Abstract
Plants have evolved and adapted to different environments. Dwarfism is an adaptive trait of plants that helps them avoid high-energy costs under unfavourable conditions. The role of gibberellin (GA) in plant development has been well established. Several plant dehydration-responsive element-binding proteins (DREBs) have been identified and reported to be induced under abiotic and biotic stress conditions. A tomato DREB gene named SlDREB, which is a transcription factor and was cloned from cultivated tomato M82, was found to play a negative role in tomato plant architecture and enhances drought tolerance. Tissue expression profiles indicated that SlDREB was expressed mainly in the stem and leaf and could be induced by abscisic acid (ABA) but suppressed by GA and ethylene. SlDREB altered plant morphology by restricting leaf expansion and internode elongation when overexpressed, and the resulting dwarfism of tomato plants could be recovered by application of exogenous gibberellic acid (GA3). Transcriptional analysis of transgenic plants revealed that overexpression of SlDREB caused the dwarf phenotype by downregulating key genes involved in GA biosynthesis such as ent-copalyl diphosphate synthase (SlCPS) and GA 20-oxidases (SlGA20ox1, -2, and -4), thereby decreasing endogenous GA levels in transgenic plants. A yeast activity assay demonstrated that SlDREB specifically bound to dehydration-responsive element/C-repeat (DRE/CRT) elements of the SlCPS promoter region. Taken together, these data demonstrated that SlDREB can downregulate the expression of key genes required for GA biosynthesis and that it acts as a positive regulator in drought stress responses by restricting leaf expansion and internode elongation.
Key words: DREB, dwarfism, ent-copalyl diphosphate synthase, gibberellin, gibberellin 20-oxidase, tomato
Introduction
Drought, high salinity, and low temperature are adverse environmental conditions that affect plant distribution (Yang et al., 2011c ). When plants encounter adverse ambiance, they exhibit a variety of responses that result in specific changes suited to the particular stress condition encountered. For example, the dwarf phenotype benefits the plant through avoidance of the high-energy cost of producing stress-tolerance proteins (Knight and Knight, 2001).
The endogenous phytohormone gibberellin (GA) is involved in plant growth and development including internode elongation, and is one of the compounds associated with the dwarf phenotype (Hedden and Kamiya, 1997). GA biosynthesis and catabolism genes in higher plants have been well characterized. Geranyl diphosphate synthase (GPS) has been reported to control the level of bioactive GA by modulating GA precursor biosynthesis in tomato (van Schie et al., 2007). Geranylgeranyl diphosphate (GGDP) is the common precursor for diterpenoids including GAs, and is converted to ent-kaurene through successive two cyclization reaction catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS). The ent-kaurene is then converted to GA12 by the two distinct P450 monooxygenases, ent-kaurene oxidase (KO) and ent-kaurenoic oxidase (KAO). GA12 can be further converted to GA53 by 13-hydroxylation. GA12 and GA53 are converted mainly to various GA intermediates (e.g. GA9 and GA20) and successively to bioactive GAs (e.g. GA1 and GA4) by GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox), respectively. GA3ox catalyses the final step in the synthesis of bioactive GAs. GA 2-oxidase (GA2ox) is the major enzyme that converts active GAs (GA1 and GA4) and their precursors (GA9 and GA20) to inactive forms (Yamaguchi, 2008; Sun, 2011).
Over the past decade, a family of transcription factors known as APETALA2 (AP2)/ethylene-responsive element binding factor (ERF) has been identified in plants. These proteins are involved in a variety of regulatory mechanisms such as fruit ripening (Chung et al., 2010), growth and development (Wilson et al., 1996), and biotic (Thara et al., 1999; Brown et al., 2003) and abiotic stresses (Liu et al., 1998; Sakuma et al., 2002; Fukao and Bailey-Serres, 2008; Li et al., 2011; Wan et al., 2011; Zhang et al., 2011; Shi et al., 2012). The AP2/ERF transcription factors are found only in the plant kingdom and are characterized by the presence of a highly conserved DNA-binding domain (Riechmann and Meyerowitz, 1998; Tournier et al., 2003). ERFs are part of the AP2/ERF superfamily, which also contains AP2 and RAV family genes and is characterized by the presence of the AP2/ERF DNA-binding domain (Sakuma et al., 2002). Based on their ERF domain, the tomato ERFs can be further divided into two subfamilies, ERF and DREB (dehydration-responsive element-binding proteins), and their ERF domain diversity is manifested at amino acid positions 14 and 19 (Sharma et al., 2010). DREBs contain a conserved AP2/ERF domain that binds specifically to the dehydration-responsive element/C-repeat (DRE/CRT) cis-acting element (core motif: G/ACCGAC) of the genes that they regulate (Busk et al., 1997; Qin et al., 2004).
DREB-binding factors play crucial roles in the regulation of abiotic and biotic responses in plants. Heterogeneous expression of a DREB gene from Leymus chinensis improved drought and salt tolerance of Arabidopsis thaliana (Xianjun et al., 2011). Similarly, expression of a DREB gene from Limonium bicolor enhanced copper tolerance in transgenic tobacco plants (Ban et al., 2011). A rice DREB gene, ARAG1, which is involved in the abscisic acid (ABA) signalling pathway, plays a role in seed germination and drought tolerance (Zhao et al., 2010). Arabidopsis DREBs play a role in the regulation of water homeostasis by regulating multiple aquaporin genes (Rae et al., 2011). The Arabidopsis DREB protein DEAR1 has an upstream regulatory role in mediating cross-talk between signalling pathways for biotic and abiotic stress responses (Tsutsui et al., 2009). Moreover, two chrysanthemum DREB genes have been reported to play a regulatory role in abiotic stress responses (Yang et al., 2009), and overexpression of OsDREB1G and OsDREB2B genes in rice significantly improved their tolerance to water deficit stress (Chen et al., 2008).
Here, we demonstrate that SlDREB encodes a transcription factor whose expression is regulated by drought, salt, and cold, and by ABA, gibberellic acid (GA3), and ethylene treatments. Our experimental results revealed that overexpression of SlDREB in tomato led to lower endogenous GA levels and consequently a dwarf phenotype consisting of restricted leaf expansion and internode elongation via downregulation of key genes involved in GA biosynthesis. Notably, SlDREB could bind specifically to the DRE/CRT elements of the SlCPS promoter region, indicating that SlDREB may directly regulate the expression of SlCPS in tomato.
Materials and methods
Plant material and plant growth regulator treatment
Tomato (Solanum lycopersicum) variety M82 and wild species Solanum pennellii (LA0716) were used to compare SlDREB nucleotide sequences between wild and cultivated species. S. lycopersicum cv. M82 was used to analyse the expression of SlDREB in different organs and the response to different plant growth regulators. Tissues from the roots, stems, leaves, flowers, and fruits of M82 plants were collected and immediately frozen in liquid nitrogen and stored at –80 °C until use. For plant growth regulator treatment, four-leaf-stage plants grown in compost plastic trays in a 16h light/8h dark regime at 25 °C in a greenhouse were sprayed with 100 µM ABA, 100 µM GA3, 100 μM ethephon (Eth; an ethylene releaser), or distilled water (control). Three leaves, one from each plant, were collected at the designated times from the different treated and untreated plants and stored as indicated above. Plant morphology was assessed by measuring the internode lengths and number of leaves, and leaf samples were taken 35 d post-inoculation (p.i.). Dwarf phenotypes were rescued by spraying with a 100 µM GA3 solution containing 0.02% Tween 20 at an interval of 3 d until run-off starting from 30 d p.i.
Isolation of SlDREB, construction of overexpression vectors, and plant transformation
In our previous studies on drought stress in tomato introgression lines (ILs), a differential expression profile of the SlDREB gene was observed between the drought-tolerant ILs and M82 (Gong et al., 2010). The tomato SlDREB gene was PCR amplified from the cDNA of M82 (forward primer: 5’-CCAATTTCCTCTCTCCCAAA-3’; reverse primer: 5’-TGTTCATGAAAATCCAAATGTCT-3’) based on the UniGene sequence (SGN-U585938). For transformation, we used the binary plasmid vector pMV2 (Yang et al., 2011a ), which carries the spectinomycin resistance gene for bacterial selection and the neomycin phosphotransferase II gene for selection of transformed plants. The binary plasmid was constructed by inserting the SlDREB cDNA between the KpnI and XbaI sites in the sense orientation driven by the cauliflower mosaic virus 35S promoter. The plasmid mediated by Agrobacterium tumefaciens strain C58 was transformed in tomato cultivar M82. After screening for regenerated shoots on selection medium containing kanamycin, the transgenic plants were further verified by PCR using genomic DNA as template and 35S forward and gene-specific reverse primers.
RNA isolation and real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, USA). DNase I (Fermentas, USA)-treated RNA was reverse transcribed using a Moloney murine leukaemia virus cDNA reverse transcription enzyme (Invitrogen) and cDNA was used for real-time RT-PCR. Real-time RT-PCR was performed with a SYBR Premix Ex Taq kit (Takara, Japan) using primers (see Supplementary Table S1 at JXB online) specific for SlDREB, SlGPS (DQ286930), SlCPS (AB015675), SlKS (AEP82778), SlKO (SGN-U583328), SlKAO (SGN-U575348), SlGA20ox1 (AF049898), SlGA20ox2 (AF049899), SlGA20ox3 (AF049900), SlGA20ox4 (EU652334), SlGA3ox1 (AB010991), SlGA3ox2 (AB010992), SlGA2ox1 (EF441351), SlGA2ox2 (EF441352), SlGA2ox3 (EF441353), SlGA2ox4 (EF441354), SlGA2ox5 (EF441355), SlGAST1 (X63093), SlDELLA (AY269087), and β-actin (SGN-U580609) transcripts as internal controls. The PCR amplification step consisted of an initial incubation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. Data were collected during the extension step, and melting-curve acquisitions and analyses were also performed on the cycler. PCR products were monitored using a LightCycler 480 (Roche, Switzerland) PCR system.
Subcellular localization assay of SlDREB
The full-length open reading frame (ORF) without the stop codon of SlDREB was PCR amplified using primers containing KpnI and BamHI restriction sites (underlined) (forward: 5’-GGTACCATGTCAA AGCGAATAAGAGAGAGTG-3’; reverse: 5’-GGATCCTTTCATCAT TTCAAAGTTGCTAAG-3’). The PCR product was purified from agarose gel, cloned into a pMD18-T vector (TaKaRa), and sequenced. The plasmid containing the correct sequence of SlDREB was digested with KpnI and BamHI and subcloned into a pCAMBIA1391 vector digested using the same restriction enzymes to create a fusion construct (pCAMBIA1391-SlDREB-GFP). Both the fusion construct (pCAMBIA1391-SlDREB-GFP) and the control vector (pCAMBIA1391-GFP) were bombarded into onion epidermal cells using a Biolistic PDS-1000 (Bio-Rad) system. The onion cells were cultured on MS medium for 24h in the dark and observed with a Leica TCSST2 confocal laser microscope.
Trans-activation activity and DRE/CRT binding assay for SlDREB in yeast
The coding region of SlDREB was PCR amplified using specific primers (forward: 5’- CCCGGGAATGTCAAAGCGAATAAGAGAGAG-3’; reverse: 5’-GTCGACTTATTTCATCATTTCAAAGTTGC-3’) containing SmaI/SalI restriction sites (underlined), corresponding to those present in the yeast expression vector pGBKT7 (Clontech, USA), to produce pBD-SlDREB. According to the manufacturer’s instructions, pBD-SlDREB, pGBKT7 (negative control), and pGBKT7-53+pGADT7-RecT (positive control) were transformed separately into the yeast strain AH109. Selection of transformants was done on SD/–Trp or SD/–Ade/–His/–Trp medium, and the trans-activation activity of each protein was evaluated according to their status of growth and the activity of X-α-Gal (5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside).
A yeast activity assay kit (Clontech) was used to investigate whether SlDREB was bound to DRE/CRT of the SlCPS promoter region. We isolated the –2500 putative promoter regions of SlCPS from the SGN database (http://solgenomics.net/) and confirmed their sequence. The full-length SlDREB was fused to the GAL4 activation domain in the vector pGADT7 digested with SmaI and BamHI to get pGAD-SlDREB. A 48bp single-stranded oligonucleotide sequence (5’-AGCTTCATGTACCGACTCCCATGTACCGACTCCCATGTACC GACTCCC-3’) and its reverse complement sequence (5’-TCGAGGG AGTCGGTACATGGGAGTCGGTACATGGGAGTCGGTAC ATGA-3’), which contains three tandem repeat copies of 5’-CATG TACCGACTCC-3’ (DRE/CRT is underlined) in the SlCPS promoter regions and cohesive termini of HindIII and XhoI was synthesized. The oligonucleotides were annealed as follows: each single-stranded oligonucleotide of 100 µM mixes at a ratio of 1:1, yielding a final concentration of 50 µM each, were heated to 95 °C for 30 s, 72 °C for 2min, 37 °C for 2min and 25 °C for 2min. After annealing, the double-stranded oligonucleotides were cloned into a pAbAi vector linearized using the restriction enzymes HindIII and XhoI, and their sequences were confirmed. The resulting pDRE-AbAi construct and p53-AbAi control vector were digested with BstBI, and the linearized plasmids were transformed into Y1H Gold yeast strain. Selection for transformants was performed on SD/–Ura medium. After PCR confirmation, the minimal inhibitory concentration of aureobasidin A (AbA) for the pDRE-AbAi yeast strain was determined. The p53-AbAi control had a minimal inhibitory concentration of 100ng ml–1 AbA (Clontech). The pGAD-SlDREB and pGADT7 vectors were transformed into the pDRE-AbAi yeast strain, while the pGAD-p53 vector was transformed into p53-AbAi containing Y1H Gold yeast strain as a positive control. A large healthy colony was picked from the yeast strains and suspended in 0.9% NaCl. The optical density at 600nm was adjusted to 0.002 (for ~2000 cells per 100 µl), and 2 µl of cells was also dotted on the SD/–Ura/–Leu medium with or without AbA to assess DNA–protein interactions. The colonies were then allowed to grow for 2–3 d at 30 °C.
Quantification of endogenous GAs
The leaves of tomato (1g) were frozen in liquid nitrogen and finely ground follow by extraction with 15ml of methanol containing 20% water (v/v) at 4 °C for 12h. Before grinding, the following labelled GAs were added as internal standards: [2H2]GA1 (1.00ng g–1), [2H2]GA3 (1.00ng g–1), [2H2]GA4 (2.00ng g–1), [2H2]GA12 (2.00ng g–1), [2H2]GA20 (2.00ng g–1), and [2H2]GA53 (4.00ng g–1). Further sample was prepared and analysed as described previously (Chen et al., 2011). In brief, the sample was first passed through a C-18 SPE cartridge (12ml, 1.5g), and the resulting eluate was evaporated under a nitrogen gas stream and redissolved in 3ml of water. The solution was acidified with 360 µl of 0.1M hydrochloric acid and extracted repeatedly with ethyl ether (10×0.5ml). The ether phases were combined, dried under nitrogen gas, and redissolved in 112 µl of acetonitrile. After the addition of 180 µl of Et3N (20 µmol ml–1) and 108 µl of 3-bromoacetonyltrimethylammonium bromide (20 µmol ml–1), the reaction solution was vortexed for 10min. The mixture was evaporated under nitrogen gas and the residue was dissolved in 30 µl of water. The prepared samples were injected using 25 kV×1min and separated by 100cm amino groups, and coated capillary electrophoresis coupled with electrospray ionization quadrupole-time-of-flight mass spectrometry was performed for analysis.
Scanning electron microscopy for leaf thickness and stomatal observation
For scanning electron microscopy observation, the third leaves from the top were collected from 2-month-old tomato plants overexpressing SlDREB and wild-type (WT) plants, all grown in a naturally illuminated glasshouse. The leaves were cut into ~0.1cm2 pieces and fixed with 2% glutaraldehyde for 24h. After washing in 0.1M cacodylate buffer, the samples were dehydrated in a graded ethanol series, dried in a desiccator (HCP-2; Hitachi), and coated with a film of gold. Observations were carried out on a JSM-6390/LV scanning electron microscope. The stomata were counted in each field of view (400×: ~0.065mm2) using an Olympus BHS/BHT System microscope (BH-2).
Results
Tomato SlDREB is a typical transcription factor
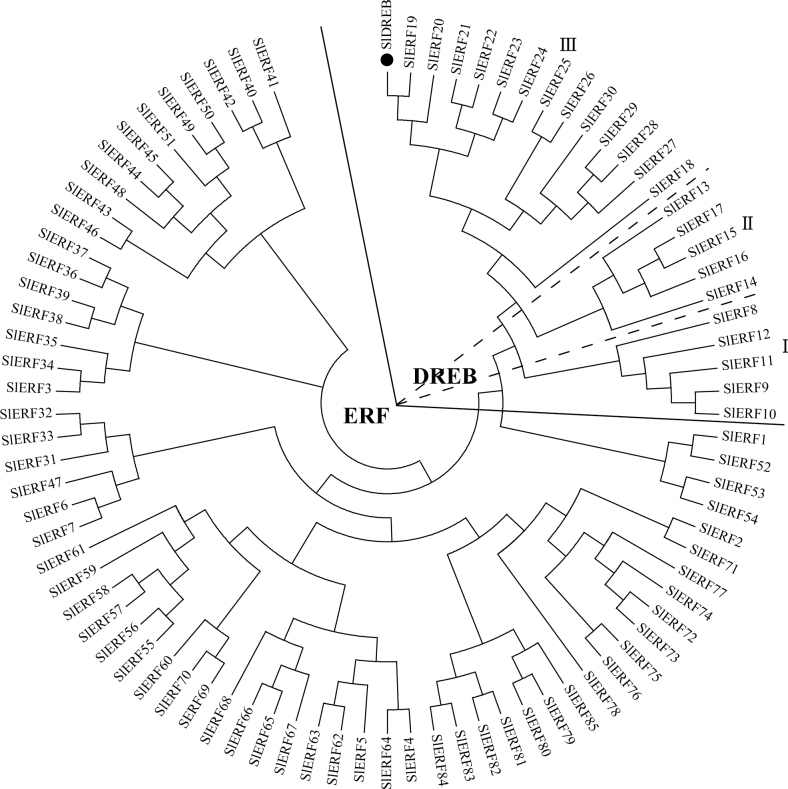
The AP2/ERF superfamily is defined by the AP2/ERF domain, which consists of ~60–70 amino acids and is involved in DNA binding. There are 112 AP2/ERFs in tomato, 85 of which are potential ERFs with a single complete AP2/ERF domain. The ERFs have been divided further into two subfamilies, ERF and DREB (Sharma et al., 2010). Based on a phylogenetic analysis based of all tomato ERFs, SlDREB was classified into the DREB subfamily (Fig. 1).
Fig. 1.
Phylogenetic analysis of tomato ERFs with SlDREB proteins. Phylogenetic analysis of the ERF domains of 85 unigenes of tomato (Sharma et al., 2010). SlDREB is indicated by a black circle. The phylogenetic tree was generated by ClustalW2 using standard parameters of the neighbour-joining method in MEGA (version 5.05).
The tomato DREB gene was previously identified as a drought-responsive gene in our microarray experiment (Gong et al., 2010). We confirmed that it is induced by drought, salt, and cold stresses (Supplementary Fig. S1 at JXB online). Using PCR, we isolated and cloned the full-length (579bp) ORF of SlDREB from the cDNA of S. lycopersicum cv. M82 and of SpDREB from S. pennellii. Comparison of their deduced protein sequences revealed that SlDREB shared 97% similarity with SpDREB (from wild tomato) and 60% similarity with StDREB (from Solanum tuberosum) but only 53% similarity with AtDREB (from Arabidopsis) (Fig. S2A at JXB online). A difference of only three amino acids—threonine, serine, and glutamic acid (in M82) in place of serine, arginine, and lysine (in S. pennellii) at positions 110, 134, and 144, respectively (Fig. S2B)—was observed between SlDREB and SpDREB.
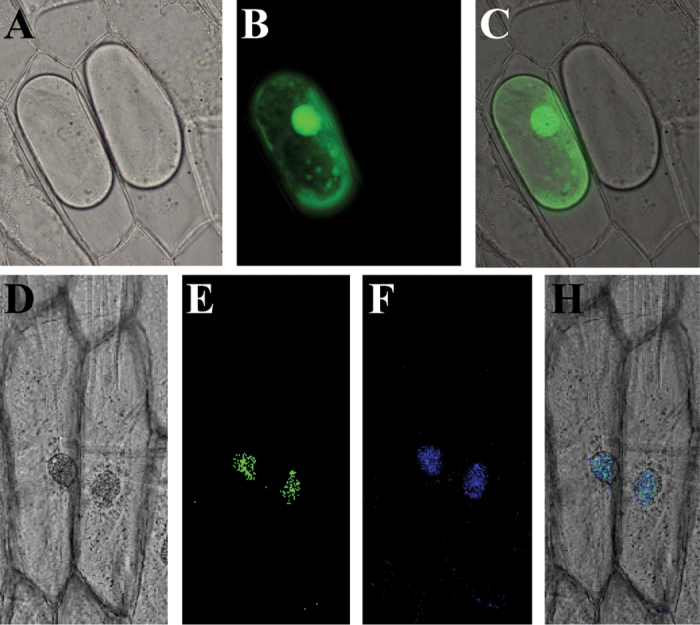
Bioinformatics analysis revealed a nuclear localization signal (positions 36–42) in the basic region of the AP2/ERF DNA-binding domain, implying that SlDREB may be localized in the nucleus. In order to verify our in silico result, subcellular localization of SlDREB was examined by monitoring GFP fluorescence in onion epidermis cells transformed with either a fusion construct (pCAMBIA1391-SlDREB-GFP) or a control construct (pCAMBIA1391-GFP). Green fluorescence signals were observed over the entire cell in onion cells transformed with the control construct (Fig. 2A–C). In contrast, fluorescence was exclusively detected in the nuclei of cells transformed with the fusion plasmid (Fig. 2D–H), implying that SlDREB is a nuclear protein.
Fig. 2.
Subcellular localization of SlDREB in onion epidermal cells. GFP and SlDREB–GFP fused constructs were expressed transiently in onion epidermal cells. Bright-field images (A, D), GFP fluorescent images (B, E), DAPI image (F), and merged images (C, H) of representative cells transformed with GFP (A–C) or the SlDREB–GFP fusion protein (D–G) are shown. (This figure is available in colour at JXB online.)
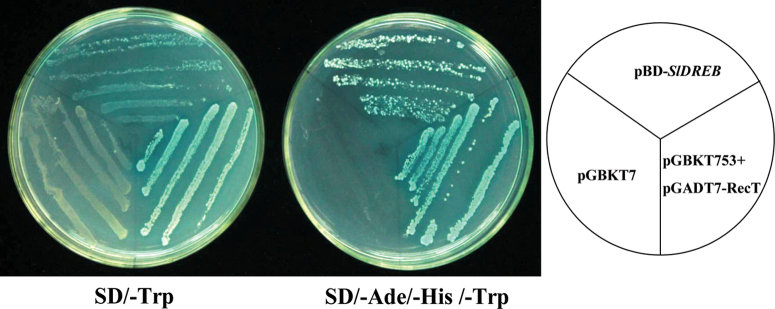
In addition to nuclear localization, transactivation activity is another defining feature of a transcription factor. We used a yeast two-hybrid system to examine the transcriptional activity of SlDREB. A GAL4 DNA-binding domain SlDREB fusion protein was expressed in yeast cells, which were then assayed for their ability to activate transcription from the GAL4 sequence. SlDREB promoted yeast growth in the absence of histidine and adenine, and showed X-α-gal activity, whilst the vector control pGBKT7 did not (Fig. 3). These data confirmed that SlDREB functions as a transcriptional activator in yeast.
Fig. 3.
Analysis of the transactivation activity of SlDREB. SlDREB and GAL4 DNA-binding domain fusion protein were expressed in the yeast strain AH109. Vectors pGBKT7 and pGBKT7-53+pGADT7-RecT were expressed in yeast as a negative and positive control, respectively. The yeast streak was cultured on SD/–Trp and SD/–Ade/–His/–Trp medium; both contained X-α-gal for assaying another yeast reporter (MEL1) gene. (This figure is available in colour at JXB online.)
SlDREB is induced by GA3 and Eth but suppressed by ABA
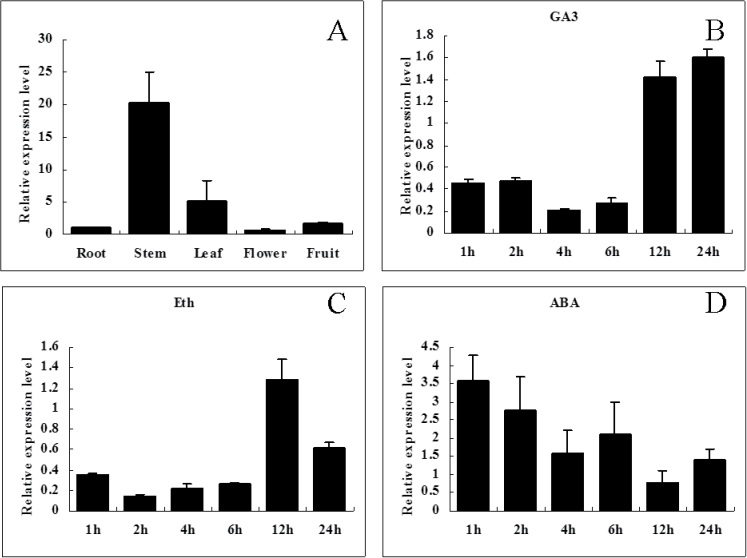
Real time-PCR detection results showed that SlDREB was highly expressed in tomato stems and leaves (Fig. 4A). By comparing the SlDREB expression levels of treated plants relative to those of untreated plants, we found that SlDREB expression was significantly suppressed by GA3 and Eth treatment for up to 6h after treatment but returned to pre-treatment levels 12h later. However, under ABA stress, SlDREB expression was induced from 1 to 6h after treatment and returned to pre-treatment levels 12h later (Fig. 4B, 4C, 4D). These expression patterns indicate that SlDREB was downregulated by GA3 and Eth treatment but upregulated by ABA application.
Fig. 4.
Real-time RT-PCR analysis of the expression of SlDREB in WT tissues (A) and in response to the phytohormones GA3 (B), ABA (C), and Eth (D). All samples were collected at the indicated time points. Expression of phytohormone-treated WT plants was compared with that in untreated plants after normalization of values with reference to the tomato β-actin gene and is presented as the relative expression level. All samples were collected at the indicated time points from three biological replicates in each treatment group. Error bars indicate the standard error (SE) of three replicates.
Transgenic plants overexpressing SlDREB have shorter internodes and increased drought tolerance
To investigate the function of SlDREB, the plasmid 35S:SlDREB was introduced into cultivated tomato M82. Transgenic plants overexpressing (OE) SlDREB were obtained after screening for regenerated shoots on selection medium containing kanamycin. The transgenic plants were analysed further by PCR using genomic DNA as template and 35S forward and gene-specific reverse primers. Twenty-three transformants (T0) regenerated from kanamycin-resistant calli contained SlDREB.
The expression level of the SlDREB gene in transgenic (T0, T1, and T2) as well as control plants was examined at the five-leaf stage by real-time PCR analyses. Expression of SlDREB was 42.84- and 19.33-fold greater in the overexpressing T2 homozygous line 4 (OE4) and overexpressing line 13 (OE13), respectively, compared with M82 control plants (Fig. 5E).
Fig. 5.
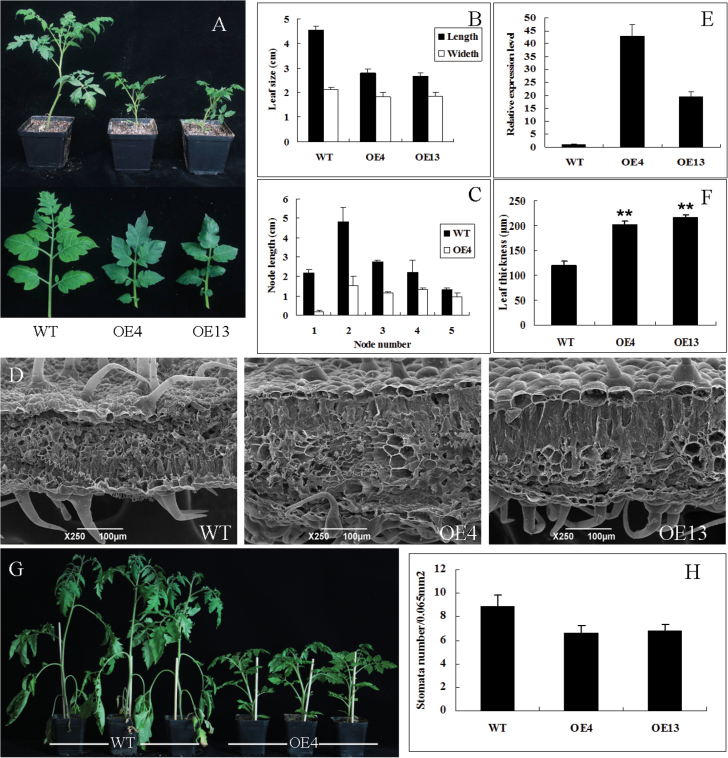
Overexpression of SlDREB restricts tomato leaf expansion and internode elongation. (A) Phenotypic appearance of seedlings (upper) and leaves (lower) of SlDREB-overexpressing tomato OE4 and OE13 lines and WT plants. (B) Leaflet size of WT and SlDREB lines OE4 and OE13. The third leaf from the top of 2-month-old transgenic and non-transgenic plants was used for leaflet size measurement; the apical leaflet was chosen from the leaf. Error bar indicates SE (n=8). (C) Internode length of 2-month-old WT and SlDREB OE4 tomato plants. Error bars indicates SE (n=8). (D) Scanning electron microscopy images showing leaf thickness. Leaves were collected from 2-month-old tomato SlDREB-overexpressing and WT plants grown in a naturally illuminated glasshouse. The apical leaflet of the third leaf from the top was collected for analysis. (E) The expression level of SlDREB in transgenic and WT control plants was examined at the five-leaf stage by real-time PCR analyses. Error bars indicates SE (n=8). (F) Graphical representation of leaf thickness. Eight individual plants were selected per line and the thickness of every leaf was measured three times. Asterisks indicate significant differences compared with WT (**P < 0.01). (G) SlDREB-overexpressing plants show drought resistance. At 40 d post-inoculation, watering was stopped for drought treatment. The photographs were taken after 15 d. Treatment was replicated three times under the same conditions (n > 8). (H) The mean number of stomata per unit area in SlDREB overexpressing and WT leaves. Stomatal number was counted in each field of view (400×, ~0.065mm2) on at least 20 microscopes and three plants. The third leaf from the top of 2-month-old transgenic and non-transgenic plants was analysed. (This figure is available in colour at JXB online.)
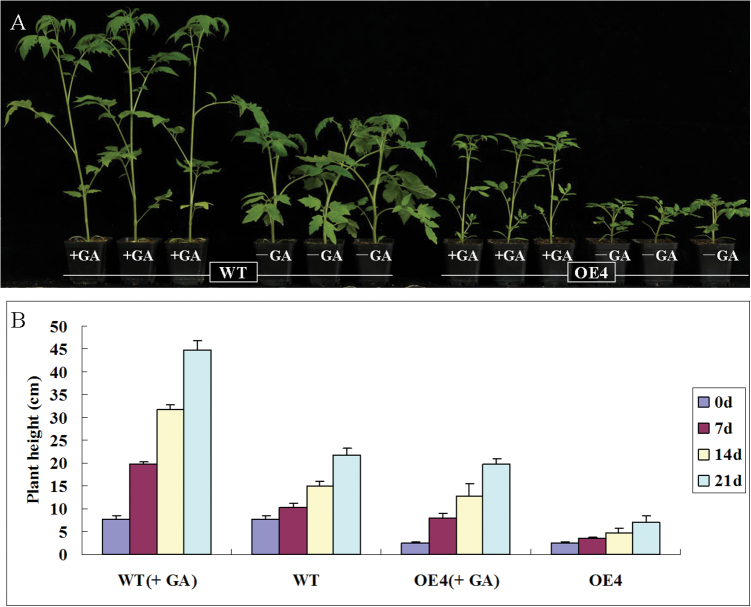
The SlDREB overexpressing transgenic lines showed dwarf phenotypes with obviously shorter internodes. For further experiments, we selected two T2 homozygous lines: OE4 and OE13 (eight individual plants of each line). The transgenic plants and WT plants were cultivated under the same conditions, and plant height was determined after 2 months. The average plant height of the control, OE4, and OE13 plants was 29.7, 11.3, and 10.6cm, respectively. Thus, OE4 and OE13 plants were 61.95% and 64.30% shorter, respectively, than the control (Fig. 5A). The leaves of the transgenic plants were also smaller: the mean length of the leaflets of the control, OE4, and OE13 lines was 4.55, 2.78 and 2.65cm, respectively. The leaf width of the transgenic plants was also slightly smaller compared with that of WT plants (Fig. 5B). Furthermore, shortening of the internode was seen in all cases, with the strongest effect on the second internode, which was shortened by a mean of 31.88% (Fig. 5C). On average, an approximate threefold decrease in internode length was observed. Spraying with 100 µM GA3 on SlDREB-overexpressing plants every 3 d rescued shoot elongation back to control levels (Fig. 6A, 6B), confirming that SlDREB-overexpressing plants indeed exhibit a classical reduced-GA phenotype (Phinney, 1956).
Fig. 6.
The dwarf phenotype of SlDREB-overexpressing plants was rescued by spraying with GA3. (A) Pictures were taken 50 d after inoculation. The dwarf phenotype was rescued by spraying GA3 every 3 d starting from 30 d after inoculation. (B) Plant height is displayed as the mean of eight plants; error bars show SE (n=8). (This figure is available in colour at JXB online.)
To investigate the effects of SlDREB on leaf structure, the leaves of 2-month-old control and SlDREB-overexpressing tomato plants were examined using scanning electron microscopy. The average leaf cross-section was 120.3 µm for control plants, while the thickest cross-section (215.45 µm) was observed in the SlDREB OE4 plant, followed by the OE13 plant (201.48 µm) (Fig. 5D, 5F).
The mean number of stomata on functional leaves of SlDREB-overexpressing plants was approximately seven compared with nine per unit area (~0.065mm2) in WT plants (Fig. 5H). This could reflect specific effects of SlDREB on tomato architecture leading to lowered transpiration and energy loss under drought stress. Consistent with this idea, the SlDREB-overexpressing plants performed very well, whilst the WT plants showed signs of wilting (Fig. 5G).
Overexpression of SlDREB downregulates GA biosynthesis genes
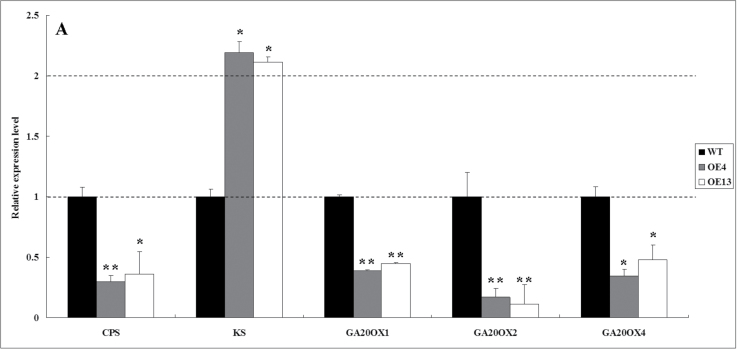
To investigate whether SlDREB altered GA metabolism, expression of GA biosynthetic genes was compared between SlDREB transgenic and M82 (control) seedlings. Two genes in the GA biosynthetic pathway, SlCPS and SlKS, showed considerable change in expression in transgenic plants. SlCPS was dramatically downregulated in SlDREB transgenic seedlings: OE4 exhibited a 3.44-fold decline in SlCPS expression compared with M82 (WT) seedlings. In contrast, expression of SlKS was 2.18-fold greater in SlDREB transgenic seedlings than in M82 WT seedlings (Fig. 7A, 7C). No significant difference in expression of other genes responsible for the early steps of GA biosynthesis, such as SlGPS, SlKO, and SlKAO, was seen between SlDREB transgenic and WT plants (Fig. 7B).
Fig. 7.
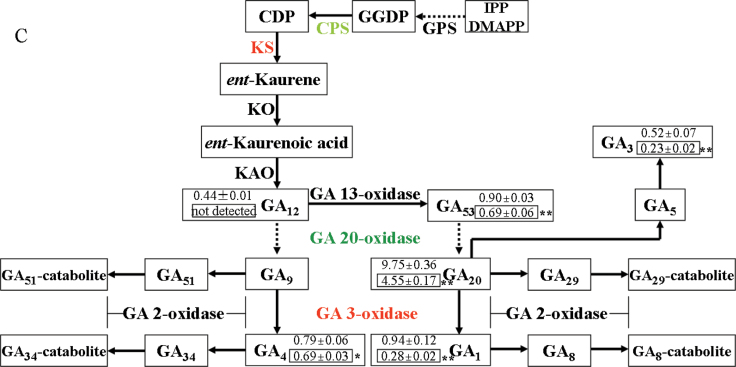
Decreased GA abundance and GA biosynthesis gene expression in SlDREB-overexpressing tomato plants. (A, B) Real-time RT-PCR analysis of the expression of tomato GA biosynthesis genes in OE4, OE13 and WT plants. *P < 0.05; **P < 0.01. Primer sequences are listed in Supplementary Table S1. All samples were collected at the indicated time points from three biological replicates in each treatment group. Error bar indicates the SE of three replicates. (C) Pathway of GA biosynthesis: green indicates significantly (P < 0.05) downregulated expression and red indicates slightly upregulated expression. Numbers indicate endogenous GA levels in the WT (upper) and transgenic plants (lower, boxed). Results that were ‘not detected’ were due to low abundance; **P < 0.01; * P < 0.05. Data are presented as means ±standard deviation from three technical replications (ng g–1 of fresh weight) using two transgenic lines. IPP, prenyldiphosphates isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPS, geranyl diphosphate synthase; GGDP, geranylgeranyl diphosphate; CPD, copalyldiphosphate; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase, KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase. (This figure is available in colour at JXB online.)
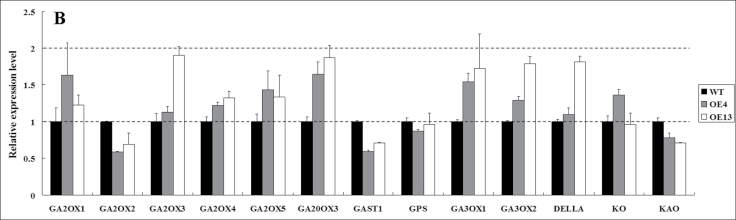
GA20oxs are also key GA biosynthetic enzymes that determine GA concentration in many plant species, and GA3oxs catalyse the final step to produce bioactive GAs (GA1, GA3, and GA4) (Hedden and Kamiya, 1997; Xiao et al., 2006; Yamaguchi, 2008). We compared transcript levels of GA20oxs and GA3oxs between transgenic plants and WT plants by quantitative RT-PCR. In transgenic plants, SlGA20ox1, SlGA20ox2, and SlGA20ox4 were sharply downregulated, but SlGA3ox1 and SlGA3ox2 were slightly upregulated when compared with their respective expression in WT plants (Fig. 7A, 7C).
To determine whether SlDREB is involved in GA inactivation, transcript levels of genes encoding GA2oxs (the main GA catabolic enzyme) in tomato (i.e. SlGA2ox1, SlGA2ox2, SlGA2ox3, SlGA2ox4, and SlGA2ox5; Serrani et al., 2008) were quantified by RT-PCR. No significant differences in expression of these genes were seen between transgenic and WT plants (Fig. 7B).
To determine further whether SlDREB affects GA signal transduction factors, the expression levels of SlDELLA, which has been reported to be a GA repressor in the GA response pathway (Sun and Gubler, 2004), and SlGAST1, a GA-responsive gene (Shi et al., 1992), were compared by quantitative RT-PCR. Expression levels of SlDELLA and SlGAST1 were not significantly different between the transgenic and WT plants (Fig. 7B).
Decreased endogenous GA abundance in SlDREB overexpressing tomato plants
As our results showed that SlDREB repressed the expression of GA biosynthesis genes, we decided to compare endogenous GA levels in SlDREB overexpressing transgenic and WT plants. As shown in the schematic representation of GA biosynthesis in Fig. 7C, endogenous levels of several GAs (GA53, GA12, GA3, GA20, GA4, and GA1) in SlDREB-overexpressing transgenic plants were significantly lower than in WT plants.
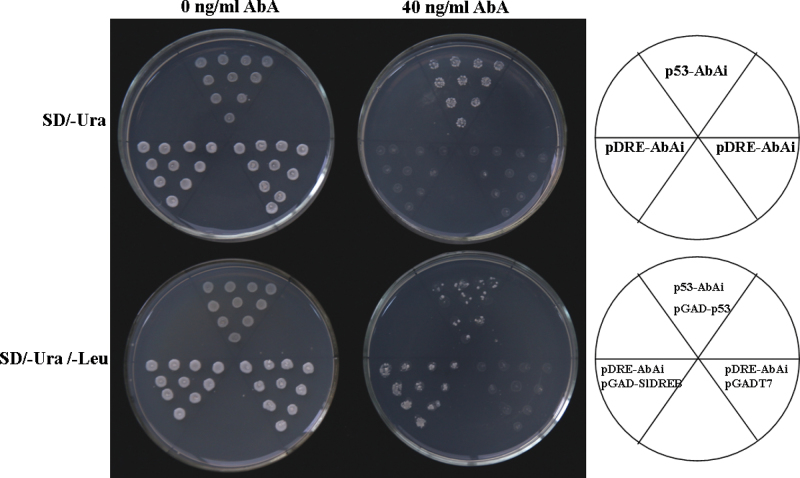
SlDREB specifically binds to the DRE/CRT elements of the SlCPS promoter region.
Previous studies have shown that DREBs can bind to the cis element of DRE/CRT (Busk et al., 1997; Qin et al., 2004). We found that the promoter region (–1928) of SlCPS contains a DRE/CRT element (Fig. S3 at JXB online). In order to investigate whether SlDREB binds to the DRE/CRT elements of the promoter region of the SlCPS gene, the full-length ORF of SlDREB was fused to the GAL4 activation domain of the vector pGADT7. The fused construct thus obtained (pGADT7-SlDREB) was co-transformed with the pDRE-AbAi construct containing triple tandem repeats of DRE/CRT from the SlCPS promoter region into the Y1H Gold yeast strain (Fig. 8). Although all yeast cells harbouring different constructs could grow on SD/–Ura without AbA, those with pDRE-AbAi did not grow in the presence of 40ng ml–1 of AbA (Fig. 8, upper panel). However, cells co-transformed with pGADT7-SlDREB and pDRE-AbAi grew normally in the presence of the minimal inhibitory concentration (40ng ml–1) of AbA. Additionally, the growth of transformants containing constructs lacking SlDREB was completely inhibited (Fig. 8, lower panel), suggesting that SlDREB could bind to the DRE/CRT cis element in yeast.
Fig. 8.
Yeast one-hybrid assays for binding of SlDREB to the DRE/CRT domains of the SlCPS promoter region. Vectors p53-AbAi+pGAD-p53 and pDRE-AbAi+pGADT7 were expressed in yeast as positive and negative controls, respectively. The minimal inhibitory concentration of AbA for the pDRE-AbAi yeast strain is 40ng ml–1. The photograph shows the growth of yeast cells on SD/–Ura or SD/–Ura /–Leu medium with (40ng ml–1) or without AbA.
Discussion
SlDREB is an AP2/ERF-binding factor and is regulated by GA, ABA, and Eth in tomato
AP2/ERF proteins have been shown to be integrators of biotic and abiotic stress responses through their interaction with cis-acting elements, the GCC box, and CRT/DRE (Knight and Knight, 2001; Sakuma et al., 2002; Brown et al., 2003; Li et al., 2011). DREB can be classified into the ERF subgroup of the AP2/ERF superfamily based on its function and group motif (Sharma et al., 2010). DREB transcription factors are involved in abiotic and biotic stress signalling pathways in plants (Agarwal et al., 2006). However, the functions of DREB in tomato remain unclear. SlDREB was isolated by screening the results of our previous microarray experiments on drought-tolerant tomato (Gong et al., 2010). SlDREB was responsive to drought, salt, and cold stresses and could be classified into subgroup III of the isolated DREBs (Figs 1 and S1). In addition, it was also shown that SlDREB is localized in the nucleus and exhibits transactivation activity in yeast (Figs 2 and 3), suggesting that SlDREB encodes a typical transcriptional factor.
It has been reported that AP2/ERF transcription factors can modulate ethylene responses (Chang and Shockey, 1999; Yang et al., 2011b). Ethylene has been found to promote GA responsiveness and shoot elongation in submergence-intolerant lines and to confer submergence tolerance in rice (Fukao and Bailey-Serres, 2008). Moreover, internode elongation of rice in deepwater requires the activity of GA and ethylene (Sauter et al., 1995). Under deepwater conditions, ethylene accumulates and triggers remarkable internode elongation via GAs (Hattori et al., 2009). It seems that GA3 and Eth provide collaboration features in the process of plant internode elongation. This is consistent with our observation that SlDREB had the same expression profile under GA3 and Eth treatment (Fig. 4B, 4C), and it is possible that SlDREB can mediate cross-talk between ethylene and GA in plant internode elongation.
GAs regulate various developmental processes including stem elongation, leaf expansion, fruit development, and seed germination (Dill et al., 2004). GA and ABA play antagonistic roles in the regulation of numerous developmental processes: GA is associated with the promotion of germination, growth, and flowering, whilst ABA acts as a competitive inhibitor of GA activity and inhibits these processes (Razem et al., 2006). This antagonistic relationship was partially confirmed by our observation that expression of SlDREB was upregulated by GA application and downregulated by ABA treatment (Fig. 4B, 4D). It appears that SlDREB functions as a balance regulator through different networking between GA and ABA signalling pathways.
SlDREB restricts leaf expansion and internode elongation by negative regulation of GA biosynthesis genes
Our results demonstrated that the dwarf phenotype of SlDREB-overexpressing plants could be attributed to decreased levels of endogenous GAs (Fig. 7C). Accordingly, dwarfism of SlDREB-overexpressing plants could be reversed by the application of GA3 (Fig. 6A, 6B). These results are in agreement with previous studies in GA-deficient mutants of various plants, including Zea mays (Fujioka et al., 1988), Arabidopsis thaliana (Magome et al., 2004), and Oryza sativa (Sakamoto et al., 2004).
To assess further whether the GA-sensitive dwarf phenotype of SlDREB-overexpressing plants was due to inhibition of transcription of GA biosynthetic genes, inhibition of GA signal transduction, and/or promotion of expression of GA-inactivating enzyme genes, we analysed the expression level of different genes involved in these pathways. Our results demonstrated that SlDREB restricted internode elongation and leaf expansion by downregulating the expression of SlCPS and SlGA20ox1, -2, and -4 (Fig. 7A), which encode key enzymes in the GA biosynthesis pathway. In fact, a series of examples showed that CPS and GA20ox genes were targeted to lead to alteration of GA level and a dwarf plant. Loss-of-function CPS mutants of several plant species, such as rice (Sakamoto et al., 2004), Arabidopsis thaliana (Sun and Kamiya, 1994), and Pisum sativum (Ait-Ali et al., 1997), have also been shown to display severe dwarf phenotypes. In contrast, Serrani et al. (2008) reported that upregulation of SlCPS, SlGA20oxs, and SlGA3ox by 2, 4-dichlorophenoxyacetic acid treatment led to higher levels of bioactive GA in tomato. GA20ox catalyses the sequential oxidation of C-20 and is responsible for the production of inactive precursors (GA9, GA20) during bioactive GA synthesis (Yamaguchi, 2008). Overexpression and downregulation of GA20ox modified plant height by altering the levels of active GAs (Coles et al., 1999; Carrera et al., 2000; Vidal et al., 2001; Fagoaga et al., 2007). No significant differences were found in the expression of other genes involved in GA biosynthesis (SlGPS, SlKO, SlKAO, SlGA20ox3, and SlGA3ox1 and -2), and the GA-inactivating enzyme (SlGA2ox1–5) between SlDREB-overexpressing lines and WT plants (Fig. 7B). On the other hand, expression of SlGA20ox3 and SlKS was slightly upregulated in transgenic plants, probably for the feedback of GA deficiency (Cowling et al., 1998). Taken together, our results show that SlDREB restricts internodal elongation and leaf expansion by downregulating the expression of key GA biosynthesis enzymes. Moreover, the expression of GA response genes (SlDELLA and SlGAST1) between SlDERB-overexpressing and WT plants was not significantly changed. This provides further evidence that SlDERB negatively regulates the internode elongation and leaf expansion processes by inhibiting GA biosynthesis and not by signal transduction.
SlDREB may directly regulate expression of SlCPS
Transcription factors bind to either enhancer or promoter regions of DNA to regulate the expression of genes (Latchman, 1997). Most transcription factors act either as activators or repressors (Ikeda et al., 2009); therefore, depending on the transcription factor, the transcription of the adjacent gene is either up- or downregulated (Gill, 2001). Transcriptional regulation is an important mechanism for controlling metabolic pathways. These pathways are generally influenced by environmental stimuli and the plant’s developmental programme (Van Moerkercke et al., 2011). As we found that expression of SlCPS and SlGA20ox1, -2, and -4 was negatively related to SlDREB expression in our study (Fig. 7A), we studied the binding of SlDREB with the promoter region of genes it is likely to regulate. Our results showed binding of SlDREB to the DRE/CRT domains of the SlCPS promoter region (Fig. 8), and thus reveal that SlDREB likely acts as a direct repressor of SlCPS in plants. We did not find DRE/CRT elements in the putative 2500bp promoter regions of SlGA20ox1, -2, and -4, which might have been downregulated in the SlDREB-overexpressing plants through other mechanisms.
SlDREB integrates environmental stress signals and reduces endogenous GA levels to avoid abiotic stress
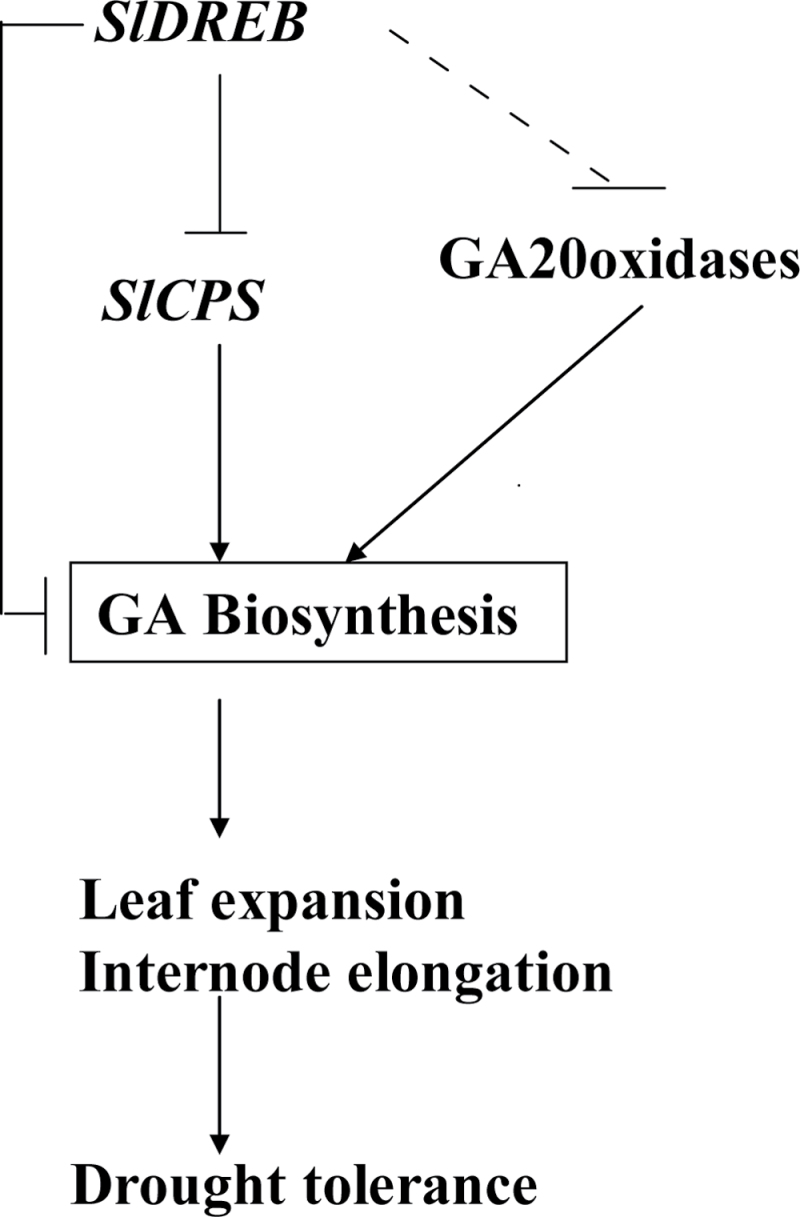
To sum up, overexpression of SlDREB in tomato plants can downregulate the expression of SlCPS and SlGA20oxs, and lowers endogenous GA levels, leading to restriction in leaf expansion and internode elongation. These effects account for the dwarfism of tomato plants and help in drought tolerance (Fig. 9). It seems that, under stress, decreased GA levels inhibit growth, allowing plants to survive better in unfavourable conditions. This hypothesis was confirmed by our results showing that overexpression of SlDREB in tomato leads to lower endogenous GA levels and consequently a dwarf phenotype, which enhances drought tolerance, and it is strengthened by the findings of (Magome et al., 2004), who also observed increased survival of GA-deficient ga1-3 mutant Arabidopsis plants exposed to salt stress and who showed that the mutants exhibited a reduced growth phenotype, which was inhibited by a reduction in GA synthesis. Thus, it can be concluded that plants need adequate GA to promote developmental changes, but limiting GA production is also necessary for them to survive under unfavourable environments. Further studies on SlDREB can provide a better understanding of how genes integrate environmental signals to restrict plant internode elongation by balancing ethylene, GA, and ABA signals.
Fig. 9.
Model of SlDREB regulation of leaf expansion and internode elongation by downregulating key genes required for gibberellin biosynthesis. SlDREB directly or indirectly downregulates the expression of SlCPS and SlGA20oxs, respectively. Transgenic plants overexpressing SlDREB have lower endogenous GA levels, leading to restriction of leaf expansion and internode elongation, which in turn increases the drought tolerance of plants.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. SpDREB expression in response to drought, salt, and cold stress factors.
Supplementary Fig. S2. Multiple alignments of the predicted protein sequence of SlDREB and DREBs from other plants.
Supplementary Fig. S3. Sequence of the promoter region of SlCPS.
Supplementary Table S1. Primer sequences used for real-time RT-PCR analysis of SlDREB and genes from the GA biosynthesis and GA response pathways in tomato.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (2009CB119002), the National High Technology R&D Program of China (2012AA100104), and the National Natural Science Foundation of China (30871712, 91017013 and 31070327).
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. 2006. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants Plant Cell Reports 25 1263–1274 [DOI] [PubMed] [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y. 1997. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A The Plant Journal 11 443–454 [DOI] [PubMed] [Google Scholar]
- Ban Q, Liu G, Wang Y. 2011. A DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco Journal of Plant Physiology 168 449–458 [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. 2003. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis Plant Physiology 132 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Jensen AB, Pagès M. 1997. Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize The Plant Journal 11 1285–1295 [DOI] [PubMed] [Google Scholar]
- Carrera E, Bou J, Garcia-Martinez JL, Prat S. 2000. Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants The Plant Journal 22 247–256 [DOI] [PubMed] [Google Scholar]
- Chang C, Shockey JA. 1999. The ethylene-response pathway: signal perception to gene regulation Current Opinion in Plant Biology 2 352–358 [DOI] [PubMed] [Google Scholar]
- Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP. 2008. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice Biotechnology Letters 30 2191–2198 [DOI] [PubMed] [Google Scholar]
- Chen ML, Huang YQ, Liu JQ, Yuan BF, Feng YQ. 2011. Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis-time of flight-mass spectrometry Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 879 938–944 [DOI] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening The Plant Journal 64 936–947 [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P. 1999. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes The Plant Journal 17 547–556 [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP. 1998. Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis Plant Physiology 117 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. 2004. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoaga C, Tadeo FR, Iglesias DJ, Huerta L, Lliso I, Vidal AM, Talon M, Navarro L, Garcia-Martinez JL, Pena L. 2007. Engineering of gibberellin levels in citrus by sense and antisense overexpression of a GA 20-oxidase gene modifies plant architecture Journal of Experimental Botany 58 1407–1420 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, Phinney BO, Takahashi N. 1988. Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L Plant Physiology 88 1367–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. 2008. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice Proceedings of the National Academy of Sciences, USA 105 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. 2001. Regulation of the initiation of eukaryotic transcription Essays in Biochemistry 37 33–43 [DOI] [PubMed] [Google Scholar]
- Gong P, Zhang J, Li H, et al. 2010. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato Journal of Experimental Botany 61 3563–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, et al. 2009. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water Nature 460 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y. 1997. Gibberellin biosynthesis: enzymes, genes and their regulation Annual Review of Plant Physiology and Plant Molecular Biology 48 431–460 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. 2009. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning Plant Cell 21 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Knight MR. 2001. Abiotic stress signalling pathways: specificity and cross-talk Trends in Plant Science 6 262–267 [DOI] [PubMed] [Google Scholar]
- Latchman DS. 1997. Transcription factors: an overview International Journal of Biochemistry and Cell Biology 29 1305–1312 [DOI] [PubMed] [Google Scholar]
- Li CW, Su RC, Cheng CP, Sanjaya, You SJ, Hsieh TH, Chao TC, Chan MT. 2011. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway Plant Physiology 156 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis Plant Cell 10 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. 2004. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor The Plant Journal 37 720–729 [DOI] [PubMed] [Google Scholar]
- Phinney BO. 1956. Growth response of single-gene dwarf mutants in maize to gibberellic acid Proceedings of the National Academy of Sciences, USA 42 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Li J, Liu Q, Li YQ, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L Plant and Cell Physiology 45 1042–1052 [DOI] [PubMed] [Google Scholar]
- Rae L, Lao NT, Kavanagh TA. 2011. Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors Planta 234 429–444 [DOI] [PubMed] [Google Scholar]
- Razem FA, Baron K, Hill RD. 2006. Turning on gibberellin and abscisic acid signaling Current Opinion in Plant Biology 9 454–459 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. 1998. The AP2/EREBP family of plant transcription factors Biological Chemistry 379 633–646 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, et al. 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice Plant Physiology 134 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression Biochemical and Biophysical Research Communications 290 998–1009 [DOI] [PubMed] [Google Scholar]
- Sauter M, Mekhedov SL, Kende H. 1995. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes The Plant Journal 7 623–632 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, Garcia-Martinez JL. 2008. Auxin-induced fruit-set in tomato is mediated in part by gibberellins The Plant Journal 56 922–934 [DOI] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. 2010. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato Molecular Genetics and Genomics 284 455–475 [DOI] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE. 1992. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato The Plant Journal 2 153–159 [PubMed] [Google Scholar]
- Shi X, Gupta S, Rashotte AM. 2012. Solanum lycopersicum cytokinin response factor (SlCRF) genes: characterization of CRF domain-containing ERF genes in tomato Journal of Experimental Botany 63 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. 2011. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants Current Biology 21 R338–R345 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F. 2004. Molecular mechanism of gibberellin signaling in plants Annual Review of Plant Biology 55 197–223 [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. 1994. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis Plant Cell 6 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thara VK, Tang X, Gu YQ, Martin GB, Zhou JM. 1999. Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate The Plant Journal 20 475–483 [DOI] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latche A, Pech JC, Bouzayen M. 2003. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element FEBS Letters 550 149–154 [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, et al. 2009. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis Journal of Plant Research 122 633–643 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Haring MA, Schuurink RC. 2011. The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias The Plant Journal 67 917–928 [DOI] [PubMed] [Google Scholar]
- van Schie CC, Ament K, Schmidt A, Lange T, Haring MA, Schuurink RC. 2007. Geranyl diphosphate synthase is required for biosynthesis of gibberellins The Plant Journal 52 752–762 [DOI] [PubMed] [Google Scholar]
- Vidal AM, Gisbert C, Talon M, Primo-Millo E, Lopez-Diaz I, Garcia-Martinez JL. 2001. The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco Physiologia Plantarum 112 251–260 [DOI] [PubMed] [Google Scholar]
- Wan L, Zhang J, Zhang H, Zhang Z, Quan R, Zhou S, Huang R. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS One. 20116:e25216. doi: 10.1371/journal.pone.0025216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. 1996. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2 Plant Cell 8 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xianjun P, Xingyong M, Weihong F, Man S, Liqin C, Alam I, Lee BH, Dongmei Q, Shihua S, Gongshe L. 2011. Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymus chinensis Plant Cell Reports 30 1493–1502 [DOI] [PubMed] [Google Scholar]
- Xiao JH, Li HX, Zhang JH, Chen RG, Zhang YY, Ouyang B, Wang TT, Ye ZB. 2006. Dissection of GA 20-oxidase members affecting tomato morphology by RNAi-mediated silencing Plant Growth Regulation 50 179–189 [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation Annual Review of Plant Biology 59 225–251 [DOI] [PubMed] [Google Scholar]
- Yang C, Li H, Zhang J, et al. 2011. a A regulatory gene induces trichome formation and embryo lethality in tomato Proceedings of the National Academy of Sciences, USA 108 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. 2011. b The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis Plant Physiology 156 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Liu XD, Chi XJ, et al. 2011. c Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways Planta 233 219–229 [DOI] [PubMed] [Google Scholar]
- Yang Y, Wu J, Zhu K, Liu L, Chen F, Yu D. 2009. Identification and characterization of two chrysanthemum (Dendronthema × moriforlium) DREB genes, belonging to the AP2/EREBP family Molecular Biology Reports 36 71–81 [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R. 2011. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis Plant Physiology 157 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Hu Y, Chong K, Wang T. 2010. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice Ann Bot 105 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.