Abstract
The persimmon fruit is a particularly good model for studying fruit response to hypoxia, in particular, the hypoxia-response ERF (HRE) genes. An anaerobic environment reduces fruit astringency by converting soluble condensed tannins (SCTs) into an insoluble form. Although the physiology of de-astringency has been widely studied, its molecular control is poorly understood. Both CO2 and ethylene treatments efficiently removed the astringency from ‘Mopan’ persimmon fruit, as indicated by a decrease in SCTs. Acetaldehyde, the putative agent for causing de-astringency, accumulated during these treatments, as did activities of the key enzymes of acetaldehyde synthesis, alcohol dehydrogenase (ADH), and pyruvate decarboxylase (PDC). Eight DkADH and DkPDC genes were isolated, and three candidates for a role in de-astringency, DkADH1, DkPDC1, and DkPDC2, were characterized by transcriptional analysis in different tissues. The significance of these specific isoforms was confirmed by principal component analysis. Transient expression in leaf tissue showed that DkPDC2 decreased SCTs. Interactions of six hypoxia-responsive ERF genes and target promoters were tested in transient assays. The results indicated that two hypoxia-responsive ERF genes, DkERF9 and DkERF10, were involved in separately regulating the DkPDC2 and DkADH1 promoters. It is proposed that a DkERF–DkADH/DkPDC cascade is involved in regulating persimmon de-astringency.
Key words: ADH, de-astringency, Diospyros kaki, ERF, ethylene, hypoxia, PDC, soluble tannins, transcriptional regulation
Introduction
Ethylene responsive factor (ERF) genes encode plant-specific transcription factors, which are located downstream of the ethylene signalling pathway (Nakano et al., 2006). ERFs are members of one of the largest transcription factor families, considered a super-family, with diverse functions in plants. Recent findings suggest that some ERF genes are important regulators of low oxygen tolerance in plants (Gibbs et al., 2011; Licausi et al., 2011a ). In fruit, ERF genes are key targets for investigating the transcriptional regulatory roles of ethylene in fruit development and ripening. Tomato LeERF2 was characterized as being a key regulator of ethylene biosynthesis (Zhang et al., 2009), and two novel ERF members, SlAP2 and SlERF6, have been shown to regulate genes involved in tomato ripening (Chung et al., 2010; Lee et al., 2012). In perennial fruit, apple CBF and kiwifruit AdERF9 transcriptionally regulate fruit cell wall-related genes (apple PG1; Tacken et al., 2010; kiwifruit AdXET5; Yin et al., 2010). However, many aspects of transcriptional regulation of fruit ripening and quality by ERF genes remain to be explored.
Recently, hypoxia-responsive ERF genes (HRE) have been characterized in Arabidopsis with HRE1 and HRE2 having partially redundant roles in increasing low oxygen tolerance (Licausi et al., 2010; Yang et al., 2011). Also, the RAP2.2, has a similar role in low oxygen signalling in Arabidopsis (Hinz et al., 2010). At least two of these ERF genes, HRE1 and RAP2.2, are involved in regulating the anaerobic fermentation-related genes, pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH) (Hinz et al., 2010; Yang et al., 2011).
In various species of fruit, anaerobic environments (low oxygen), generated by controlled atmosphere storage, have been widely applied to prolong their storage life. The persimmon fruit (Diospyros kaki) is of particular interest for anoxia-related studies, as an anaerobic environment promotes de-astringency. These fruit are unique for accumulating proanthocyanidins (also known as condensed tannins, CTs), and the fruit astringency is caused by the high content of Soluble CTs (SCTs). Although persimmons can be divided into astringent and non-astringent types (Akagi et al., 2009b ), most native cultivars grown in East Asia are of the astringent type (Yamada et al., 1994; Wang et al., 1997). Astringent-type fruit are rich in SCTs even at maturity, while the concentration of SCTs is significantly reduced during early development in fruit of the non-astringent type (Akagi et al., 2009a ). Astringent fruit tend to be rejected by consumers and artificial treatments have been developed to remove astringency, including treatments involving CO2, ethylene, and ethanol (Ikegami et al., 2007; Salvador et al., 2007), and CO2 application is usually the most effective (Salvador et al., 2007; Del Bubba et al., 2009). Furthermore, the physiological and biochemical mechanisms of CO2-driven de-astringency are much clearer than for other treatments. A high concentration of CO2 leads to anaerobic fermentation by triggering acetaldehyde metabolism (Matsuo and Ito, 1977; Pesis and Ben-Arie, 2006; Salvador et al., 2007). Acetaldehyde plays important roles in the polymerization of SCTs, converting them to insoluble condensed tannins (InSCTs) (Tanaka et al., 1994; Taira et al., 2001). CO2-driven de-astringency, therefore, is likely to occur via the modulation of acetaldehyde concentrations. Thus, studying the roles of ERF genes in regulating acetaldehyde biosynthetic genes and persimmon de-astringency is particularly important and may suggest new ways to improve the handling of this crop.
In plants, acetaldehyde can be synthesized from pyruvate and ethanol, catalysed by PDC (EC 4.1.1.1) and ADH (EC 1.1.1.1), respectively (Yamada et al., 2002; Pesis, 2005; Botondi et al., 2012). PDC and ADH have long been associated with persimmon de-astringency. During astringency loss in developing non-astringent persimmon fruit (cv. Nishimurawase), an increase in ADH enzyme activities was observed by Liu et al. (2008). The application of CO2 enhanced ADH and PDC activities during the de-astringency of ‘Saijo’ persimmons (Tamura et al., 1999). ADH and PDC enzymes are both encoded by gene families which have been studied in various plant species (Strommer, 2011), but no ADH or PDC genes have been characterized from persimmon.
In the present research, eight DkADH and DkPDC genes were isolated from persimmon fruit, based on the persimmon NCBI EST database. Transcript abundance of each of these genes was analysed in various tissues and in response to de-astringency treatments (CO2 and ethylene), in the astringent-type ‘Mopan’ persimmon fruit. DkADH and DkPDC genes specific to persimmon de-astringency were identified using principal component analysis (PCA) and were characterized by transient overexpression in persimmon leaves. By using RNA-Seq, hypoxia-responsive ERF genes were isolated and their activity in regulating target gene promoters was studied in vitro.
Materials and methods
Plant materials and treatments
‘Mopan’ (astringent-type) persimmon (Diospyros kaki) fruit were obtained from a commercial orchard at Fangshan (Beijing, China). The fruit were transported to Zhejiang University (Hangzhou, Zhejiang, China) on the second day after harvest. Uniform fruit free of visible defects were chosen for the de-astringency experiments, including CO2 and ethylene treatments.
For the ethylene treatment, the fruit were harvested in 2009, with mean colour index, firmness, and soluble solids content of 14.70, 52.31 N, and 17.88%, respectively. The fruit were divided into two lots of 180 fruit each. The control fruit were held at 20 °C free of any treatment for the same time periods as used for the ethylene-treated fruit. Continuous ethylene (100 µl l–1) in air (2 d) was applied to the second batch to remove the astringency of the fruit. For the CO2 treatment, the fruit were harvested in 2010, with mean colour index, firmness, and soluble solids content of 18.11, 43.08 N, and 18.76%, respectively. The fruit were divided into three batches (200 fruit in each) and the first two batches of fruit were treated with 95% CO2 for 1 d or 2 d, separately, according to Matsuo and Ito (1977). The third batch of fruit was stored in air at 20 °C for 4 d as the control. After treatment, the fruit were held in air at 20 °C.
All of the treatments and controls were performed with three biological replicates. At each sampling point, bulk fruit flesh samples (without skin and core) were taken from three replicates each of four fruit. The samples were frozen in liquid nitrogen and stored at –80 °C for further use.
For tissue-specific gene expression, stems, leaves, and petals were collected at full bloom, frozen in liquid nitrogen, and stored until use.
Fruit firmness and TSS
Fruit firmness and TSS (total soluble solids) were recorded at each sampling time. Fruit firmness measurements were made with a TA-XT2i texture analyser (Stable Micro Systems, UK), and the penetration measurements were carried out according to our previous report (Yin et al., 2012). Firmness was measured at two positions 90° apart at the fruit equator, after the removal of 1mm peel. Fruit TSS were measured from samples from both ends of each fruit, using a N20 digital hand-held refractometer (Atago, Japan), according to the methods described in Yin et al. (2008). Fruit firmness and TSS were measured on each of 10 fruit.
Soluble condensed tannins content
Soluble condensed tannins (SCTs) were chosen as the main de-astringency index and were also used for comparing the effectiveness of different treatments. SCTs were measured using frozen samples, according to the method described previously (Yin et al., 2012), where fruit flesh was extracted with methanol and the SCTs were measured with the Folin–Ciocalteu reagent (Sigma). The results were expressed as tannin acids equivalents g–1 fresh weight.
Acetaldehyde and ethanol
Acetaldehyde and ethanol production were measured with a gas chromatograph (Lunan Chemical Engineering Instrument Co. Ltd., model GC-14B, Shandong, China), fitted with a FID column (HP-INNOWAX). A 2g frozen fruit sample was ground in liquid nitrogen and the ground powder was mixed with 5ml saturated NaCl solution. Three ml of the mixture were transferred to 10ml air-tight vials with crimp-top caps. Before measurement, the vials were incubated at 60 °C for 1h in a water bath. Then, 0.2ml head-space gas was removed from each vial for acetaldehyde and ethanol measurement. The injector, detector, and oven temperatures were 140, 140, and 100 °C, respectively. Sec-butyl alcohol was added to each vial as an internal control. The results were calculated using standard curves for acetaldehyde and ethanol, respectively.
ADH and PDC activities
ADH and PDC enzymes were extracted and assayed according to the methods described by Botondi et al. (2012). One gram of persimmon fruit flesh was homogenized with 3ml 0.02M KH2PO4 buffer (pH 7.5) (including 2mM thiamine pyrophosphate, 2mM MgCl2, 1mM β-mercaptoethanol), on ice. The homogenate was centrifuged at 17 000 rcf for 20min at 4 °C and the supernatant was filtered with PD-10 Desalting Columns (GE Healthcare Bio-Sciences AB) and the eluate was used to measure PDC and ADH activity. Three biological replicates were performed at each sampling point.
ADH activity was measured using the increment in NADH by reading the absorbance at 340nm, in 1ml of reaction buffer [0.68ml 100mM KOH-glycine (pH 9.0), 0.07ml 11.4mM NAD+, 0.15ml 2M ethanol, 0.10ml enzyme extract].
PDC activity was measured using the decrease in NADH. The 1ml reaction mixture consisted of 0.4ml 100mM MES (pH 7.5), 0.1ml 10mM thiamine pyrophosphate, 0.1ml 10mM MgCl2, 0.1ml 2.6mM NADH, 0.1ml ADH (containing 7 kU ml–1), 0.1ml of enzyme extract, and 0.1ml 200mM pyruvate.
One activity unit (U) was defined as the change in one unit of absorbance at 340nm min–1 and the results were expressed as specific activity (mU g–1 flesh). All the reagents were the products from Sigma Aldrich Co. (USA).
RNA extraction and cDNA synthesis
Total RNA was extracted from various frozen tissues (2.0g for flesh tissues, 0.8g for leaves, 1.2g for stems, 1.6g for petals) with the method developed by Chang et al. (1993). The trace contaminating genomic DNA in total RNA was removed swith TURBO Dnase (Ambion). cDNA synthesis was initiated from 1.0 µg DNA-free RNA, using iScriptTM cDNA Synthesis Kit (Bio-Rad). For each sampling point, three biological replicates were used for RNA extraction.
Gene isolation and sequence analysis
ADH and PDC genes were isolated both using degenerate primers constructed from information for other species and also the persimmon EST database [published by Nakagawa et al. (2008) and available from NCBI (National Center for Biotechnology Information)]. The EST databases was blasted using the sequences isolated from using degenerate primers. The redundant sequences were excluded and non-redundant EST sequences were chosen for further experiments. The novel hypoxia (high CO2)-responsive ERF gene was isolated based on RNA-Seq. RNA-Seq was performed and the sequence was assembled and annotated by the Beijing Genome Institute (BGI) (Shenzhen, China). RNA isolation and the preparatory procedures were as previously reported by Feng et al. (2012). 26.7 million sequence reads were performed for each library and the sequences were compared by BLASTX to the public protein database.
Rapid amplification of cDNA ends (RACE) was chosen to amplify the 3’ end and/or 5’ end UTR (untranslated region) of ADH, PDC, and ERF, using a SMARTTM RACE cDNA Amplification Kit (Clontech). The sequences of primers used for cloning and RACE experiments are described in Supplementary Table S1 at JXB online.
The gene sequences were translated with online software (http://web.expasy.org/translate/) and were confirmed with the BLAST methods in Genbank. The deduced amino acid sequences of homologous genes in other plant species were downloaded from NCBI. Bootstrap Neighbor–Joining trees were generated with ClustalX (v 1.81), using the default parameters.
Oligonucleotide primers and real-time PCR
Oligonucleotide primers for real-time PCR analysis were designed with primer3 (v. 0.4.0, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The specificity of primers was determined by examining the melting curves and re-sequencing the PCR products. The sequences of oligonucleotide primers are described in Supplementary Table S1 at JXB online.
Real-time PCR was carried out using a CFX96 instrument (Bio-Rad). The PCR protocols were according to our previous reports, with Ssofast EvaGreen Supermix kit (Bio-Rad) (Yin et al., 2012). The relative abundance of each gene was calibrated with samples from mature fruit (for tissue specific studies) or day 0 fruit (for post-harvest experiments). Actin, a housekeeping gene, was used as the internal control (Akagi et al., 2009b ).
Transient over-expression in persimmon leaves
In order to determine the roles of DkADH and DkPDC genes in the regulation of SCTs, an unstable transformation system (transient expression) was adapted. Full-length DkADH1 and DkPDC2 coding sequences were constructed into pGreenII 0029 62-SK (Hellens et al., 2005). Both of the constructs were electroporated into Agrobacterium tumefaciens GV3101. The transient expression system (Agrobacterium culture, infiltration buffer) was according to the protocols used for tobacco in our laboratory (Espley et al., 2007; Yin et al., 2010).
Transient expression in persimmon leaves was performed with the ‘Mopan’ cultivar, the same cultivar used for astringency removal experiments. Suspensions of Agrobacterium, harbouring either DkADH1 or DkPDC2, were prepared with infiltration buffer and infiltrated into persimmon leaves, on the tree. Negative controls were included in the same leaves. Three days after infiltration, the infiltrated leaves were sampled and used for SCT analysis. 0.1g of tissue from each of the infiltrated leaves was taken for SCTs analysis, using ten single leaf replicates in total.
Genome walking and transient assay
Genome walking and a transient assay were performed according to our previous report (Yin et al., 2010). Promoters of DkADH1 and DkPDC2 were isolated with a GenomeWalker kit (Clontech), using genomic DNA from ‘Mopan’ persimmon leaves. The sequences of DkADH1 and DkPDC2 are described in Supplementary Fig. S5 at JXB online. Full-length DkERF9 was inserted into pGreen II 0029 62-SK vector (SK), while the promoters of DkADH1 (1021bp) and DkPDC2 (919bp) were inserted into pGreen II 0800-LUC vector. As the DkADH1 promoter has an endogenous NcoI site, a 12bp pGem-T easy vector sequence was incorporated between the DkADH1 promoter and luciferase reporter. All of the constructs were electroporated into Agrobacterium tumefaciens GV3101. The transient assay was performed with Nicotiana benthamiana leaves. Agrobacterium cultures were prepared with infiltration buffer (10mM MES, 10mM MgCl2, 150mM acetosyringone, pH 5.6) to an OD600 from 0.7 to 1.0. Agrobacterium culture mixtures of TFs (1ml) and promoters (100 µl) were infiltrated into tobacco leaves by needle-less syringes. Tobacco plants were grown in a greenhouse, under natural light with daylight extension to 16h. Three days after infiltration, firefly luciferase and renilla luciferase were assayed using the dual luciferase assay reagents (Promega). For each TF-promoter interaction, three independent experiments were performed (at least six replicates in each experiments).
Statistical analysis
Principal component analysis (PCA) was applied to analyse the correlations between de-astringency (soluble tannin concentrations) and gene expression with AlphaSoft version 11.0 (Alpha MOS, Toulouse, France). Software and data manipulations were according to our previous report (Zhang et al., 2011). Statistical significance of differences was calculated using Student’s t test.
Results
Isolation of novel hypoxia (high CO2)-responsive ERF genes
Four hypoxia responsive DkERF genes (DkERF1, DkERF4, DkERF5, and DkERF6) were characterized in our previous studies (Yin et al., 2012). Two novel hypoxia-responsive DkERF genes, designed as DkERF9 (JX117848) and DkERF10 (JX145122) were newly isolated after comparing RNA from 2 d CO2-treated fruit and control fruit using RNA-Seq technology. The RPKM (Reads per kb per million reads) values of DkERF9 and DkERF10 were 0 in 0 d fruit and 1.5692 and 450.2169 in 2 d CO2-treated fruit, respectively (data not shown), indicating that both DkERF9 and DkERF10 were possibly high-CO2/hypoxia-inducible. Phylogenetic analysis indicated that hypoxia responsive DkERF genes belong to different subfamilies. DkERF1 belongs to subfamily I, while DkERF4-6 belongs to subfamily VIII (according to Nakano et al., 2006). The two newly isolated hypoxia (high CO2)-responsive DkERF genes were clustered in subfamily IV (DkERF9) and VII (DkERF10) (see Supplementary Fig. S1 at JXB online). Within the six DkERF genes, only DkERF10 had a conserved MCGGAII motif at the N-end of the deduced protein.
Fruit de-astringency and ERF genes expression
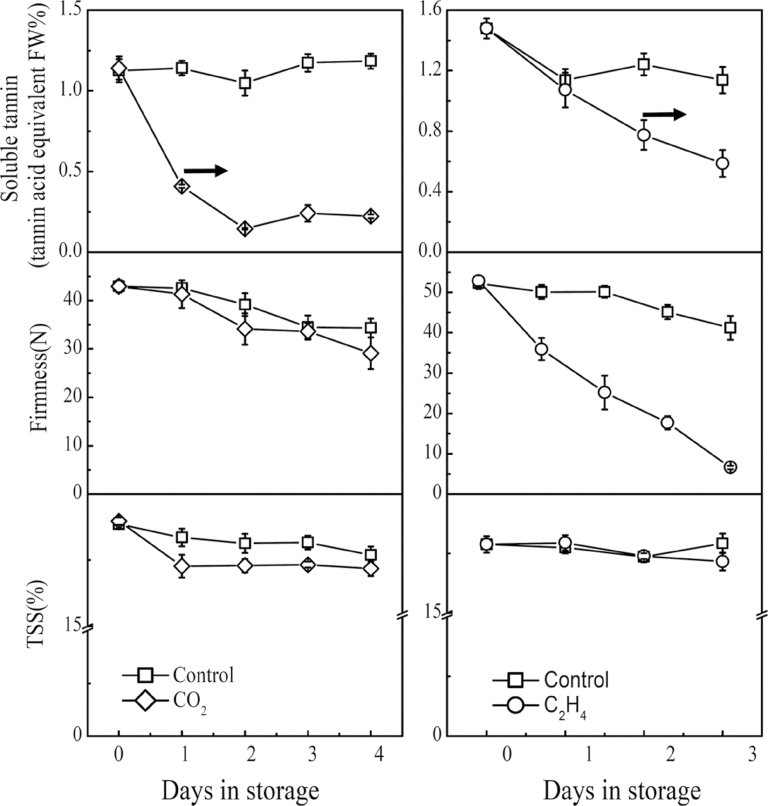
The relationship between hypoxia/high-CO2-responsive ERF genes and fruit de-astringency was evaluated in ‘Mopan’ persimmon fruit using both CO2 (95%) and ethylene (100 µl l–1) treatments. Both treatments promoted de-astringency, indicated by the decreasing concentration of SCTs (Fig. 1). However, the effects of CO2 and ethylene treatment on fruit ripening were different. Ethylene accelerated fruit texture change, and fruit softened from an initial value of 52.31 N to 14.19 N after ethylene treatment compared with 45.15 N in control fruit after 3 d in storage. By contrast, the firmness of CO2-treated fruit was similar to that of control fruit (Fig. 1). The TSS content decreased generally during both treatments, however, a larger reduction was found in CO2-treated fruit (Fig. 1).
Fig. 1.
Effects of CO2 and ethylene treatments on SCTs, firmness, and TSS of ‘Mopan’ persimmon fruit at 20 °C. Mature fruit were treated with CO2 (~95%, v/v, open diamonds, 1 d), ethylene (100 µl l–1, open circles, 2 d), and air (control, open squares), separately. The horizontal arrow represents the end day of the treatment, when the fruit were transferred to 20 °C, free of ethylene. Error bars represent SEs from three (SCTs) or 10 (firmness and TSS) biological replicates.
Acetaldehyde and ethanol, the main products of anaerobic fermentation, were also analysed in CO2- and ethylene-treated fruit. Acetaldehyde, the main compound that interacts with the SCTs to render them insoluble, was only detectable during treatment, and was undetectable once the treatments were removed (Table 1). The acetaldehyde content was much higher in CO2-treated fruit (5.94 µg g–1 at the end of 2 d continuous CO2 exposure) than ethylene-treated fruit (0.79 µg g–1 at the end of 2 d continuous ethylene exposure). Ethanol was generally detectable, although the de-astringent treatments also enhanced its accumulation several fold, and it was at its highest concentration in CO2-treated fruit (Table 2).
Table 1.
Effect of different astringency removal treatments on acetaldehyde concentration in ‘Mopan’ persimmon fruit flesh ND indicates that acetaldehyde was undetectable. Errors represent SEs from three biological replicates.
| Acetaldehyde (µg g–1 flesh weight) | |||||
|---|---|---|---|---|---|
| Days in storage | Control-1a | CO2 1d | CO2 2d | Control-2a | C2H4 2d |
| 0 | ND | ND | ND | ND | ND |
| 1 | ND | 2.51±0.21 | 2.09±0.18 | ND | ND |
| 2 | ND | ND | 5.94±0.60 | ND | 0.79±0.09 |
| 3 | ND | ND | ND | ND | ND |
| 4 | ND | ND | ND | —b | —b |
a Control-1 and Control-2 correspond to CO2 treatment and C2H4 treatments, respectively.
b —: not measured.
Table 2.
Effect of different de-astringency treatments on ethanol concentration in ‘Mopan’ persimmon fruit flesh Errors represent SEs from three biological replicates.
| Ethanol (µg g–1 flesh weight) | |||||
|---|---|---|---|---|---|
| Days in storage | Control-1a | CO2 1d | CO2 2d | Control-2a | C2H4 2d |
| 0 | 24.51±4.25 | 24.51±4.25 | 24.51±4.25 | 18.08±1.71 | 18.08±1.71 |
| 1 | 6.83±1.02 | 48.69±4.46 | 58.50±9.63 | 5.48±1.94 | 4.49±0.51 |
| 2 | 6.32±1.16 | 42.74±6.19 | 117.12±2.96 | 3.78±0.75 | 34.82±2.08 |
| 3 | 6.56±1.31 | 23.74±0.92 | 126.07±11.32 | 2.93±0.57 | 23.13±1.24 |
| 4 | 4.62±0.26 | 8.08±0.45 | 5.45±4.26 | —b | —b |
a Control-1 and Control-2 correspond to CO2 treatment and C2H4 treatments, respectively.
b —: not measured.
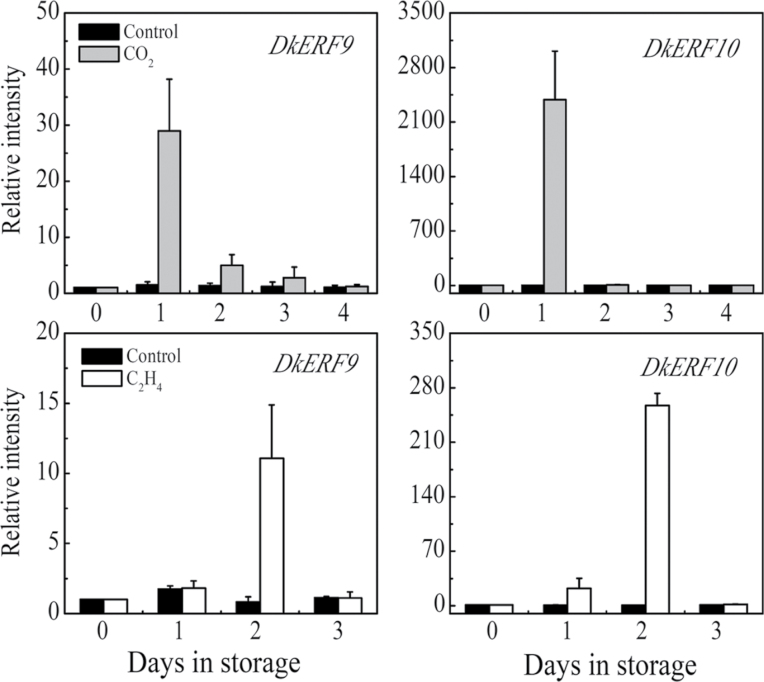
Real-time PCR analysis indicated the two newly isolated DkERF genes, DkERF9 and DkERF10, were both induced by the different de-astringent treatments (Fig. 2). Expression of DkERF10 was more responsive to these treatments than DkERF9 and the other ERFs.
Fig. 2.
Transcriptional analysis of hypoxia responsive DkERF9 and DkERF10. Transcripts of DkERF9 and DkERF10 were measured by real-time PCR in response to CO2 and ethylene treatment. Mature fruit were treated with ethylene (100 µl l–1, open circles, 2 d) and air (control, open squares), separately. A horizontal arrow represents the end day of the treatment, when the fruit were transferred to air at 20 °C. Day 0 fruit values were set as 1. Error bars indicate SEs from three biological replicates.
Isolation of anaerobic fermentation-related structural genes
Three ADH (DkADH1–3, JX117841–JX117843) and five PDC (DkPDC1–3, JX117844–JX117846; DkPDC4, DC592775; DkPDC5, JX117847) cDNAs were isolated from ‘Mopan’ persimmon fruit (astringent type). DkADH1, DkADH2, and DkPDC2 were full-length coding sequences (CDS), while others were partial CDS.
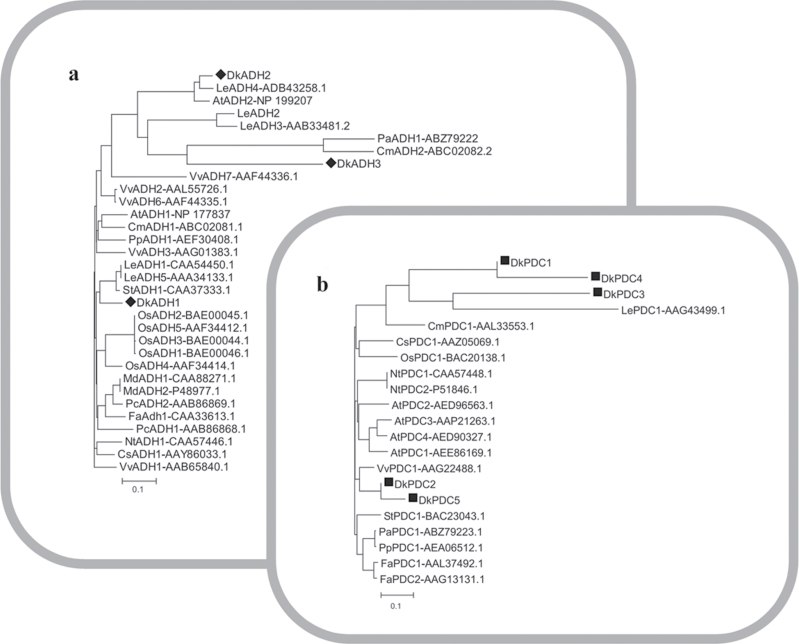
Phylogenetic analysis of the deduced amino acid sequences showed that DkADH1 clustered close to LeADH1, LeADH5, and StADH1; DkADH2 was close to AtADH2 and LeADH4; while DkADH3 was similar to PaADH1 and CmADH2 (Fig. 3). Unlike DkADH sequences, DkPDC coding sequences could be divided into two groups, with DkPDC1, DkPDC3, and DkPDC4 in one group with homology to LePDC1 and CmPDC1; while DkPDC2 and DkPDC5 were similar to each other and closely related to VvPDC1 (Fig. 3). Tissue-specific expression analysis revealed that DkADH and DkPDC genes were differentially expressed in various persimmon tissues (see Supplementary Fig. S2 at JXB online).
Fig. 3.
Phylogenetic analysis of ADH (a) and PDC (b) genes. The deduced amino acid sequences were obtained from the NBCI and the accession numbers are indicated. At, Arabidopsis thaliana; Cm, Cucumis melo; Cs, Citrus sinensis; Fa, Fragaria ananassa; Le, Lycopersicon esculentum; Md, Malus domestica; Nt, Nicotiana tabacum; Os, Oryza sativa; Pa, Prunus armeniaca; Pc, Pyrus communis; Pp, Prunus persica; St, Solanum tuberosum. The phylogenetic trees were constructed using ClustalX (v 1.81).
Enzymatic and transcriptional response of ADH and PDC during fruit de-astringency
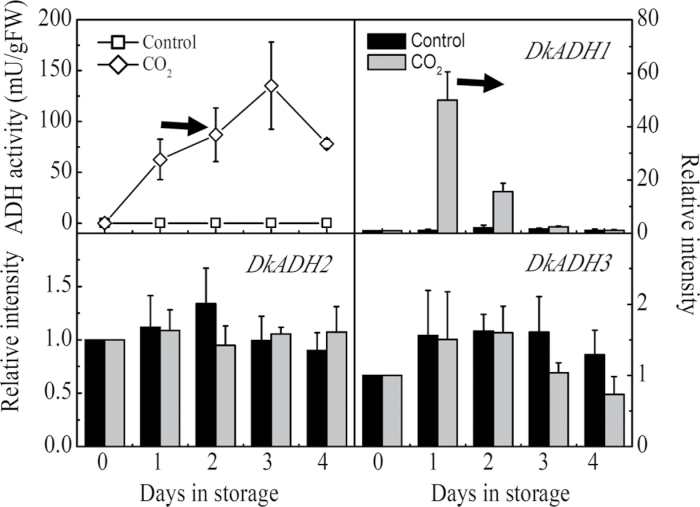
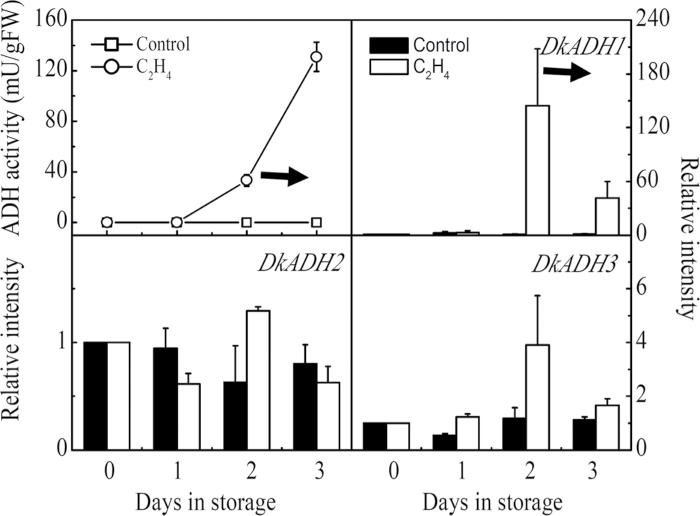
ADH activities were induced by both the 1 d CO2 and 2 d ethylene treatments. The levels in the other tissues were very high but the fruit levels were lower, but still at effective levels of activity. With the CO2 treatment, activity was induced from 1 d and reached a peak at 3 d (Fig. 4), and increased more gradually in ethylene-treated fruit (Fig. 5). Expression of DkADH1 mRNA was particularly responsive to these treatments, and paralleled the changes in enzyme activity. DkADH2 and DkADH3 expression was relatively more constant and at much lower levels. There was a stronger differential response to ethylene compared with CO2 (Figs 4, 5).
Fig. 4.
Changes in ADH enzyme activity and mRNA amounts from specific genes in response to CO2 treatment. Mature fruit were treated with CO2 (~95%, v/v, open diamonds, 1 d) and air (control, open squares), separately. The horizontal arrow represents the end day of the treatment, when the fruit were transferred to air at 20 °C. Day 0 fruit values were set as 1. Black columns and white columns represent relative mRNA abundance of the gene transcripts in control and ethylene treated fruit, respectively. Error bars indicate SEs from three biological replicates.
Fig. 5.
Changes in ADH enzyme activity and mRNA amounts from specific genes in response to ethylene treatment. Mature fruit were treated with ethylene (100 µl l–, open circles, 2 d) and air (control, open squares), separately. The horizontal arrow represents the end day of the treatment, when the fruit were transferred to air at 20 °C. Day 0 fruit values were set as 1. Black columns and white columns represent relative mRNA abundance of the gene transcripts in control and ethylene-treated fruit, respectively. Error bars indicate SEs from three biological replicates.
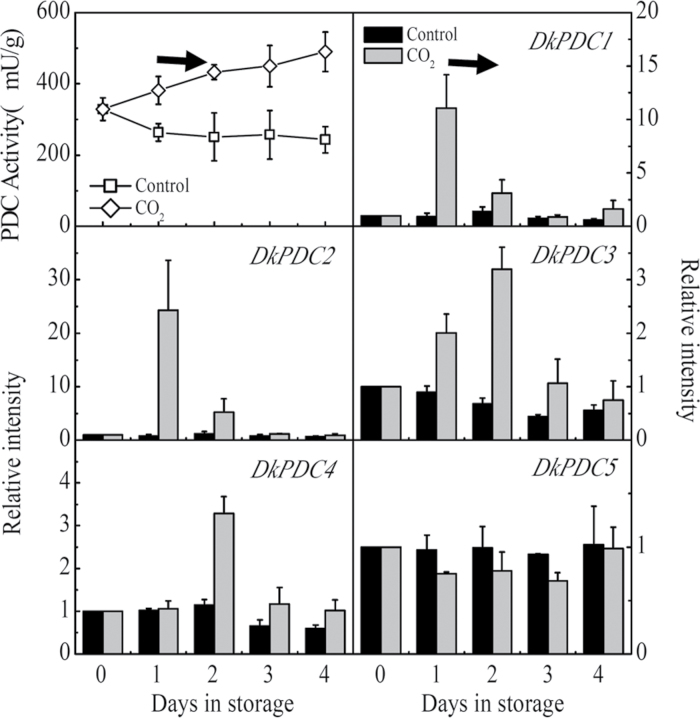
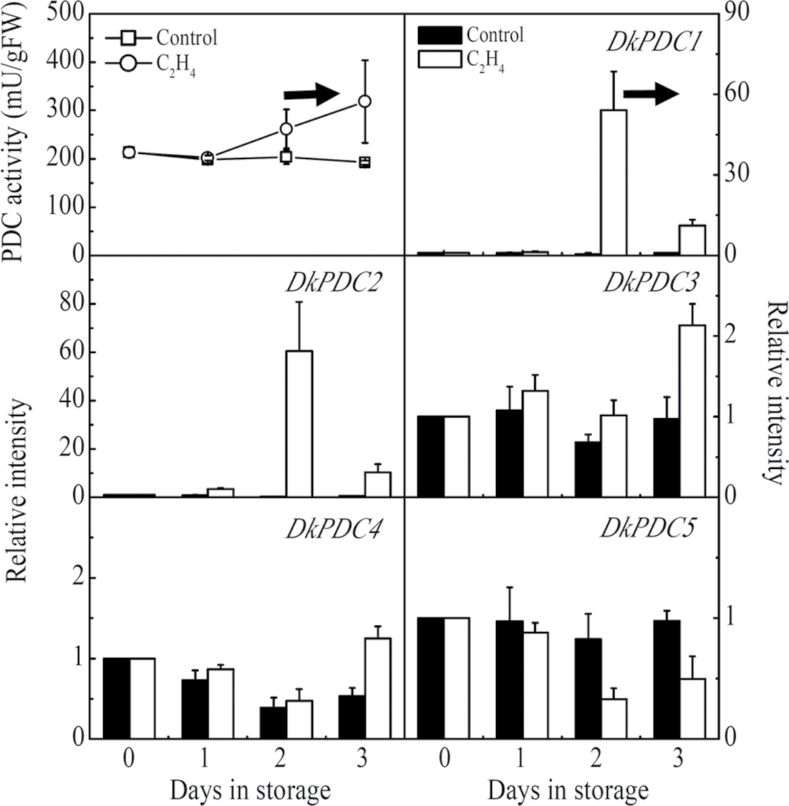
The PDC activities exhibited similar patterns, which increased in CO2-treated fruit, the increase being observed from 1 d (Fig. 6), while in ethylene-treated fruit there was a 1 d delay before PDC increased (Fig. 7). Among the PDC gene family, the transcript abundance of DkPDC1 and DkPDC2 was increased by more than 50-fold (Figs 6, 7). DkPDC3 and DkPDC4 mRNA accumulation increased only in the CO2-treated fruit and the levels were relatively lower than for DkPDC1 and DkPDC2. DkPDC5 was partially repressed by exogenous ethylene treatment (Fig. 7) The above expression patterns from the 1 d CO2 treatment (Figs 4, 6) were confirmed in the 2 d CO2-treated fruit (see Supplementary Fig. S3 at JXB online).
Fig. 6.
Changes in PDC enzyme activity and mRNA amounts from specific genes in response to CO2 treatment. Mature fruit were treated with CO2 (~95%, v/v, open diamonds, 1 d) and air (control, open squares), separately. A horizontal arrow represents the end day of the treatment, when the fruit were transferred to air at 20 °C. Day 0 fruit values were set as 1. Black columns and white columns represent relative mRNA abundance of the gene transcripts in control and ethylene-treated fruit, respectively. Error bars indicate SEs from three biological replicates.
Fig. 7.
Changes in PDC enzyme activity and mRNA amounts from specific genes in response to ethylene treatment. Mature fruit were treated with ethylene (100 µl l–1, open circles, 2 d) and air (control, open squares), separately. A horizontal arrow represents the end day of the treatment, when the fruit were transferred to air at 20 °C. Day 0 fruit values were set as 1. Black columns and white columns represent relative mRNA abundance of the gene transcripts in control and ethylene-treated fruit, respectively. Error bars indicate SEs from three biological replicates.
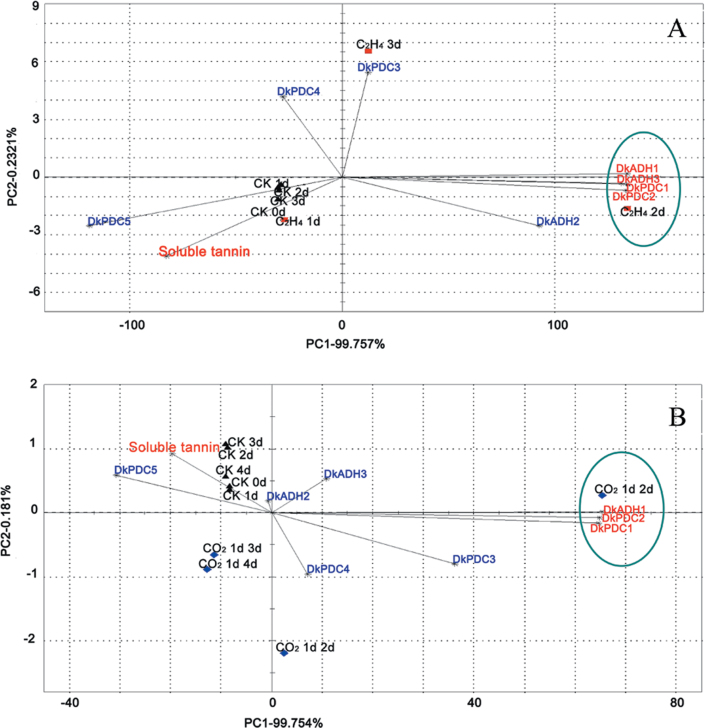
PCA analysis was performed to evaluate the relations between de-astringency and expression of ADH/PDC genes, with the mean data for soluble tannin contents and levels of gene expression. The analysis showed that, for both treatments, PC1 and PC2 accounted for over 99% of the total variation, and PC1 was dominant with 99.757% and 99.754% for ethylene and CO2 treatments, respectively (Fig. 8). As expected, de-astringency treatments showed negative correlations with soluble tannin contents. In the ethylene treatment, DkADH1, DkADH3, DkPDC1, and DkPDC2 were distributed along the negative axis of PC1, and were far from SCTs (Fig. 8). Similarly, with the CO2 treatment, DkADH1, DkPDC1, and DkPDC2 were the most distant from SCTs (Fig. 8). As PC1 was the main contributor to the total variation, these results suggest that DkADH1, DkPDC1, and DkPDC2 are most likely to be involved in de-astringency (decrease in SCTs).
Fig. 8.
PCA analysis of the relationship between DkADH and DkPDC gene expression and persimmon fruit de-astringency. Soluble tannin contents indicate the extent of fruit de-astringency. Ethylene treatment (a) and CO2 treatment (b). The mean data were used for PCA analysis.
Transient over-expression of DkADH1 and DkPDC2 in persimmon leaves
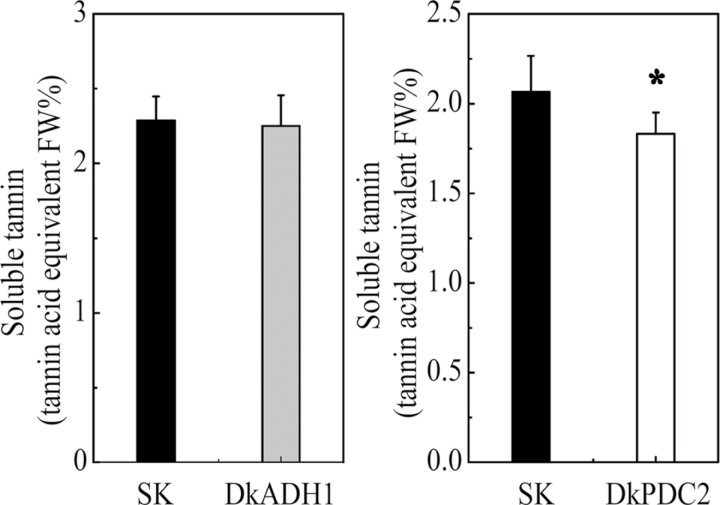
Persimmon is a perennial fruit and it takes years to generate transgenic fruit with stable transformation technologies, thus a transient overexpression system was chosen for quick gene functional analysis. DkADH1 and DkPDC2 constructs, driven by the CaMV 35S promoter, were introduced into Agrobacterium, which was then infiltrated into persimmon leaves. Analysis of SCTs in leaves indicated that the effects of DkADH1 and DkPDC2 in decreasing SCTs were different. The levels of SCTs in the leaves infiltrated with DkADH1 were similar to those in leaves infiltrated with empty vectors (SK, Fig. 9). Transient expression of DkDPC2, on the other hand, led to a small but significant (P <0.05) decrease in levels of SCTs in the leaves (Fig. 9).
Fig. 9.
Transient expression of DkADH1 and DkPDC2 in ‘Mopan’ persimmon leaves. Soluble tannins contents in leaves infiltrated with DkADH1 or DkPDC2, driven by the CaMV 35S promoter. SK represents empty vector. Error bars indicate SEs from ten biological replicates (*P <0.05).
Association and interaction of hypoxia-responsive ERFs and DkADH1 and DkPDC2
Linear correlation analysis was performed on gene expressions of DkERF genes and the target genes. Except for DkERF6, expressions of other DkERF genes were closely related with DkADH1 and DkPDC2 (see Supplementary Fig. S4 at JXB online).
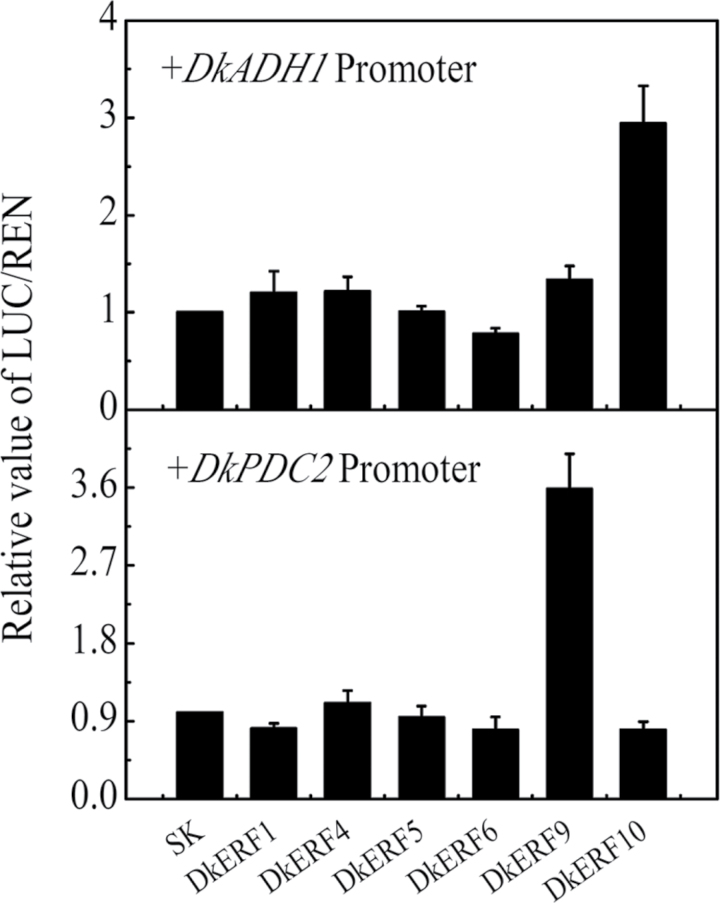
A transient assay indicated that DkERF9 and DkERF10 acted as activators, while the other five ERF genes had less effect on DkADH1 and DkPDC2 promoters (Fig. 10). Activity of the DkPDC2 promoter was induced by interaction with DkERF9, while activity of the DkADH1 promoter was only induced by DkERF10 (Fig. 10).
Fig. 10.
In vivo interaction of hypoxia-responsive ERF gene and target promoters. 100 µl samples were infiltrated into Nicotiana benthamiana leaves. Firefly luciferase and renilla luciferase were assayed 3 d after infiltration. For each TF-promoter interaction, three independent experiments were performed (at least six replicates in each experiment). The ratio of LUC/REN of the empty vector (SK) plus promoter was used as calibrator (set as 1). Error bars indicate SEs from six biological replicates.
Discussion
The majority of persimmon fruit in production are unique in that they remain astringent even at maturity, whereas astringency only occurs at the unripe stages in other fruit. Thus, the process of astringency removal is one of the most important aspects of post-harvest handling of persimmon fruit. Persimmon fruit de-astringency has been extensively studied at the physiological level, and several successful technologies for promoting astringency loss have been developed, such as CO2 and ethylene treatment. The mechanisms of action of astringency removal have mainly been studied in relation to CO2 treatment, which is thought to occur by the promotion of hypoxia-related fermentation. Despite these physiological and biochemical investigations, however, molecular insights into persimmon de-astringency are still lacking. Nevertheless, persimmon fruit provide a good model for investigating both the hypoxia response and de-astringency.
Both CO2 and ethylene treatments accelerate persimmon fruit de-astringency
By utilizing the astringent type persimmon ‘Mopan’, two de-astringency treatments, CO2 and ethylene, were compared. The results showed that CO2 was more effective than ethylene in inducing de-astringency, indicated by the decrease in SCTs (Figs 3, 34) and the increase in acetaldehyde formation (Tables 1, 12), which is in agreement with previous reports (Arnal and Del Río, 2003; Pesis and Ben-Arie, 2006; Salvador et al., 2007). This is consistent with acetaldehyde being the main compound involved in the insolubilization of SCTs.
Isolation of hypoxia-responsive ERF genes from persimmon
In model plants, HRE and RAP type ERF genes were recently shown to be the key regulators in plant responses to hypoxia (Gibbs et al., 2011; Hinz et al., 2010; Licausi et al., 2010, 2011a ; Sasidharan and Mustroph, 2011; Yang et al., 2011). Comparative analysis of the predicted amino acid sequences encoded by the HRE and RAP genes revealed that all of the five proteins had conserved N-ends, initiated by the MC domain (MCGGAII), which has recently been shown to be involved in controlling the stability of regulators of the plant oxygen-sensing mechanism (Gibbs et al., 2011; Licausi et al., 2011a ; Sasidharan and Mustroph, 2011). In persimmon, four ERF genes, DkERF1, DkERF4, DkERF5, and DkERF6 were characterized as inducible by both low oxygen and ethylene, and proposed to be de-astringency-related in our previous study (Yin et al., 2012). By utilizing RNA-Seq methodology, two additional novel hypoxia (high CO2)-responsive genes, DkERF9 and DkERF10, were isolated. However, only DkERF10, encoded the conserved MC domain. Phylogenetic analysis also indicated that DkERF10 was closely related to HRE and RAP genes, and, like them, belongs to subfamily VII. Thus, the phylogenetic and domain analyses indicated that DkERF10 had a high probability of being functionally involved in low oxygen sensing.
Real-time PCR indicated that all six DkERF genes were responsive to hypoxia/high CO2. Of these six, DkERF10 was the most responsive to the de-astringent treatment, especially to CO2 treatment (greater than a 2000-fold increase, Fig. 2). Moreover, some of hypoxia/high CO2-responsive ERF genes were also modulated by ethylene, with different patterns. DkERF4, 5, 9, and 10, were responsive to both ethylene and CO2 (present results and Yin et al., 2012 b), while DkERF1 and 6 only seemed to respond to CO2. This observation is similar to that for the hypoxia-response ERF genes in Arabidopsis, where HRE1 is both responsive to ethylene and hypoxia (Hinz et al., 2010; Yang et al., 2011), while Rap2.12 is not regulated by 1-aminocyclopropane-1-carboxylic acid (ACC) (Licausi et al., 2011b ).
Characterization of persimmon structural genes related to anaerobic fermentation
As mentioned previously, acetaldehyde is one of the main compounds that render the SCTs insoluble and cause de-astringency. Synthesis of acetaldehyde is generally catalysed by PDC, which converts pyruvate to acetaldehyde. ADH is then involved in the potentially reversible interconversion of acetaldehyde and ethanol (Strommer, 2011). In persimmon fruit, both PDC and ADH have been proposed as candidates involved in acetaldehyde formation (Yamada et al., 2002). In plants, PDC has been associated mostly with acetaldehyde formation (Nguyen et al., 2009), whereas ADH is best known for its role in the anaerobic response (Strommer, 2011), although it has also been linked in fruit to the regulation of the formation of volatile six-carbon alcohols (Speirs et al., 1998; Zhang et al., 2010). Phylogenetic analysis of persimmon DkADH and DkPDC genes revealed that the isolated genes were clustered with different homologues from Arabidopsis, tomato, tobacco, etc (Fig. 1). However, it is not possible to predict gene function simply based on phylogenetic tree analysis, particularly as the genes with high similarity were found to have different expression patterns in tissue-specific analysis (e.g. DkPDC3 and DkPDC4 were similar in sequence, but differentially expressed in various tissues).
The expression patterns of DkADH and DkPDC genes were monitored during de-astringency. The up-regulation by ethylene of DkADH1,3 and DkPDC1,2 is similar to the response of ADH and PDC genes to ethylene in other fruit, e.g. apricot PaADH (EU395433), and melon CmADH1 (DQ288986) and CmADH2 (DQ288987) (Manríquez et al., 2006; González-Agüero et al., 2009). During CO2 treatment, no matter whether for 1 d or 2 d durations (Fig. 6; see Supplementary Fig. S2 at JXB online), DkADH1, DkPDC1, DkPDC2, and DkPDC3 were transiently induced, similar to tomato LeADH2 (X77233) and tobacco TobADH1 (X81853) and TobPDC1 (X81854) genes (Longhurst et al., 1994; Bucher et al., 1995). The expression patterns of these genes have also been shown in previous reports, to coincide with the increase in related enzyme activities (Tamura et al., 1999). DkADH1, DkPDC1, and DkPDC2 were substantially more responsive to both persimmon de-astringency treatments than the other gene family members. Furthermore, the increase in expression of DkADH1, DkPDC1, and DkPDC2 occurred in parallel with the changes in ADH and PDC enzyme activities (Figs 4, 45, 46, 47), suggesting that they might be the key genes involved in persimmon de-astringency, and this suggestion is supported by the PCA results (Fig. 8). Transient expression of DkADH1 and DkPDC2 in persimmon leaves had different effects on leaf SCTs contents. Leaves infiltrated with DkPDC2 exhibited a small, but significant reduction in levels of SCTs, while DkADH1 had no affect (Fig. 9). Although further functional analysis needs to be performed with stable transformation systems, all the present results strongly indicate that DkADH1, DkPDC1, and DkPDC2 should be considered as the most important candidates for persimmon fruit de-astringency.
DkERF genes involved in the transcriptional regulation of persimmon DkADH1 and DkPDC2 and in relation to de-astringency
Transcription factors (TFs) act as the switches for regulating target genes (Riechmann et al., 2000). In model plants, expression of ADH and PDC genes is modulated by novel hypoxia-responsive ERF genes. Our results indicate that, with the exception of DkERF6 (see Supplementary Fig. S4 at JXB online), changes in transcript abundance of DkERF genes and the target genes were closely correlated. Using the transient assay, gene–gene screening indicated that only DkERF9 and DkERF10 are involved in regulating the promoters of DkPDC2 and DkADH1 target genes (Fig. 10). These results were similar to the finding obtained by RNAi methodology that Arabidopsis HRE1 is an activator of ADH1, PDC1, and PDC2 (Yang et al., 2011). Thus, DkERF9 and DkERF10 might act as the major regulators of persimmon fruit low oxygen signalling, and thus be the key regulator genes in causing de-astringency.
Interestingly, the present results indicate that two ERF genes act as switches for different targets. DkERF10 interacted with the DkADH1 promoter, while DkERF9 only induced activation of the DkDPC2 promoter. These results suggest that different ERF genes may have different targets, which is in contrast to the observations for HRE1 in Arabidopsis. This indicates that these two DkERF genes have different regulatory roles in persimmon de-astringency.
DkERF10 is a homologue of Arabidopsis HRE and RAP genes, and belongs to subfamily VII. Thus, it is not surprising that DkERF10 was characterized as a regulator of persimmon fruit hypoxia response and de-astringency. On the other hand, DkERF9 belongs to subfamily IV, which comprises the DREB subfamily. DREB type ERF genes are the main regulators of plant responses to abiotic stresses, such as drought and heat (Lata and Prasad, 2011). However, the roles of many DREB-type ERF genes have not yet been elucidated. Based on the present results, in addition to members of subfamily VII, subfamily IV ERF genes may also be involved in plant hypoxia responses.
In conclusion, both CO2 and ethylene treatments were effective in reducing ‘Mopan’ persimmon fruit astringency, by promoting the increase of acetaldehyde, ADH and PDC activities and the decrease of SCTs. Three genes, DkADH1, DkPDC1, and DkPDC2, are the most likely candidates for increasing acetaldehyde concentrations and causing de-astringency, and the role of DkPDC2 in decreasing SCTs was functionally confirmed. Two hypoxia-responsive ERF genes, DkERF9 and DkERF10, are involved in the regulation of the DkPDC2 and DkADH1 promoters, respectively. Based on the present findings, we propose that both of DkERF9 and DkERF10 are involved in the persimmon fruit anoxia response and de-astringency and increase acetaldehyde accumulation by modulating different target genes. As there are at least three hypoxia responsive ERF genes in Arabidopsis (Hinz et al., 2010; Licausi et al., 2010), additional hypoxia-responsive ERF genes may exist in persimmon fruit. Furthermore, the effects high CO2 and ethylene on the DkERF transcriptional regulation of DkADH and DkPDC genes need to be investigated further.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Sequences of the primers used for gene isolation and expression analysis.
Supplementary Fig. S1. Phylogenetic tree of ERFs.
Supplementary Fig. S2. Differential expression of DkADH and DkPDC genes in various ‘Mopan’ plant tissues.
Supplementary Fig. S3. Differential expression of DkADH and DkPDC genes in response to 2 d CO2 treatment at 20 °C.
Supplementary Fig. S4. Correlation of transcripts abundance of DkERF genes with DkADH1 and DkPDC2 expression.
Supplementary Fig. S5. Sequences of DkADH1 and DkPDC2 promoters.
Acknowledgements
We wish to thank Dr Jie Zhang for support in the collection of ‘Mopan’ fruit. This research was supported by the Special Fund for Agro-scientific Research in the Public Interest (no. 201203047), the National Natural Science Foundation of China (no. 30972059; no. 31101507), the Program for Key Innovative Research Team of Zhejiang Province (no. 2009R50036), the 111 project, and the Fundamental Research Funds for the Central Universities.
Glossary
Abbreviations:
- ADH
alcohol dehydrogenase
- AP2
APETALA2
- CBF
C-repeat-binding factor
- ERF
ethylene-response factor
- HRE
hypoxia-responsive ERF
- InSCTs
insoluble condensed tannins
- PDC
pyruvate decarboxylase
- PG
polygalacturonase
- SCTs
soluble condensed tannins
- UTR
untranslated region
- XET
xyloglucan endotransglycosylase
References
- Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, Yonemori K. 2009a. Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit Planta 230 899–915 [DOI] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K. 2009b. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit Plant Physiology 151 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal L, Del Río MA. 2003. Removing astringency by carbon dioxide and nitrogen-enriched atmospheres in persimmon fruit cv. ‘Rojo brillante’ Journal of Food Science 68 1516–1518 [DOI] [PubMed] [Google Scholar]
- Bucher M, Brander KA, Sbicego S, Mandel T, Kuhlemeier C. 1995. Aerobic fermentation in tobacco pollen Plant Molecular Biology 28 739–750 [DOI] [PubMed] [Google Scholar]
- Botondi R, Russo V, Mencarelli F. 2012. Anaerobic metabolism during short and long term storage of kiwifruit Postharvest Biology and Technology 64 83–90 [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees Plant Molecular Biology Reporter 11 113–116 [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee JM, McQuinn R, Chung JD, Klein P, Giovannoni J. 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening The Plant Journal 64 936–947 [DOI] [PubMed] [Google Scholar]
- Del Bubba M, Giordani E, Pippucci L, Cincinelli A, Checchini L, Galvan P. 2009. Changes in tannins, ascorbic acid and sugar content in astringent persimmons during on-tree growth and ripening and in response to different postharvest treatments Journal of Food Composition and Analysis 22 668–677 [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. 2007. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10 The Plant Journal 49 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Chen M, Xu Cj, Bai L, Yin XR, Li X, Allan AC, Ferguson IB, Chen KS. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genomics. 2012;13:19. doi: 10.1186/1471-2164-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, et al. 2011. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants Nature 479 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Agüero M, Troncoso S, Gudenschwager O, Campos-Vargas R, Mpya-León M, Defilippi BG. 2009. Differential expression levels of aroma-related genes during ripening of apricot (Prunus armeniaca L.) Plant Physiology and Biochemistry 47 435–440 [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. 2010. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival Plant Physiology 153 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A, Eguchi S, Kitajima A, Inoue K, Yonemori K. 2007. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit Plant Science 172 1037–1047 [Google Scholar]
- Lata C, Prasad M. 2011. Role of DREBs in regulation of abiotic stress responses in plants Journal of Experimental Botany 62 4731–4748 [DOI] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei ZJ, Tieman D, Klee H, Giovannoni J. 2012. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation The Plant Journal 70 191–204 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT. 2011a. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization Nature 479 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. 2010. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana The Plant Journal 62 302–315 [DOI] [PubMed] [Google Scholar]
- Licausi F, Weits DA, Pant BD, Scheible WR, Geigenberger P, van Dongen JT. 2011b. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability New Phytologist 190 442–456 [DOI] [PubMed] [Google Scholar]
- Liu CP, Zheng ZM, Liang YN, Li B. 2008. Relationship between activity of alcohol dehydrogenase and soluble tannin in persimmon Acta Horticulturae Sinica 35 741–746 (In Chinese, with English abstract) [Google Scholar]
- Longhurst T, Lee E, Hinde R, Brady C, Speirs J. 1994. Structure of the tomato Adh2 gene and Adh2 pseudogenes, and a study of Adh2 gene expression in fruit Plant Molecular Biology 26 1073–1084 [DOI] [PubMed] [Google Scholar]
- Manríquez D, EI-Sharkawy I, Flores FB, EI-Yahyaoui F, Regad F, Bouzayen M, Latché A, Pech JC. 2006. Two highly divergent alcohol dehydrogenases of melon exhibit fruit ripening-specific expression and distinct biochemical characteristics Plant Molecular Biology 61 675–685 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Ito S. 1977. On mechanisms of removing astringency in persimmon fruits by carbon dioxide treatment. I. Some properties of the two processes in the de-astringency Plant and Cell Physiology 18 17–25 [Google Scholar]
- Nakagawa T, Nakatsuka A, Yano K, Yasugahira S, Nakamura R, Sun N, Itai A, Suzuki T, Itamura H. 2008. Expressed sequence tags from persimmon at different developmental stages Plant Cell Reports 27 931–938 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice Plant Physiology 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Drotar AM, Monson RK, Fall R. 2009. A high affinity pyruvate decarboxylase is present in cottonwood leaf veins and petioles: a second source of leaf acetaldehyde emission? Phytochemistry 70 1217–1221 [DOI] [PubMed] [Google Scholar]
- Pesis E. 2005. The role of the anaerobic metabolites, acetaldehyde and ethanol, in fruit ripening, enhancement of fruit quality and fruit deterioration Postharvest Biology and Technology 37 1–19 [Google Scholar]
- Pesis E, Ben-Arie R. 2006. Involvement of acetaldehyde and ethanol accumulation during induced deastringency of persimmon fruits Journal of Food Science 49 896–899 [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Salvador A, Arnal L, Besada C, Larrea V, Quiles A, Pérez-Munuera I. 2007. Physiological and structural changes during ripening and deastringency treatment of persimmon fruit cv. ‘Rojo Brillante’ Postharvest Biology and Technology 46 181–188 [Google Scholar]
- Sasidharan R, Mustroph A. 2011. Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis The Plant Cell 23 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J, Lee E, Holt K, Yong-Duk K, Scott NS, Loveys B, Schuch W. 1998. Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols Plant Physiology 117 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommer J. 2011. The plant ADH gene family The Plant Journal 66 128–142 [DOI] [PubMed] [Google Scholar]
- Tacken E, Ireland H, Gunaseelan K, et al. 2010. The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening Plant Physiology 153 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira S, Ikeda K, Ohkawa K. 2001. Comparison of insolubility of tannins induced by acetaldehyde vapor in fruit of three types of astringent persimmon Journal of the Japanese Society for Horticultural Science 48 684–687 [Google Scholar]
- Tamura F, Tanabe K, Itai A, Hasegawa M. 1999. Characteristics of acetaldehyde accumulation and removal of astringency with ethanol and carbon dioxide treatments in ‘Saijo’ persimmon fruit Journal of the Japanese Society for Horticultural Science 68 1178–1183 [Google Scholar]
- Tanaka T, Takahashi R, Kouno I, Nonaka GI. 1994. Chemical evidence for the de-astringency (insolubilization of tannins) of persimmon fruit Journal of the Chemical Society, Perkin Transactions 1 20 3013–3022 [Google Scholar]
- Wang RZ, Yang Y, Li GC. 1997. Chinese persimmon germplasm resources Acta Horticulturae 436 43–50 [Google Scholar]
- Yamada M, Yamane H, Sato A, Hirakawa N, Wang RZ. 1994. Variations in fruit ripening time, fruit weight and soluble solids content of oriental persimmon cultivars native to Japan Journal of the Japanese Society for Horticultural Science 63 485–491 [Google Scholar]
- Yamada M, Taira S, Ohtsuki M, Sato A, Iwanami H, Yakushiji H, Wang RZ, Yang Y, Li GC. 2002. Varietal differences in the ease of astringency removal by carbon dioxide gas and ethanol vapor treatments among Oriental astringent persimmons of Japanese and Chinese origin Scientia Horticulturae 94 63–72 [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. 2011. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis Plant Physiology 156 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Chen KS, Allan AC, Wu RM, Zhang B, Lallu N, Ferguson IB. 2008. Ethylene-induced modulation of genes associated with the ethylene signaling pathway in ripening kiwifruit Journal of Experimental Botany 59 2097–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. 2010. Kiwifruit EIL and ERF genes involved in regulating fruit ripening Plant Physiology 153 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Shi YN, Min T, Luo ZR, Yao YC, Xu Q, Ferguson IB, Chen KS. 2012. Expression of ethylene response genes during persimmon fruit astringency removal Planta 235 895–906 [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Zhang HW, Quan RD, Wang XC, Huang RF. 2009. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco Plant Physiology 150 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shen JY, Wei WW, Xi WP, Xu CJ, Ferguson I, Chen KS. 2010. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening Journal of Agricultural and Food Chemistry 58 6157–6165 [DOI] [PubMed] [Google Scholar]
- Zhang B, Xi WP, Wei WW, Shen JY, Ferguson I, Chen KS. 2011. Changes in aroma-related volatiles and gene expression during low temperature storage and subsequent shelf-life of peach fruit Postharvest Biology and Technology 60 7–16 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.