Abstract
Autoimmune diseases are common and debilitating, but their severe manifestations could be reduced if biomarkers were available to allow individual tailoring of the potentially toxic immunosuppressive therapy required for their control. Gene expression-based biomarkers facilitating individual tailoring of chemotherapy in cancer, but not autoimmunity, have been identified and translated into clinical practice1,2. We show that transcriptional profiling of purified CD8 T cells, which avoids the confounding influences of unseparated cells3,4, identifies two distinct patient subgroups predicting long-term prognosis in two different autoimmune diseases, anti-neutrophil cytoplasmic antibody (ANCA) – associated vasculitis (AAV), a chronic, severe disease characterized by inflammation of medium and small blood vessels5, and systemic lupus erythematosus (SLE), characterized by autoantibodies, immune complex deposition and diverse clinical manifestations ranging from glomerulonephritis to neurological dysfunction6. We show that genes defining the poor prognostic group are enriched for genes of the IL7R pathway, TCR signalling and those expressed by memory T cells. Furthermore, the poor prognostic group is associated with an expanded CD8 T cell memory population. These subgroups, which are also found in the normal population and can be identified by measuring expression of only three genes, raise the prospect of individualized therapy and suggest novel potential therapeutic targets in autoimmunity.
The three clinical syndromes that make up AAV (Wegener's granulomatosis [WG], microscopic polyangiitis [MPA] and Churg-Strauss syndrome) have diverse and overlapping clinical features, including acute glomerulonephritis and granulomatous inflammation of the respiratory tract5. Mortality at five years is as high as 30%, largely owing to complications of immunosuppression7 and underlining the need for biomarkers to guide therapy. ANCA are directed against neutrophil antigens, including proteinase-3 [PR-3] and myeloperoxidase [MPO], are likely to contribute to pathogenesis8, but have not proven clinically useful as predictive biomarkers9. To detect such biomarkers we examined the transcriptomes of purified cells from 59 patients with a diagnosis of AAV (WG or MPA5) and evidence of active disease (Supplementary Methods). All responded to treatment with high dose steroid and one of a number of induction immunosuppression therapies, before maintenance therapy was commenced. Detailed clinical and laboratory information was collected prospectively for up to 1000 days (Supplementary Methods and Supplementary Tables 1 and 2). An initial cohort of 32 patients was analysed first and the subsequent 27 patients were analysed separately as a validation cohort. At enrolment, and before treatment, we purified CD4 and CD8 T cells, B cells, neutrophils and monocytes from each patient10, quantifying gene expression profiles using a genome-wide microarray platform (Mediante)11. Preliminary analysis identified distinct patterns of gene expression in each cell type, but CD8 T cell expression profiles were found to divide patients into two distinct subgroups, which prompted more intensive investigation. Abnormalities in CD8 T cells have been associated with AAV12 and they are a prominent component of vasculitic lesions, consistent with a role in pathogenesis13.
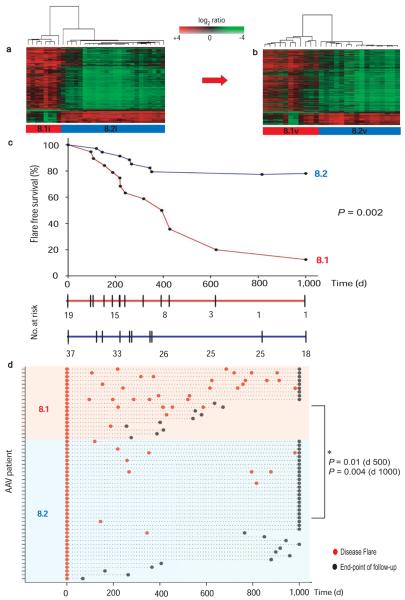
We assessed the degree of heterogeneity within the CD8 T cell gene expression dataset in the initial cohort of 32 AAV patients using multi-dimensional scaling with a global test of clustering (Supplementary Methods) and demonstrated greater “substructure” than would be expected to occur in a randomly distributed dataset (p<0.0001, Supplementary Fig. 1a). Two discrete patient subgroups best explained this substructure (Supplementary Fig. 1b, c), which was confirmed using 2 independent clustering techniques (k-means and hierarchical) and 2 additional, independent algorithms allowing both comparison between techniques (EP:NG Clustercomparison14, Supplementary Fig. 1d,e) and across multiple iterations of one technique (ConsensusCluster15, Supplementary Fig. 1b). The genes whose expression best defined the 2 subgroups (termed v8.1 and v8.2) were identified using one-way analysis of variance (ANOVA; Supplementary Table 3) and were subject to hierarchical clustering to illustrate the substructure found (Fig. 1a).
Figure 1. T cell gene expression identifies a previously unrecognized subgroup of AAV patients at increased risk of relapsing disease.
(a) Supervised hierarchical clustering using expression data from 925 genes > 2-fold significantly differentially expressed between the subgroups defined in Fig. S1 (false discovery rate (FDR) p<0.05) clusters the initial cohort of AAV patients (n = 32) into two distinct subgroups. (b) Hierarchical clustering of an independent validation cohort of AAV patients (n = 27) using expression data for the same gene list defined in (a) also identifies 2 distinct subgroups. Hierarchical clustering was performed using uncentered correlation and average-linkage. (c) Survival curve showing shorter time to first flare in v8.1 following induction therapy. Flare-free survival represented as proportion of all individuals reaching each timepoint with the numbers remaining at risk within each cohort detailed below. Significance was measured using the log-rank test. (d) Increased flare frequency in v8.1 when followed out to 1,000 days. Flare rate normalized to duration of follow-up performed at day 500 (mean flare rate 0.91/year v8.1 vs 0.17/year v8.2, p=0.01) and at day 1,000 (mean flare rate 0.81/year v8.1 vs 0.15/year v8.2, p=0.004).
We then verified the existence of these 2 subgroups in a validation cohort, which was clustered using the gene list defined in the initial cohort, identifying two discrete subpopulations which were exactly reproduced by de novo analysis of the validation cohort (Supplementary Fig. 1d, e and 2). The gene list that best differentiated the validation cohort subgroups perfectly re-clustered the initial cohort, indicating the gene lists defining the subgroups in both cohorts were highly overlapping (hypergeometric p < 1 × 10−300, Supplementary Fig. 2).
We next asked whether these expression-defined subgroups correlated with disease phenotype. There was no correlation between either v8.1 or v8.2 and technical, clinical or laboratory parameters such as disease activity score at enrolment or erythrocyte sedimentation rate (ESR), or prior or subsequent immunosuppressant therapy (Supplementary Tables 1 and 2). However, two clinical associations were found; more patients in group 8.1 relapsed after initial remission (Fig. 1c) and they had more disease flares per month of follow-up than 8.2 (> 1 relapse in 47.4% of 8.1 and 5.4% of 8.2; Fig. 1d). Thus, these hypothesis-free, unsupervised analyses allow prospective prediction of patients with relapsing disease.
The association between disease relapse and subgroups 8.1 and 8.2 was confirmed when the initial and validation cohorts were analysed independently (Supplementary Fig. 3). In patients who had experienced disease flares prior to enrolment a worse outcome was seen, as expected, but the predictive power of the gene expression signature was preserved (Supplementary Fig. 4a, b). Anti-PR3 ANCA, the best current predictor of prognosis, did not predict outcome in our cohort (Supplementary Fig. 4c), consistent with previous reports9.
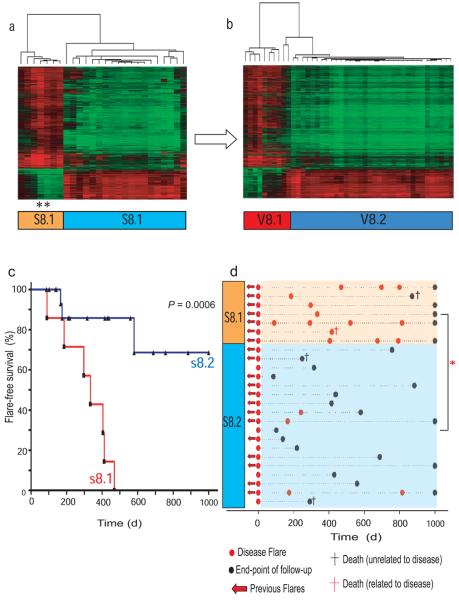
To determine if the prognosis-predicting signature was seen in other autoimmune diseases, we arrayed purified CD8 T cells from 26 patients with SLE, in which T cells are thought to be important, for example readily generating cytotoxic effector cells in active disease16. In SLE as in AAV, there is substantial disease and treatment-related morbidity and mortality17 but no clinically useful biomarkers have been identified18. All patients had active disease on enrolment and had either no previous evidence of disease or had had quiescent disease on minimal maintenance therapy (defined in Supplementary Methods). All then responded to induction immunosuppressive therapy, before weaning on to maintenance therapy (Supplementary Tables 4 and 5). Samples were processed and data analysed in an identical fashion to that described for AAV, though as a single cohort. Multidimensional scaling and consensus clustering again showed evidence of gene expression substructure (Supplementary Fig. 5a) best explained by the presence of 2 discrete patient subgroups (s8.1 and s8.2; Supplementary Figs. 5b - d). Genes defining the subgroups were identified using ANOVA, and hierarchical clustering clearly demonstrated the two groups (Fig. 2a). The gene list best defining the SLE subgroups was then used to cluster the 59 AAV patients, precisely recreating the AAV groups v8.1 and v8.2 (Fig. 2b). The converse analysis recreated the SLE groups (Supplementary Fig. 6), indicating a striking similarity in the CD8 T cell transcriptional changes defining the patient subgroups in these two distinct autoimmune diseases (hypergeometric p < 1×10−300).
Figure 2. The CD8 T cell signature that predicts prognosis in AAV defines analogous subgroups in SLE.
Consensus subgroups in a cohort of 26 SLE patients were defined by unsupervised hierarchical clustering. (a) Supervised hierarchical clustering of the SLE cohort using 1,913 genes that best define the subgroups (>2-fold statistically significant differential expression, p<0.05, FDR<0.05). One patient (black asterisk) underwent repeat analysis at the time of a second disease flare 14 months after enrolment and remained in subgroup s8.1 despite alterations in therapy. (b) Hierarchical clustering of AAV samples using expression data for the same genes as in panel (a) reproduces the same subgroups as found with de novo unsupervized clustering of the AAV dataset. Genelists defining the 2 subgroups in vasculitis (n = 1,228) and SLE (1,913) cohorts were highly overlapping (overlap = 639 genes, hypergeometric p-value <1 × 10−300). (c) Kaplan-Meier plot showing shorter time to first flare in s8.1. Significance was measured using the log-rank test. (d) Increased flare frequency in subgroup s8.2 when followed out to 1,000 days. Red asterisk = mean flare-rate normalized per unit time follow-up with Mann Whitney U test of significance performed at day 500 (flare-rate 0.86/year v 0.08/year, p = 0.0006) and at day 1,000 (flare-rate 0.81/year vs 0.09/year, p = 0.001).
As for AAV, there was a clear correlation between SLE group s8.1 and relapsing disease. All SLE group s8.1 patients had flared by 500 days after enrolment, compared to 15% of 8.2 patients (Fig. 2c), and s8.1 patients had more flares per month of follow-up than s8.2 (p=0.002, Fig. 2d). No significant association with other clinical or laboratory parameters was seen, though patients with pre-existing disease in Group 8.1 had had more immunosuppression before enrolment (Supplementary Table 5). This would be expected in a group with an increased tendency to disease relapse, and suggests that it is not a relative lack of prior therapy that has resulted in the increased flare frequency in s8.1. Induction and maintenance therapies were similar in s8.1 and s8.2 with the exception that the anti-CD20 therapy Rituximab was more frequently required in s8.1 (Supplementary Table 5). As our practice was to limit Rituximab use to refractory cases this is consistent with the poor outcome seen in this subgroup.
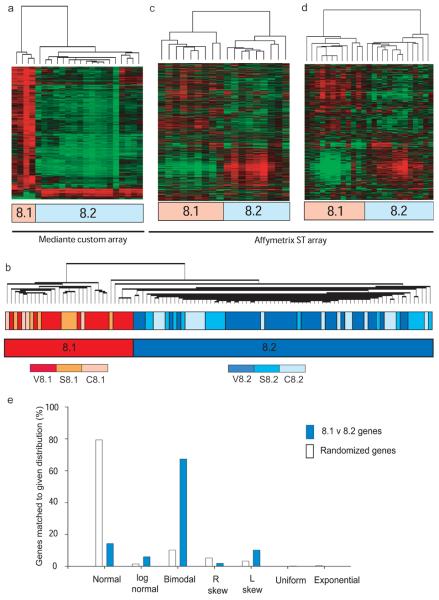
As two subgroups had been identified in two different diseases, we asked whether such heterogeneity might also exist within a population of 22 age and sex matched healthy controls analysed in an identical fashion to AAV and SLE (Supplementary Table 6). Unsupervised analysis identified 2 subgroups (c8.1 and c8.2, Fig.3a) defined by a gene list which demonstrated highly significant overlap with that defining the prognostic subgroups in both AAV and SLE (hypergeometric p<1×10−300). Genes defining the 2 subgroups in healthy individuals could be used to accurately reproduce the original subdivisions seen in both diseases (Supplementary Fig.7). When all CD8 T cell data (AAV, SLE and Control) were pooled and unsupervised hierarchical clustering performed 2 groups were again generated, divided not by diagnosis but by subgroups 8.1 and 8.2 (Fig. 3b). These findings held true independent of the array platform used, being confirmed using the Affymetrix Gene 1.0 ST array in a sample of the initial cohorts (Supplementary Fig. 8a-d), and in independent Caucasian and Singaporean control cohorts (Supplementary Table 6, Fig. 3c, d)
Figure 3. Similar subgroups can be identified in a healthy population, and the defining signature is composed of genes whose expression predominantly conforms to a bimodal distribution.
Consensus subgroups in a cohort of 22 healthy Caucasian controls, age and sex-matched to the AAV cohort, were defined by unsupervised hierarchical clustering as for the disease cohorts. (a) The 944 genes best defining these subgroups (>2-fold statistically significant differential expression, FDR p<0.05) were subject to hierarchical clustering. (b) Unsupervised hierarchical clustering of a pooled CD8 T cell expression dataset including all samples (AAV, SLE and control) reproduces subgroups 8.1 and 8.2, but shows no clustering by disease status. (c, d) CD8 T cell RNA from a cohort of 18 healthy Caucasian (c) and 25 Singaporean (d) controls was hybridised to the Affymetrix Gene 1.0 ST Array. Each cohort was clustered using data derived from 780 genes which represent the subset of the genes used in panel (a) that could be mapped onto Affymetrix feature names using common Entrez IDs. Unsupervised de novo clustering produced the same subgroups (data not shown). (e) Matched expression density distributions for genes differentially-expressed (FDR<0.05, fold-change 1.5) between patients in two randomly assigned subgroups of the AAV dataset (n=392, open bars) or subgroups 8.1 and 8.2 (n=1860, blue bars) confirming significant deviation of expression pattern from normal to bimodality (chi-square p<0.001 for bimodal versus other).
It was felt likely that the expansion of genes characterising such robust and clearly defined groups would be defined by distinct bimodal profiles rather than the extreme ends of a Gaussian distribution. When this was tested (for details see Supplementary Discussion, Supplementary Fig. 8e and Table 7) expression of most CD8 T cell genes expressed in AAV was normally distributed across the population, while those defining groups 8.1 and 8.2 predominantly fitted a bimodal distribution template, demonstrating a significant skewing away from the Gaussian distribution (p < 0.0001, Fig. 3e). The gene expression signature predicting prognosis could not be reproduced in patients during remission after initial therapy – see Supplementary Discussion.
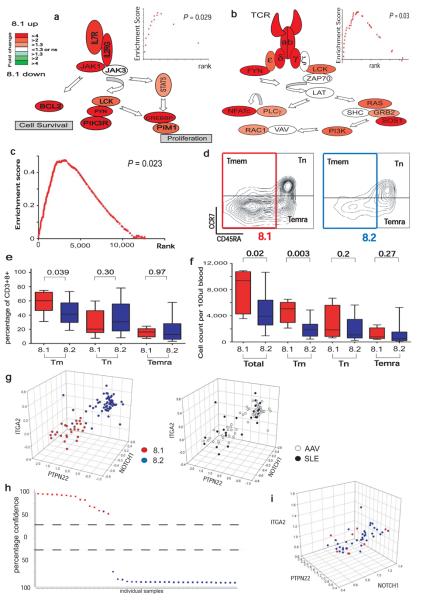
We then sought to determine if these transcription signatures were associated with biological differences that might explain their ability to predict relapsing disease. We first looked for differential expression of genes from known signalling pathways using two independently-curated pathway databases (MSigDB and Ingenuity) and two enrichment algorithms, GSEA19 and hypergeometric probability (Ingenuity20, Supplementary Methods). In v8.1 genes associated with signalling through both the T cell receptor (TCR) and the interleukin 7 receptor (IL7R) pathway were significantly upregulated (Fig. 4a, b). No enrichment was seen for other signalling pathways such as interleukin 4 (Supplementary Fig. 9a). The expression of genes differentiating subgroups 8.1 and 8.2 was validated by quantitative PCR (e.g. Supplementary Fig. 9b, c).
Figure 4. Poor prognosis in AAV correlates with over expression of mRNAs encoding proteins associated with T cell survival and memory.
(a, b) Magnitude and direction of differential gene expression between subgroups v8.1 and v8.2 for genes of (a) IL7R pathway (b) TCR signalling pathway. FDR p<0.05 and fold-change as illustrated by colour heatmap. Significance of enrichment was tested for the IL7R pathway (GSEA p=0.029, inset), Jak/Stat signalling pathway (Ingenuity, p<0.01, FDR<0.05, data not shown) and TCR signalling pathway (GSEA p=0.03, Ingenuity hypergeometric FDR p<0.05). (c) GSEA identified significant enrichment of a CD8 TEM signature in v8.1, p=0.023. (d) Memory T cell subsets of representative individuals from v8.1 (left panel) and v8.2 (right panel). Cells gated on CD3+ CD8+ showing naïve (TN, CD45RA+ CCR7+), memory (TMEM, CD45RA−) and effector memory RA (TEMRA, CCR7− CD45RA+) populations as indicated. (e, f) Boxplots showing relative proportions (e) and absolute numbers (f) of total, memory, naïve and effector memory-RA CD8 T cells in the two subgroups. (g) 3D scatterplots showing the classification of combined SLE and AAV samples using CD8 expression data from 3 genes (ITGA2, NOTCH1, PTPN22) separating patients by prognostic subgroups in both diseases (left panel) with no differentiation between diseases (right panel). mRNA expression on x, y and z axes as log2 ratio. (h) A predictive model was derived using a support vector machines algorithm on a randomized, stratified 50% subset of all samples (training set) and its performance assessed on the remaining 50% (test set), shown in (h), PPV=94%, NPV=100%. y-axis = confidence of prediction (%). (i) 3D scatterplot of the AAV cohort by the same 3-gene classifer using PBMC-derived gene expression data.
Signalling through IL7R (CD127) is vital for promoting T cell survival21, likely to be mediated by Bcl2 family-mediated inhibition of the pro-apoptotic effects of Bim22, and is critical for the expansion of effector and memory populations of CD8 T cells23. Other genes up-regulated in v8.1, including BCL2, CD27, SELL, CCR7 and CD44, are also associated with T cell memory24 and GSEA analysis using publicly available array data25 demonstrated enrichment of a human effector-memory (TEM) expression signature in subgroup v8.1 (Fig. 4c), and the same TEM signature was also associated with the 8.1 subgroup in both SLE patients and normal controls (Supplementary Fig. 9d, e). There was no enrichment of an activated T cell signature26 nor of genes known to alter expression in response to corticosteroid therapy27 (Supplementary Fig. 9f, g).
To determine if the TEM-associated signature was reflected in altered cell populations, 29 patients in remission on maintenance therapy (n = 7 v8.1; n = 22 v8.2) were re-bled between 11 and 42 months after enrolment, and their lymphocytes phenotyped. While the relationship between these results and the situation that existed at enrolment must be somewhat speculative, a clear difference in CD8 T cell populations was seen. Patients from v8.1 had an increased number of T cells in general, but of memory T cells in particular (TMEM, defined as CD3+CD8+CD45RA−; Fig. 4d-f). Most of these cells were CCR7 low, consistent with a T effector memory (or perhaps T-memory precursor effector) cell phenotype24, but the “central memory” (CCR7+ CD45RA−) cells described by Sallusto et al.28 were also significantly increased in v8.1, though not always seen as a discrete population. Correlation between TMEM numbers and the 8.1 and 8.2 subsets determined from the same sample was confirmed in 33 normal controls (Supplementary Fig. 10a-c). Attempts to classify patients into the two subgroups using only flow cytometric quantification of TMEM populations failed; TMEM populations differed between 8.1 and 8.2 but overlapped (Fig. 4e, f and Supplementary Fig. 10 b, c). Consistent with this, TN genes were also differentially expressed between 8.1 and 8.2, and TMEM genes alone cannot explain the subgroups (see Supplementary Text and Figure 10d-i).
These results taken together allow formulation of a speculative model which might explain correlation of the CD8 expression signature with clinical disease. Altered gene expression in TN cells may be imposed by germline genetic variation29, at the level of the thymus, by antigen exposure30 or by interactions with other cells31. This might in turn enhance CD8 T cell proliferation in response to antigen, at least in part via IL7R and TCR signalling32, driving the formation of an expanded memory population33 thus contributing to the TMEM component of the 8.1 signature. In the context of a response to autoantigen, such persistent autoreactive cells could have the capacity to more easily expand on restimulation, differentiating into the effector cells which drive tissue damage, manifesting clinically as a relapse of disease (Supplementary Fig. 11 and Supplementary Discussion).
To facilitate clinical translation of these prognostic expression signatures, we established that robust discrimination between 8.1 and 8.2 could be achieved using combinations of as few as 3 genes (Fig 4g, h, Supplementary Fig. 12, Supplementary Methods). These classifiers did not discriminate between diseases (Fig.4 g), and could not be replicated using RNA from un-separated PBMC (Fig. 4i).
Use of the predictive model as a prognostic biomarker may allow individualization of therapy in AAV and SLE: patients in group 8.1 may benefit from intensified maintenance therapy and follow-up, while the patients in 8.2 may need less maintenance therapy, reducing treatment-associated toxicity. A prospective clinical study will be needed to test this, but the difference in clinical outcome between 8.1 and 8.2 is at least as large as that described for clinically successful biomarkers in oncology34-36. The subgroups may also identify patient groups with differential responses to novel therapies, and may focus attention on CD8 TMEM generation as a mechanism driving disease severity in at least a subset of AAV and SLE patients. The discovery of this signature in two distinct autoimmune diseases and in a healthy population suggests that it may identify important prognostic subsets of patients not only in autoimmune diseases but also in related clinical contexts such as vaccination, infection and transplantation, and could thus help guide therapy in a range of clinical situations.
METHODS
Patients
59 AAV patients and 25 SLE patients attending or referred to the specialist vasculitis unit at Addenbrooke's hospital, Cambridge, UK between July 2004 and May 2008 were enrolled into the present study. 34 patients presenting with active disease between July 2004 and Oct 2006 composed a prospectively-defined initial vasculitis cohort, while a further 27 patients presenting between November 2006 and May 2008 composed a validation cohort, defined arbitrarily by date of presentation only. Active disease at presentation was defined by Birmingham vasculitis activity score (BVAS37) and the clinical impression that induction immunosuppression would be required. Following treatment with an immunosuppressant and tapering dose steroid therapy (Supplementary Table 3), patients were followed up monthly for up to 1000 days. Prospective disease monitoring was undertaken by clinicians blinded to gene expression findings using serial BVAS disease scoring37 and full biochemical, haematological and immunological profiling (Supplementary Table 2). At each time-point of follow-up, disease activity was allocated into one of three categories: Flare (at least 1 major or 3 minor BVAS criteria), low-grade activity (0 major and 1-2 minor BVAS criteria) or no activity (0 major or minor BVAS criteria). To differentiate between discrete flares clear improvement in disease activity was required in the form of an improvement in flare-related symptoms together with a reduction in BVAS score, a reduction in markers of inflammation (CRP, ESR), and a reduction in immunosuppressive therapy.
The SLE cohort was composed of 25 patients attending or referred to the Addenbrooke's Hospital specialist vasculitis unit between July 2004 and May 2008 meeting at least four ACR SLE criteria6, presenting with active disease (defined below) and in whom immunosuppressive therapy was to be commenced or increased. Following treatment with an immunosuppressant (Supplementary Table 5) patients were followed up monthly for up to 1000 days (Supplementary Table 4). An episode was defined as a discrete disease flare if it met all 3 of the following prospectively-defined criteria: 1. new BILAG score A or B in any system, 2. clinical impression of active disease by the reviewing physician and 3. increase in immunosuppressive therapy as a result. To differentiate between disease flares clear improvement in disease activity was required in the form of an improvement in flare-related symptoms together with a reduction in BILAG score and a reduction in immunosuppressive therapy before a further flare could be defined. Ethical approval was obtained from the Cambridge Local Research Ethics Committee.
Cell separations and microarray hybridizations
Leukocyte subsets were purified, RNA extracted10 and microarray hybridizations performed as previously described38. Custom microarrays were printed at the Centre for Microarray Resources, Department of Pathology, University of Cambridge using 50mer olignucleotide probes representing 25,342 genes and control probes11. Where indicated, later cohorts were analysed using the Human Gene 1.0 ST Array (Affymetrix) as described. Further details are given in Supplementary Methods.
Quantitative RT- PCR
Taqman Gene Expression Assays (Applied Biosystems) were performed on an ABI PRISM 7900HT instrument according to the manufacturer's instructions.
Flow cytometry
Samples were analysed on a CyAn ADP flow cytometer (Dako) using FlowJo software (Tree Star). All monoclonal antibodies were from Becton Dickinson or Abcam.
Microarray data analysis
Raw image data was extracted using Koadarray v2.4 software (Koada Technology) and probes with a confidence score >0.3 in at least one channel were flagged as present. Extracted data was imported into R where log transformation and background subtraction were performed followed by within-array print-tip Loess normalisation and between-array aquantile and scale normalisation using the Bioconductor package, Limma package. Normalized data was further analysed in Genespring v7.2 (Agilent), Cluster, Gene Pattern, BRB ArrayTools, and ExpressionProfiler:NG as described in Supplementary Methods.
Statistical analysis
Differential expression between defined phenotypes was assessed using one-way ANOVA with FDR controlled at 5%. Genes showing minimal variation between defined phenotypes were excluded from analysis using a fold change (FC) filter set at either 1.5 or 2-fold as specified. Mann Whitney U tests, Chi-square tests, and Kaplan Meier log-rank tests were performed in Prism (GraphPad Software). Overlap between differentially expressed genelists was assessed using hypergeometric probability (Genespring v7.2, Agilent). Expression density distribution profiling and differential co-expression analyses were performed in R as described in Supplementary Methods.
Classifier
A Classifier was built using a support vector machines algorithm optimized on a 50% ‘training’ subsample and validated on the remaining ‘test’ samples, generated in a randomized, stratified fashion. A predictive model was derived from the training set and applied to the test set to assess its predictive performance. Development of the predictive classifier was performed using the Genepattern 2.0 platform. Further details in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Douglas Fearon and John Todd for critical review of the manuscript, Tim Rayner, Misha Kapushesky and Menna Clatworthy for helpful discussions, Kirsty Townsend for help with patient recruitment, Hayley Woffendin and Tom Freeman for help with optimising the custom array platform and Alexander Hatton and Huzefa Ratlamwala for technical support, along with all patients and clinicians involved in enrolment and clinical follow-up. We are grateful to Elizabeth Clutterbuck, Rajeka Lazarus and the volunteers at the Oxford Vaccine Group for their help with recruitment of healthy controls. This work was supported by the NIHR Cambridge Biomedical Research Centre, the Wellcome Trust (Programme Grant number 083650/Z/07/Z), the Medical Research Council, Kidney Research UK and the National Medical Research Council of Singapore (Grant reference IRG07nov089). EFM holds a Wellcome, and LCW an MRC, Clinical Training Fellowship. KGCS is a Lister Prize Fellow and Khoo Oon Teik Professor of Nephrology at the National University of Singapore. The Cambridge Institute for Medical Research is in receipt of a Wellcome Trust Strategic Award (079895).
Footnotes
Competing Interest Statement
The authors declare no competing financial interests
References
- 1.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 2.van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 3.Bryant PA, Smyth GK, Robins-Browne R, Curtis N. Detection of gene expression in an individual cell type within a cell mixture using microarray analysis. PLoS ONE. 2009;4:e4427. doi: 10.1371/journal.pone.0004427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons PA, M. E, Rayner TF, Hatton A, Woffendin HB, Koukoulaki M, Freeman TC, Jayne DR, Chaudhry AN, Smith KGC. Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis. Ann Rheum Dis. 2009 doi: 10.1136/ard.2009.108043. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane SE, Watts RA, Shepstone L, Scott DG. Primary systemic vasculitis: clinical features and mortality. QJM. 2005;98:97–111. doi: 10.1093/qjmed/hci015. [DOI] [PubMed] [Google Scholar]
- 6.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 7.Booth AD, et al. Outcome of ANCA-associated renal vasculitis: a 5-year retrospective study. Am J Kidney Dis. 2003;41:776–84. doi: 10.1016/s0272-6386(03)00025-8. [DOI] [PubMed] [Google Scholar]
- 8.Jennette JC, Xiao H, Falk RJ. Pathogenesis of vascular inflammation by anti-neutrophil cytoplasmic antibodies. J Am Soc Nephrol. 2006;17:1235–42. doi: 10.1681/ASN.2005101048. [DOI] [PubMed] [Google Scholar]
- 9.Langford CA. Antineutrophil cytoplasmic antibodies should not be used to guide treatment in Wegener's granulomatosis. Clin Exp Rheumatol. 2004;22:S3–6. [PubMed] [Google Scholar]
- 10.Lyons PA, et al. Microarray analysis of human leucocyte subsets: the advantages of positive selection and rapid purification. BMC Genomics. 2007;8:64. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Brigand K, et al. An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 2006;34:e87. doi: 10.1093/nar/gkl485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulahad WH, Stegeman CA, Limburg PC, Kallenberg CG. CD4-positive effector memory T cells participate in disease expression in ANCA-associated vasculitis. Ann N Y Acad Sci. 2007;1107:22–31. doi: 10.1196/annals.1381.003. [DOI] [PubMed] [Google Scholar]
- 13.Lamprecht P. Off balance: T-cells in antineutrophil cytoplasmic antibody (ANCA)-associated vasculitides. Clin Exp Immunol. 2005;141:201–10. doi: 10.1111/j.1365-2249.2005.02808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrente A, Kapushesky M, Brazma A. A new algorithm for comparing and visualizing relationships between hierarchical and flat gene expression data clusterings. Bioinformatics. 2005;21:3993–9. doi: 10.1093/bioinformatics/bti644. [DOI] [PubMed] [Google Scholar]
- 15.Monti S. Consensus Clustering: A resampling-based method for class discovery and visualisation of gene expression microarray data. Machine Learning Journal. 2003;52:91–118. [Google Scholar]
- 16.Blanco P, et al. Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52:201–11. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 17.Ippolito A, Petri M. An update on mortality in systemic lupus erythematosus. Clin Exp Rheumatol. 2008;26:S72–9. [PubMed] [Google Scholar]
- 18.Illei GG, Lipsky PE. Biomarkers in systemic lupus erythematosus. Curr Rheumatol Rep. 2004;6:382–90. doi: 10.1007/s11926-004-0013-4. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ingenuity Systems www.ingenuity.com.
- 21.Buentke E, M. A, Tolaini M, Di Santo J, Zamoyska R, Seddon B. Do CD8 effector cells need IL7-R expression to become resting memory cells? Blood. 2006;108:1949–56. doi: 10.1182/blood-2006-04-016857. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini M, Belz G, Bouillet P, Strasser A. Shutdown of an acute T cell immune response to viral infection is mediated by the proapoptotic Bcl-2 homology 3-only protein Bim. Proc Natl Acad Sci U S A. 2003;100:14175–80. doi: 10.1073/pnas.2336198100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schluns KS, Lefrancois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3:269–79. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 24.Joshi NS, Kaech SM. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180:1309–15. doi: 10.4049/jimmunol.180.3.1309. [DOI] [PubMed] [Google Scholar]
- 25.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 26.Mao M, et al. T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics. 2004;83:989–99. doi: 10.1016/j.ygeno.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Galon J, et al. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J. 2002;16:61–71. doi: 10.1096/fj.01-0245com. [DOI] [PubMed] [Google Scholar]
- 28.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 29.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vezys V, et al. Memory CD8 T-cell compartment grows in size with immunological experience. Nature. 2008 doi: 10.1038/nature07486. [DOI] [PubMed] [Google Scholar]
- 31.Bevan MJ. Helping the CD8(+) T-cell response. Nat Rev Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 32.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–92. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 33.Hou S, Hyland L, Ryan KW, Portner A, Doherty PC. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature. 1994;369:652–4. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 34.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 35.van 't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 36.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 37.Stone JH, et al. A disease-specific activity index for Wegener's granulomatosis: modification of the Birmingham Vasculitis Activity Score. International Network for the Study of the Systemic Vasculitides (INSSYS) Arthritis Rheum. 2001;44:912–20. doi: 10.1002/1529-0131(200104)44:4<912::AID-ANR148>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Willcocks LC, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.