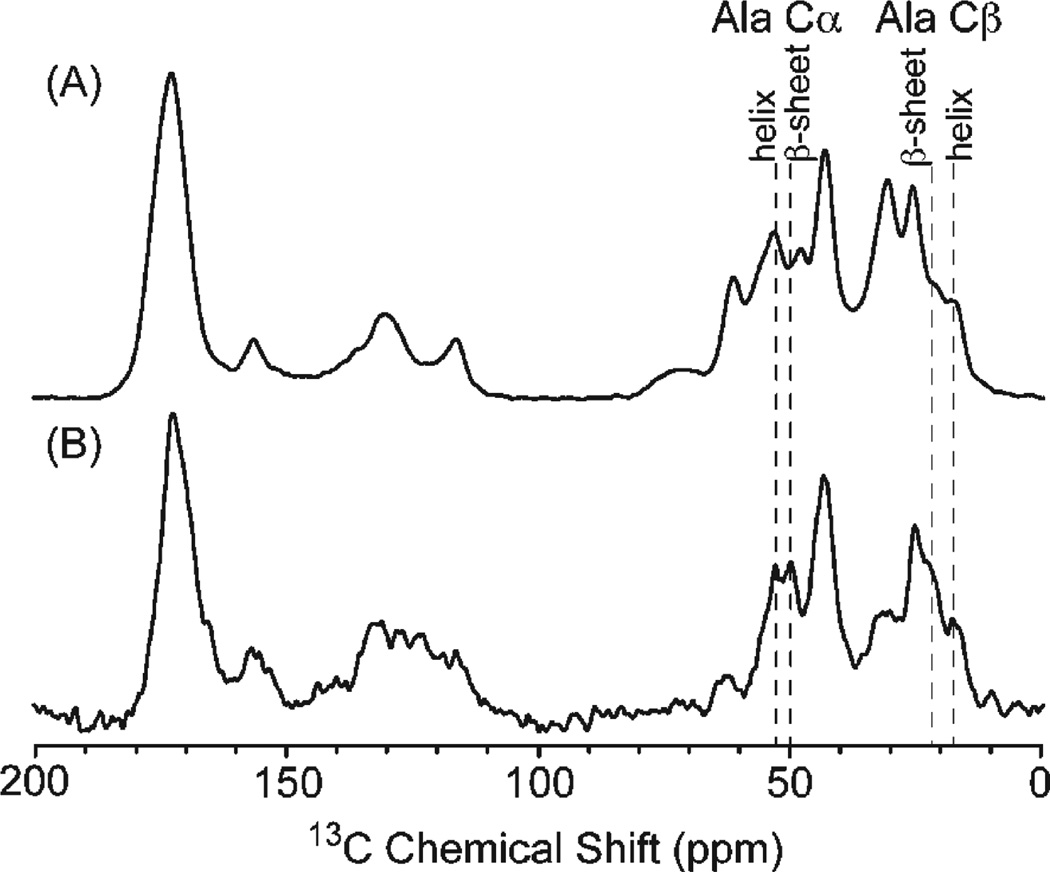
Carbon-13 CP-MAS NMR spectra were collected for the (MaSp1)24 lyophilized protein and the (MaSp1)24 fiber following postspin draw to characterize the protein secondary structure. The 13C isotropic chemical shift of the Cα and Cβ amino acid resonances is sensitive to the backbone torsion angles and, therefore, the secondary structure.38–41 This 13C chemical shift approach has been extensively used to characterize the secondary structure of the repetitive amino acid motifs in native spider silks.20,42–49 The 13C CP-MAS spectrum of the lyophilized (MaSp1)24 protein and the (MaSp1)24 fiber following postspin draw is presented in Figure 4. The overall signal-to-noise (S/N) of the lyophilized (MaSp1)24 protein is larger than the (MaSp1)24 post-spin-stretched fibers because of the larger sample size available for the lyophilized protein (~15 mg) compared with the (MaSp1)24 post-spin-stretched fibers (~4 mg). The alanine Cβ resonance displays two components at 21 and 17 ppm in both 13C CP-MAS NMR spectra that can be ascribed to alanine present in β-sheet and helical/random coil structures, respectively. Similarly, the alanine Cα resonance displays two components at 49 and 52 ppm that can be attributed to β-sheet and helical/random coil structures. Assuming similar CP efficiencies for both samples, the (MaSp1)24 synthetic fiber displays a larger fraction of the β-sheet component compared with the lyophilized (MaSp1)24 protein. This result shows that both the fiber spinning and postspin draw processing increase the β-sheet fraction and provide a molecular level understanding for the improved mechanical properties observed for the (MaSp1)24 fiber. (See Figure 3.) It also illustrates the importance of postspin drawing to drive the conversion of polyalanine from a predominantly random coil structure in the lyophilized protein starting material to a β-sheet-rich fiber following the spinning and drawing process. It should be noted that although the polyalanine β-sheet fraction is larger for the (MaSp1)24 fiber following the spinning procedure, it is not as high as that observed in native spider silks.20,43–46 This indicates that the spinning and drawing procedures can still be further optimized to produce fibers that more closely match the secondary structures observed for native spider silks.
Figure 4.
13C CP-MAS NMR spectrum of (A) (MaSp1)24 lyophilized protein and (B) (MaSp1)24 post-spin-stretched fibers. Dashed lines indicate chemical shifts of polyalanine α and β carbons in different secondary structures.
DISCUSSION
Major ampullate silk proteins of Nephila clavipes have very high molecular weights of 250–320 kDa.50,51 It is believed that protein size plays a key role in determining some fiber mechanical properties because fibers with larger protein molecules will have more intermolecular interactions and fewer protein chain ends.52 This is consistent with our results because (MaSp1)24 synthetic silk proteins are about five times smaller than native-sized proteins; the average performance in tenacity versus native fiber is five times lower. (MaSp1)24 fibers have overall superior mechanical properties versus their smaller counterpart (MaSp1)16 fibers. During our manuscript preparation, another group had successfully produced native-sized recombinant spider silk fiber with mechanical property comparable to native silk, in a metabolically engineered E. coli strain with elevated glycyl-tRNA pool.34 This report further indicates that a larger protein size contributes to better fiber mechanical properties.
In this study, the (MaSp1)24 post-spin-stretched fiber, the group of fibers that achieved the highest mechanical property, has an average of 133 MPa tensile strength, a 22% elongation, a 6 GPa Young’s modulus, and a 24 MJ/m3 toughness. The best performer in this group has a tensile strength of >230 MPa, a 30% elongation, a 6 GPa Young’s modulus, and a 47 MJ/m3 toughness. Our (MaSp1)24 post-spin-stretched fiber is comparable to the 32mer (100 KD in protein Mw) synthetic fiber produced by Xia et al. with a better average elasticity.34 Recombinant spider silk proteins based on Araneus diadematus53 and Argiope aurantia36 MaSp2 have been spun into fibers and tested. The former has similar strength to our fiber and an even higher elasticity, probably due to the GPGXX elastic module in MaSp2 protein. The latter showed inferior properties, likely due to the lack of postspin treatment. Variability seen in Figure 3B,C and Table 3 is likely caused by uneven stretching force and speed during our hand stretching.
In addition to the spider silk protein’s primary sequence and size, spinning conditions and postspin treatments are always important parts of fiber making.53 Spiders developed an impressive silk spinning system through years of evolution.7 There is still much to learn from spiders themselves. Protein concentrations in spider silk gland are 40–50%,7,54 whereas the highest protein concentration we achieved for synthetic silk protein dissolved in HFIP is ~30%. Larger size proteins appear even more difficult to dissolve in HFIP to achieve high protein concentrations.34 Moreover, unlike man-made polymers such as nylon and Kevlar, spider silk is a natural biomaterial almost completely composed of protein. It is produced and processed in a completely aqueous environment. Actually, the entire silking procedure in spider gland has been built around controlling water content.7 Finding other solvents, preferably aqueous solutions to spin fibers, as the spider does might be a key factor to achieve better synthetic fibers.
Almost all commercial synthetic fibers need some degree of postspin treatment during their production process. Postspin drawing is involved in nylon production, and heat treatment and electron bombardment increase the compressive strength of Kevlar.55,56 It is worth noting again that spiders spin their fiber by using their legs to pull silk out of their spinnerets. In contrast with the spider, our wet spinning techniques, instead of pulling, actually extrude the protein dope out of a thin needle into a coagulation bath to make fibers. Without stretching in the proper solution during spinning, protein molecules in as-spun fibers might fail to form correct secondary or tertiary structures that are critical for their mechanical properties.57 The importance of this post-spin-stretching process is well-demonstrated in this study because our stretched fibers, regardless of if they are (MaSp1)16 or (MaSp1)24, have shown noticeable improvement in mechanical property. Similar postspin draw has been done on silkworm silk fibroin,58,59 and those results closely resemble what we observe in recombinant spider silk fiber. Mechanical property enhancement was observed on both films59 and fibers58 of regenerated silkworm silk fibroin after stretching in water. On the basis of their report, molecular chain orientation in the film is increased with draw ratio. It will be interesting to perform quantitative 2H NMR in the future to see if our post-spin-stretching treatment also increases the orientation of protein molecules and promotes total crystalline content in the synthetic spider silk fiber.
As mentioned above, during the postspin stretch, we observed tiny bubbles escaping from the surface of the fiber, which indicates that holes are present in the as-spun fibers after organic solvents like HFIP or isopropanol evaporated. Those holes could prevent proper molecular interaction and crystallite orientation that hold fiber structures together. This could also explain why our as-spun fibers are soluble in pure water and our NMR data show less β-sheet formation in as-spun fibers.
Water is known as a native spider silk protein plasticizer that helps improve fiber tensile strength and stiffness by promoting silk protein crystallization.18,60 It has also been demonstrated that with a similar stretching process performed in water the strain’stress curve of native major ampullate spider silk could be predictably reproduced from supercontracted silk.61 This further suggests that the aqueous environment and stretching are essential features of the spinning process of silk fibers. During our in vitro fiber spinning, we have tried to stretch “fresh” as-spun fiber that is still soft right after spinning in the air, but without the involvement of water, post-spin-stretching in air had minimal effects on fiber mechanical properties. Also, air-dried as-spun fiber could barely be stretched in 100% isopropanol, whereas the presence of water, even as little as 10%, promoted protein structure reorganization and β-sheet formation. Water-stretched fibers showed significant tensile strength increases and were no longer soluble in water because the tightly packed hydrophobic β-sheet regions remained immobile in the presence of water.21 This change also corresponded to our NMR results because post-spin-stretched fibers formed noticeably larger amounts of β-sheets in the polyalanine region.
CONCLUSIONS
Because we are unable to farm spiders like silkworm, synthetic spider silk production is one way to make use of this exceptional biomaterial. Understanding the molecular nature of spider silk and replicating fibers comparable to native silk will continue to be the main focus for spider silk researchers. The results presented in this study confirm several important aspects of this process: first, that protein size affects mechanical properties, that is, that larger proteins produce stronger fibers; second that β-sheet formation is a key to stronger fibers, as demonstrated by our NMR results; and third, that postspin stretching improves both secondary structure and mechanical properties. Yet our results raise interesting issues for future efforts to improve the spider silk synthetic fibers.
ACKNOWLEDGMENT
This material is based on work by the Lewis laboratory supported by NIH (EB000490) and the AFOSR (FA9550-09-1-0717) and by Jeff Yarger and Greg Holland by the AFOSR (FA9550-10-1-0275) and the NSF (DMR-0805197 and CHE-1011937) for the NMR component of this project. We would also like to thank Dr. Florence Teule for the methods used for postspin stretching and Dr. Brian Cherry for help with NMR instrumentation and student training. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the AFOSR, NSF, or NIH.
REFERENCES
- 1.Vollrath F. Sci. Am. 1992;266:70–76. [Google Scholar]
- 2.Lewis RV. Acc. Chem. Res. 1992;25:392–298. [Google Scholar]
- 3.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 4.Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Trends Biotechnol. 2008;26:244–251. doi: 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Scheibel T. Microb. Cell Fact. 2004;3:14. doi: 10.1186/1475-2859-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vendrely C, Scheibel T. Macromol. Biosci. 2007;7:401–409. doi: 10.1002/mabi.200600255. [DOI] [PubMed] [Google Scholar]
- 7.Vollrath F, Knight DP. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 8.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. J. Exp. Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 9.Xu M, Lewis R. Proc. Natl. Acad. Sci. U.S.A. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinman M, Lewis RV. J. Biol. Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 11.Mita K, Ichimura S, James TC. J. Mol. Evol. 1994;38:583–592. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi CY, Shipley NH, Lewis RV. Int. J. Biol. Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 13.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 14.Garb JE, Dimauro T, Vo V, Hayashi CY. Science. 2006;312:1762. doi: 10.1126/science.1127946. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Barua B, Andersen NJ. Am. Chem. Soc. 2004;126:13679–13684. doi: 10.1021/ja047265o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palenčár P, Bleha TJ. Mol. Model. 2011 doi: 10.1007/s00894-011-0997-4. [DOI] [PubMed] [Google Scholar]
- 17.Fraser RDB, MacRae TP. Chapter 13. In Conformation in Fibrous Proteins. New York: Academic Press; 1973. [Google Scholar]
- 18.Gosline JM, Denny MW, DeMont ME. Nature. 1984;309:551–552. [Google Scholar]
- 19.Parkhe AD, Seeley SK, Gardner K, Thompson L, Lewis RV. J. Mol. Recognit. 1997;10:1–6. doi: 10.1002/(SICI)1099-1352(199701/02)10:1<1::AID-JMR338>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Simmons A, Ray E, Jelinski LW. Macromolecules. 1994;27:5235–5237. [Google Scholar]
- 21.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. J. Am. Chem. Soc. 2008;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 22.Rathore O, Sogah DY. J. Am. Chem. Soc. 2001;123:5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]
- 23.Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nakashima K, Takahashi T. Nature. 1997;387:563–564. doi: 10.1038/42391. [DOI] [PubMed] [Google Scholar]
- 24.Qin XX, Coyne KJ, Waite JH. J. Biol. Chem. 1997;272:32623–32627. doi: 10.1074/jbc.272.51.32623. [DOI] [PubMed] [Google Scholar]
- 25.Coyne KJ, Qin XX, Waite JH. Science. 1997;277:1830–1832. doi: 10.1126/science.277.5333.1830. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi CY, Lewis RV. J. Mol. Biol. 1998;275:773–784. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 27.Urry DW, Luan CH, Peng SQ. Ciba Found. Symp. 1995;192:4–22. doi: 10.1002/9780470514771.ch2. discussion 22–30. [DOI] [PubMed] [Google Scholar]
- 28.Chang DK, Venkatachalam CM, Prasad KU, Urry DW. J. Biomol. Struct. Dyn. 1989;6:851–858. doi: 10.1080/07391102.1989.10506517. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins J, Creager M, Butler E, Lewis R, Yarger J, Hol- land G. Chem. Commun. (Cambridge, U.K.) 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- 30.van Beek JD, Hess S, Vollrath F, Meier BH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lombardi S, Kaplan DJ. Arachnol. 1990:297–306. [Google Scholar]
- 32.Creager MS, Jenkins JE, Thagard-Yeaman LA, Brooks AE, Jones JA, Lewis RV, Holland GP, Yarger JL. Biomacro- molecules. 2010;11:2039–2043. doi: 10.1021/bm100399x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menassa R, Zhu H, Karatzas CN, Lazaris A, Richman A, Brandle J. Plant Biotechnol. J. 2004;2:431–438. doi: 10.1111/j.1467-7652.2004.00087.x. [DOI] [PubMed] [Google Scholar]
- 34.Xia X, Qian Z, Ki C, Park Y, Kaplan DL, Lee S. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14059–14063. doi: 10.1073/pnas.1003366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teulé F, Cooper AR, Furin WA, Bittencourt D, Rech EL, Brooks A, Lewis RV. Nat. Protoc. 2009;4:341–355. doi: 10.1038/nprot.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks AE, Stricker SM, Joshi SB, Kamerzell TJ, Middaugh CR, Lewis RV. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- 37.Earl WL, VanderHart DL. J. Magn. Reson. 1982;48:35–54. [Google Scholar]
- 38.Spera S, Bax AJ. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- 39.Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 40.Wishart DS, Sykes BD. J. Biomol. NMR. 1994:4. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 41.Wishart DS, Sykes BD. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 42.Holland GP, Lewis RV, Yarger JL. J. Am. Chem. Soc. 2004;126:5867–5872. doi: 10.1021/ja031930w. [DOI] [PubMed] [Google Scholar]
- 43.Holland GP, Jenkins JE, Creager M, Lewis RV, Yarger JL. Biomacromolecules. 2008;9:651–657. doi: 10.1021/bm700950u. [DOI] [PubMed] [Google Scholar]
- 44.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. J. Am. Chem. Soc. 2008;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 45.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Chem. Commun. 2008:5568–5570. doi: 10.1039/b812928b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins JE, Creager MS, Lewis RV, Holland GP, Yarger JL. Biomacromolecules. 2010;11:192–200. doi: 10.1021/bm9010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izdebski T, Akhenblit P, Jenkins JE, Yarger JL, Holland GP. Biomacromolecules. 2010;11:168–174. doi: 10.1021/bm901039e. [DOI] [PubMed] [Google Scholar]
- 48.Creager MS, Jenkins JE, Thagard-Yeaman LA, Brooks AE, Jones JA, Lewis RV, Holland GP, Yarger JL. Biomacro- molecules. 2010;11:2039–2043. doi: 10.1021/bm100399x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liivak O, Flores A, Lewis RV, Jelinski LW. Macromole- cules. 1997;30:7127–7130. [Google Scholar]
- 50.Sponner A, Schlott B, Vollrath F, Unger E, Grosse F, Weisshart K. Biochemistry. 2005;44:4727–4736. doi: 10.1021/bi047671k. [DOI] [PubMed] [Google Scholar]
- 51.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. PLoS ONE. 2007;2:e514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renugopalakrishnan V, Lewis RV. Bionanotechnology: Pro- teins to Nanodevices. 1st ed. Dordrecht, The Netherlands: Springer; 2006. [Google Scholar]
- 53.Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN. Science. 2002;295:472–6. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 54.Peakall DB. Nature. 1965;207:102–103. doi: 10.1038/207102a0. [DOI] [PubMed] [Google Scholar]
- 55.Sakuma Y, Rebenfeld LJ. Appl. Polym. Sci. 1966;10:637–652. [Google Scholar]
- 56.Yue CY, Sui GX, Looi HC. Compos. Sci. Technol. 2000;60:421–427. [Google Scholar]
- 57.Holland C, Terry AE, Porter D, Vollrath F. Polymer. 2007;48:3388–3392. [Google Scholar]
- 58.Zhou G, Shao Z, Knight DP, Yan J, Chen X. Adv. Mater. (Weinheim, Ger.) 2009;21:366–370. [Google Scholar]
- 59.Yin J, Chen E, Porter D, Shao Z. Biomacromolecules. 2010;11:2890–2895. doi: 10.1021/bm100643q. [DOI] [PubMed] [Google Scholar]
- 60.Work RW. Text. Res. J. 1977:650–662. [Google Scholar]
- 61.Guinea GV, Elices M, Perez-Rigueiro J, Plaza GR. J. Exp. Biol. 2005;208:25–30. doi: 10.1242/jeb.01344. [DOI] [PubMed] [Google Scholar]