Abstract
To determine if the ATP sulfurylase reaction is a regulatory step for the SO42−-assimilation pathway in plants, an Arabidopsis thaliana ATP sulfurylase cDNA, APS2, was fused to the 35S promoter of the cauliflower mosaic virus and introduced by Agrobacterium tumefaciens-mediated transformation into isolated Bright Yellow 2 tobacco (Nicotiana tabacum) cells. The ATP sulfurylase activity in transgenic cells was 8-fold that in control cells, and was correlated with the expression of a specific polypeptide revealed by western analysis using an anti-ATP sulfurylase antibody. The molecular mass of this polypeptide agreed with that for the overexpressed mature protein. ATP sulfurylase overexpression had no effect on [35S]SO42− influx or ATP sulfurylase activity regulation by S availability, except that ATP sulfurylase activity variations in response to S starvation in transgenic cells were 8 times higher than in the wild type. There were also no differences in cell growth or sensitivity to SeO42− (a toxic SO42− analog) between transgenic and wild-type cells. We propose that in Bright Yellow 2 tobacco cells, the ATP sulfurylase derepression by S deficiency may involve a posttranscriptional mechanism, and that the ATP sulfurylase abundance is not limiting for cell metabolism.
S is one of the major essential elements. It enters into the composition of the amino acids Met and Cys, and in a large variety of secondary metabolites, sulfolipids, sulfated glucides, and coenzymes (Mitchell, 1996). Plants and most of the bacteria and fungi are able to assimilate S from SO42−, whereas animals require organic S molecules as nutrients. Because of its low redox potential, SO42− is a relatively nonreactive form, and has to be activated prior to its reduction and incorporation into organic compounds (Leyh, 1993). The first step of the SO42−-activation process is catalyzed by ATP sulfurylase (ATP:sulfate adenylyl transferase, EC 2.7.7.4), which associates inorganic SO42− to ATP, resulting in the formation of APS and PPi. As this reaction is the first step in an energy-expensive sequence, ATP sulfurylase is considered to be an excellent candidate for the pathway-regulating, rate-limiting enzyme (Leustek, 1996).
APS is further phosphorylated by APS kinase using ATP, releasing PAPS. PAPS is considered to be a high-energy SO42− donor for sulfation of macromolecules in higher organisms, and can be reduced to SO32− in fungi (Thomas et al., 1992) and bacteria (Kredich, 1987). An alternative pathway has been identified recently in plants, in which SO32− formation appears to come from the reduction of APS rather than from PAPS (Gutierrez-Marcos et al., 1996; Setya et al., 1996; Hell, 1997).
In plants SO42− uptake and ATP sulfurylase activity are derepressed in response to S starvation, and both activities are repressed when SO42− availability is restored (Smith, 1975; Reuveny and Filner, 1977; Yildiz et al., 1994; Smith et al., 1995; Logan et al., 1996). To date, the regulation of SO42− uptake and ATP sulfurylase activity has been studied essentially in relation to SO42− availability as an S source (Hawkesford et al., 1993; Yildiz et al., 1996; Massonneau et al., 1997). Because SO42− activation has been considered to be a limiting step in the SO42− pathway, the reaction has been postulated to be one of the main regulatory steps of this pathway (Leustek, 1996). To address this question, we constitutively overexpressed an Arabidopsis thaliana cDNA encoding a putative chloroplastic ATP sulfurylase isoform (Logan et al., 1996) in BY2 tobacco (Nicotiana tabacum) cells. We used these cells to study the effects of three different S nutritional conditions on growth, SO42− influx, and SO42− accumulation: normal SO42− provision, S deficiency, and S nutrition bypassing the ATP sulfurylase step (using S2O32− as an S source). Comparison between wild-type cells and transformed cells indicated that the level of expression of ATP sulfurylase is not a limiting factor for growth. Our results suggest that the regulation of SO42− uptake and ATP sulfurylase activity is independent of the nature of the S source and of the abundance of the ATP sulfurylase protein.
MATERIALS AND METHODS
Culture Conditions of Isolated Tobacco Cells
BY2 tobacco (Nicotiana tabacum) cells were cultivated at 28°C in the dark on an orbital shaker at 120 rpm. The suspension was diluted to 1/50 every 7 d with a modified Murashige and Skoog medium (Nagata et al., 1992) devoid of SO42− (SO42− salts were replaced by their Cl− homologs). S was supplied as SO42− (1.5 mm K2SO4) or S2O32− (0.75 mm Na2S2O3). To achieve S-starvation conditions, 5-d-old cells were washed under sterile conditions with 3 volumes of S-less medium on a 150-μm polyamide filter (Tissages Tissus Techniques, Sailly-Saillissel, France) and grown in 1 volume of the same medium at 120 rpm for 24 h at 28°C in the dark. The composition of the solid medium for callus cultures was identical to the composition of liquid medium, except that 0.8% (w/v) agar-agar (Merck, Darmstadt, Germany) was added before sterilization. Drops (100 μL) of 7-d-old cell suspensions containing the same amount of fresh weight (40 mg) were deposited on solid medium in a Petri dish, wrapped with polyethylene film, and grown at 28°C in the dark.
Transformation of BY2 Tobacco Cells
The complete cDNA encoding APS2 (previously named ASA1), an Arabidopsis thaliana putative chloroplastic ATP sulfurylase isoform (Logan et al., 1996), was introduced between the cauliflower mosaic virus 35S promoter and the terminator of the nopaline synthase gene in place of the GUS gene of the binary vector PBI121 (Clontech, Palo Alto, CA). Agrobacterium tumefaciens LBA4404 was cultivated at 28°C into Luria-Bertani medium supplemented with 200 mg L−1 streptomycin and 150 mg L−1 rifampicin (Sigma). For transformation, an overnight 2-mL culture of A. tumefaciens was cooled in melting ice for 10 min, and then pelleted at 1500g for 5 min at 4°C. The pellet was resuspended into 100 μL of cold, sterile 20 mm CaCl2, and 2 μL of purifed plasmid suspension was added. The mixture was frozen in liquid N2, and then incubated at 37°C for 4 min. The cells were diluted into 1 mL of Luria-Bertani medium without antibiotics and allowed to recover at 200 rpm for 4 h at 28°C. The bacteria were pelleted and cultivated on solid Luria-Bertani medium supplemented with 30 mg L−1 kanamycin, 200 mg L−1 streptomycin, and 150 mg L−1 rifampicin.
A kanamycin-resistant A. tumefaciens clone was streaked on the same solid medium and an isolated colony was used for BY2 cell transformation following the procedure described by Shaul et al. (1996), except that we used 10 μg mL−1 kanamycin and 500 μg mL−1 cefotaxime (Roussel-Uclaf, Romainville, France) for primary selection. After the culture had reached the density of a 1-week-old culture, cells were diluted to 1/50 into fresh medium supplemented with 200 μg mL−1 kanamycin and 500 μg mL−1 cefotaxime. After five additional culture cycles in selective medium, the transformed cell line was considered to be established and was maintained in medium without antibiotics. All of the experiments described below were performed using medium without antibiotics.
ATP Sulfurylase Activity and Protein-Content Assay
Isolated tobacco cells corresponding to about 0.8 g fresh weight were washed with 0.2 mm CaCl2 by vacuum filtration through a 48-μm polyamide filter. The cells were frozen with liquid N2 and homogenized into a 100 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 20 mm DTT buffer at 4°C. Extracts were centrifuged at 12,000g for 15 min at 4°C, and the supernatant containing the soluble proteins was collected and stored on ice until use. The ATP sulfurylase activity of the protein extract was determined using the molybdolysis procedure described by Osslund et al. (1982). Protein content of the extract was quantified by the method of Schaffner and Weissmann (1973) using BSA as a standard.
SO42−-Content Assays
Isolated tobacco cells were washed as previously described, disrupted by incubation in 0.1 n HCl at 80°C for 30 min, and then kept at 4°C for 24 h. Total cellular SO42− was quantified by the BaCl2 turbidimetric procedure described by Tabatabai and Bremner (1970).
[35S]SO42−-Uptake Assay
The SO42−-uptake assay was performed on 5-mL aliquots of 6-d-old cultures corresponding to about 2 g fresh weight. Cell samples were deposited on a 150-μm polyamide filter, washed three times by gravity flow of 5 mL of a solution containing only the Murashige and Skoog major elements 2% (w/v) Suc and 0.2 mm K2SO4, and resuspended into the same medium. After preincubation at 120 rpm for 17 min at 28°C, 0.33 μCi of [35S]Na2SO4 (ICN Biochemicals) was added, and cells were incubated at 120 rpm for 5 min at 28°C. The radioactive medium was then removed by vacuum filtration through a 48-μm polyamide filter, and the cells were washed three times with 10 mL of ice-cold 0.2 mm CaSO4. The cell solutes were extracted by incubation into 5 mL of 0.1 n HCl for 1 h at room temperature. The radioactivity in 1 mL of the extract was quantified using a liquid-scintillation counter (Tri-Carb 2101 TR, Packard, Meriden, CT) in the presence of 3 mL of scintillation liquid (Ultima Gold, Packard).
Protein Electrophoresis and Immunoblotting
Soluble proteins were extracted as for ATP sulfurylase activity and were separated by SDS-PAGE, as described by Laemmli (1970). Electrotransfer on nitrocellulose membrane (Sartorius) was performed in a 0.22% (w/v) 3-[cyclohexylamino]-1-propanesulfonic acid and 10% (v/v) methanol buffer, and run at 35 V during 16 h. The membrane was washed into a PBS buffer containing 10 mm K2PO4, pH 7.5, 150 mm NaCl, and 0.5% (v/v) Tween for 1 h at 4°C. The membrane was then incubated in PBS buffer containing 5% (w/v) nonfat dry milk for 1 h at 22°C, and the milk excess was removed by several washes with PBS. Proteins were hybridized overnight at 22°C with a rabbit polyclonal antibody directed against the ATP sulfurylase isoform APS3 of A. thaliana. Identification of hybridized proteins was performed using an immunoblotting kit (Aurora, ICN Biochemicals) following the manufacturer's instructions.
Expression of the Results
Experiments were repeated two to three times on independent cultures of transformed and wild-type tobacco cell lines. Typical results obtained for each experiment are presented. Unless specified, ATP sulfurylase activity and protein and SO42− content data are the means of three extractions from the same culture. [35S]SO42−-uptake results are means of five extractions from the same culture. ses are calculated at the level of 0.05.
RESULTS
Effect of APS2 Overexpression on Cell Growth and ATP Sulfurylase Protein Abundance
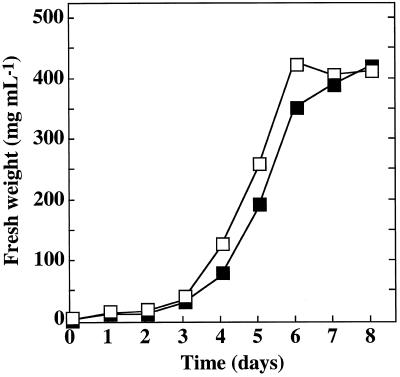
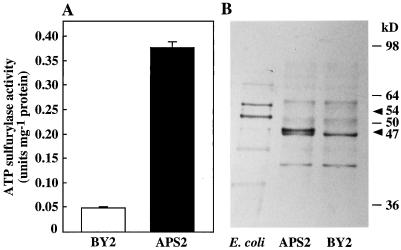
The growth kinetics and the fresh yield of the transformed APS2 cell line were identical to that of the untransformed BY2 cell line (Fig. 1). ATP sulfurylase-specific activity of 6-d-old cultures (late growing stage) of the transformed line was approximatively 8-fold that in nontransformed BY2 cells (Fig. 2A). Western analysis of protein extracts from the control BY2 line using the anti-APS3 antibody revealed a major band with a molecular mass of approximately 47 kD (Fig. 2B). In the APS2 line, this band was associated with a darker band with a molecular mass of between 47 and 50 kD. The latter band was presumably associated with the increased ATP sulfurylase activity. The same antibody recognized a 54-kD protein in the soluble protein extract of an Escherichia coli strain overexpressing APS2.
Figure 1.
Effect of ATP sulfurylase overexpression on BY2 tobacco cells grown in nonlimiting S conditions. Stationary-phase, 7-d-old cell suspensions were diluted to 1/50 with a 1.5 mm SO42−-containing medium at time 0. Each following day, culture aliquots were washed with 0.2 mm CaCl2 by vacuum filtration, and the cell fresh weight was determined. □, Nontransformed BY2 cells; ▪, transgenic APS2 cells overexpressing the APS2 cDNA.
Figure 2.
Characterization of tobacco cells overexpressing APS2. Tobacco cells were grown for 6 d on 1.5 mm SO42−-containing medium. ATP sulfurylase activity determination (A) and western analysis (B) were performed using the same soluble-protein extracts. For immunoblotting, 10 μg of soluble proteins was separated by denaturing electrophoresis on a 12% (w/v) polyacrylamide gel and hybridized with a rabbit anti-APS3 antibody. E. coli, Crude extract from an E. coli strain overexpressing the APS2 protein; BY2, extract from nontransformed tobacco cells; and APS2, extract from transgenic tobacco cells overexpressing the APS2 cDNA. Molecular mass standards are shown on the right (kD). ATP sulfurylase activities are the means of three extractions from the same culture. Arrowheads indicate ATP sulfurylase protein expressed in E. coli (54 kD) and in plants (47 kD). Bars are se at P = 0.05. Similar results were obtained from two independent experiments.
Effect of S Starvation on SO42− Uptake, SO42− Content, and ATP Sulfurylase Activity
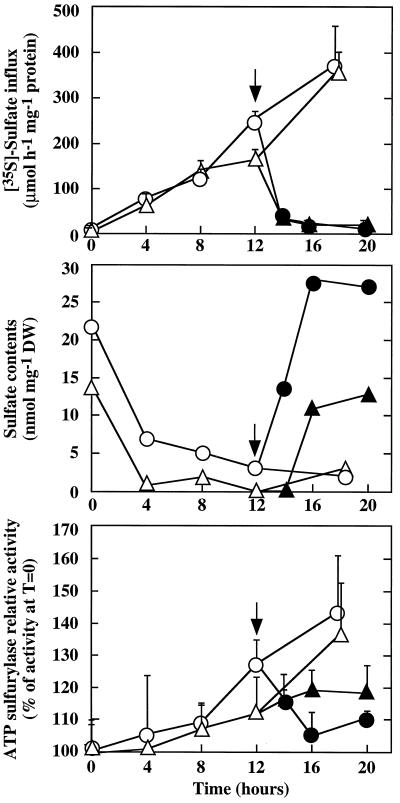
Five-day-old, nontransformed (control BY2 line) or transgenic (APS2 line) isolated tobacco cells cultivated in standard medium were washed and transferred into S-less but otherwise complete medium. SO42− uptake was derepressed progressively, correlating with a decrease of the cell SO42− content (Fig. 3). Derepression of the ATP sulfurylase activity began after 4 h of S starvation. SO42− uptake and ATP sulfurylase activity increased as long as the S deficiency was maintained. When SO42− was added to the medium, cell SO42− content increased immediately. SO42− uptake was fully repressed within 2 h, and ATP sulfurylase activity returned to its basal level within 4 h. Both BY2 control and transgenic APS2 lines reacted in similar ways.
Figure 3.
Effect of S availability on tobacco cells. Nontransformed BY2 cells (○ and •) and transgenic APS2 cells overexpressing the APS2 cDNA (▵ and ▴) were grown for 5 d in SO42− medium, then washed and cultivated in S-less medium (○ and ▵). At time 0, ATP sulfurylase activity was about 0.075 and 0.325 unit mg−1 protein, respectively, for BY2 and APS2 cells. After 12 h of treatment in S-starvation conditions, K2SO4 was added (arrow, •, and ▴) at a 1.5 mm final concentration. ATP sulfurylase activities are the means of three extractions from the same culture. [35S]SO42−-uptake results are the means of five extractions from the same culture. Bars are se at P = 0.05. Similar results were obtained from two independent experiments. DW, Dry weight.
Effect of S Source on SO42− Uptake, SO42− Content, and ATP Sulfurylase Activity
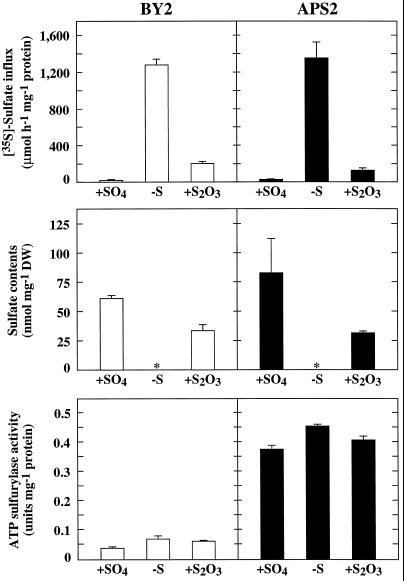
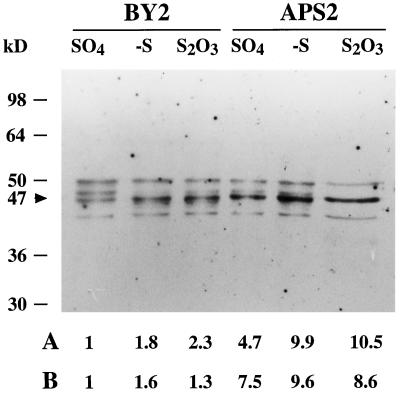
Nontransformed (control BY2 line) or transgenic (APS2 line) isolated tobacco cells were cultivated for 24 h in S-less medium or for 6 d with SO42− or S2O32− as the sole source of S. In SO42−-containing medium, SO42− uptake and ATP sulfurylase activity were repressed, and cells contained large amounts of SO42− (Fig. 4). After 24 h of S starvation, SO42− uptake and ATP sulfurylase activity were fully derepressed, in correlation with the disappearance of intracellular SO42−. In S2O3-containing medium, BY2 cells had a relatively high content of SO42−, SO42− uptake was partly derepressed, and ATP sulfurylase activity was fully derepressed. The ATP sulfurylase activity correlated with the relative abundance of the protein (Fig. 5). The APS2 line showed the same reactions as the BY2 cells to both S and SO42− deficiencies, except that the ATP sulfurylase activities were approximately 8-fold higher.
Figure 4.
SO42− uptake, SO42− contents, and ATP sulfurylase activity in tobacco cells grown in different S conditions. Nontransformed tobacco cells (BY2) and transgenic tobacco cells overexpressing the APS2 cDNA (APS2) were grown on 1.5 mm SO42− (+SO4) or 0.75 mm S2O32− (+S2O3) for 6 d, or without S (−S) for 24 h. Asterisk (*) indicates that SO42− was not detectable. ATP sulfurylase activities and SO42− contents are the means of three extractions from the same culture. [35S]SO42−-uptake results are the means of five extractions from the same culture. Bars are se at P = 0.05. Similar results were obtained from three independent experiments. DW, Dry weight.
Figure 5.
Western analysis of ATP sulfurylase in tobacco cells. Nontransformed tobacco cells (BY2) and transgenic tobacco cells overexpressing the APS2 cDNA (APS2) were grown on 1.5 mm SO42− or 0.75 mm S2O32− for 6 d, or without S (−S) for 24 h. Ten micrograms of soluble proteins was separated by denaturing electrophoresis on a 10% (w/v) polyacrylamide gel and immunoblotted with an anti-APS3 polyclonal antibody. To evaluate the ATP sulfurylase abundance, the intensity of each band (black arrowhead) was quantified using imaging software (NIH Image, National Institutes of Health, Bethesda, MD). Both the intensity of the bands and the ATP sulfurylase activity were expressed by dividing each of their values by the corresponding values of BY2 cells on SO42− medium. A, Relative intensity; B, relative activity. Molecular mass standards are shown on the left. Similar results were obtained from three independent experiments.
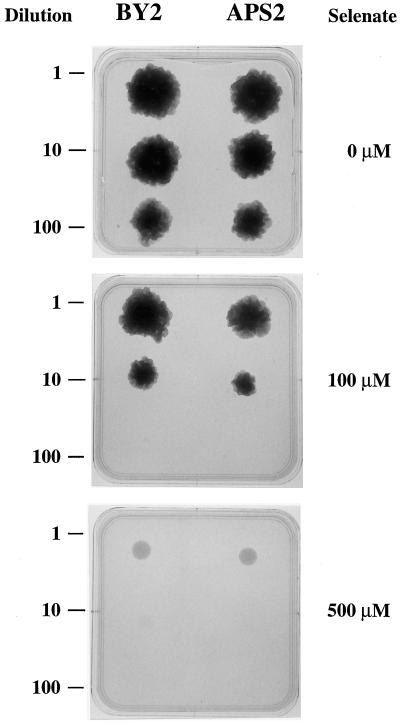
Effect of SeO42− on Callus Culture
Control BY2 and transgenic APS2 tobacco cells were grown as calli for 50 d on normal solid medium containing 0, 100, and 500 μm Na2SeO4 at three different initial dilutions for each of the cell cultures (Fig. 6). No growth differences could be noticed between the two cell lines.
Figure 6.
Effect of SeO42− on tobacco cell callus growth. Cells from 7-d-old cultures were diluted into liquid culture medium, and one drop (100 μL) of each dilution was deposited on solid culture medium containing 1.5 mm K2SO4 and 0, 100, or 500 μm Na2SeO4. Calli were grown at 28°C in the dark for 50 d. BY2, Nontransformed tobacco cells; APS2, transgenic tobacco cells overexpressing the APS2 protein. Similar results were obtained from two independent experiments.
DISCUSSION
The expression of the APS2 cDNA, an A. thaliana ATP sulfurylase, in transgenic tobacco cells was associated with an 8-fold increase in ATP sulfurylase activity (Fig. 2A) and with the appearance of a peptide that reacted with an antibody directed against the APS3 ATP sulfurylase (Fig. 2B). The relative molecular mass of this peptide (approximately 47 kD) was close to that predicted for the mature APS2 protein (Logan et al., 1996). Thus, we conclude that the transgenic cells contain increased amounts of ATP sulfurylase compared with the control cells.
After 18 h of S deprivation (Fig. 3), the relative increase of ATP sulfurylase activity was approximately the same (35–40%) in both cell lines, in spite of the 8-fold difference in the absolute value of enzymatic activities (Fig. 2A). Thus, the absolute increase in activity upon S starvation in APS2 cells was approximately 8-fold that in control cells, exceeding the activity in the latter cells in SO42−-less medium. This result implies that the transgenic ATP sulfurylase is involved in the increase in enzymatic activity upon S starvation in APS2 cells. The augmentation of the enzymatic activity upon S deficiency could be partly due to an increase in protein concentration, as suggested by the correlation between the relative enzymatic activity and immunostaining (Fig. 5). The response of the cauliflower mosaic virus 35S promoter used on the APS2 line to S-nutrition conditions is not known, but it seems unlikely that this promoter would respond to the S nutritional status in a way similar to the ATP sulfurylase promoter (Fig. 3). Therefore, the observed increase in ATP sulfurylase protein abundance in response to S starvation probably resulted from a posttranscriptional effect. An increase in the stability of the ATP sulfurylase in S-limiting conditions has been demonstrated in tobacco cells (Reuveny and Filner, 1977). Such a mechanism could be involved in the observed derepression in transgenic cells.
The kinetics of growth and SO42− uptake of the transgenic cells overexpressing the APS2 ATP sulfurylase are similar to those of wild-type cells in SO42−-containing medium, during S-starvation periods, or during return to normal nutrition (Figs. 1, 3, and 4). Since the biomass production rate is insensitive to the concentration of the ATP sulfurylase in the cells, we may infer that the cell multiplication in normal SO42− conditions is not limited by the ATP sulfurylase abundance. Several explanations for this situation can be imagined. Cell growth should not be limited by S metabolism by itself in our culture conditions. The enzyme might already be present in excess in wild-type cells, or its activity might be limited by SO42− or ATP availability. In plants the cytoplasmic concentrations of SO42− and ATP are estimated at approximately 10 and 2 mm, respectively (Cram, 1983; Roby et al., 1987). Since the Km for SO42− and ATP of the ATP sulfurylase are about 0.87 to 0.25 mm and 0.31 to 0.046 mm, respectively (Osslund et al., 1982; Renosto et al., 1993), it is unlikely that ATP sulfurylase would be limited by the availability of its substrates.
Another explanation of the insensitivity of growth to ATP sulfurylase level would be that the activity of this enzyme is strictly regulated by its product, or by those of the downstream steps of the pathway. Potent retro-inhibition of ATP sulfurylase by micromolar-range concentrations of APS is well known in fungi and higher plants (Osslund et al., 1982; Renosto et al., 1993; Foster et al., 1994). Furthermore, the ATP sulfurylase reaction toward the formation of APS is thermodynamically not favored, and is thought to depend upon the removal of the reaction products PPi and APS by pyrophosphatases and APS reductase/APS kinase, respectively (Leyh, 1993).
SeO42− can be used as a substrate by ATP sulfurylase, which results in toxic overproduction and misincorporation of selenocysteine into the proteins in place of Cys (Wilson and Bandurski, 1958; Reuveny, 1977; Cherest et al., 1997). Sensitivity to SeO42− was identical in transgenic and control tobacco cells (Fig. 6). We conclude that Se assimilation was not enhanced by the overexpression of a functional ATP sulfurylase. To investigate whether a difference between the two lines would be detected when increasing the demand for reduced S compounds such as the Cys-rich phytochelatins, which are involved in heavy-metal detoxication (Rauser, 1990; Steffens, 1990), or Met, which has been shown to have a protective effect against NaCl stress (Gläser et al., 1993; Kwon et al., 1995), calli were grown, respectively, on Cd (0, 50, and 100 μm CdCl2)- and Na (0, 100, and 500 mm NaCl)-containing solid medium. Results were very similar to those presented in Figure 6, and no difference in the sensitivity to these two compounds was noticed between the nontransformed and the transgenic cell lines (data not shown). These results would not have been expected if the ATP sulfurylase abundance was limiting to the SO42−-assimilation pathway in the nontransformed cell line.
SO42− uptake is thought to be repressed by SO42− accumulated in the cytoplasm (Smith, 1975, 1980). In both transgenic and wild-type cells, SO42− influx changed in a manner opposite to that of the whole-cell SO42− content because the latter was manipulated by various treatments (Figs. 3 and 4). We used S2O32−, a good S source for BY2 tobacco cell growth, to bypass the SO42−-activation step without causing S starvation. In yeast, S2O32− uptake is mediated presumably by the SO42−-transport system (Alonso et al., 1984). The molecule is then split into SO32− and S2−, which enter the S-assimilation pathway at the SO32− reduction and S2− incorporation steps, respectively (Thomas et al., 1992). The similar behavior of the two cell lines suggests that the level of ATP sulfurylase did not affect the level of its substrate, cytosolic SO42−. Furthermore, the SO42− level in S2O32−-grown cells was the same in both cell lines, indicating that the abundance of ATP sulfurylase did not control the equilibrium between production of SO42− (via S2O32− oxidation) and SO42− assimilation. These results indicate that in BY2 tobacco cells the amount of SO42− in the whole cell (and probably the amount of SO42− in the cytosol) is not dependent on the level of ATP sulfurylase activity.
In conclusion, the ATP sulfurylase abundance does not seem to regulate SO42− acquisition or SO42− use for growth in tobacco cells. These results suggest that this enzyme is under strict control by some products of its activity or of downstream steps of the SO42−-assimilation pathway, either by retro-inhibition or by mass-action law effects.
ACKNOWLEDGMENTS
The authors are thankful to Dr. Thomas Leustek (Rutgers University, New Brunswick, NJ) for the gift of the anti-APS3 antibodies. We also wish to thank Drs. Bruno Touraine and Anne Lappartient (Institut National de la Recherche Agronomique, Centre National de la Recherche Scientifique, Montpellier, France) for stimulating discussions about this work.
Abbreviations:
- APS
adenosine 5′-phosphosulfate
- BY2
Bright Yellow 2
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
Footnotes
This work was supported by the Institut National de la Recherche Agronomique, the Centre National de la Recherche Scientifique, and the Ecole Nationale Supérieure Agronomique de Montpellier. Y.H. was supported by a doctoral grant (no. 94123) from the Direction Générale de l'Enseignement et de la Recherche, Ministère de l'Agriculture.
LITERATURE CITED
- Alonso A, Benitez J, Diaz MA. A sulfate, sulfite and thiosulfate incorporating system in Candida utilis. Folia Microbiol. 1984;29:8–13. doi: 10.1007/BF02875902. [DOI] [PubMed] [Google Scholar]
- Cherest H, Davidian J-C, Thomas D, Benes V, Ansorge W, Surdin-Kerjan Y. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics. 1997;145:627–635. doi: 10.1093/genetics/145.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram JW. Characteristics of sulfate transport across plasmalemma and tonoplast of carrot root cells. Plant Physiol. 1983;72:204–211. doi: 10.1104/pp.72.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BA, Thomas SM, Mahr JA, Renosto F, Patel HC, Segel IH. Cloning and sequencing of ATP sulfurylase from Penicillium chrysogenum. Identification of a likely allosteric domain. J Biol Chem. 1994;269:19777–19786. [PubMed] [Google Scholar]
- Gläser K-U, Thomas D, Gaxiola R, Montrichard F, Surdin-Kerjan Y, Serrano R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993;12:3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford MJ, Davidian J-C, Grignon C. Sulphate/proton cotransport in plasma-membrane vesicles isolated from roots of Brassica napus L.: increased transport in membranes isolated from sulphur-starved plants. Planta. 1993;190:297–304. [Google Scholar]
- Hell R. Molecular physiology of plant sulfur nutrition. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- Kredich NM (1987) Biosynthesis of cysteine. In FC Neidhardt, JL Ingram, KB Low, B Magasanik, M Schaechter, HE Umbarger, eds, E. coli and S. typhimurium: Cellular and Molecular Biology. American Society of Microbiology, Washington, DC, pp 419–428
- Kwon T, Abe T, Sasahara T. Enhanced saline stress resistance in threonine and methionine overproducing mutant cell line from protoplast culture of rice (Oriza sativa) J Plant Physiol. 1995;145:551–556. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leustek T. Molecular genetics of sulfate activation in plants. Physiol Plant. 1996;97:411–419. [Google Scholar]
- Leyh TS. The physical biochemistry and molecular genetics of sulfate activation. Crit Rev Biochem Mol Biol. 1993;28:515–542. doi: 10.3109/10409239309085137. [DOI] [PubMed] [Google Scholar]
- Logan HM, Cathala N, Grignon C, Davidian J-C. Cloning of a cDNA encoded by a member of the Arabidopsis thaliana ATP sulfurylase multigene family: expression studies in yeast and in relation to plant sulfur nutrition. J Biol Chem. 1996;271:12227–12233. doi: 10.1074/jbc.271.21.12227. [DOI] [PubMed] [Google Scholar]
- Massonneau A, Cathala N, Grignon C, Davidian J-C. Effect of sulphate deficiency on the plasma membrane polypeptide composition of Brassica napus. J Exp Bot. 1997;306:93–100. [Google Scholar]
- Mitchell S (1996) Biological interactions of sulfur compounds. Taylor and Francis, London, UK
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cyt. 1992;132:1–30. [Google Scholar]
- Osslund T, Chandler C, Segel IH. ATP sulfurylase from higher plants. Purification and preliminary kynetic studies on the cabbage leaf enzyme. Plant Physiol. 1982;70:39–45. doi: 10.1104/pp.70.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauser WE. Phytochelatins. Annu Rev Biochem. 1990;59:61–86. doi: 10.1146/annurev.bi.59.070190.000425. [DOI] [PubMed] [Google Scholar]
- Renosto F, Patel HC, Martin HL, Thomassian C, Zimmerman G, Segel IH. ATP sulfurylase from higher plants: kinetic and structural characterization of the chloroplast and cytosol enzymes from spinach leaf. Arch Biochem Biophys. 1993;307:272–285. doi: 10.1006/abbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- Reuveny Z. Derepression of ATP sulfurylase by the sulfate analogs molybdate and selenate in cultured tobacco cells. Proc Natl Acad Sci USA. 1977;74:619–622. doi: 10.1073/pnas.74.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveny Z, Filner P. Regulation of adenosine triphosphate sulfurylase in cultured tobacco cells. J Biol Chem. 1977;252:1858–1864. [PubMed] [Google Scholar]
- Roby C, Martin J-B, Bligny R, Douce R. Biochemical changes during sucrose deprivation in higher plant: phosphorus31 nuclear magnetic resonance studies. J Biol Chem. 1987;262:5000. 5007. [PubMed] [Google Scholar]
- Schaffner W, Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Setya A, Murillo M, Leustek T. Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O, Mironov V, Burssens S, Van Montagu M, Inzé D. Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc Natl Acad Sci USA. 1996;93:4868–4872. doi: 10.1073/pnas.93.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Sulfate transport in tobacco cells. Plant Physiol. 1975;55:303–307. doi: 10.1104/pp.55.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Regulation of sulfate assimilation in tobacco cells. Plant Physiol. 1980;66:877–883. doi: 10.1104/pp.66.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens JC. The heavy-metal binding peptides of plants. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:553–575. [Google Scholar]
- Tabatabai MA, Bremner JM. A simple turbidimetric method of determining total sulfur in plant materials. Agron J. 1970;62:805–806. [Google Scholar]
- Thomas D, Barbey R, Henry D, Surdin-Kerjan Y. Physiological analysis of mutants of Saccharomyces cerevisiae impaired in sulphate assimilation. J Gen Microbiol. 1992;138:2021–2028. doi: 10.1099/00221287-138-10-2021. [DOI] [PubMed] [Google Scholar]
- Wilson LG, Bandurski RS. Enzymatic reactions involving sulfate, sulfite, selenate and molybdate. J Biol Chem. 1958;233:975–981. [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 1994;104:981–987. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. Sulfur availability and the SAC1 gene control adenosine triphosphate sulfurylase gene expression in Chlamydomonas reinhardtii. Plant Physiol. 1996;112:669–675. doi: 10.1104/pp.112.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]